Abstract
Cells react to viral infection by exhibiting interferon (IFN)-based innate immune responses and integrated stress responses, but little is known about the interrelationships between the two. We here report a linkage between these two host protective cellular mechanisms. We found that IRF7, the master regulator of type I IFN gene expression, interacts with ATF4, a key component of the integrated stress responses whose translation is induced by viral infection and various stresses. We have demonstrated that IRF7 upregulates ATF4 activity and expression, whereas ATF4 in return inhibits IRF7 activation, suggesting a cross regulation between the IFN response and the cellular integrated stress response that controls host innate immune defense against viral infection.
Cells react to viral infections by exhibiting innate immune responses. Central to the host innate antiviral responses is production of type I interferon (IFN), which is regulated by members of the IFN regulatory factor (IRF) family of transcription factors (1–7). Among the nine members in mammalian cells, two closely related ones, IRF3 and IRF7, have been implicated as the main regulators of type I IFN gene expression elicited by viruses (2, 4, 8–10). Although IRF3 is expressed ubiquitously and constitutively, IRF7 is expressed at low levels in most cells, but its expression is upregulated by viral infections. Despite low expression, through a positive feedback loop, IRF7 plays a dominant role in regulation of IFN induction, as evidenced by the abrogation of IFN production in most cell types of Irf7−/− but not in Irf3−/− mice (8, 9).
Host cells sense viral infection with pathogen recognition receptors such as membrane bound Toll-like receptors (TLR), cytosolic retinoic acid-inducible gene I (RIG-I) like receptors (RLR), nucleotide-binding oligomerization domain protein–like receptors, and less characterized DNA receptors DAI and AIM2 (11–13). Recognition of viral-pathogen-associated molecular patterns such as viral RNAs or DNAs by the pathogen recognition receptors triggers signaling cascades, ultimately leading to the activation of IRF3 and IRF7 that involves phosphorylation and nuclear accumulation of the two factors. Activation of ubiquitous IRF3 and the preexisting low level of IRF7 triggers initial induction of IFNβ and a subset of IFNα, followed by a positive feedback loop that allows efficient production of IFNβ and all forms of IFNα during viral infection (9, 14, 15). The secreted IFNs bind to receptors, activate the JAK-STAT pathway, and ultimately induce expression of hundreds of IFN-stimulated genes (ISGs) including PKR, RNase L, a subset of TLRs (TLR3, TLR7) and RLRs (RIG-I), and IRF7 (16). The collective effects of these ISGs allow cells to establish an antiviral state. For example, the PKR phosphorylates eIF2α, leading to global translation suppression and thus inhibition of viral replication (17, 18).
In mammalian cells, various metabolic and environmental stresses such as viral infection, perturbation of ER homeostasis (unfolded protein responses), nutrient deprivation, and reactive oxygen species induce complex cellular responses that cause phosphorylation of eukaryotic initiation factor 2α (eIF2α) (19–21). Although phosphorylation of eIF2α results in global translational suppression, it specifically increases translation of Activating Transcription Factor 4 (ATF4) through ribosomal leaky scanning of the mini open reading frames (uORFs) in the 5’-UTR of the mRNA (22–24). ATF4 belongs to the ATF/CREB (activating transcription factor/cyclic AMP response element binding protein) family of basic region–leucine zipper (bZip) transcription factors. It has been reported to function as either a transcription activator or a repressor (25). Accumulation of ATF4 induces expression of genes involved in amino-acid metabolism and transport, mitochondrial function, redox chemistry, and others that ensure supply of amino acids for protein synthesis and facilitate recovery from stress (22, 26). ATF4 is therefore thought to play a central role in cellular stress responses by initiating a feedback regulation loop to ensure the transient nature of protein synthesis inhibition (27–29).
Although the induction of type I IFN plays a key role in the control of viral infection, massive IFN production lasts only a few hours. The hosts have evolved elaborate negative regulation mechanisms to ensure that the protective response does not become excessive (30–33), but how the IFN induction process is terminated remains unclear. In the work reported here, we identified ATF4, whose expression is induced by viral infections and various stresses, as a binding partner and negative regulator of IRF7. Further studies revealed that cross regulation of the IFN response and the cellular integrated stress response mediated by IRF7 and ATF4 respectively is critical in controlling IFN induction during viral infection.
MATERIALS AND METHODS
Cells and reagents
HEK293T cells and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and antibiotics at 37°C under 5% CO2. 2ftGH, and its derivative U3A and U4A cell lines, gifts from Dr. George Stark, were cultured in DMEM supplemented with additional 1-mM sodium pyruvate. Wild-type and ATF4−/− Mouse Embryonic Fibroblasts (MEFs) were cultured similarly in DMEM supplemented with additional 55-µM β-mercaptoethanol and 1-mM nonessential amino acids instead (26). The mouse anti-Flag, anti-HA, anti-Lamin A, anti-β-actin, and rabbit anti-VSV-G antibodies were purchased from Sigma; rabbit anti-IRF7, anti-IRF3, anti-ISG15, and anti-GST antibodies were purchased from Santa Cruz Biotech; rabbit anti-PKR, anti-PERK, anti-eIF2α, and anti-phospho-eIF2α (Ser51) antibodies were purchased from Cell Signaling; mouse anti-eGFP antibody was purchased from Clontech; mouse anti-pIRF7 (Ser477/Ser479) antibody was purchased from BD Biosciences; rabbit anti-pPERK (Thr981) antibody was purchased from Biolegend; and rabbit anti-pPKR (Thr451) antibody was purchased from Biosource. Rabbit anti-ATF4 antibody was a gift from Dr. Michael Kilberg at the University of Florida. Mouse anti-ISG56 antibody was kindly provided by Dr. Ganes Sen at the Cleveland Clinic Foundation. EZview red anti-Flag M2 affinity gel beads, 3×Flag peptides, DL-homocysteine, tunicamycin, and thapsigargin were purchased from Sigma.
Plasmids
3×Flag-tagged full-length and truncation mutants were cloned by insertion of PCR-amplified fragments into pCMV-TAG3 (Stratagene). 3×HA-tagged ATF4 was cloned by PCR into pKH3 vector. GST-ATF4 constructs of amino acids 1 to 351 (full-length), 1 to 127, 1 to 90, 127 to 271, 271 to 351, and 271 to 306 were cloned by PCR into pEBG vector (provided by Dr. Yi Zhou, Florida State University). pHIV7 vector and pHIV7-GFP were kindly provided by Dr. Hengli Tang, Florida State University. pHIV7-HA-ATF4 (mouse) was generated by cloning PCR amplified fragment into pHIV7 vector (34).
Human IRF7 promoter sequence (1.7 kb) was cloned from genomic DNA into pGL3 basic vector by PCR (5’-AGCTAGTCTGGAAGTTCTTCTTC-3’; 5’-GAGCCAAGGCCATTGCTCTTC-3’). Luciferase reporter plasmids pGL3-huIFNα1, pGL3-huIFNβ, and pGL-muIFNα6 have been described previously(35); pCHOP-luc (C/EBP-LUC) was provided by Dr. Nikki Holbrook, NIA, NIH (36). pGL3-ATF4 5’UTR was kindly provided by Dr. D. Ron, New York University (23).
Yeast two-hybrid screening
A yeast two-hybrid screening was performed essentially as described previously (35). The IRF7 bait plasmid was generated by cloning of the IRF7 internal inhibitory domain (ID) aa283–466 into pAS2–1 (Clontech) in frame with GAL4 DNA-binding domain. The yeast strain Y190 carrying the plasmid pAS2–1–IRF7ID was used to screen a human lymphocyte Matchmaker cDNA library (Clontech). About one million clones were screened and plated on Leu−Trp−His− plus 25 mM 3-amino-1,2,4-triazole (3-AT) plates. The His+LacZ+ colonies were selected for sequencing and further analyses.
Immunoprecipitation (IP)
Forty-eight hours after transfection, HEK293T cells were washed with cold phosphate-buffered saline (PBS) and lysed with whole-cell lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM sodium orthovanadate [Na3VO4], 40 mM β-gylcerophosphate, 1 mM sodium fluoride, 10% glycerol, 5 mM EDTA, 5 µg/ml of aprotinin, 5 µg/ml of leupeptin, 5 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride). Cell lysates were centrifuged at 10,000 × g for 10 min at 4°C and incubated with EZview red anti-Flag M2 beads for 4 h or overnight at 4°C. After washing with lysis buffer and TBS (50 mM Tris-HCl, pH 7.4, 150 mM NaCl), proteins were eluted by incubation with 150 µg/ml 3×Flag peptide in TBS for 1 h at 4°C.
GST pull-down assay
Flag-tagged IRF7, GST-tagged ATF4 full-length, and various truncation mutants were expressed in HEK293T cells by transient transfection of the corresponding expression plasmids. GST and GST-fusion proteins were purified with glutathione sepharose beads (GE Healthcare). Equal amounts of GST and GST-tagged protein-bound glutathione beads were incubated with lysates of Flag-IRF7–expressing cells in 0.5 ml volume and rotated at 4°C for 2 h. The beads were washed twice with whole-cell lysis buffer and three times with TBS, eluted with SDS-PAGE sample buffer, and then analyzed by immunoblotting.
RNA isolation and RT-PCR
Total RNA was isolated from cells with the RNeasy Plus Mini Kit (Qiagen). First-strand cDNA was synthesized with the Cloned AMV First-Strand cDNA Synthesis Kit (Invitrogen) according to the protocols recommended by the manufacturer. The following pairs of primers were used for RT-PCR: (i) mouse IFNα (consensus primers annealing with all IFNα subtypes) sense 5’-ATGGCTAGRCTCTGTGCTTTCCT-3’, antisense 5’-AGGGCTCTCCAGAYTTCTGCTCTG-3’; (ii) mouse IFNβsense 5’-CATCAACTATAAGCAGCTCCA-3’, antisense 5’-TTCAAGTGGAGAGCAGTTGAG-3’; (iii) mouse IRF7 sense 5’-CAGCGAGTGCTGTTTGGAGAC-3’, antisense 5’-AAGTTCGTACACCTTATGCGG-3’; (iv) mouse IRF3 sense 5’-CCAGGTCTTCCAGCAGACACT-3’, antisense 5’-TAGGCTGGCTGTTGGAGATGT-3’; (v) mouse ISG56 sense 5’-ACAGCTACCACCTTTACAGC-3’, antisense 5’-TTAACGTCACAGAGGTGAGC-3’; (vi) mouse β-actin sense 5’-GGACTCCTATGTGGGTGACGAGG-3’, antisense 5’-GGGAGAGCATAGCCCTCGTAGAT-3’.
RNA interference
Hairpin-forming oligonucleotides were designed and cloned into RNAi-Ready pSIREN-Retro-Q vector (Clontech). Target sequences for IRF7 and ATF4 were siIRF7 5’-CCA AGA GCT GGT GGA ATT C-3’ and siATF4 5’-CAC TGA AGG AGA TAG GAA G-3’. Packaging of retroviruses and stable cell-line selection were performed as described previously (37).
Retrovirus
Cells stably expressing HA-ATF4 were established according to standard lentivirus expression protocols (38).
Plaque assay
Standard plaque assays were used to determine the titers of VSV as described previously (39). Briefly, HeLa or Vero cells were infected by exposure to 10-fold serially diluted VSV viruses for 1 h. The inoculum was then replaced with DMEM containing 1% methylcellulose. Twenty-four h after infection, the infected cells were fixed in 5% formaldehyde and stained with 0.1% crystal violet. All samples were assayed in duplicate, and the averages are presented.
Luciferase reporter assays
Luciferase assays were performed as previously described (35). Briefly, HEK293T cells or MEFs seeded in 24-well plates were transfected by luciferase reporter and pRL-TK internal control plasmids with Lipofectamine 2000 transfection reagent (Invitrogen). Eight h after transfection, cells were infected with 80 hemagglutinin units of Sendai viruses per well. Dual luciferase assays were performed 24 h after transfection. The relative luciferase activity was expressed as arbitrary units by normalizing firefly luciferase activity to renilla luciferase activity. Data represent the average of three independent experiments and error bars represent standard deviation. (How do you calculate the P value?)
For IRF7 promoter reporter assay, HeLa cells or MEFs in 24-well plates were transfected with a 10:1 ratio of the reporter and pRL-TK with Effectene reagents (Qiagen). Thirty-six h after transfection, dual luciferase assays were performed.
IFN ELISA
Human and murine IFNs were measured using commercial ELISA kits according to the manufacturer’s protocols (PBL Biomedical Laboratories). Briefly, 100 µl of diluted samples and the standards of known concentrations were added to each well and incubated for 1 h at RT. The wells were washed and then incubated with 100 µl of antibody solution at RT (murine IFNα for 24 h, human IFNα for 1 h). After three washes, each well was incubated with 100 µl of HRP solution at RT for 1 h. After incubation, the wells were washed three times, then incubated with 100 µl of tetramethylbenzidine substrate solution for 15 min at RT in the dark. Finally, 100 µl of stop solution were added to each well and mixed by gentle swirling. The absorbance at 450 nm was measured within 5 min. The amounts of IFNs were determined by comparison with the standard curve.
RESULTS
ATF4 binds to IRF7
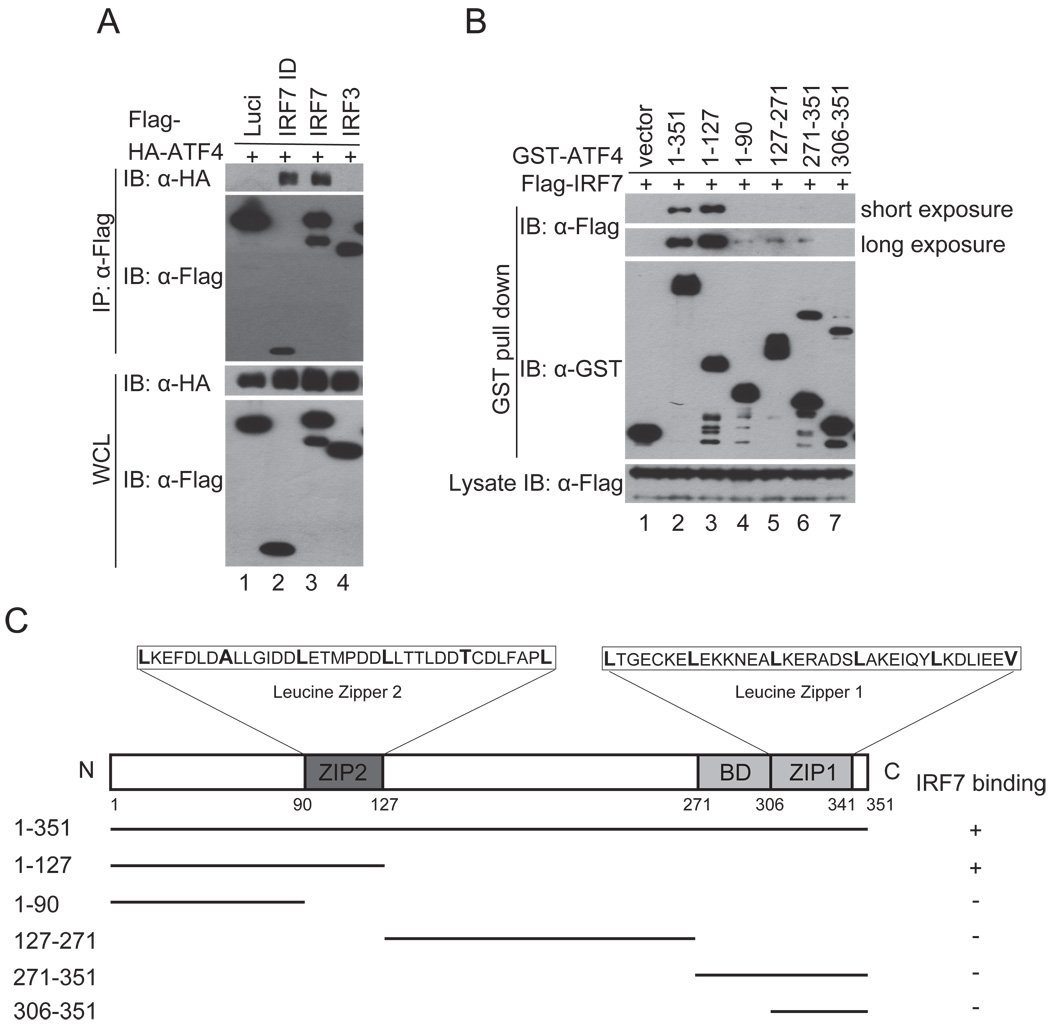
The ID of IRF7 is a binding target of several viral proteins, including KSHV ORF45, that suppress IRF7 activation (35, 40–44). This domain seems responsible for IRF7 homodimerization and heterodimerization with IRF3 and might be involved in interactions with other cellular proteins (45). To elucidate the mechanisms underlying IRF7 activation and its inactivation by KSHV ORF45, we performed a yeast two hybrid screening with the IRF7 ID domain (aa 283–466) as a bait to search for cellular proteins associated with it. The screening yielded three positive clones encoding different truncated forms of ATF4. The interaction between ATF4 and IRF7 was confirmed by coimmunoprecipitation assays. As shown in Fig. 1A, HA-ATF4 was coimmnuoprecipitated with Flag-tagged full-length IRF7 (lane 3) and the ID domain fragment (lane 2). The interaction was specific because ATF4 was not coprecipitated with luciferase (lane 1) or IRF3 (lane 4).
FIGURE 1.
Association of ATF4 with IRF7. (A) ATF4 interacts with IRF7. Flag-tagged IRF7, IRF7 ID domain, IRF3, and luciferase were cotransfected with HA-tagged ATF4 expression vectors into HEK293T cells. Lysates of the transfected cells were immunoprecipitated with anti-Flag M2 affinity beads. The IP complexes and whole cell lysates were analyzed by western blot with antibodies as indicated. (B) ATF4 interacts with IRF7 mainly through the ZIP2 domain. Flag-tagged IRF7, GST-tagged ATF4 full-length and various truncation mutants were expressed in HEK293T cells. The GST-ATF4 fusion proteins were purified with glutathione beads. After washing and blocking, the bound beads were incubated with lysates of Flag-IRF7 expressing cells. After extensive washes, the bound proteins were eluted and analyzed by immunoblotting with anti-Flag and anti-GST antibodies. (C) A schematic presentation of full-length ATF4 and its mutants. BD, basic amino acid domain.
We next mapped the regions of ATF4 that bind to IRF7 with GST pull-down assays. As shown in Fig. 1B, the N-terminal aa1–127 fragment of ATF4 bound to IRF7 as well as the full-length one does. Further truncation of the 37-aa ZIP2 domain drastically reduced the binding (compare lane 4 to lane 3). The C-terminal fragments without the ZIP2 domain bound to IRF7 weakly (aa1–90, aa127–271, and aa271–351) or did not bind at all (aa306–351). These results suggest that ATF4 interacts with IRF7 mainly through the ZIP2 region (aa90–127).
ATF4 inhibits the activation of IRF7
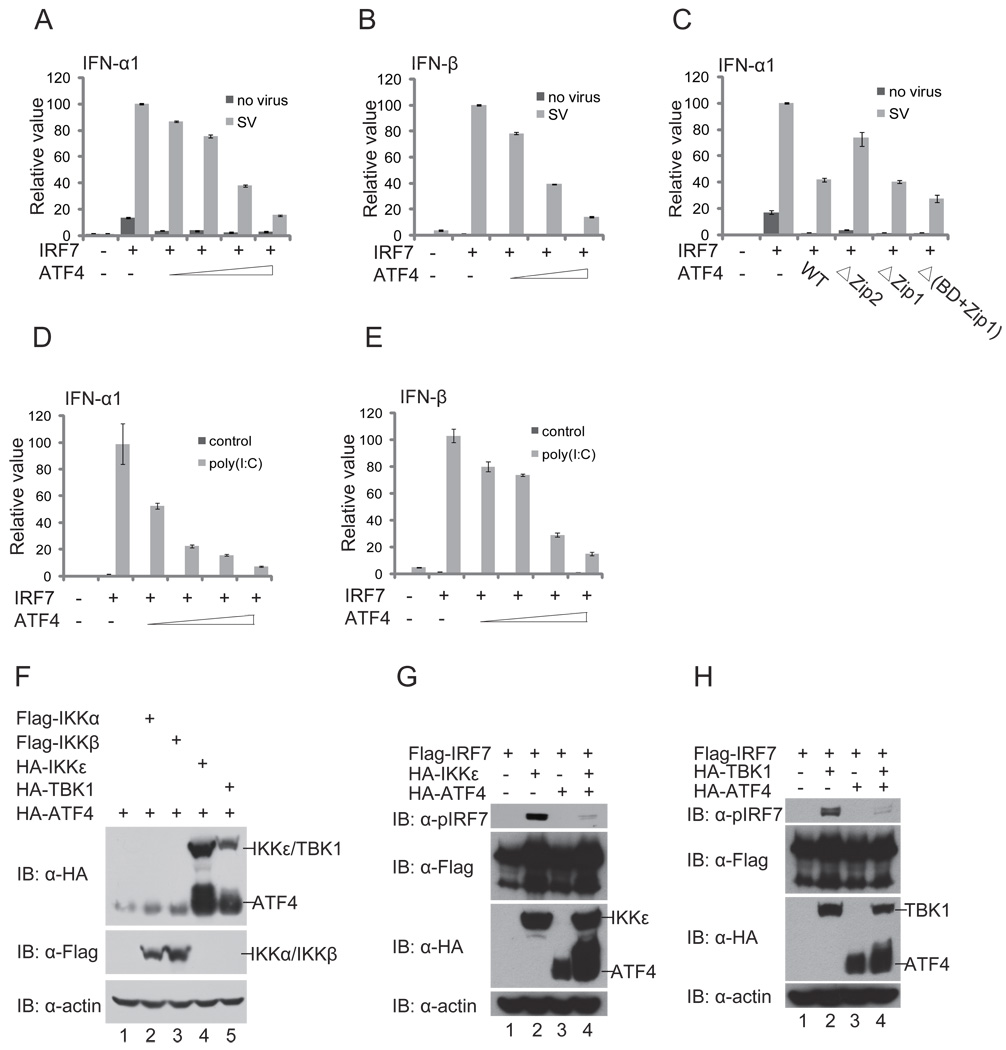
Upon viral infection, IRF7 undergoes virus-induced serine phosphorylation in its C-terminal region that stimulates protein dimerization, nuclear translocation, and cooperation with other transcriptional coactivators to induce robust expression of type I IFN genes (45). To investigate the consequence of ATF4 interaction on IRF7 function, we determined whether ATF4 affects IRF7 transactivation activity. In transient luciferase reporter assays, expression of ATF4 inhibited IRF7-induced IFNα1 and IFNβ promoter activities triggered by Sendai virus infection in a dose-dependent manner (Fig. 2A and 2B), whereas deletion of ZIP2 domain of ATF4 impaired its inhibitory activity (Fig. 2C). Moreover, ATF4 also inhibits IRF7-mediated reporter activities induced by poly(I:C) (Fig. 2D, 2E) or components in the TLR or RLR signaling pathways such as MAVS, TRIF, RIGI, TBK1, and IKKɛ (data not shown), suggesting that ATF4 inhibits IRF7 directly. These experiments demonstrate that ATF4 inhibited IRF7 activation.
FIGURE 2.
ATF4 inhibits IRF7 transactivation activity. (A) and (B) ATF4 inhibits IRF7-induced IFNα1 and IFNβ promoter activities in a dose-dependent manner. HEK293T cells were transfected with 100 ng of the IFNα1 (A) and IFNβ (B) luciferase reporter and increasing amounts (50 ng, 100 ng, 250 ng, 500 ng) of ATF4-expressing plasmids as indicated. Eight h after transfection, cells were infected with Sendai virus (SV) or left untreated as controls. Dual luciferase assay were performed at 24h after transfection. The relative luciferase activity was expressed as arbitrary units by normalizing firefly luciferase activity to renilla luciferase activity. Data represent the average of three independent experiments and error bars represent standard deviation. (C) Deletion of the Zip2 domain impairs the inhibition of IRF7 by ATF4. HEK293T cells were transfected the IFNα1 luciferase reporter and plasmids expressing wild type ATF4 or its mutants. Dual luciferase assays were performed similarly to those described above. (D) and (E) ATF4 inhibits IRF7 transactivation activities induced poly(I:C). HEK293T cells were transfected with the 100 ng of IFNα1 (D) or 100 ng of IFNβ (E) luciferase reporter, 1 µg/ml poly(I:C) and increasing amounts of ATF4-expressing plasmids (50 ng, 100 ng, 250 ng, 500 ng) as indicated. Dual luciferase assay were performed at 24 h after transfection. (F) and (G) ATF4 inhibits IRF7 phosphorylation by IKKε and TBK1. HEK293T cells were transfected with Flag-IRF7, HA-ATF4 plus HA-IKKε (F) or TBK1 (G) at the mass ratio of 10:20:1. Forty-eight h after transfection, cell lysates were analyzed by immunoblotting with anti-pIRF7 (Ser477/Ser479) phosphorylation-specific antibody and other antibodies as indicated. (H) Phosphorylation of ATF4 by IKKε/TBK1 but not IKKα/IKKβ. HEK293T cells were transfected with HA-ATF4 plus IKKα, IKKβ, IKKε, or TBK1 expression plasmids at the mass ratio of 10:1. Forty-eight h after transfection, cell lysates were analyzed by immunoblotting with antibodies as indicated.
TBK1 and IKKε, two IKK-related kinases, have been shown to phosphorylate IRF7 primarily on the residues Ser477/Ser 479 that are critical for IRF7 activation (46–48). We noticed a significant mobility shift of ATF4 on SDS-PAGE when it was coexpressed with TBK1 or IKKε, suggesting that ATF4 is phosphorylated by TBK1 and IKKε (Fig. 2F). Phosphorylation of ATF4 by TBK1 and IKKε seems to be specific because no obvious mobility shift of ATF4 was observed when IKKα or IKKβ was coexpressed (Fig. 2F). When ATF4 and IRF7 were coexpressed, ATF4 inhibited IRF7 phosphorylation of Ser477/Ser479 by TBK1 and IKKε (Fig. 2G, 2H, compare lane 4 to lane 2). Taken together, these data suggest that ATF4 negatively regulates the activation of IRF7 thus suppresses induction of IFNα an IFNβ gene expression.
Knockout of ATF4 potentiates IRF7 activation and IFN induction
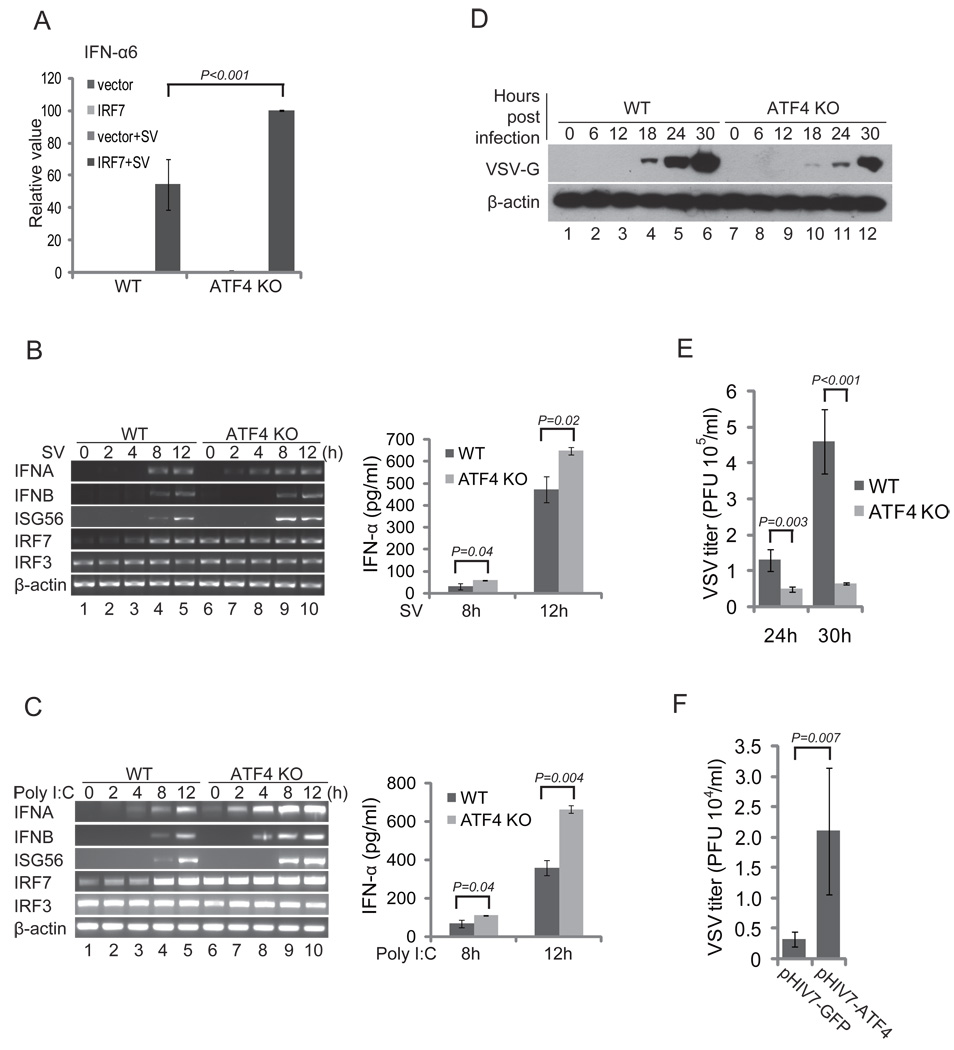
To determine whether ATF4 is involved in regulation of IRF7 under physiological conditions, we examined the effect of knockout of ATF4 on IRF7 activation and IFN induction. Transient luciferase reporter assays showed the transactivation activity of IRF7 was higher in the ATF4−/− than in wild-type MEFs, suggesting that ATF4 is a negative regulator of IRF7 (Fig. 3A). The RT-PCR assays revealed that the levels of IFNα, IFNβ, and ISG56 mRNAs induced by Sendai virus (SV) or poly(I:C) were greater in ATF4−/− than in wild-type MEFs (Fig. 3B, 3C, left panels). The differences of IFNα at protein level were confirmed by ELISA assays (Fig. 3B, 3C, right panels), indicating that ATF4 negatively regulates IRF7 activation and IFN induction. Interestingly, the basal level of IRF7 mRNA was higher in ATF4−/− than in wild-type MEFs, whereas the level of IRF3 mRNA remained the same (Fig. 3B, 3C, left panels, compare lane 6 to lane 1). These data suggest that ATF4 not only suppresses IRF7 activity but also IRF7 transcription. Therefore, lack of ATF4 leads to increase of type I IFN production.
FIGURE 3.
Knockout of ATF4 potentiates IRF7 activation and IFN production. (A) Knockout of ATF4 potentiates IRF7 transactivation activity. The wild-type and ATF4−/− MEF cells were transfected with mouse 200 ng of IFNα6 luciferase reporter plasmids. Cells were infected with SV, and dual luciferase assays were conducted as described in Fig. 1. (B) and (C) Knockout of ATF4 potentiates type I IFN induction. Wild-type and ATF4−/− MEFs were treated with Sendai virus (B) or poly(I:C) (C). Total RNAs were isolated at the times indicated, and the levels of IFNα, IFNβ, IRF7, IRF3, ISG56, and β-actin mRNA were determined by RT-PCR. Secreted IFNα in the medium collected at 8 h and 12 h were measured by ELISA. (D) and (E) Knockout of ATF4 suppresses VSV infection. Wild-type and ATF4−/− MEFs were infected with VSV at an MOI of 0.25. Cell lysates and culture medium were collected at the indicated times after infection. The lysates were analyzed by western blot for detection of VSV glycoprotein G; β-actin acted as a loading control (D). The titers in the culture medium at 24 h and 30 h after infection were determined by plaque assay (E). (F) Expression of ATF4 in MEFs potentiates VSV replication. The ATF4−/− MEFs were transduced with lentivirus vectors expressing ATF4 or GFP as a control. The transduced MEFs were infected with VSV and viral titers were determined 20 h after infection. Data in (A–C), (E), and (F) represent the average of at least three independent experiments and error bars represent standard deviation.
We next determined whether the increased type I IFN production caused by loss of ATF4 affects the susceptibility of cells to viral infection. We chose vesicular stomatitis virus (VSV) because it is sensitive to the antiviral actions of type I IFNs. We infected both wild type and ATF4−/− MEF cells with VSV and examined expression of viral proteins by western blot at various times after infection. Western blots revealed that expression level of VSV-G protein was higher in the wild-type than in ATF4−/− MEFs cells at each time point (Fig. 3D). Plaque assays revealed that loss of ATF4 reduced VSV titers by more than 10-fold (Fig. 3E). Moreover, ectopic expression of ATF4 but not GFP increased the susceptibility of the ATF4−/− MEFs to VSV infection, confirming that loss of ATF4 contributed to reduced VSV infection in ATF4−/− MEFs (Fig. 3F). Collectively, these data suggest that ATF4 is a negative regulator of IRF7 activation and IFN induction.
ATF4 inhibits transcription of IRF7
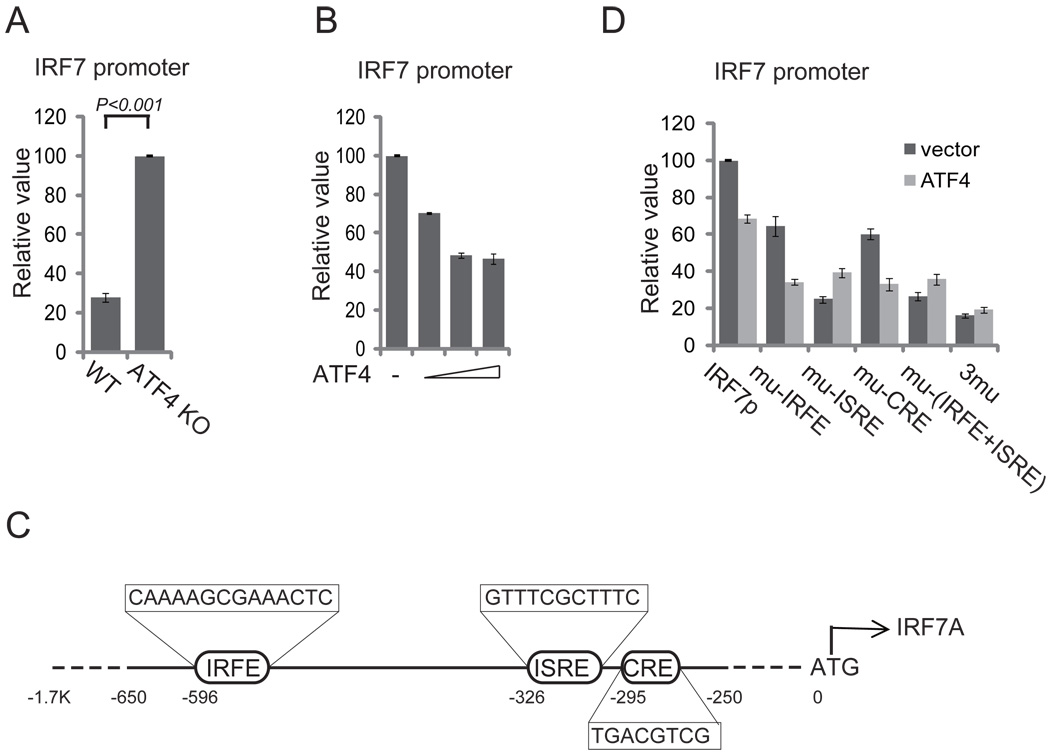
The level of IRF7 mRNA was lower in wild-type than in ATF4−/− MEFs, suggesting that ATF4 also regulates IRF7 at the level of transcription. When we cloned IRF7 promoter into pGL3 plasmid to generate a luciferase reporter and transfected it into wild type and ATF4−/− MEFs cells, IRF7 promoter activity was lower in the wild-type than in ATF4−/− MEFs cells (Fig. 4A). Moreover, overexpression of ATF4 inhibited IRF7 promoter activity in a dose-dependent manner (Fig. 4B).
FIGURE 4.
ATF4 inhibits transcription of IRF7. (A) Loss of ATF4 potentiates IRF7 promoter activity. The wild-type and ATF4−/− MEFs were transfected with 200 ng of IRF7 promoter reporter plasmid. Luciferase assays were performed 36 h after transfection. (B) ATF4 inhibits IRF7 promoter in a dose-dependent manner. HeLa cells were transfected with 200 ng of IRF7 promoter reporter and increasing amounts (50 ng, 100 ng, 250 ng) of ATF4-expressing plasmids. Luciferase assays were performed as described as above. (C) Schematic presentation of human IRF7 promoter. IRFE, ISRE, and CRE/ATF are putative regulatory elements in IRF7 promoter. (D) HeLa cells were transfected with 200 ng of IRF7 promoter reporter or the mutant constructs as depicted in (C). Luciferase assays were performed as described as above. Mutation of the ISRE (a known IRF7 binding site and positive regulatory element) but not the CRE/ATF (putative ATF4 binding site) abolishes the inhibition of IRF7 promoter by ATF4.
Several studies have shown that IRF7 positively regulates its own promoter through binding to an interferon-sensitive response elements (ISRE) and an IRF-binding element (IRFE) (49, 50). Inspection of the promoter sequence also revealed a consensus ATF/CRE element in IRF7 promoter (Fig. 4C). We wished to determine whether ATF4 regulates transcription of IRF7 through direct binding to ATF/CRE element or through interfering with IRF7 activation and thus blocking the feedback activation loop. When the wild type IRF7 promoter was used, ATF4 reduced reporter activity (Fig. 4D). As expected, mutation of IRFE and especially ISRE dramatically reduced the reporter activity, confirming that the self-regulation of its own promoter is mainly through the ISRE site (49, 50). Mutation of ATF/CRE also reduced the reporter activity, suggesting that direct binding of ATF4 to the CRE contributed little to the negative regulation of IRF7 promoter by ATF4. Negative regulation of IRF7 promoter by ATF4 was abolished only when the ISRE, the chief IRF7 positive regulation site, was mutated, suggesting ATF4 regulated IRF7 promoter mainly though interfering IRF7 activation.
IRF7 regulates ATF4 expression and activity
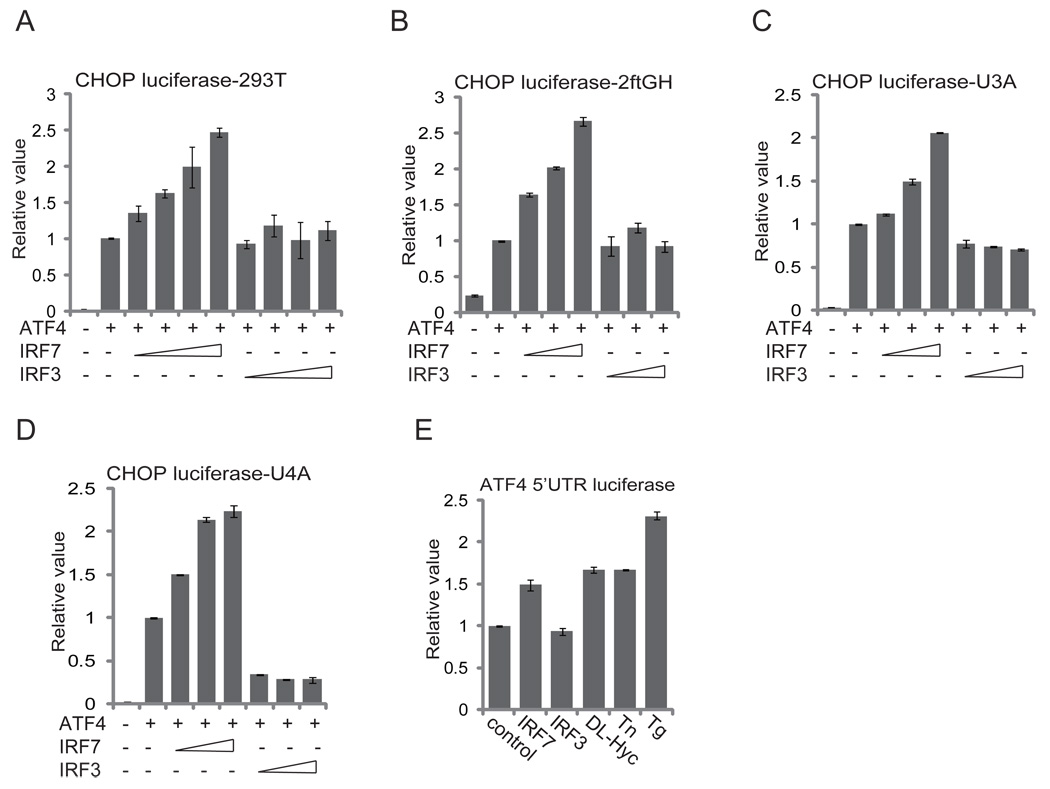
ATF4 is the key regulator of cellular responses to various stresses including viral infection and IFN signaling. Because IRF7 is induced by viral infection and triggers IFN induction, we next asked whether IRF7 affects expression and function of ATF4. In reporter assays, IRF7 but not IRF3 enhanced ATF4 transactivation activity (Fig. 5A). To determine whether IFN circuit is required for the enhancement, we repeated the assays in 2ftGH and its derivative cells in which the IFN signaling circuit is disrupted because of mutations in STAT1 (U3A) or JAK1 (U4A). IRF7 but not IRF3 increased the ATF4 activity in 2ftGH and its derivative cells, suggesting that IRF7 directly upregulates ATF4 activity in addition to the well established IFN/eIF2α-dependent mechanisms (Fig. 5B, 5C, 5D).
FIGURE 5.
IRF7 regulates ATF4 expression and activity. (A) IRF7 but not IRF3 increases ATF4 transactivation activity. HEK293T cells were transfected with 100 ng of ATF4-responsive CHOP promoter reporter and increasing amounts (50 ng, 100 ng, 250 ng, and 500 ng) of IRF7 or IRF3 expression vectors as indicated. Dual luciferase assays were performed 24 h after transfection. (B)–(D) IRF7 can increase ATF4 transactivation activity when the IFN circuit is disrupted. The above experiment was repeated in the parental 2ftGH (B) and its derivatives, U3A (STAT1) (C) and U4A (JAK1) (D) cells. (E) IRF7 upregulates translation of ATF4. MEFs were transfected with 200 ng of the ATF4 5’UTR luciferase reporter and 200 ng of IRF7 or IRF3 expression plasmids as indicated. Stress inducers DL-homocysteine (DL-Hyc), tunicamycin (Tn), and thapsigargin (Tg) were added 20 h after transfection. A dual-luciferase assay was performed 24 h after transfection. Data represent the average of at least three independent experiments and error bars represent standard deviation.
A unique feature of the 5’ UTR of ATF4 permits more efficient translation when phosphorylation of eIF2α causes global translation suppression in response to various stresses including viral infection and IFN treatment (23, 24). Expression of IRF7 increased the ATF4 5’UTR-driven luciferase reporter, whereas expression o f IRF3 did not (Fig. 5E). The level of increase caused by IRF7 was significant and comparable to that caused by treatments with various stress-inducing agents such as DL-homocysteine, tunicamycin, and thapsigargin. These results suggest that IRF7 regulates ATF4 expression and function.
Cross regulation between IFN responses and integrated stress responses
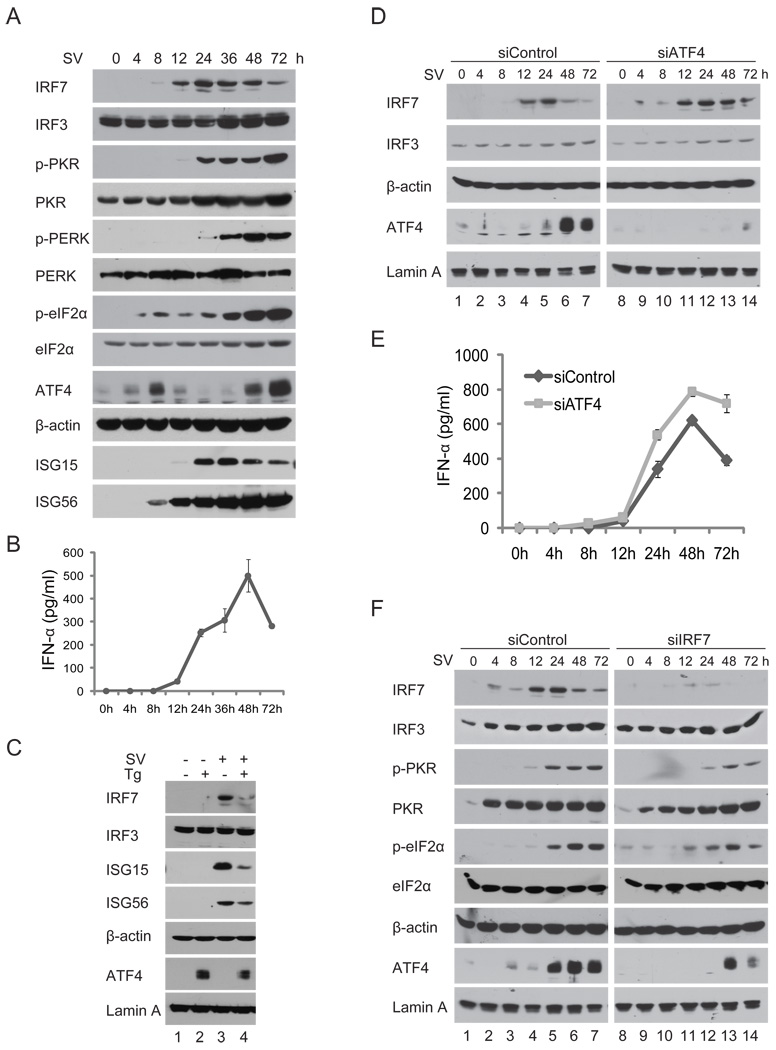
Viral infection induces IFNs and ISGs including IRF7 as well as integrated stress responses that result in phosphorylation of eIF2α and subsequent global translation suppression but increase of ATF4 translation. To seek the interrelationship between IFN and integrated stress responses, we wished to determine the expression kinetics of key components in both pathways during virus infection. Because IRF7 in MEFs is hardly detectable by western blot, we screened a number of cell lines and found A549 cells, a human lung adenocarcinoma epithelial cell line, effectively expressed both IRF7 and ATF4. As shown in Fig. 6A, IRF7 was induced by Sendai virus infection and became detectable 8 h after infection. Its expression increased over time, peaked at 24 h, then decreased. In contrast, IRF3 was expressed constitutively as expected. The kinetics of ISG15 and ISG56 expression were largely correlated with that of IFNα (Fig. 6B). Phosphorylation of eIF2α first appeared 4 h after infection and increased slightly until 8 h after infection. It remained at a low level until 24 h after infection and then increased significantly 36 h after infection and thereafter. The late increase of eIF2α phosphorylation was coincident with phosphorylation of both PKR and PERK, suggesting both these two kinase are activated during Sendai virus infection. Interestingly, ATF4 appeared to peak twice. The first peak occurred at 8 h after infection, and a late dramatic increase occurred at 48 h and 72 h after infection, when IRF7 expression began to decrease (Fig. 6A). These results confirm a reverse correlation between ATF4 and IRF7 supporting a negative regulation of IRF7 and IFN induction by ATF4.
FIGURE 6.
A cross regulation between IFN and the cellular integrated stress responses. (A) Expression kinetics of the major components of IFN and stress responses during Sendai virus infection. A549 cells were infected with Sendai virus in triplicate (160 HA units/ml). At the indicated times after infection, whole cell lysates or nuclear extract (for detection of ATF4) were prepared and analyzed by immunoblotting with indicated antibodies. Parallel samples were used to prepare RNA for RT-PCR analysis for detection of type I IFNs. (B) Activation of the integrated stress response impairs IRF7-induced expression of IFNs. A549 cells were infected with Sendai virus for 4 h and then treated with thapsigargin for another 4 h. Whole-cell lysates and nuclear extracts were prepared and subjected to immunoblotting with the indicated antibodies. (C) Knockdown of ATF4 by siRNA potentiates expression of IRF7 and IFNα. A549 cells stably transduced with siATF4 or siControl were infected with Sendai virus for time as indicated. Whole-cells lysates and nuclear extract were analyzed by immunoblotting with specified antibodies. IFNs in the culture medium were measure by ELISA. (F) Knockdown of IRF7 by siRNA reduces virus-induced cellular integrated stress responses. A549 cells stably transduced by siIRF7 or siControl were infected with Sendai virus. Whole-cell lysates and nuclear extract were analyzed by immunoblotting with the indicated antibodies.
We next determined whether artificially induction of integrated cellular stress responses affects IRF7 activation and IFN expression. We first infected A549 cells with Sendai virus and then treated the cells with stress inducing agents for 4 h and examined expression of IRF7 and ISGs by immunoblotting. As shown in Fig. 6C, immunoblotting revealed that ER stress inducer thapsigargin reduced expression of IRF7, ISG15, and ISG56 that is correlated with increased expression of ATF4 in A549 cells (Fig. 6C). Curiously, thapsigargin triggered a lower level of ATF4 induction in Sendai virus–infected cells than in mock-infected cells (Fig. 6C, compare lane 4 to lane 2); a similar observation was recently reported elsewhere (51). These experiments suggest that integrated stresses negatively regulate IFN induction.
To investigate the interrelationship between stress and IFN responses further, we knocked down expression of the key components in A549 cells by lentivirus vector-mediated siRNAs. Consistent with previous results in ATF4−/− MEFs, knockdown of ATF4 in A549 cells potentiated expression of IRF7 and increased duration of its expression (Fig. 6D, compare lanes 11–14 to lanes 4–7). Consequently expression of IFNα was increased by knockdown of ATF4 (Fig. 6E). Furthermore, knockdown of IRF7 decreased and delayed phosphorylation of PKR, reduced the level of eIF2α phosphorylation, and resulted in delayed and lower expression of ATF4 (Fig. 6F). These data confirmed the role of ATF4 in regulating IRF7 and IFN expression, and suggested that IRF7 and the IFN-PKR-eIF2α signaling cascades effectively regulate ATF4 expression (Fig. 6F). Taken together, these results suggest that IRF7 and ATF4 link the IFN-based innate immune response to the integrated stress response.
DISCUSSION
ATF4 interacts with and negatively regulates IRF7
We have identified ATF4 as a binding partner and negative regulator of IRF7. We demonstrated that overexpression of ATF4 inhibits IRF7 activation, whereas knockout of ATF4 potentiates IRF7 transactivation activity, increases IFN production, and suppresses VSV replication. ATF4 interacts specifically with IRF7 but not IRF3, mainly through the second untypical leucine zipper domain (ZIP2). The ZIP2-dependent interaction is specific to IRF7 because ATF4 interacts with most other proteins through the typical ZIP1 domain, such as zhangfei (52), mitosin/CENP-F (53), GABA(B) receptor (54), RNA polymerase 2 subunit RPB3 (55), ZIP kinase (56), and HTLV1 transactivator Tax (57). This specific interaction is required for ATF4 to inhibit IRF7 transactivation activity because deletion of ZIP2 domain impairs the inhibition. Although the detailed inhibitory mechanisms remain unclear, we found that ATF4 inhibits phosphorylation of IRF7 by TBK1 and IKKε. In addition, ATF4 inhibits transcription of IRF7 by interfering with IRF7 activation and thus disrupting the positive feedback loop. Because ATF4 down- and upregulates expression of many cellular genes, other inhibitory mechanisms may also be involved. For example, ATF4 is known to upregulate expression of 4E–BPs, negative regulators of cap-dependent translation (58). Interestingly, knockout of 4E–BPs causes a significant upregulation of IRF7 (59), indicating that 4E–BPs inhibit IRF7 translation specifically. Together, these observations suggest that ATF4 also downregulates IRF7 translation through induction of 4E–BP. Therefore, ATF4 inhibits IRF7 activation through direct protein-protein interaction and also through indirect suppression of IRF7 transcription and translation.
Negative regulation of IFN responses
Although the innate immune response is an indispensible defense against invasion by pathogens, the host must limit it, because its excessive and prolonged activation would be harmful or even fatal to the host (60). IFN induction is negatively regulated by a number of factors such as A20 (30), NLRX1 (31), SIKE (61), and Pin1 (62) that target various components in the in RLR and TLR-induced antiviral signaling. IRF activation is also negatively regulated by postactivation attenuation mechanisms. For example, IRF3 is also subject to ubiquitination-dependent proteasomal degradation (63), whereas both IRF3 and IRF7 are silenced by SUMOylation (44, 64). Furthermore, certain ISGs such as ISG56, originally thought to be involved in establishment of antiviral state, actually downregulate host antiviral responses, presumably as a mechanism for termination of IFN response (65).
Cross regulation of IFN and stress responses
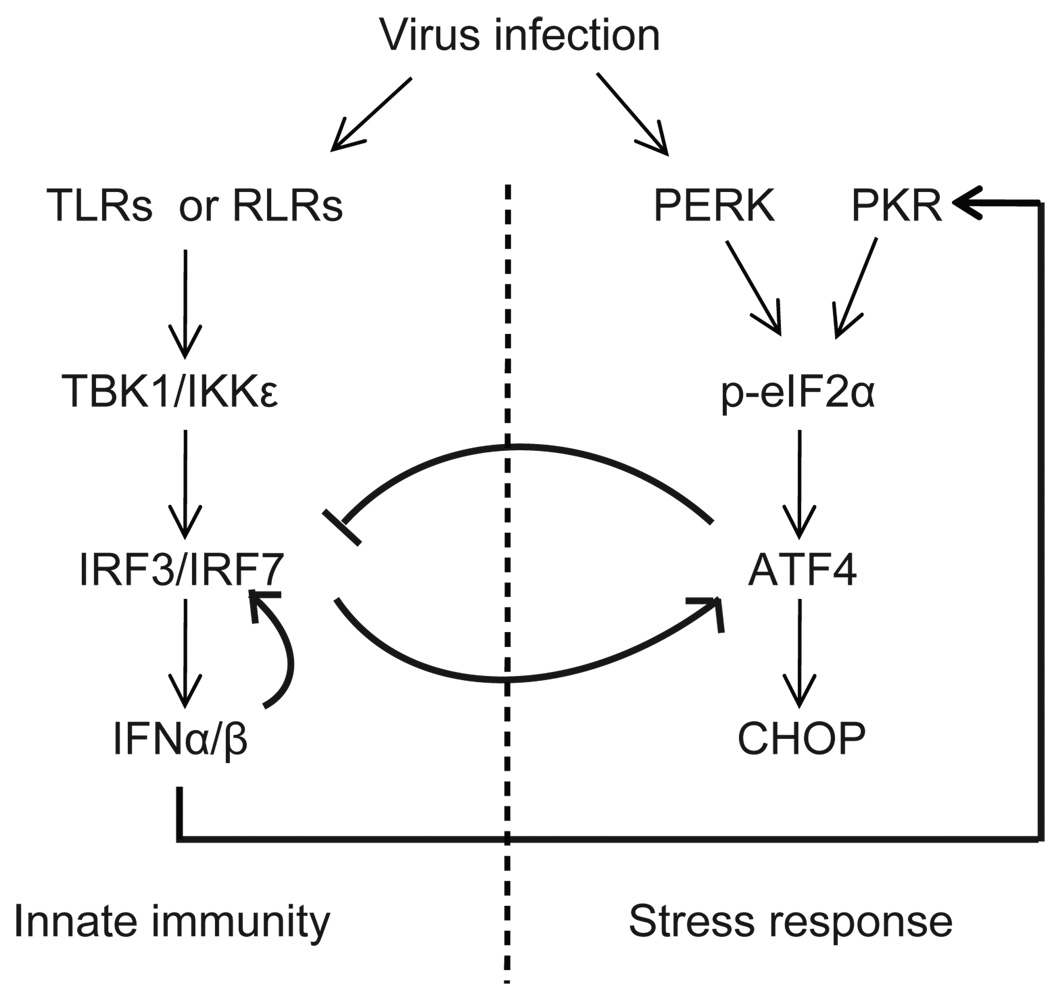
During the course of viral infection, translation of ATF4 is augmented as a result of phosphorylation of eIF2α by a group of kinases including PKR, PERK, and GCN2. These kinases are activated by dsRNA, ER stress, and amino-acid deprivation, respectively, each of which often occurs during a viral infection (66, 67). Accumulation of ATF4 not only induces genes facilitating cellular recovery but also suppresses IRF7 activation, disrupts the IFN/IRF7 positive feedback loop, and thus subsequently terminates the IFN circuit. We have demonstrated that further increase of ATF4 expression by stress-inducing agents during viral infection reduces IFN induction and IRF7 activation, that reduction of ATF4 expression by siRNA results in higher levels and longer duration of IFN induction, but that ablation of IRF7 expression reduces the level of eIF2α phosphorylation and ATF4 expression. Together these data suggest a linkage between IFN and stress responses through cross regulation by IRF7 and ATF4, the two critical regulators of these two pathways. On the basis of these data, we propose the model outlined in Fig. 7. Viral infection induces IFN expression and also activates multiple eIF2α kinases, mainly PKR, PERK, and other possible kinases. Phosphorylation of eIF2α and activation of IRF7 itself increase translation of ATF4. The increased expression level of ATF4 protein induces stress-response genes to help the cell recover but inhibits the expression and transactivation of IRF7 to terminate IFN signaling. As a result, this negative feedback loop enables host cells to terminate the IFN production and response effectively.
Figure 7.
Schematic diagram of cross regulation between IFN and integrated stress responses. Viral infection induces type I IFN expression and also activates multiple eIF2α kinases, mainly PKR and PERK. Phosphorylation of eIF2α and activation of IRF7 itself increases the translation of ATF4. The increased expression level of ATF4 protein induces stress-response genes to help cell recovery but inhibits the expression and transactivation of IRF7 to terminate IFN signaling. As a result, this negative feedback loop allows host cells to terminate the IFN responses effectively.
The ATF4/IRF7-mediated cross regulation is supported by a recent report showing that increased oxidative stress caused by aging impairs IRF7 activity, whereas reduction of the stress by antioxidant agents increases it and IFN responses (68). Because of the complex natures of the two pathways, the interrelationships between the IFN-based innate immune and integrated stress responses may not be limited to ATF4 and IRF7. For example, ATF3, a downstream target gene of ATF4 in response to ER stresses, has been identified as a negative regulator of TLR signaling (69, 70); activated PERK induced by ER stresses during viral infection promotes phosphorylation-dependent ubiquitination and degradation of IFNAR1 and thus attenuates type I IFN signaling and antiviral defenses (71).
Role of ATF4 in viral replication
Our studies revealed a novel role of ATF4 in negative regulation of IFN-based innate immune responses and thus as a potential cellular factor for better viral replications. Reovirus has been shown to induce and to benefit from an integrated cellular stress response and to require ATF4 for it replication, although the role of innate immunity was not examined (72). Influenza virus has been shown to replicate less efficiently in eIF2αS51A MEFs, in which ATF4 expression cannot be induced (73). HCMV is known to activate and modulate UPR and consequently to induce ATF4 expression (74, 75). We also found that KSHV ORF45 increases inhibition of IRF7 by ATF4 (unpublished data). These observations suggest that the cross regulation of IFN and integrated stress responses by IRF7 and ATF4 is modulated by viruses as a strategy to defeat the IFN antiviral response.
In summary, we demonstrated, for the first time, the physical and functional interactions between IRF7 and ATF4 and revealed a novel mechanism of cross regulation between the interferon and the cellular integrated stress responses.
ACKNOWLEDGMENTS
We are grateful to Nikki Holbrook, Michael Kilberg, David Levy, Rongtuan Lin, David Ron, Ganes Sen, George Stark, Hengli Tang, and Yi Zhou for kindly providing reagents. We thank members of the Zhu laboratory for reading the manuscript and for helpful discussions. We also thank Anne B. Thistle at the Florida State University for excellent editorial assistance.
This work was supported by National Institutes of Health grant R01DE016680, an FSU set-up fund, and a planning grant to F.Z.
Footnotes
DISCLOSURES
The authors have no financial conflicts of interest.
REFERENCE
- 1.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 3.Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89:744–753. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 5.Honda K, Yanai H, Takaoka A, Taniguchi T. Regulation of the type I IFN induction: a current view. Int Immunol. 2005;17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- 6.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 7.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 9.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 10.Ozato K, Tailor P, Kubota T. The interferon regulatory factor family in host defense: mechanism of action. J Biol Chem. 2007;282:20065–20069. doi: 10.1074/jbc.R700003200. [DOI] [PubMed] [Google Scholar]
- 11.Huang X, Yang Y. Innate immune recognition of viruses and viral vectors. Hum Gene Ther. 2009;20:293–301. doi: 10.1089/hum.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 13.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 15.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 17.Williams BR. Signal integration via PKR. Sci STKE. 2001:RE2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]
- 18.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Molecular Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 20.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Molecular Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 21.Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol. 2002;22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 23.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 26.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 27.Fels DR, Koumenis C. The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther. 2006;5:723–728. doi: 10.4161/cbt.5.7.2967. [DOI] [PubMed] [Google Scholar]
- 28.Ye J, Koumenis C. ATF4, an ER stress and hypoxia-inducible transcription factor and its potential role in hypoxia tolerance and tumorigenesis. Curr Mol Med. 2009;9:411–416. doi: 10.2174/156652409788167096. [DOI] [PubMed] [Google Scholar]
- 29.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, Hiscott J. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 31.Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, Lich JD, Heise MT, Chen Z, Ting JP. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 32.Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr, Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 34.Yam PY, Li S, Wu J, Hu J, Zaia JA, Yee JK. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol Ther. 2002;5:479–484. doi: 10.1006/mthe.2002.0558. [DOI] [PubMed] [Google Scholar]
- 35.Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc Natl Acad Sci USA. 2002;99:5573–5578. doi: 10.1073/pnas.082420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem J. 1999;339(Pt 1):135–141. [PMC free article] [PubMed] [Google Scholar]
- 37.Kuang E, Tang Q, Maul GG, Zhu F. Activation of p90 ribosomal S6 kinase by ORF45 of Kaposi's sarcoma-associated herpesvirus and its role in viral lytic replication. J Virol. 2008;82:1838–1850. doi: 10.1128/JVI.02119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 39.Zhu FX, Sathish N, Yuan Y. Antagonism of host antiviral responses by Kaposi's sarcoma-associated herpesvirus tegument protein ORF45. PLoS ONE. 2010;5:e10573. doi: 10.1371/journal.pone.0010573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Y, Wang SE, Hayward GS. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity. 2005;22:59–70. doi: 10.1016/j.immuni.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Buettner NVC, Martínez-Sobrido L, Weber F, Waibler Z, Kochs G. Thogoto virus ML protein is a potent inhibitor of the interferon regulatory factor-7 transcription factor. J Gen Virol. 2010;91:220–227. doi: 10.1099/vir.0.015172-0. [DOI] [PubMed] [Google Scholar]
- 42.Wu L, Fossum E, Joo CH, Inn KS, Shin YC, Johannsen E, Hutt-Fletcher LM, Hass J, Jung JU. Epstein-Barr virus LF2: an antagonist to type I interferon. J Virol. 2009;83:1140–1146. doi: 10.1128/JVI.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joo CH, Shin YC, Gack M, Wu L, Levy D, Jung JU. Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi's sarcoma-associated herpesvirus viral IRF homolog vIRF3. J Virol. 2007;81:8282–8292. doi: 10.1128/JVI.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang TH, Kubota T, Matsuoka M, Jones S, Bradfute SB, Bray M, Ozato K. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin R, Mamane Y, Hiscott J. Multiple regulatory domains control IRF-7 activity in response to virus infection. J Biol Chem. 2000;275:34320–34327. doi: 10.1074/jbc.M002814200. [DOI] [PubMed] [Google Scholar]
- 46.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 47.Paz S, Sun Q, Nakhaei P, Romieu-Mourez R, Goubau D, Julkunen I, Lin R, Hiscott J. Induction of IRF-3 and IRF-7 phosphorylation following activation of the RIG-I pathway. Cell Mol Biol (Noisy-le-grand) 2006;52:17–28. [PubMed] [Google Scholar]
- 48.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 49.Lu R, Au WC, Yeow WS, Hageman N, Pitha PM. Regulation of the promoter activity of interferon regulatory factor-7 gene. Activation by interferon and silencing by hypermethylation. J Biol Chem. 2000;275:31805–31812. doi: 10.1074/jbc.M005288200. [DOI] [PubMed] [Google Scholar]
- 50.Ning S, Huye LE, Pagano JS. Regulation of the transcriptional activity of the IRF7 promoter by a pathway independent of interferon signaling. J Biol Chem. 2005;280:12262–12270. doi: 10.1074/jbc.M404260200. [DOI] [PubMed] [Google Scholar]
- 51.Woo CW, Cui D, Arellano J, Dorweiler B, Harding H, Fitzgerald KA, Ron D, Tabas I. Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat Cell Biol. 2009;11:1473–1480. doi: 10.1038/ncb1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hogan MR, Cockram GP, Lu R. Cooperative interaction of Zhangfei and ATF4 in transactivation of the cyclic AMP response element. FEBS Lett. 2006;580:58–62. doi: 10.1016/j.febslet.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 53.Zhou X, Wang R, Fan L, Li Y, Ma L, Yang Z, Yu W, Jing N, Zhu X. Mitosin/CENP-F as a negative regulator of activating transcription factor-4. J Biol Chem. 2005;280:13973–13977. doi: 10.1074/jbc.M414310200. [DOI] [PubMed] [Google Scholar]
- 54.White JH, McIllhinney RA, Wise A, Ciruela F, Chan WY, Emson PC, Billinton A, Marshall FH. The GABAB receptor interacts directly with the related transcription factors CREB2 and ATFx. Proc Natl Acad Sci USA. 2000;97:13967–13972. doi: 10.1073/pnas.240452197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Angelis R, Iezzi S, Bruno T, Corbi N, Di Padova M, Floridi A, Fanciulli M, Passananti C. Functional interaction of the subunit 3 of RNA polymerase II (RPB3) with transcription factor-4 (ATF4) FEBS Lett. 2003;547:15–19. doi: 10.1016/s0014-5793(03)00659-8. [DOI] [PubMed] [Google Scholar]
- 56.Kawai T, Matsumoto M, Takeda K, Sanjo H, Akira S. ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol Cell Biol. 1998;18:1642–1651. doi: 10.1128/mcb.18.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reddy TR, Tang H, Li X, Wong-Staal F. Functional interaction of the HTLV-1 transactivator Tax with activating transcription factor-4 (ATF4) Oncogene. 1997;14:2785–2792. doi: 10.1038/sj.onc.1201119. [DOI] [PubMed] [Google Scholar]
- 58.Yamaguchi S, Ishihara H, Yamada T, Tamura A, Usui M, Tominaga R, Munakata Y, Satake C, Katagiri H, Tashiro F, Aburatani H, Tsukiyama-Kohara K, Miyazaki J, Sonenberg N, Oka Y. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab. 2008;7:269–276. doi: 10.1016/j.cmet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 59.Colina R, Costa-Mattioli M, Dowling RJ, Jaramillo M, Tai LH, Breitbach CJ, Martineau Y, Larsson O, Rong L, Svitkin YV, Makrigiannis AP, Bell JC, Sonenberg N. Translational control of the innate immune response through IRF-7. Nature. 2008;452:323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- 60.Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang J, Liu T, Xu LG, Chen D, Zhai Z, Shu HB. SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. EMBO J. 2005;24:4018–4028. doi: 10.1038/sj.emboj.7600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, Akira S, Yamamoto N, Lu KP, Yamaoka S. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 63.Bibeau-Poirier A, Gravel SP, Clement JF, Rolland S, Rodier G, Coulombe P, Hiscott J, Grandvaux N, Meloche S, Servant MJ. Involvement of the IkappaB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J Immunol. 2006;177:5059–5067. doi: 10.4049/jimmunol.177.8.5059. [DOI] [PubMed] [Google Scholar]
- 64.Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, Kato A, Ozato K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem. 2008;283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y, Li C, Xue P, Zhong B, Mao AP, Ran Y, Chen H, Wang YY, Yang F, Shu HB. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc Natl Acad Sci USA. 2009;106:7945–7950. doi: 10.1073/pnas.0900818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berlanga JJ, Ventoso I, Harding HP, Deng J, Ron D, Sonenberg N, Carrasco L, de Haro C. Antiviral effect of the mammalian translation initiation factor 2alpha kinase GCN2 against RNA viruses. EMBO J. 2006;25:1730–1740. doi: 10.1038/sj.emboj.7601073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 68.Stout-Delgado HW, Yang X, Walker WE, Tesar BM, Goldstein DR. Aging impairs IFN regulatory factor 7 up-regulation in plasmacytoid dendritic cells during TLR9 activation. J Immunol. 2008;181:6747–6756. doi: 10.4049/jimmunol.181.10.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, Nykter M, Shmulevich I, Aderem A. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 71.Liu J, HuangFu WC, Kumar KG, Qian J, Casey JP, Hamanaka RB, Grigoriadou C, Aldabe R, Diehl JA, Fuchs SY. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe. 2009;5:72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA. Reovirus induces and benefits from an integrated cellular stress response. J Virol. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci USA. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xuan B, Qian Z, Torigoi E, Yu D. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J Virol. 2009;83:3463–3474. doi: 10.1128/JVI.02307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Isler JA, Skalet AH, Alwine JC. Human cytomegalovirus infection activates and regulates the unfolded protein response. J Virol. 2005;79:6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]