Abstract
Alveolar macrophages are essential for clearing bacteria from the alveolar surface and preventing microbial-induced infections. It is well documented that smokers have an increased incidence of infections, in particular lung infections. Alveolar macrophages accumulate in smokers’ lungs but they have a functional immune deficit. In this study, we identify for the first time an autophagy defect in smokers’ alveolar macrophages. Smokers’ alveolar macrophages accumulate both autophagosomes and p62, a marker of autophagic flux. The decrease in the process of autophagy leads to impaired protein aggregate clearance, dysfunctional mitochondria and defective delivery of bacteria to lysosomes. This study identifies the autophagy pathway as a potential target for interventions designed to decrease infection rates in smokers and possibly in individuals with high environmental particulate exposure.
Introduction
Almost all smokers have an accumulation of macrophages in the terminal airways of the lung. This has been clearly described in bronchoalveolar fluid and on histological lung samples from smokers (1). Functional abnormalities of these cells, including a defect in killing of ingested bacteria, have been described (2, 3). Smokers’ alveolar macrophages are characterized by the presence of many vacuoles containing pigmented cargo. The types of vesicles present in alveolar macrophages from smokers have not been identified, nor is it known how they accumulate or the mechanism behind the defect in bacterial clearance.
Autophagy is an evolutionarily conserved process that captures and recycles protein aggregates, portions of the cytosol and cytoplasmic organelles (4). Autophagy dysfunction has been linked to a wide variety of diseases, including neurodegenerative diseases, cancer, and heart disease (5, 6). Changes in autophagy affect the removal of aged and defective mitochondria, protein aggregates and internalized pathogens via the lysosome. Phagocytosis and delivery of pathogens to the lysosome have both been shown to involve the autophagy process in recent publications (7). This study investigates autophagy changes in smokers’ alveolar macrophages and its functional significance.
The only studies to evaluate autophagy in smoker’s lungs come from Augustine Choi’s group. They have found increased autophagosomes in tissue from patients with COPD and in lung epithelial cells exposed to cigarette smoke extract (8, 9). The role of autophagy in smoking-induced immune dysfunction and alveolar macrophage phenotype has never been examined. Alveolar macrophages are essential for maintaining the sterility of the lung as they clear bacteria from the alveolar surface. The inability to clear bacteria increases susceptibility to pneumonia, and it is well documented that smokers have an increased incidence of pneumonia (10, 11). This study demonstrates significant accumulation of autophagosomes in smokers’ alveolar macrophages. Despite the increase in autophagosomes in alveolar macrophages of smokers, a decrease in functional autophagy is present. This autophagy defect is defined by the accumulation of protein aggregates, damaged mitochondria and decreased lysosome delivery of bacteria.
Materials and Methods
Materials
Carbon black 50 nM nanoparticles were obtained from Nanostructured & Amorphous materials Inc (#1211ER). Bafilomycin A from Streptomyces griseus was from Calbiochem (#196000). JC-1 was obtained from Guava Technologies. Antibodies used in this study were obtained from a variety of sources. Sumo 2/3 antibody was obtained from Abcam (ab3742). Phosphorylated AMPKα1 (T172, #2531), ubiquitin (#3936) and LC3b (#2775) antibodies were obtained from Cell Signaling. Horseradish peroxidase conjugated antibodies anti-rabbit (sc-2004), anti-mouse (sc-2005) and anti-goat (sc-2020) were all obtained from Santa Cruz Biotechnology. Beta actin (#A5316) antibody was obtained from Sigma Aldrich. Culture media used in experiments included serum free RPMI tissue culture media with Glutamax (Invitrogen, 61870-036) and DMEM (Invitrogen, 11095) with 10% FCS (Hyclone Fetal Bovine Serum, SH30071).
Plasmids
A plasmid expressing LC3-GFP was obtained from Addgene. The polyQ80-luciferase vector was obtained from Conrad C. Weihl, Washington University(13). The mCherry-GFP-p62 vector was obtained from Johansen Terje, University of Tromso, Norway.
Isolation of human alveolar macrophages and tissue culture cells
Alveolar macrophages were obtained from normal nonsmoking volunteers or from smokers (> 10 pack year history (a pack year is defined as 1 pack per day for one year or ~1300 cigarettes)), as previously described(14). Informed consent was obtained from each volunteer following a detailed explanation of the nature and possible consequences of the procedure. Both nonsmokers and smokers were free of any underlying disease, aside from the smokers’ smoking histories. Alveolar macrophages were cultured in RPMI 1640 media with added gentamycin. In some transfection studies, Raw 246.7 mouse macrophages (ATCC, TIB-71) were used and cultured following ATCC recommendations (Eagle’s Minimum Essential Medium with 10% fetal bovine serum).
Preparation of cigarette smoke extract (CSE)
Cigarette smoke extract was prepared as previously described(15, 16). CSE solutions were prepared using a modification of the method of Blue and Janoff (17). 10 mls of sterile serum free RPMI tissue culture media with Glutamax was drawn into a 50-ml plastic syringe. 40 ml of cigarette smoke was then drawn into the syringe and mixed with the medium by vigorously shaking them together. The smoke was expelled and the process repeated until one cigarette was used up. The generated CSE solution was filtered (0.22μm) to remove large particles (18, 19). The resulting solution was designated a 100% CSE solution and was used immediately after generation.
Protein isolation and Western analysis
Protein isolation and Western analysis were performed as previously described (20, 21).
Transfections
Primary human alveolar macrophages were transfected with LC3-GFP vector and poly Q80-luciferase vector using Invitrogen’s Neon. Briefly, 0.5 million cells were suspended in 250 ul Resuspension Buffer “T” (Invitrogen). 0.5 to 1 ug per group plasmid was added to the cell suspension before transfer to a Neon tube. Using the 100 ul Neon tip and the following parameters, 1900V, 25W, 1 pulse, the cells were electroporated. Following transfer to tissue culture wells with warmed antibiotic free medium, the cells were incubated for four hours. After replacing the serum/antibiotic free medium with complete medium, cells were incubated overnight before CSE addition. Raw 264.7 cells were transfected with polyQ80-luciferase vectors using Lipofectamine LTX and Plus Reagent (15338-100) according to the manufacturer’s instructions. Cells were cultured in 12 well plates or in two chamber coverslip slides. Plasmid (0.5 to 1 ug per condition) was suspended in 200 ul OPTIMEM with Plus reagent, mixed with the LTX reagent and allowed to incubate at room temperature for 30 minutes. After a 4 hour incubation of the cells with plasmid, 500 ul of 2× serum medium was added to the cells before overnight incubation. CSE addition or other culture manipulations were performed the next day (4-6 hours). Hela cells were transfected with the mCherry-GFP-p62 vector as described for Raw 264.7 cells.
Autophagosome sizing
To quantitate autophagosome size in alveolar macrophages from nonsmokers and smokers, TEM images were analyzed for vesicle size. TEM images were obtained through the middles of cells (verified by nuclear structure. Double-walled vesicles from each individual (3 nonsmokers and 3 smokers) were analyzed using Image J software. Briefly, circles were drawn around all double walled vesicles in a cell, identical magnifications were used for all imaging. Equal vesicle numbers were analyzed for both nonsmokers and smokers. This meant scanning more cells in the nonsmokers as smokers, as on average they contained less autophagosomes. Area was calculated for each vesicle and then standard t test analysis performed.
Long Lived Protein Degradation Assay
The effect of CSE on degradation rates of long-lived proteins was evaluated in human alveolar macrophages. Macrophages were cultured in 6 well tissue culture plates in leucine free RPMI with added 14C leucine (0.2 uCi/ml) for 8 hours. Following two PBS washes the media was changed to a chase medium with added 10 mM leucine to clear short lived proteins from the radioactive pool. Cells were incubated for 10 hours. The chase medium was replaced with fresh normal culture media (RPMI 1640 with added gentamycin) and cells exposed to either Bafilomycin A (positive control, 100 nM) or 2% CSE for 8 hours. After this third incubation, supernatants were removed and saved and cells were lysed in Western lysis buffer. Proteins were fractionated by adding 10% trichloroacetic acid (TCA, w/v) to both supernatant and lysate (1 hour 4° incubation). Precipitated proteins were isolated with a 10 minute 14,000 rpm spin. The radioactivity of the TCA soluble fraction of the supernatant (a) and TCA insoluble fraction of the cells (b) were determined via liquid scintillation counting. The percentage of long-lived proteins degraded in the culture period was determined using the formula a/a+b × 100. Data was analyzed using nonpaired Student’s t test.
ATP assay
ATP levels were monitored using CellTiter-Glo Luminescent Cell Viability Assay from Promega according to manufacturer’s instructions.
Fluorescent Microscopy
LC3-GFP and mCherry-GFP-p62 imaging was performed. At the end of the experimental period, cells were rapidly fixed (4% paraformaldehyde) and cover slipped using Vectashield mounting medium with DAPI (Vector Labs, H-1200). Images were acquired using a Leica TCS SPE confocal (truepoint-scanning spectral system) microscope with Leica LAS AF interface. Staining of endogenous LC3 and Lamp2 positive lysosomes was performed on nonsmoker and smoker alveolar macrophages. Slides were fixed in 4% paraformaldehyde and permeabilized with 0.3% saponin. Slides were blocked for 1 hour using Pierce Superblock Blocking Buffer, 37535. After washing, primary antibody (1:100 in Superblock) was added overnight, followed by secondary anti-rabbit IgG-AlexaFluor 488 (1:1000 in Superblock). Vectashield with DAPI was applied following the final wash. Images were obtained using a Leica TCS SPE confocal microscope and analysis was performed using Leica LAS AF Version 1.8.0.
Transmission electron microscopy
Samples were fixed overnight with 2.5% glutaraldehyde in 0.1 M cacodylate buffer. Post fixation was carried out for 1 hour at room temperature with a buffered 1% osmium tetroxide solution reduced with 1.5% potassium ferrocyanide. Samples were en bloc stained using 2.5% uranyl acetate. Cells were then rinsed and dehydrated. Infiltration of Spurr’s epoxy resin and acetone were carried out over several days to 100% resin and cured overnight in a 70°C oven. Sections of 100nm thickness were cut using an Ultracut E ultramicrotome (Reichert-Jung). Grids were then counterstained with 5% uranyl acetate for 12 minutes and Reynold’s lead citrate for 5 minutes. Samples were imaged using a Hitachi H-7000 transmission electron microscope.
Cargo delivery assay
Pathogen delivery to the lysosome was evaluated in chamber coverslip slides (Lab Tek) by exposing macrophages to opsonized amine-modified red fluorescent latex beads or opsonized AlexaFlour 488-tagged E.coli. at a ratio of 25 to 1. Non-phagocytosed particles were removed by washing and cells were incubated for a further 60-120 minutes. In some cases, cells were fixed and then stained for Lamp2 or LC3b. In other cases live cells were exposed to LysoTracker Red for 30 minutes at 37°C. Images were obtained using a Leica TCS SPE confocal (truepoint-scanning spectral system) microscope with Leica LAS AF interface.
Results
Cigarette smoke exposure results in accumulation of autophagic vesicles
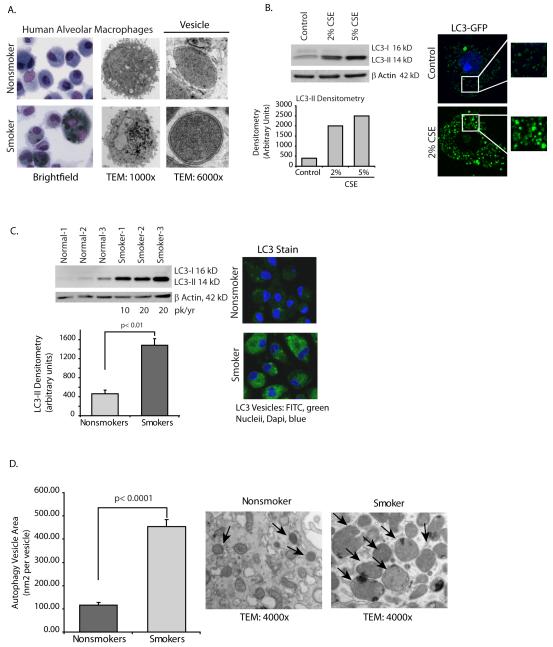
When alveolar macrophages from smokers are examined by both electron microscopy (TEM) and Wright-Giemsa stain, it is clear that smokers’ macrophages contain multiple dense cytoplasmic vesicles (Figure 1A). In this study, we identify these as autophagosomes and examine whether basal autophagy function, important for intracellular clearance of damaged proteins, organelles and intracellular pathogens, is intact.
Figure 1.
Cigarette smoke exposure in vitro and in vivo causes an increase in numbers of autophagosomes.
A. Alveolar macrophages were obtained from nonsmokers and smokers. Shown are representative TEM and Wright Giemsa stains. Higher magnification TEM image shows presence of double walled vesicles in smoker’s macrophages compared to lack of double walled vesicles in nonsmokers cells. Greater than 70% of vesicles in smoker cells had double membrane characteristic of autophagosomes.
B. Alveolar macrophages from nonsmokers were exposed to 2% or 5% CSE for five hours. Whole cell lysates were obtained and Western analysis for LC3-II performed. Equal loading is demonstrated by probing the same blot for β actin. Image is representative of 4 separate experiments. On the right is a confocal image of human alveolar macrophages transfected with LC3-GFP. After overnight incubation, some of the cells were exposed to 2% CSE for 5 hours. Confocal analysis was performed to identify GFP positive vesicles.
C. Alveolar macrophages from 3 nonsmokers and 3 smokers were analyzed for LC3-II by Western analysis of whole cell proteins. Below is a graph of densitometry results. Significance was determined using nonpaired Student’s t test. On the right is a representative image of fixed alveolar macrophages from a nonsmoker and from a smoker stained for LC3b.
D. Alveolar macrophages from nonsmokers and smokers were fixed shortly after isolation. Transmission electron microscopy (TEM) samples were prepared and images obtained. Autophagosome size was determined by analyzing area (with Image J software) of double walled vesicles in cells with slices obtained through the middle of the cell (determined by nucleus). Measurements were obtained from 3 nonsmokers and 3 smokers (12 vesicles each cell, 4 cells each person) and area of vesicle obtained by drawing circles around vesicles and performing Image j analysis. Significance was determined using nonpaired Student’s t test.
At a high magnification, the TEM images demonstrate double walled vesicles, especially in the smoker’s alveolar macrophages, consistent with autophagy vesicles(4) (Figure 1A). Greater than 70% of the vesicles observed in smokers’ alveolar macrophages possessed double membranes. Double walled vesicles were also present in the nonsmoker cells, but at low levels, consistent with normal function.
Microtubule-associated protein 1 light chain 3 (LC3), the mammalian homologue of the yeast Atg 8 protein, is a small protein that is conjugated to phosphatidylethanolamine (PE) during the formation of autophagosomes (22). It is initially synthesized in a pre-form that is cleaved at the C terminal (LC3-I) end and modified with phosphatidylethanolamine (PE) to make the mature functional protein (LC3-II). LC3-II is a reliable protein marker for mature autophagosomes and LC3-II levels correlate with numbers of autophagic vesicles (22, 23).
We first asked whether exposure to cigarette smoke extract (CSE) altered LC3 levels in human alveolar macrophages. Proteins were isolated from CSE-exposed normal alveolar macrophages and levels of endogenous LC3-II compared with LC3-I. Despite its higher molecular weight, the mature PE-conjugated form of LC3 (LC3-II) runs faster on Western gels due to its hydrophobicity and appears as a lower band. The LC3-II levels were significantly increased by exposure to CSE (Figure 1B). The right side of Figure 1B shows confocal images of alveolar macrophages transfected with a GFP expressing LC3 transgene. The transfected cells were cultured overnight and then exposed to freshly prepared CSE for 5 hours. Transfection rates were low in the primary cells but in the cells expressing the transgene, CSE exposure resulted in accumulation of LC3 positive punctate structures in alveolar macrophages (Figure 1B). The data demonstrates that in vitro exposure to CSE increases the number of autophagosomes.
Smokers’ alveolar macrophages have increased numbers of autophagic vesicles
The effect of CSE (an acute smoke exposure model) supported a pro-autophagosome accumulation effect of cigarette smoke. To analyze the effect of chronic smoke exposure as occurs in the lungs of smokers, alveolar macrophages were obtained from nonsmokers and from smokers with a >10 pack per year history. Both smokers and nonsmokers were volunteers and otherwise healthy. Figure 1C shows LC3-II Western analysis of whole cell alveolar macrophage proteins from three nonsmokers and three smokers. The blot shows significantly elevated LC3-II in the smokers’ macrophage lysates. To further examine the effect of smoking on LC3 levels, alveolar macrophages from a smoker and nonsmoker were isolated on the same day and LC3 expression analyzed by fixing the cells and staining for LC3 in both samples at once. There were significantly more LC3 positive vesicles in the smokers’ alveolar macrophages. The images are representative of four separate experiments.
A visual examination of the autophagosomes in smoker’s alveolar macrophages suggested an increase in size of the vesicles. To quantify autophagosome size, double walled vesicles in transmission electron microscopy (TEM) images were examined for vesicle area. Figure 1D shows a signficant increase in area in vesicles in smokers compared to nonsmokers. As a composite, the data in Figure 1 shows an accumulation and increase in size of autophagosomes with both acute and chronic exposure to cigarette smoke. The next experiments examine the hypothesis that the accumulation of autophagosomes in smoker’s alveolar macrophages is the result of a block in autophagic flux rather than increased functioning of the autophagosome generation machinery.
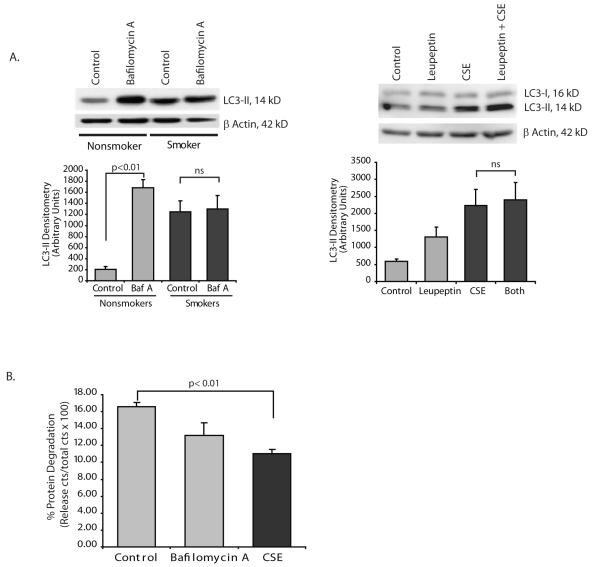
Cigarette smoke exposure blocks normal trafficking of the autophagosome to the lysosome
We examined autophagy flux in a number of ways. We first tested whether inhibition of autophagosome fusion with lysosomes altered LC3-II levels in alveolar macrophages. If inhibiting fusion of autophagosomes with lysosomes doesn’t increase LC3-II levels in smokers this would suggest that there is already a block in lysosomal fusion in the cells. Figure 2A shows that bafilomycin A (a vacuolar H(+)-ATPase inhibitor that blocks fusion of autophagosomes with lysosomes) causes an accumulation of LC3-II in alveolar macrophages from nonsmokers. In smokers, with their already high LC3-II levels, bafilomycin A had no effect. The bafilomycin data shows that in smokers’ cells, fusion is already blocked and agents that might have an effect on autophagosome/lysosome fusion can have no further effect. Leupeptin will inhibit lysosomal enzymes. In Figure 2A, alveolar macrophages were exposed to leupeptin with and without CSE. The LC3-II Western shows that CSE causes an accumulation of LC3-II that is not significantly changed by the presence of the protease inhibitor, leupeptin.
Figure 2.
Cigarette smoke exposure blocks autophagic flux.
A. Nonsmoker alveolar macrophages were exposed to bafilomycin A (100 nM) for 5 hours. Whole cell lysates were obtained and LC3-II quantified by Western analysis. The densitometry graph represents 3 separate experiments. Identical experiments were performed using leupeptin ((50 ug/ml) to inhibit lysosome function. Western anlysis and densitometry from 3 separate experiments is shown. Significance was determined using nonpaired Student’s t test.
B. Clearance of long lived proteins was determined by labeling nonsmoker alveolar macrophages with 14C leucine for 8 hours. Following a chase with cold leucine, cells were exposed to nothing (control), bafilomycin A or CSE 2%. Following an 8 hour incubation, cells and supernatants were saved. TCA precipitation isolated proteins from both samples. The percentage of long-lived proteins degraded in the culture period was determined using the formula a/a+b × 100 where a equals the TCA soluble fraction of the supernatant and b equals the TCA insoluble fraction of the cell lysate. Data was analyzed using nonpaired Student’s t test.
One measure of autophagic flux is the clearance of long lived proteins (24, 25). To evaluate the effect of CSE on protein clearance, alveolar macrophages were labelled with 14C-leucine. Short lived radioactive proteins were cleared by incubation with 10 mM cold leucine and then cells exposed to CSE or the lysosomal inhibitor bafilomycin A. The percentage of long-lived proteins degraded in the culture period was determined. Figure 2B shows that exposure to CSE decreases clearance of long lived proteins, supporting a defect in trafficking of autophagosomes to the lysosome.
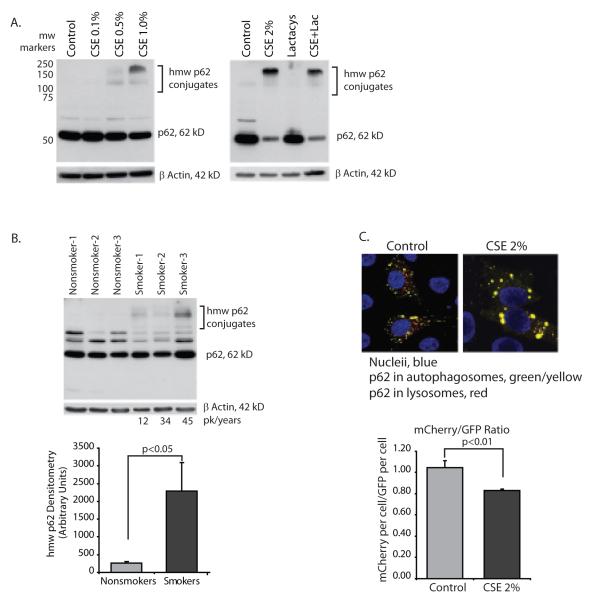
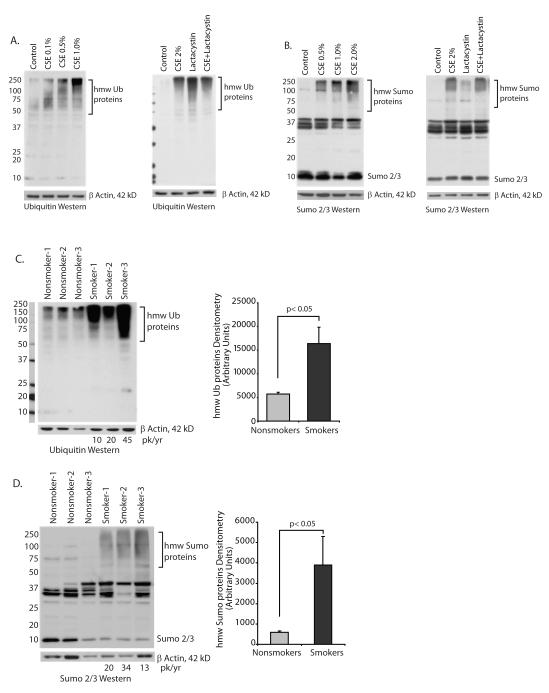
To examine autophagic flux in another manner, expression of p62 (also known as A170 or SQSTM1), an adaptor protein involved in delivery of ubiquitin bound cargo to the autophagosome (26-29) was examined. If p62 levels are increased this also would suggest a block in delivery of autophagosome cargo to the lysosome. The p62 protein (also known as SQSTM1) is a multi-functional adaptor protein with described roles in the immune system and basic cellular processes (30). The protein has a number of functional domains including a ubiquitin binding domain (UBA), an LC3 binding domain (LIR), a zinc-finger domain and a Phox domain (PB1) that mediates p62 oligomerization (28, 31). In a normal cell, p62 binds proteins with ubiquitin chains and delivers them to the autophagosome. In the process, p62 itself enters the organelle and is delivered to the lysosome with its cargo (30, 32). If autophagosome/lysosome fusion is inhibited, p62 levels will increase. Figure 3A shows that CSE exposure causes an accumulation of high molecular weight (hmw) p62 aggregates. Confirming that the accumulation of hmw p62 aggregates is a function of an autophagic flux defect and not proteasome function, we exposed alveolar macrophages to the proteasome inhibitor, lactacystin. CSE caused an accumulation of hmw p62 and lactacystin had no effect.
Figure 3.
Cigarette smoke blocks delivery of ubiquitin chaperone protein, p62 to the lysosome.
A. Nonsmoker alveolar macrophages were exposed to CSE at varying concentrations (0.1 to 1.0%) for 5 hours. Whole cell lysates were obtained and Western analysis performed for p62 protein. In the right panel, nonsmoker alveolar macrophages were exposed to lactacystin (10 uM) with and without CSE 2% for 5 hours.
B. Alveolar macrophages from 3 nonsmokers and 3 smokers were analyzed for hmw p62 aggregates by Western analysis of whole cell proteins. Densitometry reflects the p62 positive bands between 100 and 250 kD. Significance was determined using nonpaired Student’s t test.
C. Hela cells were transfected with mCherry-GFP-p62. Following an overnight incubation, cells ere exposed to 2% CSE for 5 hours. Confocal analysis was performed to identify yellow (green and red colocalization) vesicles versus red vesicles. 12 individual cells from each group were analyzed for red pixels versus green pixels (20 confocal slices per cell). Quantitation is shown in the accompanying graph. Significance was determined using nonpaired Student’s t test.
To analyze the effect of chronic smoke exposure on p62 levels in the lungs of smokers, alveolar macrophages were obtained from nonsmokers and from smokers with a >10 pack-year history of smoking. Figure 3B shows that in smokers’ macrophages, there is a significant accumulation of hmw p62 species that were not present in the lysates from nonsmokers’ macrophages.
We used a mCherry-GFP-p62 construct obtained from Terje Johansen, University of Tromso, Norway, to examine delivery of p62 to the lysosome. Because GFP is more unstable in cellular compartments with pH<6.0, a decrease in the mCherry:GFP ratio indicates decreased autophagy flux (32). We transfected Hela cells with mCherry-GFP-p62 to test the effect of CSE on p62 accumulation. Using confocal analysis, control cells showed both yellow (green and red colocalization) and red vesicles (Figure 3C). In contrast, with CSE exposure, there was significant yellow puncta with very little isolated red staining. Using image analysis software, total fluorescent units were analyzed on a per cell basis. The graph shows that with CSE exposure, the ratio of mCherry:GFP decreased, consistent with a loss of delivery of p62 to the lysosome.
Having found both accumulation of autophagosomes and decreased trafficking to the lysosome in smokers’ alveolar macrophages, we next examined systems that might be impacted by a loss of the autophagy process. Three different potential outcomes were studied: (1) clearance of protein aggregates, (2) mitochondrial function, and (3) delivery of bacteria or ingested cargo to lysosomes.
Cigarette smoke exposure causes an accumulation of protein aggregates
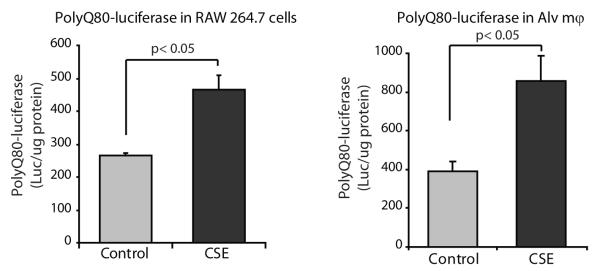
To examine accumulation of protein aggregates, we used a quantifiable luciferase reporter assay designed by Conrad Weihl (13) that measures autophagy-dependent clearance of long lived polyglutamine protein aggregates. The murine macrophage line, RAW264.7 and human alveolar macrophages were transfected with polyQ80-luciferase, allowed to rest for 24 hours, and then exposed to 2% CSE for 6 hours. Aggregate clearance was quantified as luciferase levels in CSE exposed cells compared to controls. Figure 4 demonstrates a significant decrease in aggregate clearance when polyQ80-luciferase transfected cells were exposed to CSE in all three cell types. To complement these in vitro acute exposure experiments, we analyzed accumulation of endogenous ubiquitin and sumo-conjugated protein aggregates.
Figure 4.
Cigarette smoke exposure results in cytosolic accumulation of protein aggregates.
Raw 264.7 cells and nonsmoker alveolar macrophages were transfected with a plasmid encoding firefly luciferase fused to a N-terminal polyglutamine (80) repeats (polyQ80-luciferase). Following transfection, cells were incubated overnight to allow for an accumulation of polyQ-luciferase and then exposed to 2% CSE for 6 hours. Following CSE exposure, cells were lysed and luciferase assayed. Data represent mean ±SEM (n=3). Significance was determined using nonpaired Student’s t test.
Cigarette smoke exposure causes an accumulation of high molecular weight (hmw) aggregates of ubiquitin and sumo modified proteins
Aggregate clearance in alveolar macrophages was also studied by examining accumulation of hmw ubiquitin conjugates. As originally described, ubiquitin modification of proteins is linked to degradation by the proteasome. More recently, ubiquitin modification has also been linked to clearance by the autophagy/lysosomal pathway (30). Clearance of monoubiquitinated proteins, ubiquitin-conjugated misfolded and aggregated proteins as well as signal related K63 ubiquitin chains have been linked to autophagy (33-36). Figure 5A illustrates analysis of alveolar macrophages from nonsmokers exposed to increasing doses of CSE for 5 hours. The blot demonstrates a dose-dependent increase in hmw ubiquitin-bound proteins. We examined the role of the proteasome by incubating cells with the proteasome inhibitor, lactacystin. Proteasome inhibition also causes a buildup of ubiquitin conjugates, consistent with the important role of the proteasome in clearing ubiquitinated proteins. However, in the CSE-exposed cells, more of the ubiquitin staining bands are at the very top of the gel, suggesting that CSE ubiquitin accumulation is not necessarily a function of changes in proteasome function.
Figure 5.
Cigarette smoke exposure causes an accumulation of high molecular weight (hmw) ubiquitin and sumo-conjugated proteins.
A. Ubiquitin analysis: The accumulation of hmw ubiquitin conjugates was assessed in alveolar macrophages from nonsmoker’s exposed to 0.1 to 1.0% CSE for 5 hours. Western analysis for ubiquitin was done on whole cell lysates. In the experiment shown on the right, nonsmoker alveolar macrophages were exposed to lactacystin (10 uM) with and without 2% CSE for 5 hours. Images are representative of three separate experiments.
B. Sumo analysis: The accumulation of hmw sumo conjugates was assessed in alveolar macrophages from nonsmoker’s exposed to 0.1 to 1.0% CSE for 5 hours. Western analysis for sumo 2/3 was done on whole cell lysates. In the experiment shown on the right, nonsmoker alveolar macrophages were exposed to lactacystin (10 uM) with and without 2% CSE for 5 hours. Images are representative of three separate experiments.
C. Ubiquitin analysis: Alveolar macrophages from 3 nonsmokers and 3 smokers were analyzed for hmw ubiquitin conjugates by Western analysis of whole cell proteins. Denstitometry reflects the ubiquitin positive bands between 75 and 250 kD. Significance was determined using nonpaired Student’s t test.
D. Sumo analysis: Alveolar macrophages from 3 nonsmokers and 3 smokers were analyzed for hmw sumo conjugates by Western analysis of whole cell proteins. Densitometry reflects the sumo positive bands between 75 and 250 kD. Significance was determined using nonpaired Student’s t test.
Because it was hard to distinguish the proteasome effects from the autophagy/lysosome effects, we also analyzed sumo-conjugated hmw complexes (37-39). Sumo is a ubiquitin-like protein that also binds lysines via an isopeptide bond and does not target substrates to the proteasome (40). There are three sumo isoforms, 1, 2 and 3. Sumo 1 binds to substrate lysines as a monomer. Sumo 2 and 3, like ubiquitin, can make long sumo chains. We used an antibody specific for Sumo 2/3 to determine if cigarette smoke exposure altered clearance of sumo-conjugated substrates. Figure 5B shows an accumulation of sumo positive aggregates in CSE exposed alveolar macrophages. In contrast to the ubiquitin conjugated proteins, lactacystin had very little effect on sumo-conjugate accumulation. As an aggregate, this data suggests that cigarette smoke exposure caused an accumulation of hmw aggregates that should normally be cleared by the autophagy/lysosome system.
To examine the in vivo effect of cigarette smoking on the accumulation of hmw ubiquitin-bound and sumo-bound proteins, we analyzed baseline lysates from nonsmokers’ alveolar macrophages and smokers’ alveolar macrophages for ubiquitin-linked proteins and sumo-linked proteins. Figure 5C shows a significant increase in the amount of hmw ubiquitin bound proteins in the smokers’ macrophages. The smoking histories of the three individuals consisted of 10, 20 and 45 pack years. To examine hmw sumo aggregates, lysates from nonsmokers and smokers (pack years of 24, 13 and 34 years) were examined by Western analysis for sumo 2/3. Figure 5D shows an accumulation of hmw species in cell lysates from smokers. As a composite, the data in Figure 5 demonstrates that in vivo cigarette smoke exposure causes an accumulation of protein aggregates and blocks the normal cycling of ubiquitin or sumo 2/3 conjugated proteins. This is consistent with a loss of autophagy function.
Cigarette smoke exposure disrupts normal mitochondrial architecture and function
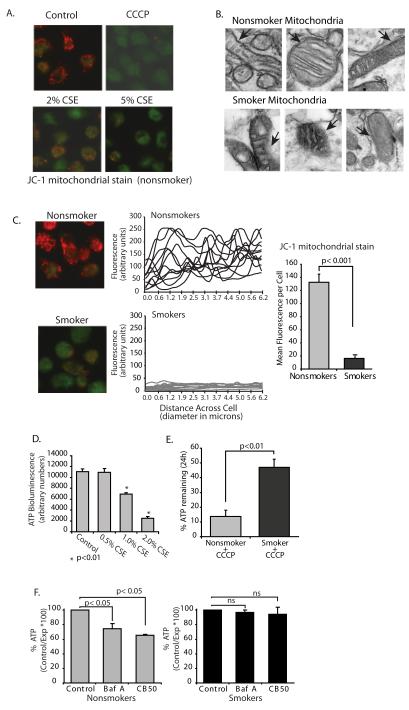
Autophagy is the only known mechanism for turning over large organelles such as mitochondria (41). In the case of mitochondrial turnover by the autophagosome system, the process is known as mitophagy. It is required for mitochondrial homeostasis (42). If the process of autophagy in smokers’ macrophages is nonfunctional, then mitochondrial dysfunction is possible sequelae. We first examined mitochondrial membrane potential (mitΔΨ) in CSE-exposed alveolar macrophages. Mitochondrial membrane potential is determined by the electrochemical gradient across the inner mitochondrial membrane and the fluorescent cationic dye, JC-1, monitors mitΔΨ. Figure 6A shows normal alveolar macrophages exposed to the mitochondrial uncoupler, CCCP, or CSE for 4 hours. JC-1 accumulates in healthy mitochondria as red aggregates. The dye remains in the cytosol in a green monomeric form with loss of mitΔΨ. Both CSE and CCCP caused a loss of the red aggregated JC-1 form. We asked if smokers’ alveolar macrophages had damaged mitochondria in the cytosol. Figure 6B shows TEM images from both smokers’ and nonsmokers’ alveolar macrophages. The smokers’ cells were characterized by the presence of damaged mitochondria, which is consistent with an inability to clear old and/or damaged mitochondria. Arrows identify mitochondrial membrane that appears compromised in the smokers cells.
Figure 6.
Cigarette smoke exposure results in loss of mitochondrial electron transport chain function and accumulation of defective mitochondria.
A. Human alveolar macrophages were cultured (1×106/ml in 2 chamber microscope slides) with and without 2-5% CSE or the uncoupler (CCCP). At the end of the incubation period, the mitochondrial stain JC-1 was added as described in the methods. Red/orange stain denotes intact mitochondria with no disruption of membrane potential. Green staining denotes loss of mitΔΨ.
B. TEM was performed on smoker and nonsmoker macrophages. Shown are mitochondria demonstrating damaged mitochondria in the smoker’s cells. Arrows in images point to intact (nonsmoker) and damaged (smoker) mitochondrial membranes.
C. Nonsmoker and smoker macrophages were allowed to adhere to 2 chamber slides and stained with JC-1. Quantitation was performed as described in the methods and individual cell levels of red fluorescence from two separate experiments are shown in the line graphs and as a composite in the bar graph.
D. Normal alveolar macrophages were cultured (96 well plate) with varying concentrations of CSE (0.5%-2.0%). ATP levels were measured using a chemiluminescence assay system. Data is presented as arbitrary luminescent units and is a composite of three experiments. Significance was determined using nonpaired Student’s t test.
E. Nonsmoker and smoker alveolar macrophages were exposed the uncoupler, CCCP, for 24 hours. ATP levels with and without the uncoupler were obtained and percent remaining ATP calculated. Data is a composite of three smokers and three nonsmokers. Significance was determined using nonpaired Student’s t test.
F. Nonsmoker alveolar macrophages were exposed to the specific inhibitor of vacuolar-type H+-ATPase, bafilomycin A (100 nM) or carbon black 50 nM nanoparticles for 5 hours. ATP levels were measured using a chemiluminescence assay system. Data is presented as arbitrary luminescent units and is a composite of three experiments. Significance was determined using nonpaired Student’s t test.
Smokers’ and nonsmokers’ alveolar macrophages were tested for mitΔΨ to examine the long-term effect of cigarette smoke exposure. In order to control for staining differences, bronchoalveolar lavages were performed on both smokers and nonsmokers on the same day. Cells from both subjects were loaded onto two chamber coverslip slides and stained with JC-1. Figure 6C shows red mitochondrial staining in the nonsmoker alveolar macrophage and green staining indicative of loss of mitΔΨ in the smoker’s cell. Random cells from each of three smokers and nonsmokers were analyzed for red fluorescent levels. The data shows a significant loss of mitΔΨ in smokers’ alveolar macrophages.
Cigarette smoke exposure decreases oxidative respiration
We next examined functional consequences of a loss in mitochondrial function. We have shown previously that the majority of nonsmokers’ alveolar macrophage ATP comes from the mitochondria (21). We asked if acute exposure to CSE would lead to a loss of ATP. The graph in Figure 6D shows decreasing ATP levels with increasing doses of CSE. It is possible (and not inconsistent with the literature) that long term smoke exposure would lead to reliance on anaerobic glycolysis for smokers’ alveolar macrophage energy generation. We addressed the question of whether long term smoking led to a shift in ATP production from oxidative phosphorylation to glycolysis. We exposed smokers’ and nonsmokers’ alveolar macrophages to CCCP for 4 hours. CCCP is a mitochondrial uncoupler that rapidly dissipates the proton gradient and stops mitochondrial ATP production. The nonsmoker alveolar macrophages were much more sensitive to the ATP-depleting effects of CCCP than were the smoker macrophages (Figure 6E). This is consistent with an increase in anaerobic glycolysis (mitochondrial independent) ATP production in smokers’ alveolar macrophages.
We do not propose that loss of mitochondrial ATP production is entirely due to a loss in autophagic flux. However, lack of clearance of damaged organelles would set up a feedback loop, preventing mitochondrial recovery (42). To investigate whether blocking autophagic flux alone would damage mitochondria in alveolar macrophages, we treated normal alveolar macrophages with bafilomycin A to block autophagosome lysosome fusion. We also added carbon black nanoparticles (a process that would mimic the accumulation of particulates from cigarette smoke) that would stress the fusion process by accumulating in autophagosomes. ATP was used as a measure of mitochondrial injury. Both bafilomycin A and carbon black 50 nm nanoparticles caused a drop in ATP in nonsmokers’ alveolar macrophages (Figure 6F), showing that inhibiting autophagosome/lysosome fusion caused mitochondrial dysfunction. There was no effect of either bafilomycin A or carbon particles in the smoker macrophages, suggesting a decreased reliance on oxidative phosphorylation in smoker macrophages. The data in Figure 6 show that both in vitro and in vivo smoke exposure leads to accumulation of defective mitochondria and results in the subsequent loss of mitochondrial derived ATP. In addition, we show that in nonsmoker macrophages, inhibiting fusion of the autophagosome with the lysosome decreases mitochondrial ATP production.
Cigarette smoke exposure blocks delivery of phagocytosed cargo to lysosomes
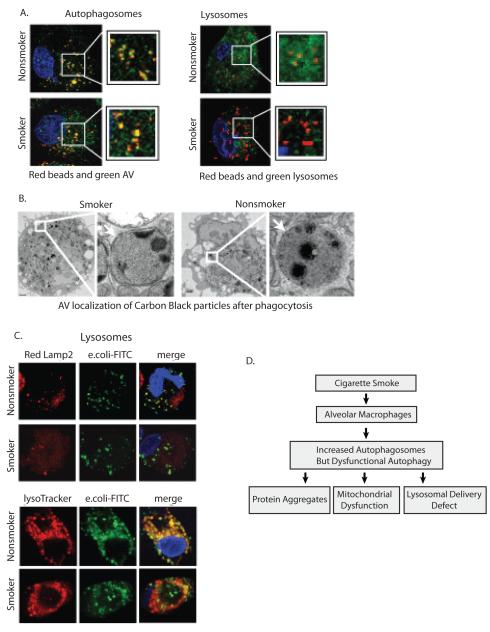
Though first described as a process that improves cellular survival in conditions of nutrient depletion, recent work suggests an important role for autophagy in the immune system (43, 44). Xenophagy refers to the autophagic process that targets intracellular pathogens to the lysosome via an autophagosome (5, 45-47). We first examined the delivery of opsonized fluorescent latex beads to either the autophagosome or the lysosome in nonsmoker and smokers’ macrophages. Nonsmoker and smokers’ alveolar macrophages were cultured on coverslip chamber slides and exposed to a 25:1 solution of opsonized 0.5 micron amine-modified latex beads for 1 hour. The amine-modified beads are routinely used in phagocytosis assays and easily opsonized. Non-internalized beads were washed off and the cells incubated for a further hour before fixing and staining for either autophagosomes (LC3) or lysosomes (lamp2a). Figure 7A demonstrates the colocalization of the red beads with green autophagosomes in macrophages from both smokers and nonsmokers. However, only the nonsmoker cells showed co-localization of beads and lysosomes (Lamp2a staining).
Figure 7.
Cigarette smoke exposure impairs delivery of cargo to lysosomes.
A. Alveolar macrophages from nonsmokers and smokers were placed in culture. Following a 3 hour stabilizing period, cells were exposed to 0.5 micron latex beads for 60 minutes. Nonphagocytosed beads were washed off and cells were incubated for a 2 hour chase period. Cells were then fixed and stained for either LC3b (autophagosomes) or Lamp2 (lysosomes).
B. Alveolar macrophages from nonsmokers and smokers were placed in culture. Following a 3 hour stabilizing period, cells were exposed to 50 nM carbon black nanoparticles for 60 minutes. Nonphagocytosed nanoparticles were washed off and cells were incubated for a 2 hour chase period. Cells were then fixed and TEM analysis performed. Shown is a representative cell demonstrating carbon black particles in double walled vesicles.
C. Alveolar macrophages from smokers and nonsmokers were cultured on a coverslip chamber slide. Cells were exposed to AlexaFlour 488-E.coli for 30 minutes, nonphagocytosed bacteria was washed off, a 1 hour chase incubation followed before staining the cells with Lysotracker Red DND-99 or fixing and staining for Lamp2 with an Alexa 568 secondary antibody. The images are representative of three separate experiments. The photomicrographs show red lysosomes, green E. coli and yellow merged image of E. coli in a lysosome.
D. The diagram outlines the conclusions from this study. Prolonged cigarette smoke exposure leads to accumulation of nonfunctional autophagosomes by blocking lysosomal trafficking. The block in autophagic flux leads to decreased clearance of protein aggregates, mitochondrial dysfunction and reduced delivery of phagocytosed bacteria to the lysosome.
To test the delivery of other types of phagocytosed cargo to the autophagosome, alveolar macrophages were exposed to opsonized 50nm carbon nanoparticles for 1 hour. The cells were then fixed and TEM studies performed. Figure 7B shows that in both nonsmoker and smoker macrophages, carbon black nanoparticles end up in double walled vesicles characteristic of autophagosomes.
We examined delivery of E. coli particles to lysosomes in nonsmoker versus smoker macrophages. As in earlier comparison stains, smoker and nonsmoker cells were seeded onto the same 2 chamber coverslip slide. Alveolar macrophages were exposed to Alexa Fluor 488 tagged E.coli particles for 1 hour. This was followed by a chase period for the particles to move to lysosomes. For lysosomal staining, a red Lysotracker dye was added to live cells or fixed cells were stained for Lamp2. Fluorescent images were obtained. It is important to note that the smokers’ alveolar macrophages underwent less phagocytosis, consistent with existing literature. However, the smokers’ cells did phagocytose some particles, which allowed us to ask whether internalized particles were delivered to the lysosome. The images in Figure 7C show that the e. coli particles are delivered to the lysosome in the nonsmoker alveolar macrophages. This was not true in the smoker cells where green (E. coli) and red lysosomes do not co-localize. These data suggest that the normal delivery of pathogen cargo to the lysosomes by autophagosomes is disrupted in smokers’ alveolar macrophages.
Discussion
This study’s major finding reveals that chronic inhalation of cigarette smoke causes a profound defect in autophagy/lysosomal function in human alveolar macrophages (Figure 7D). This was evidenced by lack of clearance of aggregated proteins and hmw ubiquitinated proteins, accumulation of abnormal mitochondria with altered function, and loss of cargo delivery to lysosomes. The same alterations could be triggered in alveolar macrophages from nonsmokers acutely exposed to cigarette smoke in vitro. This defect in autophagy/lysosomal function was not due to fewer autophagosomes since smokers’ macrophages had significantly more of these vesicles than nonsmokers’ macrophages. The data are consistent with a loss of autophagic flux, leading to an accumulation of autophagosomes and defects in autophagy clearance functions.
A relationship between smoking and autophagy has been elegantly described by Augustine Choi’s group. They found increased autophagosomes in epithelial tissue from patients with COPD compared to normal tissue (8). This work showed increased expression and activation of autophagy regulator proteins including LC3B, beclin 1, Atg5 and Atg7. A similar increase in autophagosomes was found in the lungs of mice subjected to chronic cigarette smoke inhalation and in lung epithelial lines exposed to cigarette smoke extract (CSE) (8). The increase in generation of autophagosomes was reversed by overexpressing the oxidant stress protein, heme oxygenase 1 (9). In another study, they found that CSE caused an increase in autophagosomes in Beas2b human bronchial epithelial cells (9). The CSE-induced autophagy defect served a pro-apoptotic function in cigarette smoke-exposed cells. Both of the studies from Choi and colleagues demonstrate concurrent up-regulation of both autophagy and apoptosis. Their findings might explain the loss of peripheral lung tissue and apoptosis that is characteristic of emphysema. Our findings with alveolar macrophages differ from those of Choi et al in that the work from Choi’s group shows increased autophagy function, while our study found decreased autophagy function in alveolar macrophages. It is likely that our observations of decreased autophagy function in macrophages relate to the increased phagocytic capacity of alveolar macrophages which makes them differentially susceptible to cigarette smoke exposure. One of the main functions of alveolar macrophages is phagocytosis of inhaled material including particulates in cigarette smoke, making their exposure to cigarette smoke quite different from exposure of airway epithelium. In fact we found that ingestion of carbon black particles by alveolar macrophages induces a similar defect as cigarette smoke. It is likely that the different effects on the autophagy process lie in the very different functions of lung epithelium and alveolar macrophages.
It is interesting to speculate that the increase in autophagy seen by A. Choi’s group is the same as the increase in autophagy associated with programmed cell death recently reported as essential for presentation of phoshatidylserine (PS) on the cell surface (48). It is possible that autophagy in dying epithelial cells contributes to externalization of phosphatidylserine while a lack of autophagy in alveolar macrophages contributes to a lack of clearance of dead cell from the alveolar surface. Both of these effects on the process of autophagy could contribute to smoking-related diseases.
This study demonstrates a significant accumulation of autophagosomes in smokers’ alveolar macrophages. This was also demonstrated in in vitro models using macrophages from nonsmokers acutely exposed to CSE. Our data suggests that these structures accumulate because they are not being turned over via fusion with lysosomes. In this regard, smokers’ macrophages contain a large amount of cigarette smoke particulates that appear to be within the autophagosomes. Montgomery et. al. showed that loading macrophages with poorly digestible material resulted in a defect in autophagy/lysosomal function (49). Our data supports a defect in the delivery of autophagosomes to the lysosome in smokers’ alveolar macrophages.
Long term in vivo cigarette smoke exposure led to a buildup of hmw ubiquitin and sumo complexes in smokers’ alveolar macrophages. We believe this build-up is linked to a block in the process of autophagy. Recent studies have shown that autophagy, as well as the proteasome, clear ubiquitin bound substrates (50). Three proteins that bind both ubiquitin and LC3, p62/SQTSM1, NBR1 and NDP52 have been identified (51). They are thought to act as chaperones, delivering ubiquitinated proteins to the autophagosome (30, 32). In the present study, cigarette smoke exposure led to an increase in sequestration of p62 in hmw complexes.
Mitophagy refers to the clearance of damaged mitochondria by autophagy. The importance of autophagy in clearing damaged mitochondria has been convincingly shown in recent studies (52-54). The effect of smoking-induced mitochondrial dysfunction in alveolar macrophages could be due to a global effect on autophagy or could be due to an effect on one of the adaptor proteins shown recently to target damaged mitochondria to the autophagosome. In mammalian cells, the cytosolic protein Parkin is selectively recruited to damaged mitochondria (55). Recruitment of Parkin and the PTEN-induced putative kinase-1 (PINK1) to damaged mitochondria leads to mitophagy and clearance of mitochondria. Another adaptor protein involved in mitophagy is the protein Nix. Nix forms a complex with LC3 and GABARAP-L1 that mediates mitochondrial clearance (56). It will be interesting to see if smoking alters expression or function of either Parkin or Nix. It is clear from our data that in smokers’ alveolar macrophages, there is a loss of mitochondrial ATP generating capacity (consistent with mitochondrial dysfunction) and an increase in damaged mitochondria.
Autophagy has a number of described immune functions including homeostasis of B, T and other immune cells, activation or dampening of pro-inflammatory processes and clearance of pathogens that reach the cytosol (45). Xenophagy is the term coined to cover the subset of autophagy involved in pathogen clearance (43, 57). Bacteria that reach the cytosol are targeted to nascent LC3 containing phagophores by adaptor proteins that have an LC3-interaction domain (LIR). For example, Listeria lacking ActA that escapes to the cytosol becomes covered with ubiquitin. It is then recognized by p62 and targeted to the autophagosome (58). Salmonella that escapes from damaged vacuoles has been shown to colocalize with polyubiquitin complexes, which are delivered to autophagosome via p62 and LC3 (59). Much of the early work on xenophagy has been performed by Beth Levine’s group, including an elegant study in nematodes showing an in vivo role for xenophagy in Salmonella Typhimurium clearance (46). In our study, E. coli particles that are phagocytosed by smokers’ alveolar macrophages are not delivered to the lysosome. This data is consistent with a defect in xenophagy with chronic cigarette smoke exposure.
This study makes two new contributions to our knowledge of the effects of cigarette smoke in the lung. The first is the demonstration of a basic biological mechanism whereby cigarette smoke alters the process of autophagy. The second contribution addresses the clinical issue of recurrent infections in people who smoke. Obviously, the best treatment is to stop smoking. However, chronic obstructive pulmonary disease (COPD) is the fourth leading cause of death in the U.S. and cigarette smoke exposure accounts for more that 80-90% of U.S. COPD cases (60). With the understanding that there is a defect in pathogen clearance due to altered autophagy, it is possible that pharmaceutical agents that target autophagy could decrease smoking-related infection rates and subsequently healthcare costs.
Acknowledgements
The authors thank Chris Weihl, Washington University, for sharing the polyQ80-luciferase vector and Johansen Terje, University of Tromso, Norway for sharing the mCherry-GFP-p62 vector.
Footnotes
Bibliography
- 1.Niewoehner DE, Kleinerman J, Rice DB. Pathologic changes in the peripheral airways of young cigarette smokers. The New England journal of medicine. 1974;291:755–758. doi: 10.1056/NEJM197410102911503. [DOI] [PubMed] [Google Scholar]
- 2.Green GM. Cigarette smoke: protection of alveolar macrophages by glutathione and cysteine. Science (New York, N.Y. 1968;162:810–811. doi: 10.1126/science.162.3855.810. [DOI] [PubMed] [Google Scholar]
- 3.Laurenzi GA, Guarneri JJ, Endriga RB, Carey JP. Clearance of Bacteria by the Lower Respiratory Tract. Science (New York, N.Y. 1963;142:1572–1573. doi: 10.1126/science.142.3599.1572. [DOI] [PubMed] [Google Scholar]
- 4.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science (New York, N.Y. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 8.Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, Dhir R, Landreneau RJ, Schuchert MJ, Yousem SA, Nakahira K, Pilewski JM, Lee JS, Zhang Y, Ryter SW, Choi AM. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS ONE. 2008;3:e3316. doi: 10.1371/journal.pone.0003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HP, Wang X, Chen ZH, Lee SJ, Huang MH, Wang Y, Ryter SW, Choi AM. Autophagic proteins regulate cigarette smoke-induced apoptosis: protective role of heme oxygenase-1. Autophagy. 2008;4:887–895. doi: 10.4161/auto.6767. [DOI] [PubMed] [Google Scholar]
- 10.Nuorti JP, Butler JC, Farley MM, Harrison LH, McGeer A, Kolczak MS, Breiman RF. Cigarette smoking and invasive pneumococcal disease. Active Bacterial Core Surveillance Team. The New England journal of medicine. 2000;342:681–689. doi: 10.1056/NEJM200003093421002. [DOI] [PubMed] [Google Scholar]
- 11.Drannik AG, Pouladi MA, Robbins CS, Goncharova SI, Kianpour S, Stampfli MR. Impact of cigarette smoke on clearance and inflammation after Pseudomonas aeruginosa infection. American journal of respiratory and critical care medicine. 2004;170:1164–1171. doi: 10.1164/rccm.200311-1521OC. [DOI] [PubMed] [Google Scholar]
- 12.Woods A, Azzout-Marniche D, Foretz M, Stein SC, Lemarchand P, Ferre P, Foufelle F, Carling D. Characterization of the role of AMP-activated protein kinase in the regulation of glucose-activated gene expression using constitutively active and dominant negative forms of the kinase. Molecular and cellular biology. 2000;20:6704–6711. doi: 10.1128/mcb.20.18.6704-6711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju JS, Miller SE, Jackson E, Cadwell K, Piwnica-Worms D, Weihl CC. Quantitation of selective autophagic protein aggregate degradation in vitro and in vivo using luciferase reporters. Autophagy. 2009;5:511–519. doi: 10.4161/auto.5.4.7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monick MM, Carter AB, Hunninghake GW. Human alveolar macrophages are markedly deficient in REF-1 and AP-1 DNA binding activity. The Journal of biological chemistry. 1999;274:18075–18080. doi: 10.1074/jbc.274.25.18075. [DOI] [PubMed] [Google Scholar]
- 15.Nyunoya T, Monick MM, Klingelhutz A, Yarovinsky TO, Cagley JR, Hunninghake GW. Cigarette smoke induces cellular senescence. American journal of respiratory cell and molecular biology. 2006;35:681–688. doi: 10.1165/rcmb.2006-0169OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nyunoya T, Monick MM, Klingelhutz AL, Glaser H, Cagley JR, Brown CO, Matsumoto E, Aykin-Burns N, Spitz DR, Oshima J, Hunninghake GW. Cigarette smoke induces cellular senescence via Werner’s syndrome protein down-regulation. American journal of respiratory and critical care medicine. 2009;179:279–287. doi: 10.1164/rccm.200802-320OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blue ML, Janoff A. Possible mechanisms of emphysema in cigarette smokers. Release of elastase from human polymorphonuclear leukocytes by cigarette smoke condensate in vitro. The American review of respiratory disease. 1978;117:317–325. doi: 10.1164/arrd.1978.117.2.317. [DOI] [PubMed] [Google Scholar]
- 18.Panayiotidis MI, Stabler SP, Allen RH, Ahmad A, White CW. Cigarette smoke extract increases S-adenosylmethionine and cystathionine in human lung epithelial-like (A549) cells. Chemico-biological interactions. 2004;147:87–97. doi: 10.1016/j.cbi.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Carnevali S, Petruzzelli S, Longoni B, Vanacore R, Barale R, Cipollini M, Scatena F, Paggiaro P, Celi A, Giuntini C. Cigarette smoke extract induces oxidative stress and apoptosis in human lung fibroblasts. American journal of physiology. 2003;284:L955–963. doi: 10.1152/ajplung.00466.2001. [DOI] [PubMed] [Google Scholar]
- 20.Hansdottir S, Monick MM, Hinde SL, Lovan N, Look DC, Hunninghake GW. Respiratory epithelial cells convert inactive vitamin D to its active form: potential effects on host defense. J Immunol. 2008;181:7090–7099. doi: 10.4049/jimmunol.181.10.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monick MM, Powers LS, Barrett CW, Hinde S, Ashare A, Groskreutz DJ, Nyunoya T, Coleman M, Spitz DR, Hunninghake GW. Constitutive ERK MAPK activity regulates macrophage ATP production and mitochondrial integrity. J Immunol. 2008;180:7485–7496. doi: 10.4049/jimmunol.180.11.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr., Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy Reduces Acute Ethanol-Induced Hepatotoxicity and Steatosis in Mice. Gastroenterology. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogier-Denis E, Houri JJ, Bauvy C, Codogno P. Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. The Journal of biological chemistry. 1996;271:28593–28600. doi: 10.1074/jbc.271.45.28593. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature cell biology. 12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 27.Moscat J, Diaz-Meco MT. To aggregate or not to aggregate? A new role for p62. EMBO reports. 2009;10:804. doi: 10.1038/embor.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin H. W. t., Kyei GB, Johansen T, Vergne I, Deretic V. Delivery of Cytosolic Components by Autophagic Adaptor Protein p62 Endows Autophagosomes with Unique Antimicrobial Properties. Immunity. 32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Molecular cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 31.Moscat J, Diaz-Meco MT, Albert A, Campuzano S. Cell signaling and function organized by PB1 domain interactions. Molecular cell. 2006;23:631–640. doi: 10.1016/j.molcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. The Journal of biological chemistry. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 33.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. The Journal of cell biology. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, Ho MW, Troncoso J, Gygi SP, Lee MK, Dawson VL, Dawson TM, Lim KL. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Human molecular genetics. 2008;17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- 36.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. The Journal of biological chemistry. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 37.Hemelaar J, Borodovsky A, Kessler BM, Reverter D, Cook J, Kolli N, Gan-Erdene T, Wilkinson KD, Gill G, Lima CD, Ploegh HL, Ovaa H. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Molecular and cellular biology. 2004;24:84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hochstrasser M. Biochemistry. All in the ubiquitin family. Science (New York, N.Y. 2000;289:563–564. doi: 10.1126/science.289.5479.563. [DOI] [PubMed] [Google Scholar]
- 39.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nature cell biology. 2000;2:E153–157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 40.Ulrich HD. The fast-growing business of SUMO chains. Molecular cell. 2008;32:301–305. doi: 10.1016/j.molcel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 41.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. The Journal of biological chemistry. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell host & microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 45.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, Levine B. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 49.Montgomery RR, Webster P, Mellman I. Accumulation of indigestible substances reduces fusion competence of macrophage lysosomes. J Immunol. 1991;147:3087–3095. [PubMed] [Google Scholar]
- 50.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Molecular cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivanov S, Roy CR. NDP52: the missing link between ubiquitinated bacteria and autophagy. Nat Immunol. 2009;10:1137–1139. doi: 10.1038/ni1109-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. Faseb J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 53.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Archives of biochemistry and biophysics. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of cell biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proceedings of the National Academy of Sciences of the United States of America. 107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO reports. 11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deretic V. Autophagy in infection. Current opinion in cell biology. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, Kakizuka A, Sztul E, Chakraborty T, Sasakawa C. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nature cell biology. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 59.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 60.Le A, Zielinski R, He C, Crow MT, Biswal S, Tuder RM, Becker PM. Pulmonary epithelial neuropilin-1 deletion enhances development of cigarette smoke-induced emphysema. American journal of respiratory and critical care medicine. 2009;180:396–406. doi: 10.1164/rccm.200809-1483OC. [DOI] [PMC free article] [PubMed] [Google Scholar]