Abstract
Myeloid differentiation factor 2 (MD-2) is a secreted glycoprotein that assembles with Toll-like receptor 4 (TLR4) to form a functional signaling receptor for bacterial lipopolysaccharide (LPS). In this study we have identified a novel alternatively spliced isoform of human MD-2, termed MD-2 short (MD-2s), which lacks the region encoded by exon 2 of the MD-2 gene. Similar to MD-2, MD-2s is glycosylated and secreted. MD-2s also interacted with LPS and TLR4, but failed to mediate LPS-induced NF-κB activation and interleukin-8 production. We show that MD-2s is upregulated upon IFN-γ, IL-6 and TLR stimulation and negatively regulates LPS-mediated TLR4 signaling. Furthermore, MD-2s competitively inhibited binding of MD-2 to TLR4. Our study therefore pinpoints a mechanism that may be employed to regulate TLR4 activation at the onset of signaling and identifies MD-2s as a potential therapeutic candidate to treat human diseases characterized by an overly exuberant or chronic immune response to LPS.
INTRODUCTION
Detection of microbial pathogens and instigation of an appropriate innate and subsequent adaptive immune response is highly reliant on Toll-like receptors (TLRs) (1). TLR4 is one of the most widely studied of the family and recognizes a varied repertoire of ligands, such as heat shock protein 60 (2), respiratory syncytial virus fusion protein (3) and lipopolysaccaride (LPS) (4–8) a major component of the outer membrane of Gram-negative bacteria. Host sensitivity to LPS is enhanced by the accessory proteins, LPS-binding protein (9) and CD14 (10), but for LPS recognition to occur, TLR4 requires the co-receptor myeloid differentiation factor (MD)-2 (11–13). Upon LPS binding, a receptor multimer composed of two copies of the TLR4-MD-2-LPS complex is formed (14), which triggers a downstream signaling cascade, culminating in the activation of transcription factors such as nuclear factor (NF)-κB and the interferon regulatory factors (IRFs), which in turn induce various immune and inflammatory genes.
Tight regulation of TLR4 signaling is imperative in order to prevent an overactivated immune response that could contribute to the pathogenesis of autoimmune, chronic inflammatory and infectious diseases, such as diabetes (15), asthma (16) and sepsis (4). One method of downregulating TLR4 signaling involves the production of inhibitory isoforms, by alternatively splicing specific genes encoding essential signaling components. To date, several such splice variants have been identified, examples include smTLR4 (17, 18), myeloid differentiation factor 88S (MyD88S) (19), TRAM adaptor with GOLD domain (TAG) (20) and murine MD-2B (21).
Here we report the identification and characterization of a novel alternatively spliced isoform of human MD-2, that we have called MD-2 short (MD-2s). Similar to full-length MD-2, this protein was glycosylated and secreted. However, despite its ability to interact with TLR4 and LPS, MD-2s failed to mediate NF-κB activation and interleukin 8 (IL-8) production following LPS exposure. We also determined that MD-2s competitively inhibited binding of full length MD-2 to TLR4 and identified this short isoform as a negative regulator of LPS-mediated TLR4 activation. We show that MD-2s is an inducible protein that may function as a negative feedback inhibitor. Our results therefore define a novel mechanism used by human isoform of MD-2 that may curtail excessive activation of the innate immune response at the initiation of the TLR4 signal transduction pathway.
METHODS
Cell culture and biological reagents
Immortalized HMECs were cultured in MCDB-131 medium, supplemented with 10% heat-inactivated fetal bovine serum, 2 mM glutamine, and 100 μg/ml penicillin and streptomycin. The HEK293 cell line, mouse RAW 264.7 macrophage cell line, mouse aortic endothelial cells (MAEC) were cultured in Dulbecco’s modified Eagle’s medium, supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 2 mM glutamine. LPS (TLRGrade; Alexis). Biotin-LPS (Ultrapure, Invivogen).
Real-time PCR (RT-PCR)
Total cellular RNA was isolated from cells using the RNeasy mini kit (Qiagen) and treated with DNase. RNA from human lung, pancreas, thymus, kidney, spleen, liver, heart, and placenta was purchased from Ambion. Following reverse transcription with Omniscript cDNA synthesis kit (Qiagen), PCR analysis was performed using primers specific for human MD-2 (5′-ATGTTACCATTTCTGTTT-3′, 5′-CTAATTTGAATTAGGTTG-3′) or mouse MD-2 (5′-TCTGCAACTCCTCCGATG-3′, 5′-GGCGGTGAATGATGGTGA-3′). The PCR was performed using Taq DNA polymerase (Invitrogen). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a loading control. For quantitative RT-PCR the following primer and probe set was used to detect MD-2s: 5′-ATT GGG TCT GCA ACT CAT CC-3′, 5′-TTC TTT GCG CTT TGG AAG AT-3′ and 5′-CAC CTA CTG TGG GAG AGA TTT AAA GCA-3′. The comparative cycling threshold method (ΔΔCT) was used for relative quantification compared to untreated samples after normalization with GAPDH expression.
Immunoprecipitation and Immunoblotting
HEK293 cells were seeded into 100 mm dishes (1.5×106) 24 h prior to transfection. Transfections were performed using lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. For co-immunoprecipitations, 4 μg of each construct was transfected. For competition experiments where the effect of increasing MD-2s expression on complex formation between MD-2 and TLR4 was examined, 3 μg of each signaling molecule expression plasmid was transfected in the presence of increasing amounts (10 μg or 15μg) of the MD-2s expression plasmid. The total amount of DNA in each sample was kept constant by using empty vector cDNA. Cells were harvested 24 h following transfection in 600 μl of lysis buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40 containing protease inhibitor cocktail, and 1 mM sodium orthovanadate). For immunoprecipitations, the indicated antibodies were incubated with the cell lysates for 2 h or overnight at 4°C. Subsequently, Trueblot™ IgG beads were added and the samples were incubated at 4°C for 1 h. The immune complexes were then washed and the associated proteins were eluted from the beads by boiling in 35 μl of sample buffer, and then fractionated by SDS-PAGE. For immunoblotting, primary antibodies were detected using horseradish peroxidase–conjugated secondary antibodies, followed by enhanced chemiluminescence (Amersham Biosciences).
Reporter gene assays
HEK293 cells were transiently transfected with the expression vectors noted in combination with constructs encoding the NF-κB-luciferase reporter gene, and the phRL-TK reporter gene to normalize for transfection efficiency. In all cases, total DNA concentration was kept constant by supplementation with empty vector control. Following overnight incubation, cells were stimulated for 6 h with 50 ng/ml LPS, lysed and then luciferase activity was measured. For the inhibition studies, HEK293 cells were transfected with a constant amount (3 μg) of MD-2 expressing plasmid alone or in combination with increasing concentrations of a plasmid expressing MD-2s (5 μg, 10 μg, 20 μg). In all cases, total DNA concentration was kept constant by supplementation with empty vector control. Supernatants derived from these cells (5 ml final volume) were then transferred (100 μl per well) to HEK293 cells stably transfected with TLR4 and a NF-κB reporter gene. These supernatant-treated cells were then stimulated with LPS (5 ng/ml) for 6 h, lysed and then luciferase activity was measured. Data are shown as mean ± S.D. of three or more independent experiments and are reported as a percentage of LPS-stimulated NF-κB promoter activity.
Enzyme-linked immunosorbent assay (ELISA) of human IL-8
After LPS stimulation, supernatants were collected and the production of human IL-8 was measured with an ELISA kit following the manufacturers directions (R&D Systems).
Flow-cytometric analysis
HEK293T cells were retrovirally transduced to generate a cell line stably expressing TLR4-mCitrine. TLR4-mCitrine cells were seeded into 12-well dishes and transfected with 300 ng of plasmid encoding Myc-tagged MD-2 or MD-2s using GeneJuice lipofection reagent (Novagen) and cultured overnight. Cells were dislodged by scraping, washed once with cold PBS and incubated with a 1:20 dilution of anti-Myc Alexa 647-conjugated antibody (AbD Serotec) at 4 °C for 20 min. Cells were washed four times with PBS and fluorescence was assessed with an LSR II flow cytometer (BD Biosciences) running FACS Diva software (BD Biosciences). Data was processed using FlowJo software v8.6.3 (Tree Star).
Confocal imaging
TLR4-mCitrine cells were cultured on glass-bottom confocal dishes (MatTek) coated with collagen. After 24 h of culture, cells were transfected with up to 300 ng of plasmid encoding myc-tagged MD-2 or MD-2s using GeneJuice lipofection reagent (Novagen) and cultured for approximately 24 h. Cells were stained with anti-myc Alexa-647 antibody (AbD Serotec) in culture medium at 4 °C for 20 min. Cells were gently washed three times with PBS and culture medium was replaced. Cells were subsequently imaged at room temperature using an SP2 AOBS confocal laser scanning microscope (Leica Microsystems) running LCS software (Leica Microsystems).
RESULTS
Identification of a novel human MD-2 splice variant
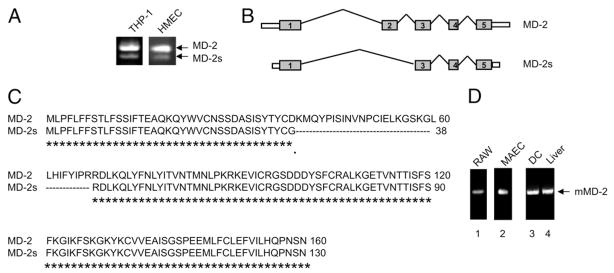
We examined the expression profile of MD-2 in human monocytic THP-1 cells by RT-PCR analysis and detected two cDNA products, suggesting that the human MD-2 gene is alternatively spliced (Fig. 1a). RT-PCR amplification using cDNA derived from human dermal microvascular endothelial cells (HMECs) also yielded two fragments. Sequencing of the larger cDNA fragment determined that it corresponded to the published sequence of full-length human MD-2. Sequence analysis of the smaller cDNA fragment, subsequently referred to as MD-2s, revealed that this was a novel splice variant of human MD-2, lacking the region encoded by exon 2 (Fig. 1b and 1c). The open reading frame of MD-2s consists of 390 bp, which translates into a predicted protein of 130 amino acids. Excision of exon 2 resulted in an in-frame deletion of 90 bp and an amino acid substitution (D38G) at the junction between exons 2 and 3. The existence of MD-2s was further confirmed given that a full-length mRNA sequence, deposited in the NCBI database, corresponded to MD-2s (Accession code: BM918324). Table I depicts the 3′ and 5′ splice-site sequences at the intron-exon boundaries for MD-2 and MD-2s.
Figure 1. Identification of a splice variant of human MD-2, which lacks exon 2.
(a) RT-PCR with human MD-2 specific primers on RNA isolated from THP-1 cells or HMECs. (b) A schematic representation depicting the mature human MD-2 protein shown above the alternatively spliced MD-2s isoform. MD-2s is generated by skipping exon 2 (90 base pair deletion). An amino acid substitution, D38G, also occurs at the junction between exons 2 and 3. (c) ClustalW alignment of MD-2 and MD-2s. The amino acids that are common to both proteins are indicated underneath with an “*”. The D38G amino acid substitution is indicated underneath with a “.”. (d) RT-PCR with murine MD-2 specific primers on RNA isolated from RAW 246.7 cells, MAEC, murine derived dendritic cells and liver tissue.
TABLE I.
Intron-exon boundaries of the human MD-2 gene.
 |
The intronic and exonic sequences are in lowercase and uppercase letters respectively. The splice-acceptor sites are in bold underlined letters. The single letter amino acid translations are located below the middle nucleotide of the codon.
We also examined the expression profile of murine MD-2 by performing RT-PCR on cDNA derived from the murine monocytic cell line, RAW246.7, and mouse aortic endothelial cells (MAECs). Using primers specific for murine MD-2, only the larger RT-PCR product was detected (Fig. 1d, lanes 1 and 2 respectively). To confirm that the absence of the smaller fragment was not specific to the murine cell lines selected, we also amplified cDNA from murine bone marrow derived dendritic cells and liver tissues obtained from C57BL/6 mice. Again only the larger cDNA fragment was observed, suggesting that MD-2s is not expressed in mice (Fig. 1d, lanes 3 and 4). Furthermore, of the four murine MD-2 splice variants that have been deposited in the NCBI database, exon 2 is transcribed in all isoforms identified.
To determine why the human but not the mouse MD-2 gene alternatively skips exon 2, we compared the gene structures and sequences of the two species. Both the human and murine MD-2 genes are organized into five exons and four introns, and each species encodes a predicted full-length MD-2 protein of 160 amino acids. Alignment of the human and mouse genomic regions revealed that the exons and coding sequences were well conserved. However, analysis of the non-coding regions revealed a number of differences. In particular, it was noted that intron 1 of human MD-2 is composed of 13241 base pairs, while the mouse has 3064. The longer intron 1 may increase the probability for alternative lariat formation during the splicing process of human MD-2. In addition, after comparing the sequences at the 3′ end of intron 1, we found that murine MD-2 has more pyrimidines than its human ortholog, which may lead to a more stable lariat formation in murine MD-2, thereby preventing alternative splicing of exon 2 in this species.
Expression and regulation of human MD-2s
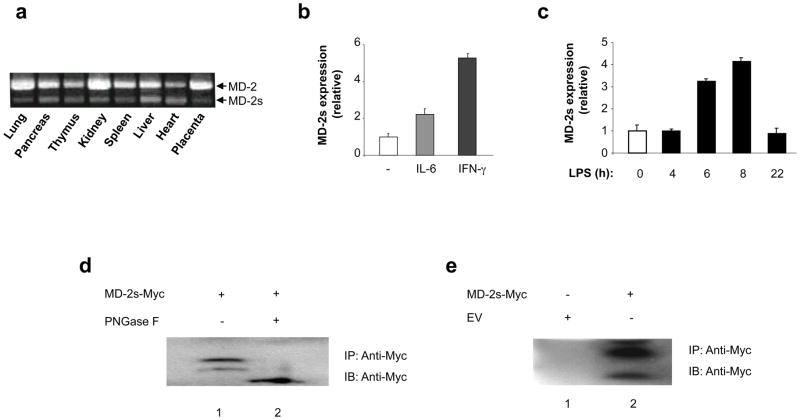
To further characterize the expression profile of human MD-2s, we performed RT-PCR analysis on a variety of human tissues. As shown in Fig. 2a, MD-2s mRNA was expressed in all tissues examined. We also observed that although the ratio between MD-2 and MD-2s varies in different tissues, full-length MD-2 is the predominant form detected. Previous studies have indicated that MD-2 mRNA is upregulated following exposure to interferon-gamma (IFN-γ) (22, 23) or IL-6 (13). We therefore investigated what effect these stimuli would have on MD-2s mRNA levels. RNA was extracted from THP-1 cells stimulated with IFN-γ- or IL-6, and quantitative RT-PCR was performed with primers specific for MD-2s and the reference gene GAPDH. Notably, we observed induction of MD-2s mRNA following treatment with both IL-6 and IFN-γ (Fig. 2b). We next examined what effect LPS stimulation would have on MD-2s expression. Interestingly, MD-2s expression was also upregulated in response to LPS (Fig. 2c). This suggests that MD-2s may play an important regulatory role during immune responses and TLR signaling.
Figure 2. Expression and regulation of MD-2s.
(a) MD-2 and MD-2s were amplified by RT-PCR from the indicated human tissues. (b) Quantitative RT-PCR analysis of RNA from untreated, IL-6-, and IFN-γ-treated THP-1 cells using primers specific for MD-2s. Expression is normalized to the reference gene GAPDH and is presented relative to that of untreated controls. (c) RT-PCR analysis of RNA from THP-1 cells treated with LPS for 4, 6, 8, and 22 hours. Primers specific for MD-2s were used. Expression is normalized to the reference gene GAPDH and is presented relative to that of untreated controls. (d) Myc tagged proteins were immunoprecipitated (IP) with an anti-Myc antibody from cell lysates prepared from HEK293 cells transiently transfected with a plasmid encoding Myc-MD-2s. Immunoprecipitates were either left untreated (lane 1) or treated with PNGase F (lane 2). Samples were subsequently analyzed by SDS-PAGE and immunoblotted (IB) with an anti-Myc antibody. (e) Myc-MD-2s was immunoprecipitated from culture supernatants that were obtained from HEK293 cells transiently transfected with a plasmid encoding Myc-MD-2s. Samples were subsequently analyzed by SDS/PAGE and immunoblotted with an anti-Myc antibody. Mock transfected culture supernatants were used as a negative control.
MD-2s is glycosylated and secreted
In order to further characterize MD-2s, we amplified the smaller RT-PCR fragment and cloned it into an expression vector encoding for the Myc tag. When this plasmid was transfected into human embryonic kidney (HEK293) cells, overexpressed MD-2s migrated with different electrophorectic mobilities following SDS-PAGE analysis (Fig. 2d, lane 1). MD-2 displays a similar expression profile due to N-linked glycosylations at positions Asn26 and Asn114 (24). These residues are still present in MD-2s, but given that it lacks 30 amino acids, its tertiary structure was likely to be different than that of MD-2, which could result in occlusion of these known glycosylation sites. Nevertheless, we found that following treatment with the Peptide-N-glycosidase F (PNGase F), the slowest migrating forms of MD-2s were no longer evident in the PNGase F-treated sample (Fig. 2d, lane 2). Thus, MD-2s is also a glycoprotein.
MD-2 belongs to the MD-2-related lipid recognition (ML) family, the signature sequence of which is a secretion signal (25). This signal is located at the N-terminus of MD-2, and studies have shown that MD-2 is secreted (26, 27). MD-2s also contains this leader sequence. We therefore examined if MD-2s exists as a stably secreted protein. Myc-tagged proteins were immunoprecipitated from culture supernatants collected from HEK293 cells transiently expressing MD-2s-Myc. As shown in Fig. 2e lane 2, a secreted product corresponding to MD-2s was detected.
MD-2s fails to induce NF-κB activation following LPS stimulation
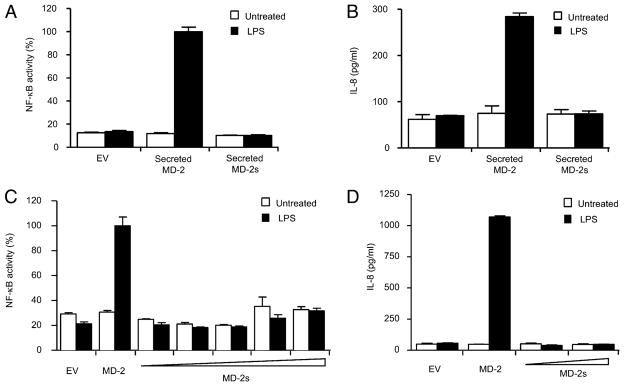
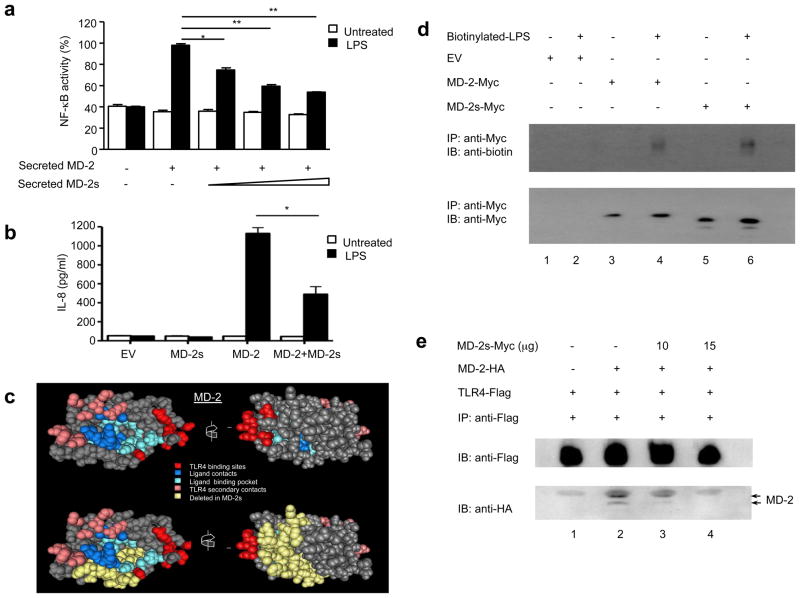
We investigated the requirement for MD-2s in LPS mediated NF-κB activation. It is known that the soluble form of full length MD-2 confers LPS responsiveness to cells expressing TLR4 (26, 28, 29), we therefore investigated if soluble MD-2s was also bioactive. Culture supernatants were collected from HEK293 cells transiently expressing control vector, MD-2, or MD-2s, and incubated with HEK293 cells stably transfected with TLR4 and an NF-κB-dependent luciferase reporter gene. Consistent with published results, soluble MD-2 conferred the ability of these reporter cells to respond to LPS by inducing both NF-κB activation and IL-8 secretion (Fig 3a and 3b, respectively). In stark contrast the secreted form of MD-2s could not activate either of these responses (Fig. 3a and 3b). We next assessed the capacity of MD-2s to mediate signal transduction triggered by LPS following transient transfection of the MD-2s expression vector. HEK293TLR4 cells stably expressing the NF-κB-dependent luciferase reporter gene were transiently transfected with plasmids encoding CD14 in combination with either MD-2 or MD-2s, and subsequently stimulated with LPS. Although full length MD-2 activated NF-κB and lead to secretion of IL-8 in the presence of LPS, overexpressed MD-2s remained inactive, even at higher concentrations (Fig. 3c and 3d). Taken together, these results suggest that it is essential for MD-2 to contain the region encoded by exon 2 in order to mediate LPS-induced TLR4 signaling.
Figure 3. MD2-s fails to mediate LPS-dependent NF-κB activation and IL-8 secretion.
(a) HEK293 cells stably transfected with TLR4 and a NF-κB reporter gene were treated with supernatants containing either MD-2 or MD-2s. Cells were left untreated or stimulated with LPS (250 ng/ml) for 6 h. Mean relative stimulation of luciferase activity ±S.D. for a representative experiment, each performed in triplicate, is shown. (b) Supernatants were collected and measured for IL-8 secretion. (c) HEK293 cells stably transfected with TLR4 and a NF-κB reporter gene were transiently transfected with plasmids encoding for CD14, and either MD-2 (25 ng), or MD-2s (wedge represents 25–200 ng). 24 h later, cells were left untreated or stimulated with LPS (50 ng/ml) for 6 h. Mean relative stimulation of luciferase activity ±S.D. for a representative experiment, each performed in triplicate, is shown. (d) Supernatants were also collected and measured for IL-8 secretion.
MD-2s interacts with TLR4
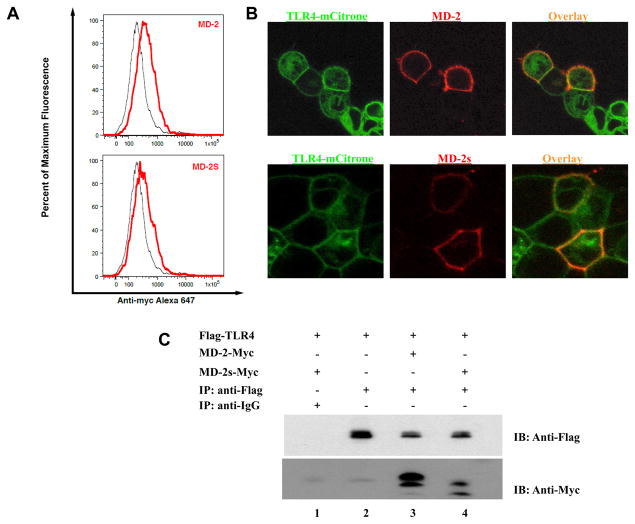
Although MD-2 lacks transmembrane and intracellular regions, it can be membrane-bound through its association with the extracellular portion of TLR4 (30). Given that MD-2s failed to mediate LPS signaling, we next asked whether this might be due to an inherent inability to interact with TLR4. To address this possibility, we first investigated if MD-2s was located on the surface of TLR4 expressing cells. We transiently transfected TLR4-mCitrine expressing cells with plasmids encoding either MD-2 or MD-2s and performed flow cytometric analysis. We observed that MD-2s was bound to the cell surface of TLR4 expressing cells (Fig. 4a). We then examined the cellular distribution of MD-2s with respect to TLR4 by confocal microscopy. We found that MD-2s exhibited the same cellular distribution as MD-2 and colocalized with TLR4 on the cell surface (Fig. 4b). We next assessed if MD-2s was anchored to the membrane via association with TLR4. We observed that MD-2s immunoprecipitated together with TLR4 (Fig. 4c, lane 4). Furthermore, as has been reported with MD-2, both the non-glycosylated and glycosylated form of MD-2s immunoprecipitated with TLR4. This indicates that although MD-2s failed to confer LPS responsiveness to cells in the absence of MD-2, this cannot be attributed to an inability to associate with TLR4.
Figure 4. MD-2s colocalizes and immunoprecipitates with TLR4.
(a) Cells stably expressing TLR4-mCitrine were transiently transfected with plasmids encoding for full-length MD-2 (upper panels) or MD-2s (lower panels) and surface-stained with anti-myc Alexa 647-conjugated antibody. Cells were analyzed by flow cytometry and gated to select single cells and the TLR4-positive population. Specific staining of TLR4 cells expressing MD-2 (upper panel, red line) or MD-2S (lower panel, red line) could be distinguished from nonspecific staining of untransfected cells (black line). (b) Cells stably expressing TLR4-mCitrine (green) were transiently transfected with plasmids encoding wild-type MD-2 (upper panels) or MD-2s (lower panels). 24 h later, cells were incubated with an anti-Myc antibody and cross-liked with Alexa 647-conjugated anti-mouse polyclonal secondary antibody. Co-localization of TLR4 and MD-2 isoforms on the cell surface is visualized in yellow. (c) HEK293 cells were transiently transfected with a plasmid encoding for Flag-tagged TLR4 (lanes 1–4) and co-transfected with either a Myc-tagged MD-2 (lane 3) or a Myc-tagged MD-2s (lanes 1 and 4) expressing plasmid. Co-immunoprecipitation experiments were then performed using an anti-Flag antibody (lanes 2–4) or an IgG isotype control antibody (lane 1). Samples were fractionated by SDS-PAGE and immunoblotting with an anti-Myc antibody was performed. Experiment shown is representative of 4 separate experiments.
MD-2s interacts with LPS and inhibits TLR4 signaling by preventing the formation of an active MD-2-TLR4 complex
Having determined that MD-2s cannot reconstitute LPS signaling in cells lacking MD-2, we next investigated the effect of soluble MD-2s on TLR4 activation. HEK293 cells stably expressing TLR4 and an NF-κB-dependent luciferase reporter gene were incubated with supernatants collected from HEK293 cells that contained a constant amount of MD-2 but increasing concentrations of MD-2s. We noted that soluble MD-2s blocked NF-κB activation in response to LPS in a dose-dependent manner (Fig. 5a). We also observed that overexpressed MD-2s inhibited LPS-induced IL-8 secretion (Fig. 5b). These findings indicate that MD-2s functions as a negative regulatory protein in the TLR4 signaling pathway, suggesting that MD-2s may be used as a therapeutic or preventative agent for modulating an overactivated MD-2-TLR4 immune response.
Figure 5. MD-2s inhibits LPS-induced NF-κB activation by preventing the formation of an active MD-2-TLR4 complex and interacting with LPS.
(a) HEK293 cells stably transfected with TLR4 and a NF-κB reporter gene were treated with supernatants containing either MD-2 or MD-2s as depicted (see Methods for details) and left untreated or incubated with LPS (5 ng/ml). Mean relative stimulation of luciferase activity ± S.D. for a representative experiment from three separate experiments, each performed in triplicate, is shown. (b) HEK293 cells, stably transfected with TLR4 and a NF-κB reporter gene, were transiently transfected with plasmids expressing CD14 and MD-2 (25 ng) in combination with, or without, an MD-2s expressing vector (100 ng). Cells were left untreated or incubated with LPS (50 ng/ml). Supernatants were subsequently collected and measured for IL-8 secretion. (c) Structural analysis of MD-2. Structural features representing the TLR4 binding sites (red); the ligand contacts (dark blue); the ligand binding pocket (light blue) and the TLR4 secondary contacts (pink) are illustrated. The residues deleted in MD-2s (yellow) are also shown. (d) HEK293 cells were transiently transfected with plasmids encoding for Myc-tagged MD-2 (lanes 3 and 4) or a Myc-tagged MD-2s (lanes 5 and 6) expressing plasmid and treated with biotinylated LPS as indicated. Co-immunoprecipitation experiments were then performed using an anti-Myc antibody. Samples were fractionated by SDS-PAGE and immunoblotting with an anti-biotin (upper panel) or anti-Myc antibody (lower panel) was performed. (e) HEK293 cells were transiently transfected with plasmids encoding for Flag-tagged TLR4 (lanes 1–4) in combination with a HA-tagged MD-2 (lanes 2 to 3) and increasing concentrations of Myc-tagged MD-2s (lanes 3 and 4). Co-immunoprecipitation experiments were then performed using an anti-Flag antibody. Samples were fractionated by SDS-PAGE and immunoblotting with an anti-Flag antibody (upper panel) or anti-HA antibody (lower panel) was performed.
We next analyzed the published structural models of MD-2 to ascertain the structural effect of deleting exon 2. MD-2 consists of a β-cup fold with two anti-parallel β-sheets, one composed of six β-strands (numbered 1/2/9/8/5/6) and the other of three (numbered 3/4/7) (31, 32). The missing exon 2 of MD-2s encodes the first two β-strands of the three-stranded β-sheet (β3 and β4 strands) (Fig. 5c). The hinges connecting β-strands 2 to 3 and 4 to 5 are also partially lost. Furthermore, the disulphide bond between Cys25 and Cys51, which assists in closing the MD-2 cavity and stabilizing the cup-like structure, is disrupted. The β6 and β7 strands that line the entrance to the deep hydrophobic cavity are still encoded by the mRNA of MD-2s.
Elucidation of the TLR4-MD-2-LPS complex revealed that LPS binding instigates the formation of a receptor multimer which is composed of two copies of the complex arranged symmetrically (14). Eleven residues are involved in the main dimerization interface of the TLR4-MD-2-LPS complex, of these 10 are still present in MD-2s. However, deletion of exon 2 in MD-2 implied that the ligand-binding pocket of MD-2s may be severely disrupted, suggesting that it may be unable to bind LPS efficiently. To address this possibility, HEK293 cells were transiently transfected with plasmids encoding either MD-2-Myc or MD-2s-Myc. Culture supernatants were then incubated with biotinylated-LPS and Myc-tagged proteins were immunoprecipitated. In agreement with previous studies (33–35), secreted MD-2 bound readily to LPS (Fig. 5d, lane 4). Interestingly, under similar conditions, an interaction between secreted MD-2s and LPS was also detected (Fig. 5d, lane 6), suggesting that soluble MD-2s may sequester LPS from binding to the MD-2-TLR4 complex and thus diminish TLR4 signal transduction.
Given that MD-2s inhibits LPS-induced TLR4 signaling, but still interacts with LPS and TLR4, we hypothesized that it may compete with MD-2 to form an interaction with TLR4. To investigate this possibility, we increased the expression level of MD-2s-Myc, but kept the concentration of the constructs for HA-MD-2 and Flag-TLR4 constant. As expected, both the glycosylated and non-glycosylated forms of MD-2 immunoprecipitated with TLR4 in HEK293 cells transiently transfected with plasmids encoding both proteins (Fig. 5e, lane 2). However, overexpression of MD-2s competitively inhibited binding of MD-2 to TLR4 (Fig. 5e, compare lanes 3 and 4 to lane 2), indicating that MD-2s downregulates TLR4 signaling by inhibiting the formation of an active MD-2-TLR4 signaling complex.
DISCUSSION
An inappropriately mounted or dysregulated immune response can cause considerable morbidity and mortality in a number of diseases. One such example is sepsis, which is among the most common causes of death in the United States, with over 750,000 cases presenting annually, of which more than one-quarter are fatal (36). Excessive inflammation is the hallmark of a number of related infectious pathologies as well, including sepsis, acute respiratory distress syndrome (ARDS) and multiple organ failure (37). LPS derived from bacterial sources can contribute to these diseases, and does so by interacting with MD-2 and TLR4. To circumvent an overactivated host immune response to LPS, it is imperative that TLR4 signal transduction be tightly regulated, but the precise molecular mechanisms by which this is accomplished are only partly understood.
Here we further elucidate the complexities involved in averting a prolonged and dysregulated immune response to LPS by the identification of a naturally occurring alternatively spliced isoform of human MD-2, which we have termed MD-2s. We report that human MD-2s is generated by skipping exon 2 of the MD-2 gene, which leads to an in-frame deletion of 30 amino acids spanning positions 39–69, and one amino acid substitution (D38G). Under similar conditions and using primers specific to the murine MD-2 gene, we could not detect a corresponding murine splice variant. A prior study reported an alternatively spliced version of murine MD2 (MD-2B) (21), which lacks the first 54 bases of exon 3 and downregulates LPS signaling. However, there are no data provided as to whether it is secreted or inducible and the human relevance is unknown (see Table II for comparison between MD-2s and MD-2B).
TABLE II.
Comparison of the MD-2 splice variants, MD-2s and MD-2B.
| MD-2s (human) | MD-2B (murine) | |
|---|---|---|
| Deleted region | Exon 2 | 54bp of Exon 3 at 5′ end |
| Glycosylated | Yes | N.D. but contains multiple bands |
| Secreted | Yes | N.D. |
| Mediate LPS-induced NF-κB activation | No | No |
| Upregulated | Yes | N.D. |
| Immunoprecipitates with TLR4 | Yes | Yes |
| Immunoprecipitates with LPS | Yes | N.D. |
| Inhibits LPS signaling | Yes | Yes |
| Inhibits TLR4 surface expression | No | Yes |
| Inhibits formation of the MD-2-TLR4 complex | Yes | Yes, on the cell surface |
Our results identify MD-2s as an important negative regulatory component of the TLR4 signaling pathway. The mRNA expression profile of MD-2s in human tissues revealed that it is ubiquitously expressed, suggesting that this isoform may perform a widespread role in modulating TLR4-mediated responses. Previous studies have shown that IFN-γ exerts anti-inflammatory responses by inducing specific secreted inhibitors, examples include the IL-1 receptor antagonist (IL-1Ra) (38) and IL-18 binding protein (IL-18BP) (39). Both molecules suppress the activity of IL-1 and IL-18 respectively. Many LPS-inducible negative regulators have also been identified, such as smTLR4 and MyD88s. We determined that MD-2s is upregulated in response to IFN-γ, IL-6 and LPS, indicating that MD-2s may be a key component involved in the negative regulation of TLR4 signaling.
Similar to full-length MD-2, MD-2s is a secreted glycoprotein. However, MD-2s overexpression failed to trigger NF-κB activation and IL-8 secretion following LPS treatment, indicating the importance of exon 2 for MD-2 function. Importantly, we also observed that MD-2s negatively regulated both NF-κB activation and IL-8 secretion following LPS stimulation, suggesting that MD-2s may be used as a therapeutic or preventative agent for modulating endotoxemia. Whereas the murine isoform, MD-2B, inhibited TLR4 from being expressed on the cell surface (21), MD-2s is anchored to the cell surface of TLR4 expressing cells, and both proteins localize together on the cell membrane. We also determined that MD-2s and TLR4 immunoprecipitated together. This may have been predicted, given that MD-2s retains most of the residues reported to be essential in mediating a MD-2-TLR4 interaction, with the exception of I66 and R68 (31, 40). In addition, MD-2s immunoprecipitated with LPS. This is consistent with a study demonstrating that a 15 residue peptide fragment of MD-2, encompassing the F126 loop (positions 119–132), still binds LPS (41). Additional studies have verified that residues within this fragment of MD-2 are essential for LPS binding (34, 40, 42). This region is preserved in MD-2s and most likely confers the ability of MD-2s to associate with LPS. Furthermore, although the hydrophobic pocket of MD2-s is predicted to be disrupted, with the exception of K58 all the MD-2 residues that have been shown to be involved in the main dimerization interface of the TLR4-MD-2-LPS complex, are preserved in MD-2s. Whilst it appears that these residues are sufficient to form an effective interaction between MD-2s and LPS, the binding affinity is most likely affected.
Several studies have shown that negative regulators can control TLR signal transduction by inhibiting the formation of active signaling complexes, including the recently identified splice variant TAG (20), IRF-4 (43), RP105 (44) and the IL-1Ra (45–47). Based on our results, we propose that MD-2s functionally modulates TLR4 signaling by inhibiting the formation of an active MD-2-TLR4 signaling complex. In addition, it is possible that MD-2s may behave like a decoy co-receptor by binding LPS and TLR4 to form a non-functional complex that does not activate NF-κB, thereby negatively regulating signaling.
Collectively our results define an important mechanistic role for MD-2s in modulating the LPS-TLR4 signal transduction pathway at the initial phase of activation. MD-2s therefore represents a prospective target for pharmacological intervention and development of new therapeutic and preventive strategies for sepsis and other diseases resulting from an over-exuberant MD2-TLR4-induced immune response.
Acknowledgments
We sincerely thank P. Sun for technical assistance; K. Miyake (Tokyo University) for the plasmid encoding Flag MD-2; F.J. Candal (Center for Disease Control and Prevention) for the HMECs, and Terence M. Doherty (Cedars-Sinai Medical Center) for editorial assistance.
Supported by the National Institutes of Health Grants (HL-66436 and AI-058128 to MA).
The abbreviations used are
- TLR
Toll-like receptor
- MD-2
Myeloid differentiation factor 2
- LPS
lipopolysaccharide
References
- 1.Brikos C, O’Neill LA. Signalling of toll-like receptors. Handb Exp Pharmacol. 2008:21–50. doi: 10.1007/978-3-540-72167-3_2. [DOI] [PubMed] [Google Scholar]
- 2.Ohashi K, Burkart V, Flohe S, Kolb H. Cutting Edge: Heat Shock Protein 60 Is a Putative Endogenous Ligand of the Toll-Like Receptor-4 Complex. J Immunol. 2000;164:558–561. doi: 10.4049/jimmunol.164.2.558. [DOI] [PubMed] [Google Scholar]
- 3.Kurt-Jones EA, Popova L, Kwinn L, Haynes LM, Jones LP, Tripp RA, Walsh EE, Freeman MW, Golenbock DT, Anderson LJ, Finberg RW. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- 4.Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: Mutations in Tlr4 Gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 5.Poltorak A, Smirnova I, He X, Liu MY, Van Huffel C, McNally O, Birdwell D, Alejos E, Silva M, Du X, Thompson P, Chan EK, Ledesma J, Roe B, Clifton S, Vogel SN, Beutler B. Genetic and physical mapping of the Lps locus: identification of the toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol Dis. 1998;24:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 6.Qureshi ST, Lariviere L, Leveque G, Clermont S, Moore KJ, Gros P, Malo D. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4) J Exp Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 8.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 9.Schumann RR, Leong SR, Flaggs GW, Gray PW, Wright SD, Mathison JC, Tobias PS, Ulevitch RJ. Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 10.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 11.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamann L, Kumpf O, Muller M, Visintin A, Eckert J, Schlag PM, Schumann RR. A coding mutation within the first exon of the human MD-2 gene results in decreased lipopolysaccharide-induced signaling. Genes Immun. 2004;5:283–288. doi: 10.1038/sj.gene.6364068. [DOI] [PubMed] [Google Scholar]
- 13.Tissieres P, Dunn-Siegrist I, Schappi M, Elson G, Comte R, Nobre V, Pugin J. Soluble MD-2 is an acute-phase protein and an opsonin for Gram-negative bacteria. Blood. 2008;111:2122–2131. doi: 10.1182/blood-2007-06-097782. [DOI] [PubMed] [Google Scholar]
- 14.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 15.Kolek MJ, Carlquist JF, Muhlestein JB, Whiting BM, Horne BD, Bair TL, Anderson JL. Toll-like receptor 4 gene Asp299Gly polymorphism is associated with reductions in vascular inflammation, angiographic coronary artery disease, and clinical diabetes. Am Heart J. 2004;148:1034–1040. doi: 10.1016/j.ahj.2004.05.049. [DOI] [PubMed] [Google Scholar]
- 16.Eisenbarth SC, Piggott DA, Huleatt JW, Visintin I, Herrick CA, Bottomly K. Lipopolysaccharide-enhanced, Toll-like Receptor 4-dependent T Helper Cell Type 2 Responses to Inhaled Antigen. J Exp Med. 2002;196:1645–1651. doi: 10.1084/jem.20021340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwami KI, Matsuguchi T, Masuda A, Kikuchi T, Musikacharoen T, Yoshikai Y. Cutting edge: naturally occurring soluble form of mouse Toll-like receptor 4 inhibits lipopolysaccharide signaling. J Immunol. 2000;165:6682–6686. doi: 10.4049/jimmunol.165.12.6682. [DOI] [PubMed] [Google Scholar]
- 18.Jaresova I, Rozkova D, Spisek R, Janda A, Brazova J, Sediva A. Kinetics of Toll-like receptor-4 splice variants expression in lipopolysaccharide-stimulated antigen presenting cells of healthy donors and patients with cystic fibrosis. Microbes Infect. 2007;9:1359–1367. doi: 10.1016/j.micinf.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Janssens S, Burns K, Tschopp J, Beyaert R. Regulation of interleukin-1-and lipopolysaccharide-induced NF-kappaB activation by alternative splicing of MyD88. Curr Biol. 2002;12:467–471. doi: 10.1016/s0960-9822(02)00712-1. [DOI] [PubMed] [Google Scholar]
- 20.Palsson-McDermott EM, Doyle SL, McGettrick AF, Hardy M, Husebye H, Banahan K, Gong M, Golenbock D, Espevik T, O’Neill LAJ. TAG, a splice variant of the adaptor TRAM, negatively regulates the adaptor MyD88-independent TLR4 pathway. Nat Immunol. 2009;10:579–586. doi: 10.1038/ni.1727. [DOI] [PubMed] [Google Scholar]
- 21.Ohta S, Bahrun U, Tanaka M, Kimoto M. Identification of a novel isoform of MD-2 that downregulates lipopolysaccharide signaling. Biochem Biophys Res Commun. 2004;323:1103–1108. doi: 10.1016/j.bbrc.2004.08.203. [DOI] [PubMed] [Google Scholar]
- 22.Bosisio D, Polentarutti N, Sironi M, Bernasconi S, Miyake K, Webb GR, Martin MU, Mantovani A, Muzio M. Stimulation of toll-like receptor 4 expression in human mononuclear phagocytes by interferon-gamma: a molecular basis for priming and synergism with bacterial lipopolysaccharide. Blood. 2002;99:3427–3431. doi: 10.1182/blood.v99.9.3427. [DOI] [PubMed] [Google Scholar]
- 23.Abreu MT, Arnold ET, Thomas LS, Gonsky R, Zhou Y, Hu B, Arditi M. TLR4 and MD-2 expression is regulated by immune-mediated signals in human intestinal epithelial cells. J Biol Chem. 2002;277:20431–20437. doi: 10.1074/jbc.M110333200. [DOI] [PubMed] [Google Scholar]
- 24.Ohnishi T, Muroi M, Tanamoto K. N-linked glycosylations at Asn(26) and Asn(114) of human MD-2 are required for toll-like receptor 4-mediated activation of NF-kappaB by lipopolysaccharide. J Immunol. 2001;167:3354–3359. doi: 10.4049/jimmunol.167.6.3354. [DOI] [PubMed] [Google Scholar]
- 25.Inohara N, Nunez G. ML -- a conserved domain involved in innate immunity and lipid metabolism. Trends Biochem Sci. 2002;27:219–221. doi: 10.1016/s0968-0004(02)02084-4. [DOI] [PubMed] [Google Scholar]
- 26.Visintin A, Mazzoni A, Spitzer JA, Segal DM. Secreted MD-2 is a large polymeric protein that efficiently confers lipopolysaccharide sensitivity to Toll-like receptor 4. Proc Natl Acad Sci U S A. 2001;98:12156–12161. doi: 10.1073/pnas.211445098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato K, Morrison AM, Nakano T, Tashiro K, Honjo T. ESOP-1, a secreted protein expressed in the hematopoietic, nervous, and reproductive systems of embryonic and adult mice. Blood. 2000;96:362–364. [PubMed] [Google Scholar]
- 28.Kennedy MN, Mullen GE, Leifer CA, Lee C, Mazzoni A, Dileepan KN, Segal DM. A complex of soluble MD-2 and lipopolysaccharide serves as an activating ligand for Toll-like receptor 4. J Biol Chem. 2004;279:34698–34704. doi: 10.1074/jbc.M405444200. [DOI] [PubMed] [Google Scholar]
- 29.Teghanemt A, Widstrom RL, Gioannini TL, Weiss JP. Isolation of monomeric and dimeric secreted MD-2. Endotoxin.sCD14 and Toll-like receptor 4 ectodomain selectively react with the monomeric form of secreted MD-2. J Biol Chem. 2008;283:21881–21889. doi: 10.1074/jbc.M800672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akashi S, Shimazu R, Ogata H, Nagai Y, Takeda K, Kimoto M, Miyake K. Cutting edge: cell surface expression and lipopolysaccharide signaling via the toll-like receptor 4-MD-2 complex on mouse peritoneal macrophages. J Immunol. 2000;164:3471–3475. doi: 10.4049/jimmunol.164.7.3471. [DOI] [PubMed] [Google Scholar]
- 31.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Ohto U, Fukase K, Miyake K, Satow Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 33.Mullen GE, Kennedy MN, Visintin A, Mazzoni A, Leifer CA, Davies DR, Segal DM. The role of disulfide bonds in the assembly and function of MD-2. Proc Natl Acad Sci U S A. 2003;100:3919–3924. doi: 10.1073/pnas.0630495100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visintin A, Latz E, Monks BG, Espevik T, Golenbock DT. Lysines 128 and 132 enable lipopolysaccharide binding to MD-2, leading to Toll-like receptor-4 aggregation and signal transduction. J Biol Chem. 2003;278:48313–48320. doi: 10.1074/jbc.M306802200. [DOI] [PubMed] [Google Scholar]
- 35.Viriyakosol S, Tobias PS, Kitchens RL, Kirkland TN. MD-2 binds to bacterial lipopolysaccharide. J Biol Chem. 2001;276:38044–38051. doi: 10.1074/jbc.M105228200. [DOI] [PubMed] [Google Scholar]
- 36.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 37.Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Micro. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 38.Kline JN, Fisher PA, Monick MM, Hunninghake GW. Regulation of interleukin-1 receptor antagonist by Th1 and Th2 cytokines. Am J Physiol Lung Cell Mol Physiol. 1995;269:L92–98. doi: 10.1152/ajplung.1995.269.1.L92. [DOI] [PubMed] [Google Scholar]
- 39.Paulukat J, Bosmann M, Nold M, Garkisch S, Kampfer H, Frank S, Raedle J, Zeuzem S, Pfeilschifter J, Muhl H. Expression and Release of IL-18 Binding Protein in Response to IFN-{gamma} J Immunol. 2001;167:7038–7043. doi: 10.4049/jimmunol.167.12.7038. [DOI] [PubMed] [Google Scholar]
- 40.Re F, Strominger JL. Separate functional domains of human MD-2 mediate Toll-like receptor 4-binding and lipopolysaccharide responsiveness. J Immunol. 2003;171:5272–5276. doi: 10.4049/jimmunol.171.10.5272. [DOI] [PubMed] [Google Scholar]
- 41.Mancek M, Pristovsek P, Jerala R. Identification of LPS-Binding Peptide Fragment of MD-2, a Toll-Receptor Accessory Protein. Biochemical and Biophysical Research Communications. 2002;292:880–885. doi: 10.1006/bbrc.2002.6748. [DOI] [PubMed] [Google Scholar]
- 42.Teghanemt A, Re F, Prohinar P, Widstrom R, Gioannini TL, Weiss JP. Novel roles in human MD-2 of phenylalanines 121 and 126 and tyrosine 131 in activation of Toll-like receptor 4 by endotoxin. J Biol Chem. 2008;283:1257–1266. doi: 10.1074/jbc.M705994200. [DOI] [PubMed] [Google Scholar]
- 43.Negishi H, Ohba Y, Yanai H, Takaoka A, Honma K, Yui K, Matsuyama T, Taniguchi T, Honda K. Negative regulation of Toll-like-receptor signaling by IRF-4. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Divanovic S, Trompette A, Atabani SF, Madan R, Golenbock DT, Visintin A, Finberg RW, Tarakhovsky A, Vogel SN, Belkaid Y, Kurt-Jones EA, Karp CL. Negative regulation of Toll-like receptor 4 signaling by the Toll-like receptor homolog RP105. Nat Immunol. 2005;6:571–578. doi: 10.1038/ni1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hannum CH, Wilcox CJ, Arend WP, Joslin FG, Dripps DJ, Heimdal PL, Armes LG, Sommer A, Eisenberg SP, Thompson RC. Interleukin-1 receptor antagonist activity of a human interleukin-1 inhibitor. Nature. 1990;343:336–340. doi: 10.1038/343336a0. [DOI] [PubMed] [Google Scholar]
- 46.Carter DB, Deibel MR, Dunn CJ, Tomich CSC, Laborde AL, Slightom JL, Berger AE, Bienkowski MJ, Sun FF, McEwan RN, Harris PKW, Yem AW, Waszak GA, Chosay JG, Sieu LC, Hardee MM, Zurcher-Neely HA, Reardon IM, Heinrikson RL, Truesdell SE, Shelly JA, Eessalu TE, Taylor BM, Tracey DE. Purification, cloning, expression and biological characterization of an interleukin-1 receptor antagonist protein. Nature. 1990;344:633–638. doi: 10.1038/344633a0. [DOI] [PubMed] [Google Scholar]
- 47.Eisenberg SP, Evans RJ, Arend WP, Verderber E, Brewer MT, Hannum CH, Thompson RC. Primary structure and functional expression from complementary DNA of a human interleukin-1 receptor antagonist. Nature. 1990;343:341–346. doi: 10.1038/343341a0. [DOI] [PubMed] [Google Scholar]