Abstract
To investigate the cell cycle checkpoint response to aberrant S phase-initiation, we analyzed mutations of the two DNA primase subunit genes of Schizosaccharomyces pombe, spp1+ and spp2+ (S. pombe primase 1 and 2). spp1+ encodes the catalytic subunit that synthesizes the RNA primer, which is then utilized by Polα to synthesize the initiation DNA. Here, we reported the isolation of the fission yeast spp1+ gene and cDNA and the characterization of Spp1 protein and its cellular localization during the cell cycle. Spp1 is essential for cell viability, and thermosensitive mutants of spp1+ exhibit an allele-specific abnormal mitotic phenotype. Mutations of spp1+ reduce the steady-state cellular levels of Spp1 protein and compromised the formation of Polα–primase complex. The spp1 mutant displaying an aberrant mitotic phenotype also fails to properly activate the Chk1 checkpoint kinase, but not the Cds1 checkpoint kinase. Mutational analysis of Polα has previously shown that activation of the replication checkpoint requires the initiation of DNA synthesis by Polα. Together, these have led us to propose that suboptimal cellular levels of polα–primase complex due to the allele-specific mutations of Spp1 might not allow Polα to synthesize initiation DNA efficiently, resulting in failure to activate a checkpoint response. Thus, a functional Spp1 is required for the Chk1-mediated, but not the Cds1-mediated, checkpoint response after an aberrant initiation of DNA synthesis.
INTRODUCTION
Upon perturbation of DNA replication or after DNA damage, eukaryotic cells have the checkpoint mechanism to arrest or delay the cell cycle progression to maintain genome integrity (Hartwell and Weinert, 1989; Hartwell and Kastan, 1994; Weinert, 1997; Carr, 1998; Weinert, 1998). In fission yeast, Schizosaccharomyces pombe, six proteins named “checkpoint Rads” function in both the replication and the DNA damage checkpoints (Caspari and Carr, 1999). Five of these six checkpoint Rad proteins are evolutionarily conserved from yeast to man (Caspari et al., 2000). In response to replication stress and/or DNA damage, the checkpoint Rad proteins signal to at least two downstream checkpoint kinases, Cds1 and Chk1, to regulate the cell cycle machinery and delay or arrest the cell cycle (Murakami and Okayama, 1995; O'Connell et al., 1997; Boddy et al., 1998; Lindsay et al., 1998; Rhind and Russell, 1998; Zeng et al., 1998; Furnari et al., 1999). When S phase is blocked or DNA damage is inflicted in S phase, Cds1 is phosphorylated, and this phosphorylation correlates to an increase in its kinase activity (Lindsay et al., 1998). In response to DNA damage during late S phase or G2, Chk1 is phosphorylated. It is not yet clear whether Chk1 protein phosphorylation correlates with activation of Chk1 kinase activity. However, Chk1 protein phosphorylation does correlate with cell cycle arrest in response to damage or replication perturbation (Walworth and Bernards, 1996; Martinho et al., 1998). Phosphorylation of Cds1 and Chk1 are both dependent on the kinase domain of the Rad3 checkpoint Rad protein (Bentley et al., 1996; Lindsay et al., 1998; Martinho et al., 1998). In vitro work suggests that activated Cds1 and Chk1 kinase can phosphorylate two regulators of the mitotic inducer Cdc2, Wee1, and Cdc25 (Furnari et al., 1997; Boddy et al., 1998; Zheng et al., 1998; Lopez-Girona et al., 1999). A number of replication mutants are synthetic lethal in cds1Δ background but not in chk1Δ background (Bhaumik and Wang, 1998; Tan and Wang, 2000). Furthermore, deletion of cds1+ lowers the semipermissive temperature and exacerbates the mutation rate of several thermosensitive replication mutators (Liu et al., 1999). These data suggest that Cds1 allows the cells to tolerate the perturbation and to recover from the stress in these replication mutants to prevent accumulation of further damage of the genome, thus contributing to the mitotic checkpoint (Lindsay et al., 1998; Rhind and Russell, 1998).
We are interested in understanding how the two checkpoint effector kinases, Cds1 and Chk1, respond to aberrant initiation of S phase. In eukaryotic cells, an evolutionarily conserved four-subunit enzyme complex, DNA polymerase α-primase, is the principal enzyme that initiates DNA synthesis (Wang, 1996). The primase in this enzyme complex synthesizes an RNA primer. Polymerase α can only utilize the RNA primer synthesized by primase of greater than or equal to seven nucleotides in length to initiate the DNA synthesis (Kuchta et al., 1990). Primase in the Polα–primase complex is a heterodimeric enzyme. The catalytic subunit of primase, named p49 in mammalian cells and PRI1 in budding yeast, synthesizes the RNA primer. The second subunit, named p58 in mammalian cells, PRIII in budding yeast, and Spp2 in fission yeast, is required for coupling the RNA primer synthesis by primase catalytic subunit with Polα for initiation DNA (iDNA) synthesis (Tan and Wang, 2000).
We isolated the gene and cDNA of S. pombe polymerase α, polα+, and the primase catalytic subunit and the coupling subunit named, spp1+ and spp2,+ for S. pombe primase 1 and 2, respectively. We have analyzed the mutational effects of polα+ and spp2+ on the cell cycle checkpoint responses (Bhaumik and Wang, 1998; Tan and Wang, 2000). Mutational studies of fission yeast polα+ indicate that cells with polαΔ enter mitosis with 1C DNA content, and cells harboring a catalytically dead but physically intact Polα in the Polα–primase complex enter mitosis inappropriately. These findings indicate that the catalytic activity of Polα is a prerequisite for activation of the S-M phase checkpoint response (Bhaumik and Wang, 1998). Analysis of spp2+ thermosensitive mutants has indicated that mutations of spp2+ affect the stability of the Polα–primase complex. Maintaining a stable Polα–primase complex by Spp2 protein is required for the activation of the Cds1-mediated intra–S phase checkpoint (Tan and Wang, 2000).
In this report, we analyzed the mutational effect of the primase catalytic subunit, spp1+, on the checkpoint kinase response. We found that mutations of spp1+ reduced the cellular mutant protein's steady-state levels and compromised the proper Polα–primase complex formation. In contrast to mutations of spp2, upon cell cycle arrest the aberrant mitotic phenotype of spp1 mutants is due to failure to induce Chk1 phosphorylation, not due to inability to activate Cds1 kinase. On the basis of the results of this study and previous in vitro kinetic studies of primase by others (Kuchta et al., 1990), we propose how mutations of spp1+ might affect the Chk1-mediated S-M phase checkpoint response.
MATERIALS AND METHODS
Strains and Genetic, Molecular, and Cell Biology Techniques
All strains used in this study were derived from the wild-type strain 972 h−. Unless stated otherwise, cells were grown in either YE5S-rich media, or EMM minimal media containing required supplements as described (Moreno et al., 1991). Standard crosses were performed on minimal media lacking ammonium chloride, as previously described (Gutz et al., 1974). Mutants were isolated by either random spore or tetrad analysis, where appropriate. Standard molecular biology techniques were performed as described in Maniatis et al. (1982). Transformation of fission yeast was achieved by the lithium acetate method of Kanter-Smoler et al. (1994) or Bahler et al. (1998). For survival analysis of spp1 mutants, cells were grown in rich media to a density of 2 × 106/ml at the permissive temperature of 26°C and then were shifted to the nonpermissive temperature of 36.5°C. At indicated times after the temperature shift, defined aliquots of cells were removed, diluted, and plated in duplicate on rich media. Surviving colonies were allowed to grow for 4 days at 26°C, and survival was expressed as a percentage of the number of cells surviving at time zero. For cytological analysis, cells were fixed in 70% ethanol and stained with the DNA-specific dye 4′,6-diamidi-2′-phenylindole (DAPI) as previously described (Uchiyama et al., 1997).
Growth curves of spp1 mutants were determined in rich media at the indicated temperature, with a starting cell density of 2 × 106/ml. Four hundred-microliter samples were removed hourly and fixed in 1.6 ml of formolsaline. Samples were diluted 10-fold in Isoton (Becton-Dickinson, Mountain View, CA), and cell number was determined using a Coulter counter. For hydroxyurea (HU) block experiments, HU was added to cultures growing in rich media to a final concentration of 11 mM. For Figure 6, A and B, a further 11 mM HU was added to the culture immediately before the temperature shift from 26 to 36.5°C. Synchronous cell cultures using cdc25-22 was performed by growing cells to a density of 3.5 × 106 in rich media and then shifting to the nonpermissive temperature of 36.5°C for 3.5 h. Release was performed by rapid cooling to 26°C, and samples were analyzed at the indicated times. Synchrony by HU arrest was achieved by growth to 4 × 106 cells/ml in rich media followed by incubation in 11 mM HU for 3.5 h at 32°C. Cells were filtered, washed with an equal volume of prewarmed media, and reinoculated in fresh media. Arrest points from nitrogen-starved cells were determined by starving cells for 14 h in minimal media lacking ammonium chloride at 26°C, increasing the temperature to 36.5°C for 1 h, and then adding ammonium chloride to the cultures. FACS analysis was performed using cells fixed in 70% ethanol as previously described (Sazer and Sherwood, 1990).
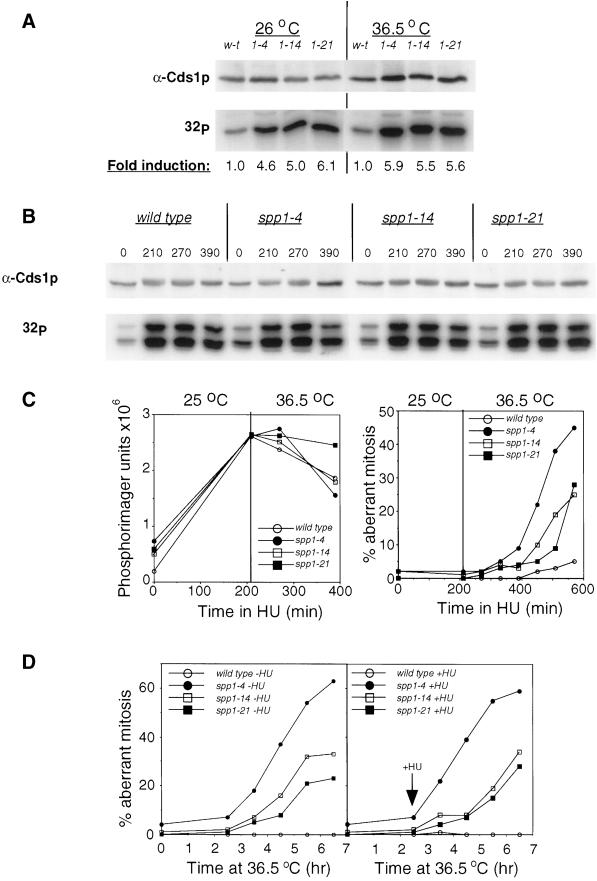
Figure 6.
Cds1 kinase response in the spp1 mutants. (A) Cds1 kinase activity in spp1 mutants. Extracts were prepared from wild-type (w-t) and the spp1 mutants at either the permissive temperature (26°C) or after incubation at the nonpermissive temperature (36.5°C) for 3.5 h. Cds1 kinase was immunoprecipitated from cell extract and assayed for kinase activity using MBP as substrate as described in MATERIALS AND METHODS. Top: a control blot of 1/30th of the extract used for immunoprecipitation; bottom: autoradiograph of the Cds1 kinase activity of the immunoprecipitates. Fold induction was calculated using the activity of the wild-type control at the respective temperature as 1.0 and was quantified using Phosphorimager software. (B) Cds1 kinase activity induced by hydroxyurea (HU) arrested wild-type and spp1 mutant cells. Cell cultures were synchronized with 11 mM HU for 3.5 h at 26oC after which fresh HU (11 mM) was added to each culture, and cultures immediately were transferred to a water bath for further incubation at the nonpermissive temperature of 36.5oC. Cell samples were removed at the indicated times and lysed. Cds1 protein was immunoprecipitated and assayed for Cds1 kinase activity. Top: a control blot with anti-Cds1 antibodies of 1/30th of the cell extract used for immunoprecipitation; bottom: autoradiograph of Cds1 kinase activity using MBP as substrate. (C) Left: quantification of the Cds1 kinase activity shown in B; right: percentage of cells displayed aberrant mitosis determined by microscopic examination of DAPI-stained cells at the indicated times. (D) The percentage of cells displaying aberrant mitosis in wild-type and spp1-arrested cells at 36.5°C, either in the absence (left) or presence of 11 mM HU (right) added 2.5 h after shift to the nonpermissive temperature. Cells were scored by DAPI staining of ethanol-fixed samples taken at the indicated times.
Construction of spp1Δ Strains
A genomic clone of spp1+ was isolated by colony hybridization from the pURSP1 library (Barbet et al., 1992). Briefly, a known fragment of spp1 was amplified by PCR from the genomic library using primers DG1 (5′-ATGCCCAAGCCGATTTGAAGTTGGA-3′) and DG2 (5′-CGGATCTTGGTCTTCAAGGACAA-3′). This 500-bp fragment was radiolabeled with [α-32P]dCTP as described (Maniatis et al., 1982) and used to probe a bacterial library that had been transformed with the SP1 library. Two clones were isolated to single colonies by repeated probing, isolation, dilution, and reprobing. Both were found by restriction mapping to contain a 3-kb fragment that contains the entire spp1 open reading frame (ORF) and ∼1 kb of sequence 5′ to the ORF, and 500 bp 3′ to the ORF (Figure 1B). The 1.5-kb ClaI-ClaI fragment (that contains all but the first 4 codons, and extends 16 bp past the Stop codon) was excised, and a ClaI-AccIII linker was inserted (5′-CGTATTCCGGAATA-3′). This derivative was used to insert the ura4+ gene that had been excised from the plasmid pUD18 (Barbet et al., 1992). The resulting construct was excised from the pUR vector with KpnI-PstI, and the linear fragment was used to transform the h+/h− strain KG23 (ade6-M210/ade6 M216 leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1). Cells were selected for uracil prototrophy, and accurate integration was confirmed by Southern blot analysis. Two correct isolates were induced to sporulate by incubation on minimal media containing adenine and lacking ammonium chloride. Dissection of the resulting tetrads produced no viable uracil prototrophs, confirming that Spp1 is essential for cell viability. To examine the terminal phenotype of spp1-deleted germinating spores, the spp1+/spp1Δ diploid was induced to sporulate as above, and spores were treated overnight with zymolyase to remove the ascus wall and any residual vegetative cells. Resulting spores were washed three times with water and inoculated into minimal media lacking uracil at a density of ∼4 × 106/ml. Cultures were incubated at 32°C, and samples were removed and fixed in 70% ethanol for FACS and cytological analysis at indicated times. The wild-type control for this experiment was strain PN1842 (ade6-M210/ade6-M216 ura4-D18/ura4+ leu1-32/leu1-32 his3-D1/his3+ h+/h−) and was treated in parallel to the spp1Δ strain.
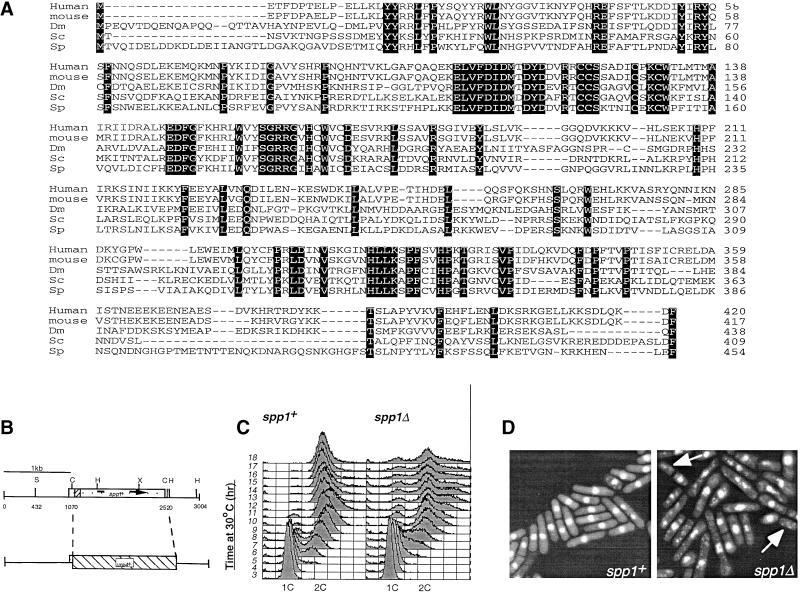
Figure 1.
Identification and deletion of the fission yeast Spp1. (A) Alignment of the amino acid sequence of fission yeast Spp1 with the homologous proteins from human, M. musculus (mouse), D. melanogaster (Dm), and S. cerevisiae (Sc). Sequences and residues that are identical to Spp1 are shaded in black. (B) Deletion of the open reading frame (ORF) of spp1+ was achieved by replacing the ClaI-ClaI fragment that contains the majority of the ORF with the ura4+ gene. S, SalI; C, ClaI; H, HindIII; X, XbaI. This linear construct was transformed into a wild-type h+/h− diploid, and uracil prototrophs were selected. (C) FACS analysis of germinating spores arising from either spp1−/spp1+ (right) or spp1+/spp1+ (left) diploids. Time points were taken hourly after the inoculation of spores in minimal media containing nitrogen, but lacking uracil. (D) Morphology of either spp1+ (left) or spp1− (right) cells 18 h after germination in nitrogen-containing media lacking uracil. Cells displaying the “cut” phenotype are indicated with an arrow.
Immunofluorescence Studies
An spp1-GFPKan strain expressing Spp1 tagged at the C-terminus with the Green Fluorescent Protein (GFP) was constructed by the PCR-based method recently described by Bahler et al. (1998). The spp1-GFPKan PCR product was transformed into the strain PN670 (h+/h− ade6-M216/ade6-M210), and transformants were selected for kanamycin resistance. Successful transformants were induced to sporulate and germinated on rich media containing kanamycin. Western blotting of total protein extracts by anti-Spp1 antibody confirmed the loss of the wild-type Spp1 protein (52 kDa) and the appearance of the Spp1-GFP fusion protein (∼80 kDa). For immunostaining with anti-GFP (gift from K. Sawin) and antitubulin (anti-Tat1, gift from K. Gull) antibodies, cells were grown in rich media, and ∼2.5 × 108 cells were harvested per sample, either by centrifugation or filtration. Cells were fixed by the addition of 10 ml of −80°C methanol and processed for immunofluorescence essentially as described (Hagan and Hyams, 1988). Secondary antibodies (Alexa; Molecular Probes, Eugene, OR) were used at 1:1000 dilution, and cells visualized using a Hamamatsu (Bridgewater, NJ) cooled CCD camera and software.
Isolation of Temperature-sensitive spp1 Mutants
Temperature-sensitive mutant alleles of the spp1 gene were constructed by the method described by Tatebayashi et al. (1998). The ura4+ gene was inserted into the 3′ClaI site of the genomic spp1+ clone in pUR19 (described in Figure 1B). The resulting 4.7-kb construct was excised by digestion with KpnI and PstI, gene-cleaned, and used as a PCR template for PCR mutagenesis using the primers Spp1–843F (5′-GCACAGAGTTTGATACGTATCTCG-3′) and Spp1-12975R (5′-CAAATCAATCTGATAGTGAATTGG-3′). Reactions were performed in 100 μl volume using 10 μl of 10× Taq buffer, 10 μl of 2.5 mM dNTPs, 40 pM primer 1, 40 pM primer 2, 10 ng of template DNA, and 1 μl of Taq polymerase. MnCl2 was added to the reaction mixture at 0.1, 0.2, and 0.3 mM final concentration, and PCR was performed over 30 rounds of 95°C for 1 min, 56°C for 1 min, and 72°C for 5 min. Reaction products were pooled, extracted with phenol:chloroform, ethanol-precipitated, and used to transform yeast strain DG501 (ade6–704 leu1-32 ura4-D18 h−). After 4 days' growth on selective minimal media at 26°C, ∼3500 uracil prototrophs were replica-plated to rich media containing phloxin B and incubated overnight at 36.5°C. Temperature-sensitive colonies were identified and selected for further analysis. All three mutants presented in this article were confirmed to be stable uracil prototrophs, with the spp1:ura4 construct integrated at the spp1 locus, as judged by Southern blot analysis. Backcrossing to the parental strain DG501 was used to confirm the cosegregation of uracil prototrophy with the temperature-sensitive phenotype. Finally, the temperature sensitivity of all three was fully rescued by transformation with spp1+ in pREP81.
Generation of Anti-Spp1 Polyclonal Antibodies
The spp1 cDNA was isolated from the S. pombe cDNA library in pREP3 (Norbury and Moreno, 1997) by colony hybridization, using the 1.1-kb HindIII fragment as a probe, essentially as described as above. The isolated cDNA was then used as a template in a PCR reaction using primers Spp1 NdeI (5′-GACGTTAATTCATATGACTGTTCAAAT-3′) and Spp1 SalI (5′- ATTATTACTGTCGATTAACGGAA-3′). The resulting products were cloned into pREP1, and two isolates sequenced in their entirety. Both were found to be identical to the sequence contained within the fission yeast sequence database cosmid c6B12 and had correctly spliced the 84-bp intron located at +75 of the ORF (where A of the ATG is +1). One was subcloned into the bacterial expression vector pET16b (Promega, Madison, WI) and transformed into competent BL21pLysS E. coli. His-Spp1 expression was induced by the addition of 1 mM isopropyl-thiogalactopyranoside to a midlogarithmically growing culture at 30°C for 4 h. Cells were lysed by freeze/thawing in a hypotonic buffer (50 mM Tris, pH 8.0, 2 mM EDTA), and his-tagged Spp1 was purified according to the manufacturer's protocol in 6 M guanidinium hydrochloride (his-Spp1 is insoluble). Eluted protein from the Ni3+ resin was dialyzed overnight with 50 mM Tris-HCl and 8 M urea, and the protein was further purified by extraction from a 9% SDS-polyacrylamide gel slice. This gel slice (∼300 μg) was used as an antigen for immunization into rabbits. Polyclonal sera was affinity-purified by coupling ∼1 mg of his-Spp1 to Affigel (Bio-Rad, Hercules, CA) in 50 mM HEPES, pH 7.9, 8 M urea according to the manufacturers protocol. Sera were incubated with the his-Spp1 beads overnight at 4°C, and antibodies were eluted by serial elution with 100 mM glycine (pH 2.5) and 100 mM triethylamine (pH 11.5) into 1 M Tris-HCl (pH 7.5) as described in Lane and Harlow (1988).
Immunoblot and Immunoprecipitation
To detect expression of Polα, Spp1, and Spp2 in cells, 10 ml of cell culture incubated at either permissive or restrictive temperature were grown for 5 h to a cell density of 107/ml and harvested. Cells extracts were prepared by glass bead disruption of cells in a lysis buffer containing 250 mM KCl, 150 mM HEPES, pH 7.9, 1 mM EDTA, 10% glycerol, and a mixture of protease inhibitors. For immunoblot analysis, 1.5 μg of total cell extracts were fractionated on a 8% SDS gel, transferred onto a membrane, and probed with either anti-Polα (Park et al., 1993) at 1:5000 dilution, anti-Spp2 (Tan and Wang, 2000) at 1:1000 dilution, or anti-Spp1 as described above at 1:500 dilution overnight at 4°C.
The Polα–primase immnocomplex was precipitated from cell lysates with anti-Polα antibody immobilized on Protein-A agarose. Crude cell extract proteins (150 μg) were incubated with 10 μl of Protein-A agarose in the above lysis buffer for 1 h at 4°C with end-to-end rotation. The immunocomplex was then collected, washed three times with the lysis buffer, resuspended in 30 μl of SDS sample loading buffer, and fractionated on SDS gel, followed by immunoblotting with antibodies against Polα, Spp2, and Spp1.
Chk1 Mobility Shift Assays
Protein extraction of fission yeast was performed by glass bead disruption in buffer containing 250 mM NaCl, 50 mM HEPES, pH 7.9, 80 mM β-glycerophosphate, 5 mM EDTA, 0.1% NP-40, and 10% glycerol. Thirty-five micrograms of total protein was analyzed for the Chk1 mobility shift by boiling in SDS sample buffer and was separated on an 8% SDS acrylamide gel (200:1 acrylamide:bisacrylamide) followed by immunoblotting with the anti-hemagglutinin (HA) mouse monoclonal antibody 12CA5.
Cds1 Protein Kinase Assays
Protein kinase assays were performed on Cds1 as previously described (Lindsay et al., 1998). Briefly, total protein was prepared from 25 OD595 units of cells by glass bead disruption in lysis buffer (as above). Soluble protein was obtained by centrifugation (13,000 rpm, 10 min), and Cds1 was immunoprecipitated from 1 mg of soluble protein in 300 μl volume, using 2 μl of affinity-purified rabbit anti-Cds1 antibody and 10 μl of Protein A agarose (5:1 slurry) (Lindsay et al., 1998). As a control for equal starting material, 1/30th of the sample was removed before immunoprecipitation and analyzed by Western blotting, because we were unable to quantify the immunoprecipitation (Cds1 comigrates with the IgG heavy chain at ∼50 kDa). Immunoprecipitates were washed five times with lysis buffer and an additional three times with kinase buffer (10 mM HEPES, pH 7.9, 75 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT). Kinase reactions were performed at 30°C for 15 min in a 20 μl volume using 5 μg of myelin basic protein (MBP) as a substrate in the presence of 5 μCi of [γ-32P]ATP and 100 μM ATP. Reactions were terminated by boiling in an equal volume of 2× SDS sample buffer, and products were analyzed by 12% SDS PAGE. Gels were stained with Coomassie Brilliant Blue, fixed in 40% methanol and 10% acetic acid, dried under vacuum, and exposed to film.
RESULTS
Isolation of spp1+ Gene
To facilitate the characterization of the DNA polymerase α holoenzyme and interacting proteins, a biochemical purification of the fission yeast DNA polymerase α catalytic subunit was developed (Davis and Wang, unpublished results). A protein of approximate molecular weight of 50 kDa that reproducibly copurified with Polα catalytic subunit was identified by mass spectrometry to contain residues that were homologous to the p49/PRI1 primase catalytic subunit from human and yeast. This protein also identified an identical peptide sequence within the fission yeast genome sequence project with the coding region residing in chromosome I (cosmid c6B12). Further characterization of this region identified the entire ORF of the fission yeast homologue of mammalian primase p49 and budding yeast primase PRI1, which we have named Spp1 (S. pombe primase 1). This sequence displays 41, 40, 41, and 38% sequence identity to the p49/PRI1 from S. cerevisiae, human, mouse, and Drosophila, (Lucchini et al., 1987; Bakkenist and Cotterill, 1994; Stadlbauer et al., 1994), respectively (Figure 1A), and differs only in its extended N-terminal, which is conserved with the Drosophila homologue. Colony hybridization of a S. pombe genomic library (Barbet et al., 1992) was used to isolate a genomic clone that contained the entire ORF.
Spp1 Is Essential for Growth
The ClaI-ClaI fragment that contains all but the first four codons of the spp1+ ORF was replaced with the ura4+ selectable marker (Figure 1B). This construct was transformed into an h−/h+ heterothallic diploid strain, and transformants selected for uracil prototrophy. Resulting colonies were induced to sporulate, and tetrad dissection failed to produce any viable haploid ura+ colonies, indicating that in fission yeast spp1+ is essential for vegetative growth.
To investigate the terminal morphology of cells harboring spp1Δ, a spore germination experiment was performed. FACS analysis of germinating spores in media that selected for the spp1::ura4+ allele demonstrated that spp1Δ spores were retarded in S-phase progression in comparison to a wild-type control (Figure 1C, compare the 8-h time point). However, unlike cells deleted for polα+, which arrest with a 1C DNA content (Bhaumik and Wang, 1998), spp1Δ germnating spores were able to complete S phase, and exhibited a 2C DNA content. Examination of the cell morphology demonstrated that the cells had prevented mitotic entry and displayed predominantly elongated cell morphology, with <20% of cells displaying the abnormal mitotic nuclear phenotype (Figure 1D, arrows indicate “cut” cells). This is similar to the germinating spores harboring the deletion of spp2 (Tan and Wang, 2000), but in contrast to polαΔ germinating spores, which have ∼60% of the germinating spores displaying abnormal mitotic phenotype (Bhaumik and Wang, 1998). Because cells with spp1Δ divided several times and then died, the observed cdc phenotype and 2C DNA profile of spp1Δ most likely were due to the carryover of residual Spp1 protein from the diploid.
Spp1 Localizes in the Nucleus and Is Constitutively Expressed throughout the Cell Cycle
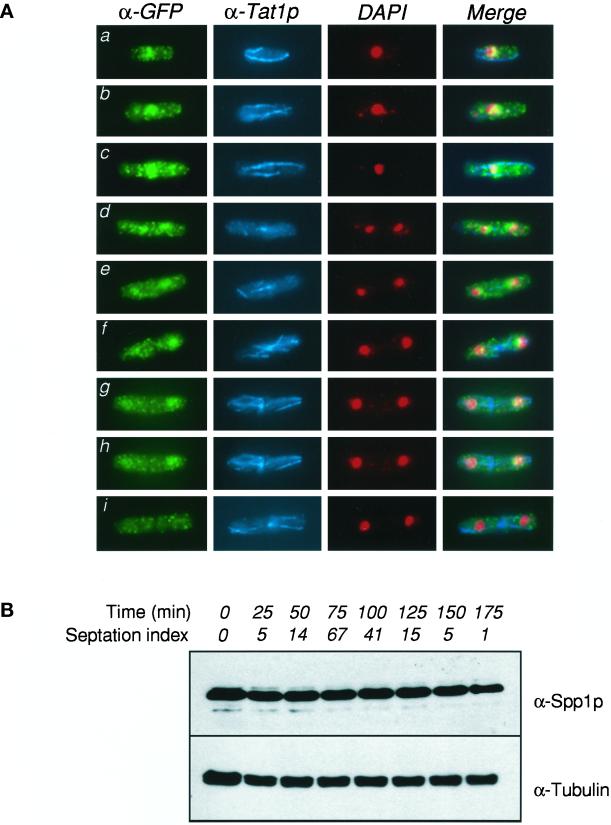
To investigate the cellular localization and expression of Spp1 during the cell cycle, we constructed a wild-type strain expressing an Spp1-GFP protein fusion, where the spp1-gfp gene fusion is integrated and under the control of the endogenous spp1+ promoter. Exponentially growing cells were examined for the presence of the Spp1-GFP fusion by fixation and immunoflourescence with anti-GFP antibody (Figure 2A). As an indicator of cell cycle stage within individual cells, samples were costained with antibodies to Tat1, which recognizes tubulin (Figure 2A). Typical G2 cells (a, b) displayed a punctate cytoplasmic staining that is characteristic of the polyclonal anti-GFP antibody on untagged strains (data not shown). A concentration of signal toward the middle of the cell, in a region coincidental with the nuclear material, was observed. A merge of the anti-GFP and DAPI signals, however, suggested a concentration of Spp1-GFP in peri-centric nuclear regions, and such separation of DAPI and GFP signals was more obvious in longer cells (c), which are presumably late G2/premitotic cells. The difference between nuclear DAPI-stained material and the GFP signal was more apparent when mitotic cells were examined (d, e; note the mitotic spindle), where it was seen to trail the condensed mitotic chromosomes to the poles of the dividing cell. As the nuclei decondensed and returned to the centers of the newly divided cells (f, g, h; note the post-anaphase array), the GFP signal again colocalized with the DAPI-stained material. We consistently observed, however, that a high proportion of binucleate cells displayed much-reduced GFP staining compared with a uninucleate or mitotic cell (i), despite efficient staining with anti-Tat1 antibodies. These observations of differential cellular staining were further confirmed using synchronous cells obtained by release from a cdc25-22 block.
Figure 2.
Immunolocalization of Spp1 in asynchronous cells. (A) Localization of Spp1-GFP (α-GFP), tubulin (α-Tat1p), and DAPI-stained nuclei (DAPI) in an exponential population of Spp1-GFP–expressing cells. Cells at different stages of interphase and mitosis are shown. (B) The expression of Spp1 protein is not cell cycle regulated. Total protein was prepared from cell samples at 25-min intervals after release from a cdc25-22 block, as described in MATERIALS AND METHODS, and probed with anti-Spp1 polyclonal antisera (top), or anti-Tubulin (bottom) as a loading control. The septation index is given as an estimate for cell synchrony and approximate time of S phase.
Because the levels of anti-GFP signals were seen to vary somewhat during the cell cycle, we tested whether this reflected a difference in the expression of total Spp1 protein during the cell cycle. Using polyclonal anti-Spp1 antibodies on protein samples obtained from cells released from a cdc25-22 arrest, we found that the Spp1 protein was expressed at a constant level throughout the cell cycle (Figure 2B). We, therefore, consider it likely that the antigenic site recognized by the anti-GFP antibody shown in Figure 2A is transiently masked in bincucleate cells, possibly as a result of protein–protein and/or protein–DNA interactions.
Isolation of Temperature-sensitive Mutants of spp1
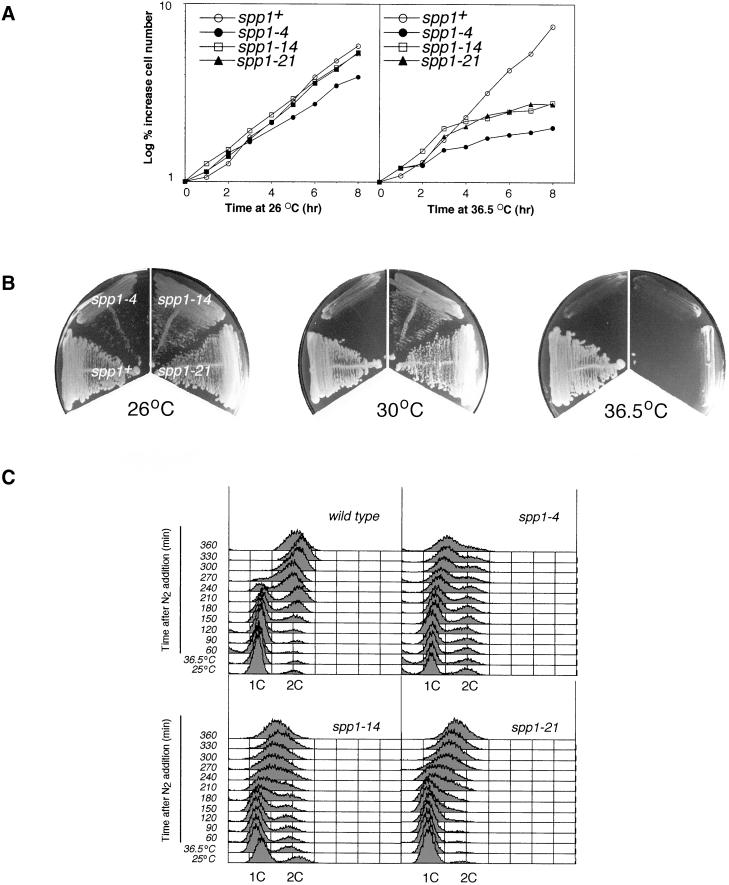
To investigate the effect of spp1 mutation on the cell cycle checkpoint response, we isolated a panel of thermosensitive mutants of spp1+ as described in MATERIALS AND METHODS. Three representative mutants are described here, two of which, spp1-14 and spp1-21, displayed near wild-type growth rates at the permissive temperature (Figure 3A, left). The third, spp1-4 was found to exhibit an ∼20% slower growth rate at 26°C (3 h 40 min versus 3 h at 26°C in rich media; Figure 3A, left). Importantly, all three were found to cease cell division in the first cell cycle after shift to the nonpermissive temperature (Figure 3A, right). These three mutants were found to differ in their temperature sensitivity, with spp1-4 being the most temperature sensitive, followed by spp1-14 and spp1-21. For example, at 30°C, spp1-4 was unable to form colonies, and spp1-14 grew poorly, whereas spp1-21 grew well (Figure 3B). As predicted from the growth rate analysis, all three mutants were unable to form colonies at 36.5°C (Figure 3B). When cells were synchronized by nitrogen starvation and released into the nonpermissive temperature, wild-type cells completed replication and exhibited a 2C DNA profile 270 min after being released into rich media. In contrast, majority of the three spp1 mutant cells, as expected, arrested in mid–S phase with a 1.5 C DNA content (Figure 3C). The is consistent with the fact that leading strand DNA synthesis, which only requires Spp1 to initiate at the origin, contributes to the DNA content profile.
Figure 3.
Temperature-sensitive spp1 mutants. (A) Growth curve of wild-type and spp1-4, spp1-14, and spp1-21 mutants in rich media at either the permissive (left) or nonpermissive (right) temperature. Cell numbers were determined hourly using a Coulter counter. (B) Temperature sensitivity of the spp1 mutants at 26, 30, and 36°C. Cells were streaked onto rich media and allowed to grow for 4 days at the indicated temperature. (C) FACS analysis of spp1+ (wild-type), spp1-4, spp1-14, and spp1-21 mutants released from nitrogen-starved G1-arrested cells by addition of ammonium chloride and incubation at the nonpermissive temperature.
Characterization of the Temperature-sensitive Mutant Alleles of spp1
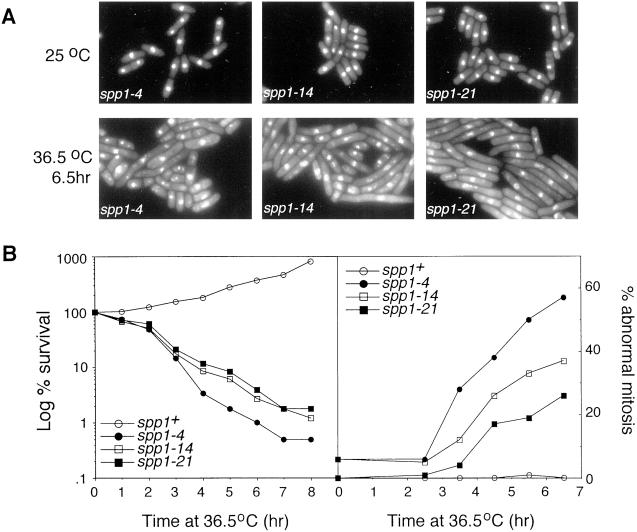
We further characterized the terminal morphology of the spp1 thermosensitive mutants at the nonpermissive temperature. Surprisingly, we found that the phenotype of the spp1 mutants at the nonpermissive temperature was allele specific (Figure 4). Although all cells appeared morphologically wild type at 26°C, incubation at 36.5°C produced a high percentage of spp1-4 cells with an aberrant mitotic phenotype, whereas a high percentage of spp1-21 cells displayed a cdc phenotype. Mutant spp-14 had some cells that exhibited aberrant mitotic phenotype, yet the majorities were cdc in appearance (Figure 4A; quantified in 4B, right). Despite this difference in terminal morphology, all three mutants lost viability with approximately the same kinetics when asynchronous cells were shifted to the nonpermissive temperature (Figure 4B, left). Finding that spp1-21 lost viability with kinetics similar to spp1-4 suggests that the ability to prevent premature mitosis in spp1-21 does not contribute to its ability to recover from S-phase arrest at the restrictive temperature. Furthermore, it is noteworthy that even in the case of spp1-4, which displayed the highest percentage of cells with abnormal mitosis, the survival at which point cells started to display an abnormal mitotic phenotype (∼2.5 h after shift to 36.5°C) was already 30%. These data suggest that despite the phenotypic differences between spp1-4 and spp1-21 at the nonpermissive temperature in terms of checkpoint arrest, the primary cause of lethality is most likely due to an inability to recover DNA synthesis.
Figure 4.
Analysis of the spp1 mutants at the nonpermissive temperature. (A) Microscopic analysis of DAPI-stained spp1-4, spp1-14, and spp1-21 cells at either the permissive temperature (top) or after 6.5 h incubation at the nonpermissive temperature (bottom). (B) Survival curve of spp1 mutants at the nonpermissive temperature (left), determined by the percentage survival of cells after indicated times at 36.5°C in rich media. The percentage of cells displaying abnormal nuclear morphology (right) was determined by microscopic examination of DAPI-stained cells at the indicated times.
Analysis of Spp1 Mutant Protein
We have previously found that thermosensitive mutations of spp2 significantly destabilize the Polα–primase complex and that the aberrant mitotic phenotype observed in spp2 mutants correlates to the severity of Polα–primase instability (Tan and Wang, 2000). We thus tested whether mutations of spp1+ will also compromise the Polα–primase complex stability and whether the observed allele-specific abnormal mitotic phenotype correlates to the severity of the Spp1 mutation-induced Polα–primase complex defect.
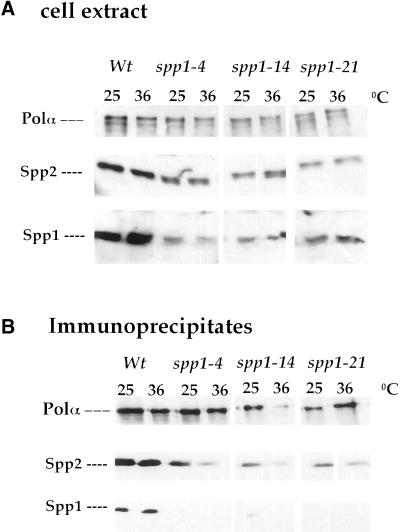
We first tested the Spp1 protein levels in the spp1 mutants and compared with that of the wild-type cells. Cell extracts were prepared from wild-type cells and the three spp1 mutants after incubation at the permissive or restrictive temperature for 5 h and probed with antibodies against Polα, Spp1, or Spp2 (Figure 5A). At either temperature, all three spp1 mutants had lower levels of Spp1 protein compared with wild-type extract (Figure 5A). Furthermore, mutant spp1-4 at 36°C had lower levels of Spp1 protein than at 25°C and also slightly lower levels than that of spp1-14 and spp1-21. In contrast, spp1-21 had comparable levels of Spp1 protein at either 25 or 36°C. Thus, all three spp1 mutants have reduced steady-state levels of the Spp1 mutant protein.
Figure 5.
Analysis of the Spp1 mutant proteins. (A) Levels of Spp1 protein in wild-type and spp1 mutants at the permissive and the restrictive temperature. Total cell extracts prepared from cells grown for 5 h at 25 or 36°C were prepared and analyzed by immunoblotting as described in MATERIALS AND METHODS. (B) Analysis of the association of mutant-Spp1 proteins with the Polα immunocomplex. Polα immunocomplex was analyzed by immunoblotting with antibodies against Polα, Spp2, and Spp1 as described in MATERIALS AND METHODS.
We next tested the association of Spp1 with Polα-Spp2 in these three spp1 mutants. Wild-type and spp1 mutant cells incubated at either 25 or 36°C for 5 h were tested for coimmunoprecipitation of Spp1 with Polα-Spp2 by anti-Polα antibody. The Polα immunocomplexes were then probed with antibodies against Polα, Spp1, and Spp2 (Figure 5B). In wild-type cells at either the permissive or the restrictive temperature, Spp1 coimmunoprecipitated with Polα and Spp2 as an intact Polα–primase complex. Surprisingly, all three spp1 mutants had barely detectable Spp1 protein coprecipitated with Polα at either the permissive or the restrictive temperature. These results thus suggest that suboptimal levels of Spp1 proteins in all three spp1 mutants (Figure 5A) compromised the Polα–primase complex subunit association, especially the association of Spp1 protein with Polα-Spp2. Given the fact that spp1-4 had lower amount of Spp1 mutant protein expressed at 36°C than at 25°C and that in spp1-14 and spp1-21, this suggests that Spp1 protein of spp1-4 mutant has a more severe defect than the Spp1 mutant proteins in spp1-14 and spp-21.
It is important to mention that mutations of spp1 also affected the ability of Spp2 protein to associate with Polα. Comparison of the ratio of Polα protein levels to Spp2 protein levels at 25 to 36°C indicated that mutation of spp1-4 allele reduced the affinity between Polα and Spp2 protein (Figure 5B; see Spp2 level in spp1-4 lane at 36°C). In contrast, the ratio of Polα to Spp2 protein levels in spp1-14 and spp1-21 at 25 and 36°C did not exhibit a significant difference.
Together, these experiments indicate that thermosensitive mutations of spp1+ cause a decrease of the cellular steady-state levels of Spp1-mutant protein. This may compromise the cellular levels of Polα–primase complex formation. Importantly, at the restrictive temperature Spp1 mutant protein of spp1-4 seems to have more severe defects than the Spp1 mutant proteins in spp1-14 and spp1-21, and an spp1-4 mutant also exhibits a more severe aberrant mitotic phenotype.
Cds1 Responses of the spp1 Mutants
Finding that the spp1 thermosensitive mutants exhibit allele-specific aberrant mitotic phenotype at the restrictive temperature (Figure 4A) led us to investigate the checkpoint response of these mutants. We were unable to isolate double mutants of the spp1 mutants with any of the checkpoint rad deletion mutants or the cds1Δ mutants. This is in common with the thermosensitive mutants of polα (Bhaumik and Wang, 1998) and spp2 (Tan and Wang, 2000). This finding is thought to be due to a requirement for Cds1 function in coordinating S phase in the presence of a perturbed replication complex. Because, in our hands, overexpression of cds1+ by thiamine derepression of the nmt81 or nmt41 promoter causes severe cell cycle delay, we have been unable to study the terminal phenotype of any spp1 cds1Δ double mutant with pREP switch-off strains. However, we were able to measure the relative activity of the Cds1 kinase using MBP as a substrate at the permissive and nonpermissive temperature (Figure 6A). At 26°C, we observed an increase of four- to sixfold in Cds1 activity over wild-type levels, suggesting that 26°C was semipermissive for the spp1 mutants' growth and that these spp1 mutants required Cds1 kinase function at this temperature for viability, because complete loss of Cds1 in spp1 mutants would be a lethal event. At the nonpermissive temperature, levels of Cds1 were increased slightly relative to the levels observed at 26°C (even in wild-type), but they were still only approximately sixfold over wild-type levels at 36°C. Importantly, there was no correlation between the spp1 arrest-induced Cds1 kinase activity and the subsequent cell-cycle checkpoint defect of the respective spp1 mutants, suggesting that, in cells arrested by spp1 mutation, Cds1 kinase activity does not play a major role in preventing mitosis at the nonpermissive temperature.
Cds1 kinase activity is highly activated after exposure of wild-type cells to HU (Lindsay et al., 1998). We have found that temperature-sensitive mutations of spp2 exhibit an allele-specific defect of HU-induced Cds1 kinase activation, but not the spp2 arrest-induced Cds1 kinase activation (Tan and Wang, 2000). Upon finding that the Cds1 kinase activity was induced in all three of the spp1 mutants after arrest at 36.5°C, we investigated a potential role for Spp1 in the activation and/or maintenance of the HU-induced Cds1 kinase activity. At 26°C, HU-induced Cds1 kinase activity strongly in wild-type cells and all three of the spp1 mutants (Figure 6B; compare 210-min time point to 0 min). Upon incubation at the nonpermissive temperature for 3 h (390-min time point), the HU-induced Cds1 kinase activity in all three mutants was found to decay, with kinetics indistinguishable from wild-type (Figure 6C, left graph). These data indicate that the spp1 mutants are defective neither in the HU-induced Cds1 kinase activation nor in the maintenance of HU-induced Cds1 kinase activity after activation. It is noteworthy that despite the high levels of HU-induced Cds1 kinase activity in the spp1 mutants, all three spp1 mutants began to enter inappropriate mitosis after ∼2- to 2.5-h incubation at the restrictive temperature. Greater than 40% of spp1-4 and nearly 30% of both spp1-14 and spp1-21 entered aberrant mitosis after prolonged incubation at the nonpermissive temperature (Figure 6C, right graph). Therefore, despite high levels of Cds1p activity, induced by HU, spp1 mutants entered mitosis with kinetics approximately equal to those observed in the absence of HU at the nonpermissive temperature (see Figure 6, A and C).
Having determined that Cds1 kinase can be fully activated by HU in the spp1 mutants, we then investigated whether HU could rescue the abnormal mitotic phenotype when cells were presynchronized at the spp1 arrest point. As shown in Figure 6D, the addition of HU to spp1 cells that had been arrested for 2.5 h at the nonpermissive temperature had no effect on the kinetics of the accumulation of the aberrant mitotic phenotype. Therefore, the HU-activated Cds1 kinase activity is unable to suppress the abnormal mitotic phenotype associated with spp1 mutants, irrespective of whether HU is added before, or subsequent to, arrest at the nonpermissive temperature.
Chk1 Response of the spp1 Mutants
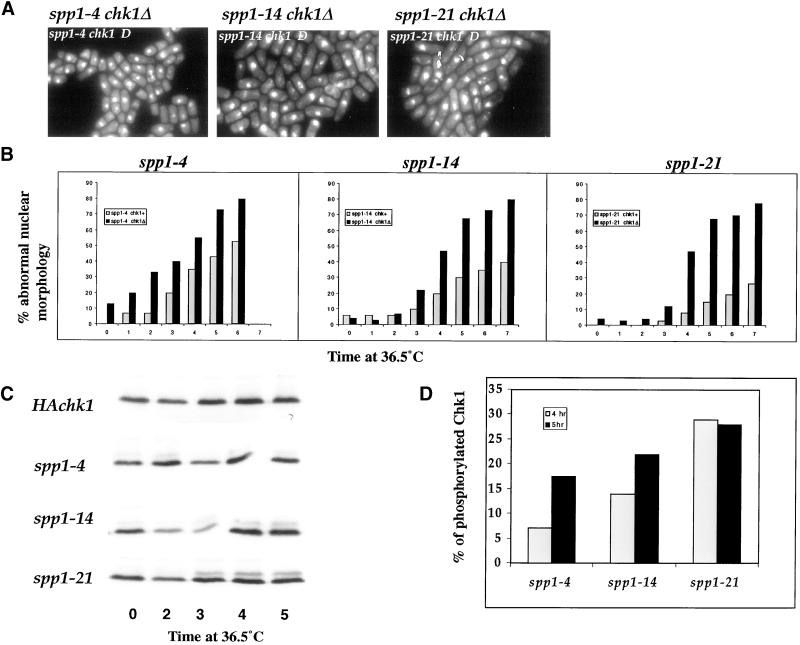
Finding that Cds1 kinase does not play a major role in preventing aberrant mitotic entry of the spp1 mutants, we investigated the Chk1 response in these mutants. In contrast to cds1Δ, all three spp1 mutants were viable in chk1Δ background at the permissive temperature. Similar to polα and spp2 thermosensitive mutants (Bhaumik and Wang, 1998; Tan and Wang, 2000) as well as other temperature-sensitive cdc mutants that arrest S-phase progression, spp1 chk1Δ double mutants displayed a severe mitotic checkpoint defect in comparison to respective single mutants when arrested at the nonpermissive temperature (Figure 7A). We then compared the kinetics of each spp1 mutant entering aberrant mitosis in Chk1+ or Chk1Δ background at 36.5°C (Figure 7B). As shown in Figure 7B, all three spp1 mutants had a substantially higher percentage of cells that entered aberrant mitosis in the chk1Δ background than in the chk1+ background throughout 6-h incubation at 36.5°C. Mutant spp1-21 showed the greatest difference between the chk1+ and chk1Δ background, whereas spp1-4 had the least difference. A strain expressing a three-HA epitope–tagged Chk1 protein has previously been shown to undergo a phosphorylation-dependent mobility shift after DNA damage and S-phase progression arrest, using temperature-sensitive cdc mutants, and this has been used as a biochemical marker for Chk1 activation (Walworth and Bernards, 1996). We thus examined the ability of the spp1 mutants to induce phosphorylation of Chk1 at the nonpermissive temperature (Figure 7C). After incubation at the restrictive temperature for 3 h, the phosphorylated form of Chk1 was observed in spp1-21, and slightly lower levels of phosphorylated Chk1 were detectable in spp1-14. In contrast, spp1-4 had barely detectable levels of phosphorylated Chk1 (Figure 7C). To ascertain that the observed differences of Chk1 phosphorylation in these three mutants were not due to uneven loading of the cell extracts, we quantified the percentage of phosphorylated Chk1 in total Chk1 detected in each mutant after 4 and 5 h incubations at 36°C (Figure 7D). The quantification was performed with an exposure of the gel in which the HA-tagged Chk1 signals were in linear range. The percent of Chk1 in spp1-4 being phosphorylated was 7.2 and 17.5% at 4 and 5 h, respectively. Mutant spp1-14 had 14 and 22% of the total Chk1 phosphorylated, whereas spp1-21 had 29 and 28.5% of Chk1 phosphorylated. Thus, there is a correlation between the aberrant mitotic phenotype of the spp1 mutants and a lower ability to induce and/or maintain the phosphorylated form of HA-Chk1.
Figure 7.
Chk1 response in the spp1 mutants. (A) Photomicrographs of DAPI-stained spp1 chk1Δ double mutants after incubation at the nonpermissive temperature for 6 h. (B) Comparison of the percentage of abnormal nuclear morphology in spp1 mutant cells at 36°C in chk1+ (gray bar) or chk1Δ (black bar) background. Logarithmic-growth spp1 mutants in either chk1+ or chk1Δ background were shifted to 36°C, and cell samples were removed at the indicated time, stained by DAPI, and scored for the percentage of cells that displayed an abnormal mitotic nuclear phenotype. (C) Status of HA-tagged Chk1 phosphorylation in wild-type (HAchk1) and the spp1 mutant strains after incubation at the nonpermissive temperature for the indicated times. (D) Percent of phosphorylated Chk1 in each spp1 mutants after 4 and 5 h shifted to 36.5°C. Gray and black bars represent the percent of phosphorylated Chk1 after 4 and 5 h at 36.5°C, respectively.
DISCUSSION
To understand the checkpoint response after the aberrant initiation of S phase, we have analyzed how mutation of each subunit component of the Polα–primase complex could affect the cell cycle. In this study, we analyzed how mutation of the catalytic subunit of primase Spp1 induces the response of cell cycle checkpoint kinases. We found that thermosensitive mutations of spp1+ caused a decrease of the cellular Spp1 protein level and compromised the formation of Polα–primase complex, particularly the affinity of Spp1 protein to the Polα-Spp2. Analysis of these spp1 mutants on the checkpoint kinase responses suggests that the aberrant mitotic phenotype is due to a failure to properly phosphorylate Chk1, not activation of Cds1. Furthermore, the strong induction of Cds1 kinase activity using HU cannot suppress the aberrant mitotic entry of the spp1 mutants, suggesting that Cds1 does not play a major role in preventing the spp1 mutants' aberrant mitotic entry. We discuss these findings below and propose a model of how these spp1 mutants might fail to activate Chk1 and enter aberrant mitosis.
Analysis of spp2+ has shown that mutations of spp2+ affect the stability of Polα–primase complex and the HU-induced Cds1 kinase activation, but not the Cds1 kinase induced by early S-phase arrest by either spp2 mutants or polα mutants (Tan and Wang, 2000). These thus suggest that Cds1 response to initiation arrest by either Spp2 or Polα mutation is different from the Cds1-mediated intra–S phase checkpoint response induced by HU arrest. The Cds1 response to S-phase arrest induced by either initiation enzyme mutation or HU is also distinguished from the S-M phase checkpoint response that requires the catalytic activity of Polα (Tan and Wang, 2000). Here, we found that Cds1 kinase is activated in cells arrested in early S phase by spp1 mutants, similar to that of spp2 mutants (Figure 6A). In contrast to spp2 mutants, at the restrictive temperature, the HU-induced Cds1 kinase levels observed in all three of the spp1 mutants are essentially wild-type like, regardless of their DNA structure checkpoint defect (Figure 6, B and C). Furthermore, the kinetics of aberrant mitotic entry of spp1 mutants at the restrictive temperature in the presence or absence of HU is similar (Figure 6D). These experiments suggest that the Cds1 kinase activation induced by early S-phase arrest by spp1 mutants or by HU does not play a major role in preventing inappropriate mitotic entry of these mutants. These results also suggest that Cds1 activation induced by HU for maintaining the intra–S-phase checkpoint may not require the function of Spp1.
We have previously found that cells with polα+ deletion enter inappropriate mitosis with 1C DNA content and that activation of the S-M phase checkpoint requires the catalytic function of Polα (Bhaumik and Wang, 1998). We showed in this study that mutations of spp1+ cause a reduction in the steady-state levels of mutant Spp1 protein (Figure 5A). All mutant-Spp1 protein showed substantially decreased affinity to Spp2 and Polα (Figure 5B). Moreover, at the restrictive temperature, association of Spp2 to Polα was also compromised in all spp1 mutants, suggesting that absence of Spp1 protein could either alter the structure of Spp2 or affect the affinity between Spp2 and Polα. Thus, the formation of an optimally functional Polα–primase complex may require the presence of all these three subunits in proper stoichiometric ratio. As shown in Figure 5B, at the restrictive temperature, spp1-4 had significantly lower levels of Spp2 protein coprecipitated with Polα than that at 25°C. This suggests that Spp1 proteins in spp1-4 may be more severely compromised than the Spp1 protein of spp1-14 and spp1-21. In vitro kinetic studies have shown that there is a stringent RNA primer length requirement for Polα to use as primer. Only RNA primers synthesized by Spp1 of greater than or equal to seven nucleotides can be efficiently utilized by Polα to synthesis the iDNA (Kuchta et al., 1990). Suboptimal cellular levels of Polα–primase complex due to severely reduced levels and/or defective Spp1 protein in spp1-4 would have lower efficiency to synthesize RNA primers that can be utilized by Polα for initiation-DNA synthesis. A moderately compromised Polα–primase complex in spp1-21 would have relatively higher efficiency than spp1-4 in synthesizing optimal length RNA primers, albeit with reduced efficiency in comparison to wild-type cells. We have previously shown that the catalytic function of Polα to synthesize an iDNA structure is required for activation of the replication checkpoint. Thus, cells with polα mutant alleles that fail to synthesize an initiate DNA structure enter inappropriate mitosis and exhibit aberrant mitotic phenotype. Mutant alleles of polα that cause a stalled replication structure induce Chk1 phosphorylation and displaying a cdc phenotype (Bhaumik and Wang, 1998). At the restrictive temperature, a severely compromised Polα–primase complex in spp1-4 may have relatively low efficiency to synthesize an optimal size RNA primer that is utilizable by Polα to synthesize an iDNA. Thus, spp1-4 mutant cells fail to activate Chk1 and have a high percentage of the cells displaying aberrant mitotic phenotype (Figure 4B). The mildly comprised Polα–primase complex of spp1-21 may be able to synthesize sufficient levels of optimal size RNA primers to allow iDNA structure synthesis by Polα. Therefore, the spp1-21 cells are able to activate Chk1 and exhibit a cdc phenotype (Figure 4B). We thus propose that the differential activation of Chk1 in spp1 mutants (Figure 7) is the consequential effect of each mutant's capability to efficiently synthesize RNA primers of an appropriate length for Polα to utilize for synthesis of the iDNA structure.
The molecular details of how the aberrant initiation of DNA synthesis activates a checkpoint response are not yet clear. Our systematic mutational analysis of the three critical genes involved in initiation, polα+, spp2+, and spp1+, have provided evidence suggesting the following: (i). The catalytic function of Polα to synthesize an iDNA structure is a prerequisite for the activation of the S-M phase checkpoint (Bhaumik and Wang, 1998). (ii). A normal Spp2 to properly couple Spp1 with Polα to form a stable initiation enzyme complex is required for the activation of the Cds1-mediated, intra–S-phase checkpoint response (Tan and Wang, 2000). (iii). An optimal size RNA primer synthesized by Spp1 that can be efficiently utilized by Polα to synthesize an iDNA structure is required for the activation of Chk1 response to prevent inappropriate mitotic entry.
ACKNOWLEDGMENTS
We thank members of the Wang and Nurse laboratory for helpful discussions during the course of the study. This study was supported by a grant CA54415 from the National Cancer Institute, of the National Institutes of Health, to T.W. D.J.F.G. is a recipient of a Wellcome International Traveling Fellowship (046754/Z/96/JMW/LEC/CG).
REFERENCES
- Bahler J, Wu JQ, Longtine MS, Shah NG, 3rd, AM, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile P.C.R-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Cotterill S. The 50-kDa primase subunit of Drosophila melanogaster DNA polymerase alpha: molecular characterization of the gene and functional analysis of the overexpressed protein. J Biol Chem. 1994;269:26759–26766. [PubMed] [Google Scholar]
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for the use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Bhaumik D, Wang TS-F. Mutational effect of fission yeast Polα on cell cycle events. Mol Biol Cell. 1998;9:2107–2123. doi: 10.1091/mbc.9.8.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinase Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- Carr AM. Analysis of fission yeast DNA structure checkpoints. Microbiology. 1998;144:5–11. doi: 10.1099/00221287-144-1-5. [DOI] [PubMed] [Google Scholar]
- Caspari T, Carr AM. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochemie. 1999;81:173–181. doi: 10.1016/s0300-9084(99)80050-9. [DOI] [PubMed] [Google Scholar]
- Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hoffmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;74:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari B, Blasina A, Boddy MN, McGowan CH, Russell P. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol Biol Cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari B, Rhind N, Russell P. cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King RC, editor. Handbook of Genetics. I. New York: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Kanto-Smoler G, Dahlkvist A, Sunnerhagen P. Improved method for rapid transformation of intact Schizosaccharomyces pombe cells. Biotechniques. 1994;16:798–800. [PubMed] [Google Scholar]
- Kuchta RD, Reid B, Chang LM. DNA primase. Processivity and the primase to polymerase alpha activity switch. J Biol Chem. 1990;265:16158–65. [PubMed] [Google Scholar]
- Lane D, Harlow E. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Lindsay HD, Griffiths DJF, Edwards R, Murray JM, Christensen PU, Walworth N, Carr AM. S-phase specific activation of Cds1 kinase defines a subpathway of the checkpoint response in S. pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu VF, Bhaumik D, Wang TS-F. Mutator phenotype induced by aberrant replication. Mol Cell Biol. 1999;19:1126–1135. doi: 10.1128/mcb.19.2.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14–3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- Lucchini G, Francesconi S, Foiani M, Badaracco G, Plevani P. Yeast DNA polymerase–DNA primase complex; cloning of PRI 1, a single essential gene related to DNA primase activity. EMBO J. 1987;6:737–742. doi: 10.1002/j.1460-2075.1987.tb04815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Martinho RG, Lindsay HD, Flaggs G, DeMaggio AJ, Hoekstra MF, Carr AM, Bentley NJ. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- Norbury C, Moreno S. Cloning cell cycle regulatory genes by transcomplementation in yeast. Methods Enzymol. 1997;283:44–59. doi: 10.1016/s0076-6879(97)83006-6. [DOI] [PubMed] [Google Scholar]
- O'Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Francesconi S, Wang TS-F. Cell cycle expression of two replicative DNA polymerases α and δ from Schizosaccharomyces pombe. Mol Biol Cell. 1993;4:145–157. doi: 10.1091/mbc.4.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S, Sherwood SW. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- Stadlbauer F, Brueckner A, Rehfuess C, Eckerskorn C, Lottspeich F, Forster V, Tseng BY, Nasheuer HP. DNA replication in vitro by recombinant DNA-polymerase-alpha-primase. Eur J Biochem. 1994;222:781–793. doi: 10.1111/j.1432-1033.1994.tb18925.x. [DOI] [PubMed] [Google Scholar]
- Tan S, Wang TS-F. Analysis of fission yeast primase defines the checkpoint responses to aberrant S phase initiation. Mol Cell Biol. 2000;20:7853–7866. doi: 10.1128/mcb.20.21.7853-7866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K, Katoa J, Ikeda H. Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics. 1998;148:49–58. doi: 10.1093/genetics/148.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama M, Galli I, Griffiths DJF, Wang TS-F. A novel mutant allele of Schizosaccharomyces pombe rad26 defective in monitoring S phase progression to prevent premature mitosis. Mol Cell Biol. 1997;17:3103–3115. doi: 10.1128/mcb.17.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- Wang TS. Cellular DNA polymerases. In: DePamphilis ML, editor. DNA Replication in Eukaryotic Cells. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 461–493. [Google Scholar]
- Weinert T. A DNA damage checkpoint meets the cell cycle engine. Science. 1997;277:1450–1451. doi: 10.1126/science.277.5331.1450. [DOI] [PubMed] [Google Scholar]
- Weinert T. DNA damage and checkpoint pathways: Molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Forbes KC, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Forbes KC, Wu A, Moreno S, Piwinca-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]