Abstract
The human pathogen Candida albicans undergoes a well-defined switch between two distinct cell types, named “white” and “opaque.” White and opaque cells differ in metabolic preferences, mating behaviors, cellular morphologies, and host interactions. Each cell type is stable through many generations; switching between them is rare, stochastic, and occurs without any known changes in the primary sequence of the genome; thus the switch is epigenetic. The white-opaque switch is regulated by a transcriptional circuit, composed of four regulators arranged in a series of interlocking feedback loops. To understand how switching occurs, we investigated the order of regulatory changes that occur during the switch from the opaque to the white cell type. Surprisingly, changes in key transcriptional regulators occur gradually, extending over several cell divisions with little cell-to-cell variation. Additional experiments, including perturbations to regulator concentrations, refine the signature of the commitment point. Transcriptome analysis reveals that opaque cells begin to globally resemble white cells well before they irreversibly commit to switching. We propose that these characteristics of the switching process permit C. albicans to “test the waters” before making an all-or-none decision.
Introduction
The architecture of transcriptional circuits often determines how organisms orchestrate developmental programs and how they respond to environmental cues. For example, circuit characteristics such as cooperativity and feedback can determine whether responses are graded or bistable and whether a transcriptional pattern can be directly inherited by descendent cells. Understanding the behavior of complex transcriptional circuits has important implications for many areas of biology, including the differentiation of stem cells into adult tissues, the response of cells to environmental perturbations, and the ability of cells to “remember” their cell type through repeated cell divisions.
A well defined system for examining how transcriptional patterns can be inherited is found in the human commensal yeast Candida albicans. Although a normal resident of the gastrointestinal tract of healthy humans, C. albicans is also the most prevalent fungal pathogen in humans, causing a variety of infections, particularly in immunocompromised individuals. C. albicans can switch between two genetically identical but phenotypically distinct types of cells, each of which is inherited through many generations (Slutsky et al., 1987; Soll et al., 1993; Bennett et al., 2003; Johnson, 2003; Lockhart et al., 2003; Lohse and Johnson, 2009; Soll, 2009; Morschhäuser, 2010). These two cell types, referred to as white and opaque, are distinguished by the differential regulation of approximately one-tenth of the genome (Lan et al., 2002; Tsong et al., 2003) resulting in distinct cellular and colony morphologies (Slutsky et al., 1987), metabolic preferences (Lan et al., 2002), mating behaviors (Miller and Johnson, 2002), and interactions with the host immune system (Kvaal et al., 1997; Kvaal et al., 1999; Geiger et al., 2004; Lohse and Johnson, 2008). Each cell type is stable through many generations, with switching between the two cell types estimated to occur every 104 generations (Rikkerink et al., 1988). Thus the switch is epigenetic in the classical sense of the word: it produces a heritable change of phenotype without a change in the nucleotide sequence of the genome. Although switching is stochastic, environmental cues can affect the frequency of switching events in one direction or the other. These cues include temperature (Rikkerink et al., 1988), oxidative stress (Kolotila and Diamond, 1990), anaerobic conditions (Dumitru et al., 2007; Ramírez-Zavala et al., 2008), and carbon dioxide (Huang et al., 2009).
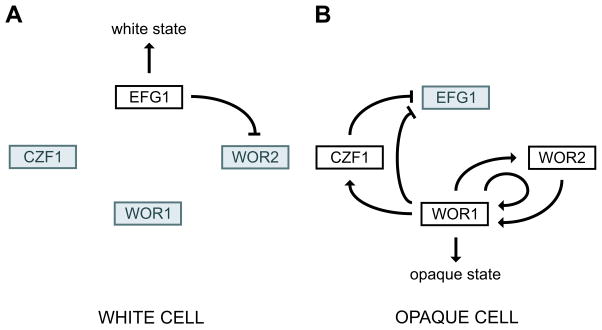
White-opaque switching offers many experimental advantages for studying the inheritance of transcriptional patterns. First, it takes place on well defined laboratory medium and requires no input from other cells or tissue. Second, each cell type is stable through many generations, so pure cultures of each cell type can easily be obtained. Third, the switching is reversible. Finally, C. albicans can be easily manipulated genetically (for example, genes can be easily deleted, over-expressed, or tagged), and many of the key regulators of switching have been identified. White-opaque switching is controlled by a core circuit of four transcriptional regulators arranged in multiple interlocking feedback loops, shown in Figure 1 (Huang et al., 2006; Srikantha et al., 2006; Zordan et al., 2006; Vinces and Kumamoto, 2007; Zordan et al., 2007). It has been hypothesized that this transcriptional network determines many of the characteristics of switching: according to the model, the circuit is largely inactive in the white state and this constitutes the default state (Figure 1a). Switching from the white to the opaque state is postulated to occur when the circuit becomes excited, and the series of positive feedback loops ensures that the circuit remains active (Figure 1b). According to the hypothesis, molecules of the regulators are passed on following cell division; the concentrations of these regulators in the progeny cells would then be sufficiently high to keep the circuit active, and the progeny cells would thereby stay opaque for many generations. Switching from the opaque to the white state would occur when the activity of the circuit decays, perhaps due to a stochastic decrease of one or more of the key regulators below a critical threshold level.
Figure 1.
Working model of the white-opaque regulatory circuit and its activity in white and opaque cells. (a) In white cells, EFG1 represses WOR1 indirectly through WOR2 to maintain white cell identity. (b) In opaque cells, WOR1, WOR2, and CZF1 establish a series of positive feedback loops, maintaining opaque cell identity and repressing EFG1. Up-regulated genes and active relationships are indicated in black. Down-regulated genes are indicated in gray. Arrows and bars represent activation and repression respectively. Figure adapted from Zordan et al., 2007 (Zordan et al., 2007).
Circuits composed of a series of transcriptional regulators arranged in feedback loops are common in biology. For example, eye development in flies ((Czerny et al., 1999), reviewed in (Silver and Rebay, 2005)) and muscle development in mammals ((Molkentin and Olson, 1996) reviewed in (Tapscott, 2005)) are specified by such circuits. Although the circuit summarized in Figure 1 can account, at least in broad outline, for the stability of the white and opaque states, it does not describe how a switching event occurs. This is an important general question because the characteristics of switching determine the stability of the two states; for example, if the two states are heritable through many generations (as they are in C. albicans), there must be a barrier to frequent switching. In C. albicans, it is known that concentrations of the four principle regulators of Figure 1 must change during a switching event: EFG1 (orf19.610) transcript levels are higher in white cells than in opaque cells and WOR1 (orf19.4884), WOR2 (orf19.5992), and CZF1 (orf19.3127) transcript levels are higher in opaque cells than white cells (Lan et al., 2002; Tsong et al., 2003); however, we do not know the order of these changes or the concentration of the regulators at the commitment point, the point at which a switching event becomes irreversible. In addition, on a single cell basis, it is not known whether white-opaque switching is a gradual process-- in which different sets of genes are activated sequentially-- or whether it is an abrupt, irreversible phenomenon. Finally, hundreds of genes are differentially expressed between white and opaque cells, and we do not know the order of changes in expression of these genes, or how they correspond to changes in the key regulators depicted in Figure 1.
Using a combination of fluorescence microscopy, western blotting, RT-qPCR, and microarray analysis, we describe the order of events during a switching event and develop a model of switching, one that has implications for other switching systems and for C. albicans’ ability to survive in a human host.
Results
Synchronized opaque-to-white switching
To effectively study the opaque-to-white switch using biochemical approaches, a majority of cells must be made to undergo this switch at approximately the same time. We chose to examine the opaque-to-white switch using a physiological cue: a temperature shift from 25°C to 37°C. Although several other experimental manipulations can cause an entire population of cells to switch (ectopic expression of a key transcriptional regulator, for example), we chose temperature for several reasons. The effect of temperature has been well characterized (Rikkerink et al., 1988) and it likely represents a physiological signal, one encountered in C. albicans’ warm blooded animal hosts. Although the average internal body temperature of C. albicans’ human host is 37°C, C. albicans experiences a range of temperatures in different niches; for example, skin surfaces are significantly cooler. Rikkerink et al. showed that the temperature induced switch was not instantaneous: following a shift from 20°C to 34°C of a suspended culture of opaque cells, approximately 6 hours passed before a significant number of opaque cells had committed to the white cell type; before that time if the temperature was lowered, the cells could be “rescued” and would remain opaque. The commitment point was determined experimentally by plating cells from the 34°C culture and incubating the plates at 25°C, the temperature where opaque cells are stable. Approximately 6 hours after the temperature was raised, roughly half the cells (all of which started off as opaque) had committed to the white state; that is, they formed white colonies even if the temperature was lowered back to 25°C. At later times, higher fractions of white colonies were observed and by approximately 28 hours the transition was complete with all plated cells giving rise to white colonies (Rikkerink et al., 1988). We will use the term “50% commitment point” to refer to the time by which half the opaque cells in a culture had committed to the white state following the temperature shift.
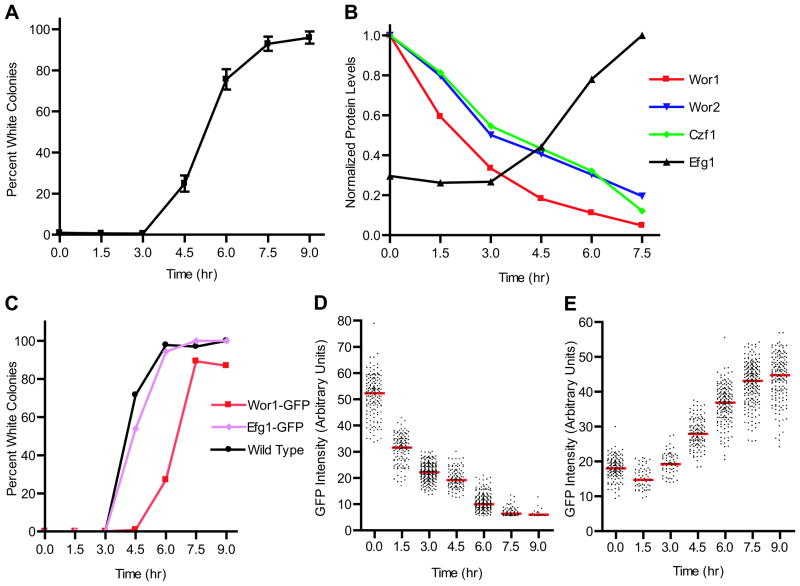
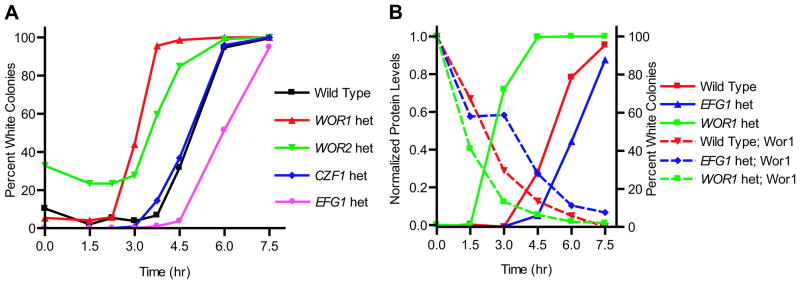
We compared the concentrations of the four regulators of the white-opaque regulatory circuit shown in Figure 1 to the commitment point. We also followed mRNA levels across the entire genome over the time course of switching. Briefly, a population of pure opaque cells was grown in suspension at 25°C, shifted to 37°C, and then plated at 25°C at various time points to monitor the percent commitment at each time point. Aliquots of the same samples were harvested for protein and mRNA measurements. In the experiments reported here, we observed significant numbers of white colonies by 4.5 hours. By 6 hours the majority of cells plated gave rise to white cells, and by 7.5 hours virtually all colonies were white (Figure 2a). The 50% commitment point is therefore 4.5 to 5 hours for the conditions used in this work. Similar results were observed in multiple independent experiments (see error bars in Figure 2a).
Figure 2.
Changes in protein levels of Wor1, Wor2, Czf1, and Efg1 during the opaque to white switch. (a) Commitment point. Opaque cells in suspended culture were shifted from 25°C to 37°C at T=0 hr. At each time point, an aliquot of cells was collected and plated at room temperature to determine the percentage of cells that had switched to the white cell type. Data points reflect the mean of six experiments (except for the 9 hour time point which reflects 5) and error bars represent the standard error of the mean. (b) Levels of the four regulatory proteins as determined by western blotting. As described in the methods, protein levels were normalized to the maximum levels for each regulator. For Wor1, Wor2, and Czf1 the maximum level occurs in opaque cells, while Efg1 is maximally expressed in white cells. Data points reflect the mean of four (Wor1 and Efg1) or three (Wor2 and Czf1) experiments; error bars for each protein level are shown in Supplemental Figure S1. (c) Commitment point for the experiments in (d) and (e); the percentage of colonies with the white colony phenotype is plotted for wild type, Wor1-GFP, and Efg1-GFP containing strains. (d) Single cell measurements of Wor1. Wor1-GFP levels in individual cells were quantitated as described in the methods. The red line represents the median value at each time point and each dot represents a single cell. (e) Single cell measurements of Efg1; symbols are as described in (d).
Graded-- not switch-like-- changes in regulatory proteins and transcripts
To follow levels of the regulatory proteins of Figure 1 on a population level over the time course of the opaque-to-white switch, cells were harvested at different time points after the temperature shift, lysed, and analyzed by western blotting (Figure 2b and Supplemental Figure S1). We first consider the activators of the opaque state (Wor1, Wor2, Czf1), which are expressed at higher levels in opaque cells than white cells. Levels of these proteins showed clear decreases within 90 minutes of the 25°C to 37°C temperature shift. By 4.5 hours-- the 50% commitment point-- levels of Wor1 had decreased to approximately 20% of its starting concentration and those of Wor2 and Czf1 to approximately 40% of their starting levels. At later time points, well past the commitment point, the levels of these proteins had further decreased to approximately the same levels observed in stable white cells. Efg1, the white-enriched regulator, showed a nearly reciprocal but delayed response. Its levels had increased less than 2-fold by the 4.5 hour commitment point, with significant increases in Efg1 levels observed only when the majority of cells were past the commitment point.
Western blots of cell cultures give the average characteristics of the population but do not reveal the behavior of single cells. To observe switching at the single-cell level, we tagged each of the four regulatory proteins with GFP and observed individual cells undergoing switching by fluorescence microscopy. C. albicans is a diploid, so both copies of a given regulator were tagged in order to ensure that the protein being followed was actively participating in the switch. Cells containing the tagged constructs exhibited switching behavior comparable to wild-type strains, although the tagged Wor1 resulted in a slight delay, relative to the untagged control strain, in reaching the commitment point (Figure 2c). Fluorescence levels were quantitated using a custom MATLAB script. Due to low fluorescence levels, it was not possible to reliably track Czf1 levels. For the other three proteins, we observed gradual, uniform changes in the levels in individual cells rather than all-or-none behaviors (Figures 2d and 2e, Supplemental Figure S2). In other words, the cells showed intermediate levels of the regulatory proteins across the time course as opposed to mixed populations of low and high expressors. Overall, the trends were similar to those observed in the bulk culture by western blotting. These results clearly show that individual cells undergo a gradual and uniform transition from the opaque to the white state. As noted above, in the Wor1-tagged strain the commitment point was slightly delayed (from 4.5 to 6 hours); we do not believe this delay complicates our interpretation because the results are very similar to those observed by western blotting of an untagged strain.
White and opaque cells exhibit numerous observable differences; many of these are attributable to the large number of genes (approximately 400) that are differentially regulated at least two-fold between the two states (Lan et al., 2002; Tsong et al., 2003). To examine the temporal order of the large change in gene expression that occurs during the opaque-to-white switch, we isolated mRNA at different time points following the 25°C to 37°C temperature shift of a culture of opaque cells. In the same experiment, aliquots of cells were also analyzed for the commitment point as described above. Four independent experiments were performed to ensure the reproducibility and statistical significance of the results. As controls for this experiment, four independent time courses were performed with populations of white cells that were subjected to the same 25°C to 37°C temperature shift. Switching does not occur here, allowing us to separate changes induced solely by temperature from those resulting from the opaque to white switch.
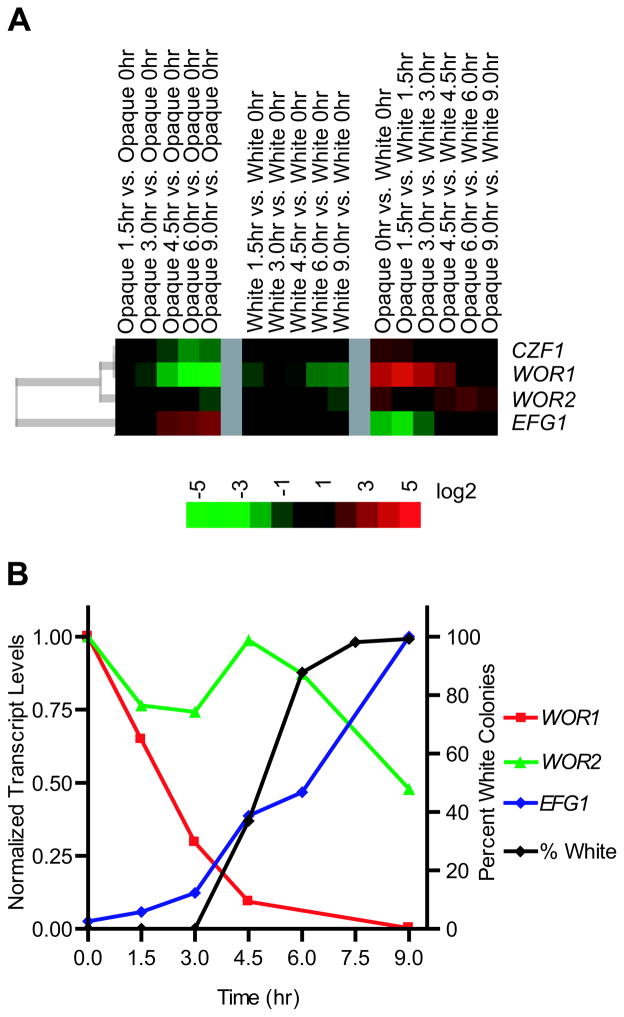
We first consider changes in the mRNA levels of WOR1, WOR2, CZF1, and EFG1 (Figure 3a). For WOR1 and CZF1, the mRNA and protein levels declined simultaneously. For WOR2, a lag in the mRNA decrease relative to the protein decrease was observed. For EFG1, the mRNA levels increased before the protein levels, as might be expected for an increase in expression levels. To confirm these observations, we monitored mRNA levels of WOR1, WOR2, and EFG1 in opaque cells by RT-qPCR (Figure 3b). Although the magnitude in changes observed using RT-qPCR data was greater (likely due to the more limited dynamic range of microarray analysis), the trends remained the same. Thus, both the mRNA and protein measurements indicate that changes to the core regulatory circuit during the opaque to white switch are relatively slow and gradual, occurring over a period of several hours.
Figure 3.
Transcript levels of WOR1, WOR2, CZF1, and EFG1. (a) Microarray transcript data, with the median of four independent experiments shown. The first block represents changes in opaque cells during the time course, normalized to the starting (T=0 hr) opaque cells. The second block highlights the changes due to temperature alone by comparing a time course performed on white cells (which do not switch) to the starting (T=0 hr) culture of white cells. The last block shows the profile of the opaque cell cultures at each time point, compared to the white cell cultures at the same time point. The color key is based on a log2 scale. (b) Levels of WOR1, WOR2, and EFG1 transcripts as determined by RT-qPCR (left axis). The levels of each transcript are normalized to their maximum levels in opaque (WOR1, WOR2) or white (EFG1) cells. The right axis displays the percentage of colonies with the white colony phenotype for the same experiment.
Transcriptome reprogramming
We now turn to the behavior of the remainder of the C. albicans transcriptome during the temperature-induced opaque-to-white switch. We first note that a few transcripts annotated as part of the heat shock or stress responses changed in both white and opaque cells after the temperature shift, but most did not. The lack of general heat shock and stress responses probably reflects the use of 37°C, a modest change in temperature. We also noted additional expression changes that occurred in both the white and opaque populations, likely involved in a general response to a subtle temperature change. The data for these genes are available in supplemental materials (Supplemental Table S1), and we will not discuss them further as they are not specific to the switching event. It is also worth noting that the 25°C to 37°C temperature shift can trigger hyphal growth (Sudbery et al., 2004), this response can explain some of the gene expression changes observed in both white and opaque cells.
Based on the four matched time courses, we developed a high-confidence set of 64 white- and 145 opaque-enriched genes that we followed during the switch from opaque to white (Figure 4; Supplemental Table S2). Our criteria for inclusion in this set was a greater than two-fold mRNA difference between white and opaque cells at 25°C (the T=0 time point) in at least 3 of the 4 replicate time courses, with the differences all in the same direction. The smaller number of white and opaque enriched genes relative to previous sets (Lan et al., 2002; Tsong et al., 2003) largely reflects the increased stringency of our criteria. For example, many additional white and opaque-enriched genes consistently differed by less than two-fold in expression; because it is difficult to meaningfully track such subtle changes across a time course we excluded them from our analysis, focusing instead on higher-magnitude changes. Significance Analysis of Microarrays for this gene set is discussed in the supplemental materials.
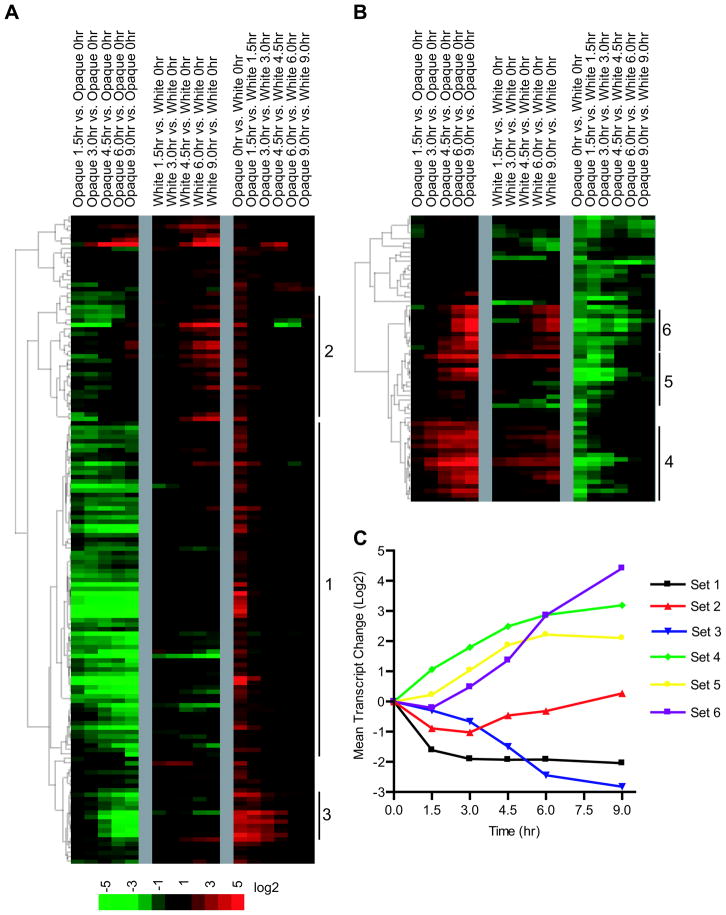
Figure 4.
Behavior of 209 white- and opaque-enriched genes over the time course of switching. The three column blocks are described in Figure 3a, with the median value from the four experiments shown for each gene. (a) Changes in the 145 opaque-enriched genes during the time course. (b) Changes in the 64 white-enriched genes during the time course. The color key for (a) and (b) is based on a log2 scale. (c) The six gene sets were binned and the mean transcriptional change at each time point for each set was determined in opaque cells. Sets of genes (1, 2, 3, etc.) are described in more detail in the text. Sets 1 and 2 are the opaque-enriched genes that are rapidly down-regulated upon the temperature shift. Set 3 is the 11 gene WOR1/CZF1 cluster of opaque-enriched genes. Set 4 contains the white-enriched “early” genes. Set 5 is white-enriched “middle” genes, and Set 6 is the white-enriched “late” gene group. Data are from the same set of experiments shown Figure 3a.
The opaque-enriched genes in our analysis cluster into distinct groups based on the timing of their expression change during the opaque-to-white switch (Figure 4a). For example, a large group of 63 opaque-enriched genes were fully down-regulated within the first 90 minutes following the temperature shift, well before the 50% commitment point was reached. These mRNAs remained at this level for the remainder of the time course (Figure 4a; Set1, Supplemental Table S3). A second group of 29 genes was down-regulated in a similarly rapid manner but, subsequently, these genes were slightly up-regulated (Figure 4a; Set2, Supplemental Table S3). These two sets of genes comprise roughly 65% of the opaque-enriched gene set; thus opaque cells transition to a “white-like” expression state well before the 50% commitment point (4.5 hours in these experiments) is reached. Significantly, the mRNAs of the key opaque regulators WOR1, WOR2, and CZF1 decrease hours later than these 63- and 29-gene sets. Other classes of opaque-enriched genes are discussed in the Supplemental Materials and Supplemental Table S4, as is a comparison of these genes.
White-enriched genes also fall into several temporal clusters (Figure 4b) which, in many respects, show reciprocal behavior to the opaque clusters. Mirroring the rapid down-regulation of many opaque-enriched genes, at least a quarter of the white enriched genes (‘early genes’) approach their final levels well before the commitment point is reached (Figure 4b; Set4). A second cluster of genes (“middle” genes), including EFG1, change more slowly but are also up-regulated before the cells reach the 50% commitment point (Figure 4b; Set5). Finally, a third group of 11 genes (“late” genes) is not up-regulated until after the commitment point is reached (Figure 4b; Set6). A summary of the behavior of each set of genes is included in Figure 4c.
In summary, the behavior of the transcriptome during the temperature-induced switch from opaque to white reveals that large changes occur well before the cells reach the commitment point. Not only do the cells lose much of their opaque signature prior to commitment, but they also up-regulate expression of many white-enriched genes. Levels of the key transcriptional regulators themselves change only after this initial transformation has taken place, thus lagging behind most of the transcriptome changes.
Perturbing the commitment point
Thus far we have described, during the course of switching, the changes in levels of the four known switching regulators (Wor1, Wor2, Czf1, Efg1) and the behavior of the transcriptome relative to the 50% commitment point-- the time at which 50% of the cells have passed the “point of no return.” We now consider the molecular events that coincide with commitment. Although post-translational modifications could play important roles in the switch (Hnisz et al., 2009), the simplest model holds that the commitment point is caused by changes in Wor1, Wor2, Czf1, and/or Efg1 levels. To examine this possibility, we perturbed levels of each regulator and monitored the effect on the commitment point. Due to the extensive feedback loops in the switching circuit (see Figure 1), it is not possible to drastically change the level of one regulator without changing the others as well. We therefore turned to more subtle changes; more specifically, we lowered the gene dosage of each regulator by knocking out one of the two gene copies. The commitment point in these heterozygous strains was then determined experimentally.
The largest effect was observed with the WOR1/wor1 heterozygote: the 50% commitment point was reached more quickly than with the control strain (less than 3 hours for the heterozygote versus 5 hours for the wild type) (Figure 5a). Heterozygous strains constructed for the other regulators showed less pronounced effects: removing one copy of CZF1 had very little effect on the commitment point while removing one copy of WOR2 decreased the time to commitment by about one hour (Figure 5a). Removal of one copy of EFG1 delayed the commitment point by about one hour, from 5 to 6 hours (Figure 5a). It is worth noting that each of the heterozygous strains had altered frequencies of white-to-opaque switching (data not shown), suggesting that small changes in regulator levels affect stochastic switching as well as temperature induced switching.
Figure 5.
Effects of deleting one copy of WOR1, WOR2, CZF1, or EFG1 on the commitment point. (a) Temperature-induced switching from opaque to white cell types for the wild type strain and for heterozygous deletions of the four known regulators. The percentage of colonies with the white colony phenotype is plotted for each strain. The 50% commitment points for these strains are 5 hours (wild type and CZF1 het), 3 hours (WOR1 het), and 6 hours (EFG1 het). As the WOR2 heterozygous strain had a consistent background of 20–30% white cells, we have defined the commitment point for this strain as the point at which half the opaque colonies, or 60–65% of the total colonies, are white (approximately 4 hours). All temperature shift experiments in (a) were performed in parallel on the same day. (b) Wor1 protein levels in wild type, WOR1 heterozygous, and EFG1 heterozygous strains. Western blotting data (left axis, dashed lines) and percent commitment (right axis, solid lines) are plotted for Wor1 in wild type (red), WOR1 heterozygous (green), and EFG1 heterozygous (blue) strains. Data points represent the mean of two experiments.
We further investigated the one hour delay in the commitment point in the EFG1/efg1 strain by tracking the levels of Wor1 in this strain. As shown in Figure 5b, the time required for Wor1 to decline by 80% is increased by roughly one hour in the EFG1/efg1 strain. Deleting both copies of EFG1 resulted in even slower decreases in Wor1, whose levels dropped by less than 50% after 9 hours (Supplemental Figure S3a). We also performed the complementary experiment, tracking Efg1 levels in a WOR1/wor1 heterozygote. Although deleting one copy of WOR1 advanced the commitment point by two hours, we observed only a small increase (<10%) in Efg1 at the commitment point (Supplemental figure S3b). We note that this increase in Efg1 levels is much less than that observed in a wild type cell at commitment. Thus, the commitment point shows a higher correlation with Wor1 levels than with Efg1 levels. To determine the effect of deleting one copy of Wor1 on its absolute levels, we used normalized western blotting and flow cytometery. We found that the level of Wor1 in the WOR1/wor1 opaque cells was roughly 70–80% that of the WOR1/WOR1 control strain (data not shown). We also note that Wor1 levels had dropped by more than 80% in less than 3 hours in this strain (Figure 5b).
These data, taken together, are consistent with the decline in Wor1 levels triggering commitment, although we cannot rule out more complicated models. We note that the commitment point in these different strains corresponds to an 80% decline in Wor1 levels.
Transcriptional Signature of Commitment
What are the characteristics of the commitment point on a genome-wide scale? To answer this question, we compared the transcriptional profiles of a wild-type strain and a WOR1/wor1 strain at the commitment point. As described above, the WOR1/wor1 strain reaches commitment more than 2 hours earlier than the wild type strain, so, in principle, this experiment distinguishes commitment per se from other effects (e.g. nutrient depletion, heat shock, ect.) of the experimental manipulations.
This analysis, described in greater detail in the Supplemental Text, identified a set of 36 genes whose mRNA expression levels track with the commitment point in both strains (Supplemental Figure S4, Supplemental Table S5). Since levels of these genes change more rapidly in the WOR1 heterozygote compared to wild type cells, these genes appear linked to the commitment process itself. Most of these genes (27/36) are white or opaque-enriched genes that appear to be structural genes, transporters, or genes of unknown function. Significantly, this gene set includes three transcriptional regulators—WOR1, CZF1, and EFG1—thus supporting the model that changes in concentration of these regulators are responsible for the switching commitment point. Of these three, WOR1 levels have changed the most at commitment, consistent with the idea that a decline in this protein underlies commitment.
Discussion
The human commensal yeast C. albicans can switch between two distinct cell types, white and opaque. Switching between the two cell types is rare, with each cell type being stable for hundreds of generations. A transcriptional regulatory circuit responsible for the regulation of the two cell types has been recently described (Figure 1) which can account, in part, for the stability of the two cell types but does not illustrate the order of events during a switch. In this paper we address the following questions: Is the switch gradual or does it display rapid, all or none behavior? What is the order of events when cells switch from opaque to white? What features of switching stabilized the two states (i.e. what prevents rapid switching back and forth)? What triggers the switch? Do properties of the switch provide specific advantages to the organism?
Temperature induced opaque-to-white switching is a gradual and synchronous process
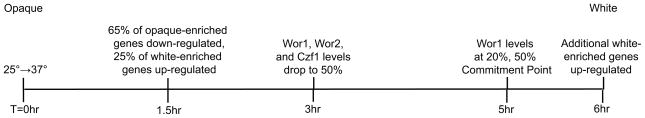
It has previously been established that temperature-induced opaque-to-white switching is a slow process, requiring approximately 5–6 hours for 50% of cells to reach the commitment point-- the point at which switching has become irreversible-- and approximately 12 hours for full commitment of the entire population (Rikkerink et al., 1988). But what happens during this time? We found that the levels of the four regulatory proteins shown in Figure 1 (Wor1, Wor2, Czf1, and Efg1) change in a gradual, ordered manner during this time period (Figure 2b and Supplemental Figure S1). These gradual changes were observed in both bulk populations of cells and in individual cells. A summary of events following the temperature shift is presented in Figure 6.
Figure 6.
Order and timing of events following temperature shift of an opaque population.
We were initially surprised by the smooth nature of the opaque-to-white transition and by the lack of variation in regulatory protein levels between individual cells. Based on the bistable nature of the switch circuitry and the existence of time points where only a fraction of cells had committed to switching, we expected to observe two clearly differentiated sub-populations of cells and few transitional intermediates in a switching population. Instead, the analysis revealed that the levels of regulatory proteins changed gradually in individual cells with very few outliers, let alone more than one distinct population at any time point. It is possible that the gradual decline in the regulators prevents frequent switching and thereby contributes to the stability of the two types of cells.
Given the gradual changes observed for the regulators, how can the all-or-none nature of the commitment point on a single cell level be explained? We found that small perturbations of the levels of Wor1, Wor2, and Efg1 (but not Czf1) changed the commitment point. Because (1) perturbation of Wor1 had the largest effects, (2) it is the regulator whose concentration changes the most between white and opaque cells, and (3) changes in its levels appear to track the best with commitment in the various strains examined, we propose that the decline in Wor1 levels is the critical event in switching. However, because the transcriptional network controlling switching is highly interconnected (that is, each regulator controls every other regulator), it is currently difficult to definitively test this proposal.
We do not know why Wor1 levels decrease slowly in response to a temperature shift, but a simple model can account for this: if the temperature shift blocks new WOR1 synthesis, then Wor1 levels gradually decline simply because new mRNA synthesis and translation fail to keep up with degradation rates and with dilution due to cell growth and division. This idea is supported by two observations described in this work: (1) the decline in Wor1 protein and mRNA levels track together (Figures 2b and 3) and (2) the 25°C to 37°C temperature shift reduces WOR1 mRNA levels in both white and opaque cells (Figure 3a) even though the level of expression of WOR1 is much greater in opaque cells. As first noted by Bergen et al. (Bergen et al., 1990) and Srikantha et al. (Srikantha et al., 2006), following the temperature shift, the cells continue to divide for at least two rounds of cell division before the commitment point is reached. Multiple rounds of cell division following the temperature shift also occurred in the experiments reported here (data not shown). Thus, reducing new WOR1 synthesis, coupled with dilution due to cell growth and division, can easily account for the gradual decrease in WOR1 levels following the temperature shift. In addition, Wor1 is known to activate its own transcription in opaque cells (Huang et al., 2006; Zordan et al., 2006) and so a continual decline in levels could eventually break this positive feedback loop and lead irreversibly to the white state. Regardless of its exact mechanism, the slow decline in Wor1 documented here would create a time window of several hours during which the cells are clearly changing in response to temperature, but have not yet committed to switching. As described below, we believe this time window has important implications for the switching program.
Opaque cells phenotypically transition to “white-like” cells before the switch becomes heritable
We were surprised to find that a massive transformation of the opaque cell transcriptional program into a “white-like” program occurred shortly after the temperature shift and several hours before the commitment point was reached. Srikantha et al. previously reported that the transcript levels of two opaque-enriched genes (OP4 (orf19.4934) and SAP1 (orf19.5714)) decreased prior to the commitment point (Srikantha et al., 2000); our results show that this response encompasses approximately 65% of all opaque-enriched genes (which are down-regulated) and 25% of the white-enriched genes (which are up-regulated). Thus, immediately following the temperature shift, opaque cells switch their transcriptome program to that resembling white cells and, only if the temperature shift is maintained for several more hours, do they make these changes irreversible by committing to the switch.
What advantage might be conferred by changing the transcriptome in advance of the switch? One possibility, of course, is that this strategy is not adaptive at all; that is, this feature of the switch was a non-selected by-product of the evolutionary history of the switch. However, changing the transcriptome in advance of the switching event may, at least on paper, offer certain advantages. The white-opaque switch has long been hypothesized to allow C. albicans to better colonize a range of environments in the host; consistent with this idea, a major class of genes that is differentially regulated between the two cell types encodes metabolic enzymes (Lan et al., 2002). Rapidly changing from an opaque transcriptional program to a white program before the switching mechanism locks in could allow opaque cells to “test the waters” before switching. For example, if a temperature shift is only temporary, the cells can quickly return to the opaque state. Such a temporary separation of the bulk of the transcriptome from its control circuit may allow opaque cells to sample a new environment, committing to switching only when a favorable match occurs between the gene expression profile and the environment. This idea can account for why, in the absence of oxygen and/or the presence of carbon dioxide, increased temperature fails to induce the opaque-to-white switch (Dumitru et al., 2007; Ramírez-Zavala et al., 2008; Huang et al., 2009). It remains to be seen if this temporary separation of a regulatory circuit and the transcriptome is a strategy used by other microbial pathogens to respond to changes in the host environment.
Similarity to other cell differentiation programs
On the surface, white-opaque switching resembles other examples of cell differentiation where cells remember their differentiated state through many rounds of cell division without environmental cues. For example, the gradual nature of the switch and the relatively long time lag before the commitment point is reached are reminiscent of certain features of the formation of induced pluripotent stem (iPS) cells from differentiated tissues (see (Scheper and Copray, 2009) and (Hochedlinger and Plath, 2009) for reviews). Both processes appear to be controlled by a series of transcriptional regulators, each of which controls the others, arranged in a complex network that includes multiple feedback loops. Moreover, each has a relatively long time delay (up to 2 weeks for iPS cells) with the transcriptome changing in a stepwise fashion.
The slow, gradual nature of iPS cell reprogramming is often attributed to slow changes in methylation/demethylation or in chromatin structure. However, a systematic investigation of histone modifying enzymes and chromatin remodelers in C. albicans indicated relatively small changes in white-opaque switching rates in comparison to the effects of the transcriptional regulators (Hnisz et al., 2009). We have proposed that the slow decay of a key transcriptional regulator is sufficient to explain the delay and its drop below a threshold level sufficient to explain the commitment point of the switch. Whether this type of framework applies to other cases of cell differentiation remains to be determined.
Experimental procedures
Strains and primers used in this paper are listed in Supplemental Tables S6 and S7 respectively.
Media
Cells were grown in synthetic complete media supplemented with 2% glucose and 100μg/mL uridine (SD+aa+Urd).
Plasmids
Plasmids for GFP tagging were based on the SAT1 flipper cassette (Reuss et al., 2004). Briefly, the last 500bp of a gene (excluding the stop codon) was PCR amplified with a 5′ SphI site. C. albicans optimized GFP was PCR amplified with a 3x Gly linker and a 3′ XhoI site. The first 500bp immediately 3′ of the open reading frame (ORF) was PCR amplified between NotI and AatII sites. These three fragments were combined in a further round of PCR, resulting in a SphI-ORF-GFP-XhoI-NotI-3′ flank-AatII fragment that was digested with SphI and AatII and ligated into pUC19 (NEB). Plasmids were sequenced to ensure matching to the sequence listed at the Candida Genome Database (http://www.candidagenome.org/). Plasmids with correct sequences were digested with XhoI and NotI, then ligated to a similarly digested SAT1 flipper cassette from pSFS2A. Plasmids were then digested with SphI and AatII for transformation into C. albicans.
Strains
All strains are derivatives of SC5314. Creation of the a/a selected –Leu –His starting strain RZ47 has been described previously (Zordan et al., 2006). C. dublieniensis LEU2 and C. maltosa HIS1 were added back to this strain at the LEU2 locus. The white and opaque strains in this background were used as wild type, independent isolates of this strain were gifts from Aaron Hernday and Quinn Mitrovich and have been described previously (Mitrovich et al., 2007). The EFG1, WOR1, WOR2, and CZF1 heterozygous deletion strains and the efg1/efg1 deletion strain were made using the His or Leu knockout cassettes (Noble and Johnson, 2005) in the RZ47 background and were a gift from Aaron Hernday and Rebecca Zordan. GFP tagging of Wor1, Wor2, Czf1, and Efg1 was accomplished using the plasmid designs described above. Strains were transformed with linearized plasmids and selected for growth on 200 μg/mL nourseothricin (clonNAT, WERNER BioAgents, Jena, Germany). Insertion was verified with PCR against both flanks and the marker was then flipped out by growth in Yeast Peptone Dextrose (YPD) media supplemented with 2% Maltose for 6–24 hours. Cells were then plated on YPD plus 25 μg/mL nourseothricin to identify small colonies which had lost the nourseothricin resistance marker. Marker excision was then verified by PCR. The second copy of the gene was then tagged by repeating this process, performing additional PCR checks to ensure that untagged copies of the gene did not remain in the cell.
Temperature Shift Assay
Room temperature overnight cultures were started from single colonies with no sectors on SD+aa+Urd plates. Cultures were diluted back to OD600=0.02 in the morning and allowed to grow back to OD600=0.1–0.2 at room temperature. Cells were then diluted (OD600=0.00012 for opaques and OD600=0.00002 for whites) in 500mL SD+aa+Urd in 2.8L flasks and grown at 25°C overnight. When cultures reached OD600=0.1, 0 hr samples were removed and the remaining culture was transferred to pre-warmed 37°C flasks and swirled in a 37–40°C water bath for 5 minutes to ensure the temperature quickly reached 37°C. Cultures were then placed in a 37°C shaking incubator. Modifications of this assay for single cell experiments (Figure 2c–e, Supplemental Figure S2) and some heterozygous strain assays (Figure 5a) are described later. Modifications of this assay for the commitment microarray analysis are described in the Supplemental Text.
Samples were then harvested at 1.5, 3, 4.5, 6. 7.5, and 9 hour time points. Samples for microarray analysis and qPCR were harvested by centrifugation and frozen in liquid nitrogen. Samples for western blotting were centrifuged, resuspended in 1mL PBS, transferred to 2mL tubes, pelleted, decanted, and frozen in liquid nitrogen. Samples for microscopy were centrifuged, washed 2x with PBS, and placed on slides for analysis. At each time point, cells were also plated on room temperature SD+aa+Urd plates which were scored for colony phenotype 5–7 days later to determine percent switching. Between 100 and 500 cells were plated at each time point, generally 100 cells were plated on each of two plates.
Quantitative RT-PCR
Sample preparation, RT-qPCR, and data analysis were performed as previously described (Zordan et al., 2006). Normalization was versus PAT1 (orf19.3792), as described in that reference.
Microarrays
Total RNA was extracted from samples via buffered phenol extractions. Following cDNA synthesis, samples were dye coupled to both Cy3 and Cy5 in separate reactions. A pooled reference was created from the Cy5 labeled fractions. Equal amounts of Cy3 labeled sample and Cy5 labeled reference were hybridized overnight to custom designed Agilent 8*15k Microarrays (AMADID #020166) containing a minimum of 2 independent probes for each ORF. Following hybridization, slides were processed and then scanned on a Genepix 4000A or 4000B Axon Instrument Scanner (Axon/Molecular Devices, Sunnyvale, CA USA). Arrays were gridded using GenePix Pro version 5.1. Global Lowess normalization analysis was performed for each array using a Goulphar script (Lemoine et al., 2006) for R (The R Foundation for Statistical Computing). Normalized data was then transformed before collapsing all points for an ORF to the median value using a custom java script written by Oliver Homann. Temperature shifted opaque samples were transformed versus a pre-shift opaque sample to determine how genes had changed relative to their starting values. Temperature shifted white cells compared to a pre-shifted white population allowed us to control for general temperature responses as well as entry into stationary phase at late time points. Transformations of an opaque sample at a time point versus a white sample at the same time point were also performed. Data shown in Figures 3a and 4 represent the median values from the four experiments. Transformations used for commitment characterization are discussed in the Supplemental Materials.
Microarray data were clustered using Cluster version 3.0 (de Hoon et al., 2004) and visualized using Java TreeView Version 1.1.3 (Saldanha, 2004). The microarray data has been uploaded to Array Express (http://www.ebi.ac.uk/microarray-as/ae/), the accession number for the time course experiments is E-MEXP-2705 and the accession number for the commitment analysis experiments is E-MEXP-2834.
Western Blotting
Extracts were prepared in Urea Lysis buffer (Ubersax et al., 2003). 10μL of each sample was run on an SDS-PAGE gel and then analyzed by Western Blotting. Blots were analyzed with rabbit affinity-purified antibody generated against synthetic peptides unique to each protein from the various genes (Bethyl Laboratories Inc., Montgomery TX). Wor1 antibodies have been previously described (Zordan et al., 2006). Wor2 antibodes were raised against the sequences CSAVINRVSVADLLK and CYSPNSPYSLPTRPSN. Efg1 antibodies were raised against the sequences CQANQSASTVAKEEK and CYGQYNAPGKNQNTPA. Czf1 antibodies were raised against the sequences CKVLRGIVEYRSK and CLPSNVSPPNSRAVPT. To reduce non-specific binding, the Wor2, Efg1, and Czf1 antibodies were additionaly purified as follows. Lysates of C. albicans strains deleted for the respective genes were bound to activated Sepharose 4B. A mixture of both antibodies for each protein were then incubated with the resin for 12–18 hours before elution. Elution fractions with high protein concentrations were pooled for further use, creating a mixture of both antibodies against a given protein. Rat α-tubulin (no. ab1616; Abcam, Cambridge, MA) was used as a loading control. Secondary antibodies were goat α-rabbit IrDye800 (611-132-122, Rockland Immunochemicals Inc., Gilbertsville, PA) and goat α-rat AlexaFluor680 (A-21096, Invitrogen, Carlsbad, CA). Blots were scanned on a Oddessey Imaging System scanner (LI-COR Biotechnology, Lincoln, NE) using both the 700 and 800 channels. Protein levels in each lane were first normalized versus the tubulin loading control, then normalized to the maximum value (set to 1) for each protein. Data shown for the four proteins in Figure 2b and Supplemental Figure S1 represent the mean of four (Wor1 and Efg1) or three (Wor2 and Czf1) independent experiments and the error bars in Supplemental Figure S1 reflect the standard error of the mean.
Single Cell Fluorescence and Data Analysis
The temperature shift assay was modified as follows for the single cell experiments. GFP strains were grown overnight at room temperature, diluted to OD600=0.2, and allowed to grow at room temperature. At OD600=0.6, samples were harvested, an equal amount of pre-warmed 37°C SD+aa+Urd was added, and samples were swirled in a 37°C water bath for 5 minutes. At each time point, samples were washed twice with PBS and imaged on a Zeiss Axiovert 200M microscope (Carl Zeiss, Oberkochen, Germany). Constant GFP exposure times were used for each strain during an assay. The Rhodamine channel was used to detect autofluoresence. Images were analyzed using a custom MATLAB (The MathWorks Inc., Natick, MA) script which determined mean GFP fluorescence levels for each cell. Experiments with the heterozygous strains shown in Figure 5a also used this general temperature shift assay approach, although without the microscopy.
Supplementary Material
Acknowledgments
The authors thank Aaron Hernday, Rebecca Zordan, and Quinn Mitrovich for strains. The authors thank Chris Cain, Sarah Foss, Aaron Hernday, Chris Baker, Anita Sil, Hiten Madhani, and Oliver Homann for valuable comments on this manuscript. MBL was supported by a National Science Foundation Graduate Research Fellowship. Research was supported by grants from the National Institutes of Health (RO1 AI49187) and the Ellison Foundation (ID-SS-0628-04) to ADJ.
References
- Bennett RJ, Uhl MA, Miller MG, Johnson AD. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol. 2003;23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen MS, Voss E, Soll DR. Switching at the cellular level in the white-opaque transition of Candida albicans. J Gen Microbiol. 1990;136:1925–1936. doi: 10.1099/00221287-136-10-1925. [DOI] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Dumitru R, Navarathna DH, Semighini CP, Elowsky CG, Dumitru RV, Dignard D, et al. In vivo and in vitro anaerobic mating in Candida albicans. Eukaryot Cell. 2007;6:465–472. doi: 10.1128/EC.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger J, Wessels D, Lockhart SR, Soll DR. Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infect Immun. 2004;72:667–677. doi: 10.1128/IAI.72.2.667-677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Schwarzmüller T, Kuchler K. Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Mol Microbiol. 2009;74:1–15. doi: 10.1111/j.1365-2958.2009.06772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Srikantha T, Sahni N, Yi S, Soll DR. CO(2) regulates white-to-opaque switching in Candida albicans. Curr Biol. 2009;19:330–334. doi: 10.1016/j.cub.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. The biology of mating in Candida albicans. Nat Rev Microbiol. 2003;1:106–116. doi: 10.1038/nrmicro752. [DOI] [PubMed] [Google Scholar]
- Kolotila MP, Diamond RD. Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicans WO-1. Infect Immun. 1990;58:1174–1179. doi: 10.1128/iai.58.5.1174-1179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaal C, Lachke SA, Srikantha T, Daniels K, McCoy J, Soll DR. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun. 1999;67:6652–6662. doi: 10.1128/iai.67.12.6652-6662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaal CA, Srikantha T, Soll DR. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect Immun. 1997;65:4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan CY, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, Agabian N. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci U S A. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine S, Combes F, Servant N, Le Crom S. Goulphar: rapid access and expertise for standard two-color microarray normalization methods. BMC Bioinformatics. 2006;7:467. doi: 10.1186/1471-2105-7-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Daniels KJ, Zhao R, Wessels D, Soll DR. Cell biology of mating in Candida albicans. Eukaryot Cell. 2003;2:49–61. doi: 10.1128/EC.2.1.49-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS One. 2008;3:e1473. doi: 10.1371/journal.pone.0001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD. White-opaque switching in Candida albicans. Curr Opin Microbiol. 2009;12:650–654. doi: 10.1016/j.mib.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, Tuch BB, Guthrie C, Johnson AD. Computational and experimental approaches double the number of known introns in the pathogenic yeast Candida albicans. Genome Res. 2007;17:492–502. doi: 10.1101/gr.6111907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci U S A. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhäuser J. Regulation of white-opaque switching in Candida albicans. Med Microbiol Immunol. 2010;199:165–172. doi: 10.1007/s00430-010-0147-0. [DOI] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Zavala B, Reuss O, Park YN, Ohlsen K, Morschhäuser J. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 2008;4:e1000089. doi: 10.1371/journal.ppat.1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Rikkerink EH, Magee BB, Magee PT. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Scheper W, Copray S. The Molecular Mechanism of Induced Pluripotency: A Two-Stage Switch. Stem Cell Rev. 2009;5:204–223. doi: 10.1007/s12015-009-9077-x. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Rebay I. Signaling circuitries in development: Insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR. Why does Candida albicans switch? FEMS Yeast Res. 2009;9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- Soll DR, Morrow B, Srikantha T. High-frequency phenotypic switching in Candida albicans. Trends Genet. 1993;9:61–65. doi: 10.1016/0168-9525(93)90189-O. [DOI] [PubMed] [Google Scholar]
- Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, et al. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Tsai LK, Daniels K, Soll DR. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J Bacteriol. 2000;182:1580–1591. doi: 10.1128/jb.182.6.1580-1591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Vinces MD, Kumamoto CA. The morphogenetic regulator Czf1p is a DNA-binding protein that regulates white opaque switching in Candida albicans. Microbiology. 2007;153:2877–2884. doi: 10.1099/mic.0.2007/005983-0. [DOI] [PubMed] [Google Scholar]
- Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.