Abstract
Synovial joints enable smooth articulations between different skeletal elements and are essential for the motility of vertebrates. Despite decades of extensive studies of the molecular and cellular mechanisms of limb and skeletal development, the molecular mechanisms governing synovial joint formation are still poorly understood. In particular, whereas several signaling pathways have been shown to play critical roles in joint maintenance, the mechanism controlling joint initiation is unknown. Here we report that Osr1 and Osr2, the mammalian homologs of the odd-skipped family of zinc finger transcription factors that are required for leg joint formation in Drosophila, are both strongly expressed in the developing synovial joint cells in mice. Whereas Osr1−/− mutant mice died at midgestation and Osr2−/− mutant mice had only subtle defects in synovial joint development, tissue-specific inactivation of Osr1 in the developing limb mesenchyme in Osr2−/− mutant mice caused fusion of multiple joints. We found that Osr1 and Osr2 function is required for maintenance of expression of signaling molecules critical for joint formation, including Gdf5, Wnt4 and Wnt9b. In addition, joint cells in the double mutants failed to upregulate expression of the articular cartilage marker gene Prg4. These data indicate that Osr1 and Osr2 function redundantly to control synovial joint formation.
Keywords: joint formation, joint fusion, limb development, Odd-skipped, Osr1, Osr2, synovial
Introduction
The successful adaptation of vertebrate animals to a wide variety of ecological niches depended largely on the formation and function of synovial joints. Synovial joints facilitate smooth articulation between two or more skeletal elements and thus play essential roles in vertebrate movement. The tissues that make up the synovial joint include articular cartilages of the opposing skeletal elements, ligaments, synovium, and the fibrous capsule. These tissues are prone to injury and malfunction during natural aging, resulting in impaired mobility. Moreover, synovial joints are often targets of diseases, such as osteoarthritis and inflammatory rheumatoid arthritis, which are the most common causes of disability in the adult human population (Elders, 2000). Despite the obvious importance of synovial joints, however, there is currently very limited understanding of the molecular mechanisms of joint development.
During embryogenesis, the limb skeleton initiates by condensation and differentiation of mesenchymal cells to form cartilage, which further undergoes endochondrial ossification to form bone. The various long bones of the limb do not initiate as discrete individual elements, but rather initially form as continuous condensed mesenchyme that is subsequently divided into distinct elements by segmentation. For example, the humerus, radius and ulna rudiments initially form as an uninterrupted Y-shaped prechondrogenic mesenchymal condensation marked by high levels of expression of Type II collagen (Col2) (Hinchliffe and Johnson, 1980; Craig et al., 1987; Lizarraga et al., 2002). The first overt sign of joint development is the appearance of the so-called interzone, consisting of densely packed flattened mesenchymal cells that provide a clear demarcation between the adjacent cartilaginous elements (Holder, 1977; Mitrovic, 1978; Pacifici et al., 2005). It has been proposed that initiation of joint formation involves inhibition of the interzone progenitor cells from differentiating into chondrocytes by downregulating or blocking Col2 expression (Lizzarraga et al., 2002; Später et al., 2006; Pitsillides and Ashhurst, 2008).
One of the earliest markers of joint interzone cells identified is Gdf5, a member of the TGF-β family of signaling molecules. Mice lacking Gdf5 exhibited multiple joint fusions, particularly involving interphalangeal joints and the carpal or tarsal bones (Storm et al., 1994, 1996). Joint defects are also associated with mutations in GDF5 in humans (Francis-West et al., 1999b; Buxton et al., 2001). Genetic lineage tracing studies showed that the Gdf5-expressing interzone cells gave rise to most joint tissues including articular cartilage but contributed little to the growth plate cartilage (Koyama et al. 2008). However, application of exogenous Gdf5 protein to the developing chick or mouse limb mesenchyme stimulated cartilage development (Storm and Kingsley, 1999), indicating that Gdf5 signaling does not specify joint cell fate and that cellular responses to Gdf5 signaling may be cell-type specific. Joint development failed to initiate in mice lacking Noggin, a secreted BMP antagonist (Brunet et al., 1998). Noggin mRNA is expressed throughout the prechondrogenic mesenchymal condensation, in both the cartilage anlagen and the presumptive joint cells. In addition to lacking joints, the Noggin−/− mutant mice exhibited gross skeletal hyperplasia, suggesting that failure of joint formation in these mutant mice was due to increased recruitment of the prechondrogenic limb mesenchyme into cartilage formation (Brunet et al., 1998). Indeed, implantation of agarose beads soaked with Bmp2 or Bmp4 adjacent to developing chick digital joints caused partial joint fusion (Archer et al., 2003).
Another signaling pathway that plays critical roles in joint development is the Wnt/β-catenin pathway. Hartmann and Tabin (2001) showed that Wnt9a (formerly Wnt14) mRNA was expressed early during interzone formation in the developing chick limb and that misexpression of Wnt9a induced ectopic expression of several interzone cell molecular markers including Gdf5 and Chordin. Moreover, Wnt9a misexpression in primary chick sternal chondrocytes caused downregulation of Col2 and upregulation of Col3 (Später et al., 2006), mimicking the molecular events during interzone formation. Expression of either Wnt9a or a constitutively active form of β-catenin driven by the Col2a1 gene promoter in transgenic mice also caused ectopic expression of joint cell markers in the developing limbs (Guo et al., 2004). However, targeted disruption of Wnt9a did not affect joint induction but resulted in synovial chondroid metaplasia in some joints (Später et al., 2006). Although the Wnt4−/−Wnt9a−/− double mutant newborn mice showed fusion of some carpal and tarsal elements, the fusion occurred after initiation of the joint interzones. Moreover, tissue-specific inactivation of β-catenin in the early developing limb mesenchyme did not block joint interzone formation (Später et al., 2006), indicating that other mechanisms are involved in joint specification.
Previous studies have shown, in addition to the selected signaling molecules of the Gdf and Wnt families, that several transcription factor genes, including Cux1, Erg, Osr1 and Osr2, are also expressed in highly restricted patterns during synovial joint formation (Dhordain et al., 1995; Lizarraga et al., 2002; Stricker et al., 2006; Iwamoto et al., 2007; Koyama et al., 2008). Cux1, encoding a large homeodomain-containing DNA-binding protein, is highly expressed at sites of incipient joint formation during limb development in chick (Lizarraga et al., 2002). Misexpression of Cux1 mRNA suppressed chondrocyte differentiation in limb mesenchyme cultures. However, overexpression of Cux1 did not induce expression of known joint markers and mice deficient in Cux1 did not have limb developmental defects (Lizarraga et al., 2002; Ellis et al., 2001). Thus, the role of Cux1 in joint development is unclear. Erg, encoding a member of the Ets family of transcription factors (Sharrocks, 2001), is co-expressed with Gdf5 in the incipient joint cells during mouse limb development and exogenous Gdf5 protein induced Erg mRNA expression in the developing limb mesenchyme (Iwamoto et al., 2007). Transgenic mice overexpressing Erg mRNAs throughout the cartilaginous skeleton did not induce ectopic joint formation but rather blocked chondrocyte maturation toward hypertrophy, suggesting that Erg may be an important factor downstream of Gdf5 in the differentiation and maintenance of articular cartilage (Iwamoto et al., 2007). Osr1 and Osr2 are the vertebrate homologs of the Drosophila odd-skipped family of zinc finger proteins (So and Danielian, 1999; Lan et al., 2001; Gao et al., 2009). In Drosophila, all four odd-skipped family genes, odd, sob, drm, and bowl, are expressed in a segmental pattern in the developing leg imaginal discs. Ectopic expression of odd, sob, or drm, induced invaginations in the leg disc epithelium and morphological changes in the adult leg that are characteristic of joint cells (Hao et al., 2003). Osr1 and Osr2 mRNAs exhibit partially overlapping expression in the early limb bud and both are expressed at high levels in the incipient joints during limb development in chick and in mice (Stricker et al., 2006), suggesting that this family of transcription factors may be involved in joint cell fate specification in vertebrates as well. Since most mouse embryos lacking Osr1 died in midgestation (Wang et al., 2005), we have generated mice with limb-specific inactivation of Osr1 and demonstrate in this report that Osr1 and Osr2 function partially redundantly to control synovial joint formation in mice.
Materials and Methods
Mouse strains and breeding strategies
The Osr1+/− and Osr2+/− mice, which carry in-frame fusion of the lacZ gene, have been described previously (Lan et al., 2004; Wang et al., 2005). Mice carrying a conditional Osr1 allele (Osr1f) were generated by inserting an FRT-flanked neo expression cassette together with a loxP sequence in the first intron and another loxP sequence in the second intron of the Osr1 gene. The Osr1f/f homozygous mice appear normal and Cre-mediated germ line deletion of the loxP-flanked exon 2 recapitulated the Osr1 null mutant mouse phenotypes (Lan et al., submitted). Other mouse strains used include the β-catenin conditional mice (Catnbf/f) (Brault et al., 2001), the Osr2IresCre mice (Lan et al., 2007), and the Prrx1-cre transgenic mice (Logan et al., 2002).
To generate mice lacking both Osr1 and Osr2 in the developing limb mesenchyme, Osr1f/f homozygous mice were crossed with Osr2+/− mice to generate the Osr1f/+Osr2+/− mice, which were then crossed with Osr1f/f mice to generate Osr1f/fOsr2+/−mice. Prrx1-cre transgenic mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Prrx1-cre mice were bred to the Osr1f/fOsr2+/− mice to generate Osr1f/+Osr2+/−Prrx1-cre mice. No gross morphological defects were found in these mice. Timed mating was set up between Osr1f/+Osr2+/−Prrx1-cre mice and Osr1f/fOsr2+/− mice to generate the Osr1f/fOsr2−/−Prrx1-cre mutant embryo for analysis.
To inactivate β-catenin in the developing limb joint cells, Catnbf/f mice were crossed to Osr2IresCre/+ mice to generate Catnbf/+Osr2IresCre/+ mice, which were then crossed to Catnbf/f mice to generated Catnbf/fOsr2IresCre/+ mutant embryos for analysis.
Skeletal preparations and histology
Timed-mating was set up for embryos with desired genotypes. Embryos were collected at E17.5. Tail DNA was extracted and genotypes of the embryos were determined by allele-specific PCR. Skeletal preparations were performed following the protocol described previously (Martin et al., 1995). For histology analysis, embryos were fixed in Bouin’s fixative, dehydrated through graded ethyl alcohols, embedded in paraffin, sectioned at 7-μm thickness and stained with alcian blue, hematoxylin and eosin.
Detection of LacZ expression and section in situ hybridization
Timed mating was set up for embryos at desired stages. Tail DNA was extracted for genotyping with allele-specific PCR. X-gal staining of whole-mount embryos and cryostat sections were performed as described (Hogan et al, 1994). Sections were counterstained with eosin. For in situ hybridization, embryos were fixed in 4% paraformaldehyde in PBS overnight at 4 °C, washed with PBS, dehydrated through graded ethyl alcohols, embedded in paraffin and sectioned at 7-μm thickness. Section in situ hybridization was carried out as previously described (Zhang et al., 1999) with digoxygenin-labeled antisense RNA probes.
Dual color whole mount LacZ staining and in situ hybridization
E11.5 and E12.5 Osr1+/− and Osr2+/− embryonic limbs were dissected and fixed on ice for 2 hours with 4% paraformaldehyde in PBS and stained at 37°C for 2 hours in Salmon-gal (Biosynth International, Naperville, IL) staining solution. After Salmon-gal staining, samples were washed with PBS, post-fixed with 1% paraformaldehyde in PBS and processed for whole-mount in situ hybridization as described previously (Jiang et al, 1998).
Results
Expression patterns of Osr1 and Osr2 overlap in most developing synovial joints and Osr2−/− mice have mild joint defects that correlate with its distinct expression
It has been reported that Osr1 and Osr2 mRNAs are both expressed in the developing joint tissues (Stricker et al., 2006). Since Osr1−/− mutant embryos died in midgestation (Wang et al., 2005), we first examined the Osr2−/− mutant mice, which die shortly after birth due to craniofacial defects (Lan et al., 2004), for possible joint defects. We found that all Osr2−/− mutant mice had abnormally fused tarsal elements in the ankle region (Fig. 1A, B) while other joints in the limb appeared normal.
Fig. 1.
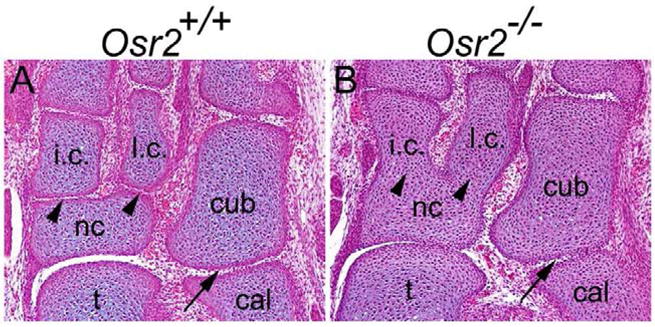
Osr2−/− mutant mice have fused tarsal elements. (A) In the E17.5 wildtype embryo, tarsal elements are separated by synovial joints. (B) In the E17.5 Osr2−/− embryo, both intermediate cuneiform and lateral cuneiform are fused with the navicular bone (arrowheads in B). Cuboidal and calcaneus bones in Osr2−/− embryos remain separate (arrow in B). cal, calcaneus bone; cub, cuboid bone; i.c., intermediate cuneiform; l.c., lateral cuneiform; nc, navicular bone; t, talus bone.
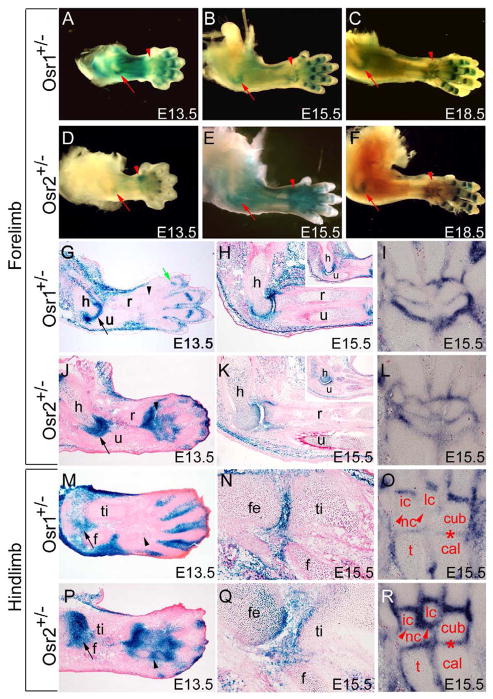
We investigated the expression patterns of Osr1 and Osr2 during limb development in detail using the Osr1+/− and Osr2+/− mice, which carry targeted in-frame fusion of the lacZ reporter gene to the Osr1 and Osr2 genes, respectively (Lan et al., 2004; Wang et al., 2005). From E13.5 to E18.5, both Osr1 and Osr2 are strongly expressed in the developing synovial joints, including interphalangeal, metacarpal-phalangeal, elbow and knee joints (Fig. 2A–F). While Osr1 is more broadly expressed that Osr2 in the developing limb mesenchyme surrounding the skeletal elements at E13.5, including inter-digital mesenchyme (Fig. 2A, D, G, J, M, R), expression of Osr1 was much weaker than that of Osr2 in the carpal and tarsal regions at this stage (Fig. 2G, J, M, P). At E15.5, while both Osr1 and Osr2 mRNA expression were clearly detected in the developing joint cells in between the carpal elements in the wrist (Fig. 2I, L), strong Osr2 mRNA expression was detected in the developing joints between the tarsal elements but little Osr1 mRNA was expressed in that region (Fig. 2Q, R). The differential expression of Osr1 and Osr2 in the developing tarsal joints correlated very well with the joint defects in the Osr2−/− mutant mice (Fig. 1B). Together with the strong overlapping expression patterns in most developing joint tissues, these data suggest that Osr1 and Osr2 may play critical but partially redundant roles in synovial joint development.
Fig. 2.
Expression patterns of Osr1 and Osr2 during synovial joint development. (A–F) Whole-mount X-gal staining of the forelimbs of E13.5 to E18.5 Osr1+/− and Osr2+/− embryos. Both Osr1 and Osr2 are strongly and highly specifically expressed in the developing interphalangeal, metacarpal-phalangeal, and elbow joints (arrows in A–F). In the carpal region, expression of Osr1 is weak (arrowhead in A) as compared with that of Osr2 (arrowhead in D) at E13.5. At E15.5 and E18.5, both Osr1 and Osr2 are strongly expressed in the carpal joints. (G, H) X-gal-stained sections of the forelimbs of E13.5 and E15.5 Osr1+/− embryos, respectively. Black arrow points to the elbow joint, green arrow points to the developing metacarpophalangeal joint in the digit, and black arrowhead points to the carpal region. (I) Section in situ hybridization of E15.5 wildtype embryos detected Osr1 mRNA expression in the carpal joints. (J, K) X-gal stained sections of the forelimbs of E13.5 and E15.5 Osr2+/− embryos, respectively. Strong lacZ activity was detected in between the carpal elements (arrowhead in J) and in the elbow joint (arrowhead in K). (L) Section in situ hybridization detected Osr2 mRNA expression in the carpal joint cells in the E15.5 wildtype embryos. (M, N) X-gal stained sections of the hindlimbs of E13.5 and E15.5 Osr1+/− embryos, respectively. (P, Q) X-gal stained sections of the hindlimbs of E13.5 and E15.5 Osr2+/− embryos, respectively. Expression of Osr1 (arrowhead in M) is much weaker than that of Osr2 (arrowhead in P) in the tarsal region at E13.5. However, both were expressed strongly in the knee joint at E13.5 and E15.5. (O, R) Section in situ hybridization detected difference in expression of Osr1 (O) and Osr2 (R) in the developing tarsal joints at E15.5. Strong Osr2 mRNA expression was detected in the developing joints between the tarsal elements (R) while little Osr1 mRNA was detected in between intermediate cuneiform and navicular bones and in between lateral cuneiform and navicular bones (arrowheads in O). Both Osr1 and Osr2 mRNAs were detected in between the cuboidal and calcaneus bones (asterisk in O and R). cal, calcaneus bone; cub, cuboid bone; f, femur; fe, femur; h, humerus; i.c., intermediate cuneiform; l.c., lateral cuneiform; nc, navicular bone; r, radius; t, talus bone; ti, tibia; u, ulna.
Defects in synovial joint development in Osr1f/fOsr2−/−Prrx1-cre double mutant mice
Since Osr1−/− mutant mice died at midgestation due to heart defects, we crossed the Osr1f/f mice with Prrx1-cre transgenic mice, which express the Cre recombinase throughout the developing limb mesenchyme as early as E9.5 (Logan et al., 2002), and generated Osr1f/fPrrx-cre mice to examine the role of Osr1 in limb joint development. We found that the Osr1f/fPrrx-cre mice, lacking Osr1 gene product in the developing limb mesenchyme, did not have obvious defects in synovial joint development (data not shown). To investigate further the roles of Osr1 and Osr2 in synovial joint development, we generated and analyzed limb development in Osr1f/fOsr2−/−Prrx1-cre mice. The Osr1f/fOsr2−/−Prrx1-cre mutant mice had the open-eyelid phenotype as found in Osr2−/− mutants (Lan et al., 2004). The wrist and ankle regions of the limbs in the double mutant were abnormally shaped, causing the limbs to wrap in front of the trunk (data not shown). Skeletal preparations of the E17.5 Osr1f/fOsr2−/−Prrx1-cre and control embryos showed multiple joint defects in the double mutants: the humerus was fused with both the ulna and radius at the elbow region while the tibia was fused with the fibula at both the proximal and distal ends (Fig. 3, A–H). In addition, the double mutant mice exhibited delay in mineralization of digital bones (Fig. 3B, D). Histological sections further confirmed the fusion of the elbow joints and revealed additional joint defects in the wrist and ankle regions (Fig. 3, I–P). In the wrist region, the carpal elements were fused together in the double mutants (Fig 3N). In the ankle region, in addition to the fusion between intermediate cuneiform (i.c.) and navicular (nc) bones and between lateral cuneiform (l.c.) and nc seen in the Osr2−/− mutants, the double mutants exhibited fusion between cuboidal (cub) and calcaneous (cal) bones (Fig. 3P). The interphalangeal and metacarpophalangeal joints were also disorganized in the double mutants in comparison with the control littermates (Fig. 3, Q–R). These data confirm that Osr1 and Osr2 play critical and partially redundant roles in synovial joint development.
Fig. 3.
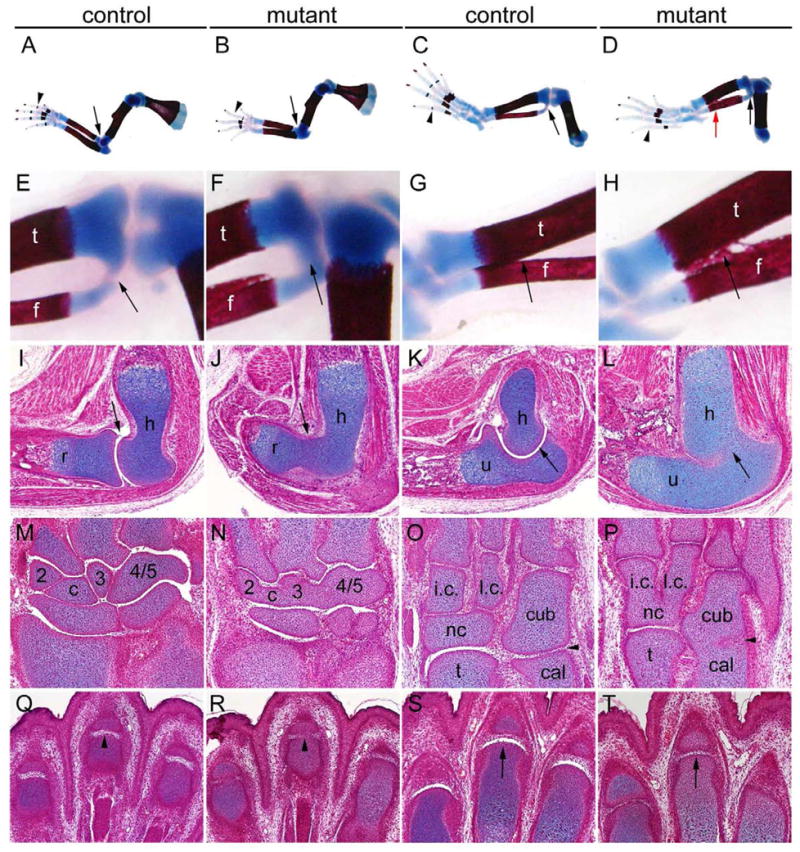
Joint defects in Osr1f/fOsr2−/−Prrx1-cre double mutant mice. (A–D) Alizarin red and alcian blue stained skeleton preparations of E17.5 limbs of Osr1f/fPrrx1-cre (A, C) and Osr1f/fOsr2−/−Prrx1-cre mutant (B, D) embryos. The double mutants showed delay in mineralization of phalanges (arrowheads in B and D), fusion of the elbow joints (arrow in B), and fusion between tibia and fibula in both the distal (red arrow in D) and proximal (black arrow in D) regions. (E–H) High magnification views of proximal and distal regions of the tibia and fibula in control (E, G) and mutant (F, H) hindlimbs. Arrows point to the aberrant fusion between these two elements in the mutants and corresponding regions in the control littermates. (I–L) Osr1f/fPrrx1-cre embryo had normal synovial joint development in the elbow (arrows in I and K) while in Osr1f/fOsr2−/−Prrx1-cre mutant embryo humerus is fused to radius (arrow in J) and to ulna (arrow in H). (M, N) Carpal elements are demarcated clearly by the synovial joints in E17.5 Osr1f/fPrrx1-cre littermate (M), while they are fused together in the E17.5 Osr1f/fOsr2−/−Prrx1-cre mutant embryo (N). (O, P) Tarsal elements are separated by synovial joints in the E17.5 Osr1f/fPrrx1-cre embryo (O), while the intermediate cuneiform and lateral cuneiform were fused respectively with the navicular bone in the Osr1f/fOsr2−/−Prrx1-cre mutant (P). In addition, the cuboidal and calcaneus bones were also fused in Osr1f/fOsr2−/−Prrx1-cre mutant embryo (arrowhead in P). (Q–T) Histological sections through the interphalangeal (Q, R) and metacarpophalangeal (S, T) joint regions in the E17.5 forelimbs of control (Q, S) and Osr1f/fOsr2−/−Prrx1-cre mutant (R, T) embryos. Arrowheads in Q and R point to corresponding interphalangeal joints, whereas arrows in S and T point to the corresponding metacarpophalangeal joints in the control and mutant littermates. cal, calcaneus bone; cub, cuboid bone; h, humerus; i.c., intermediate cuneiform; l.c., lateral cuneiform; nc, navicular bone; t, talus bone; r, radius; u, ulna.
Osr1 and Osr2 are expressed in the presumptive joint cells prior to and are critical for interzone formation
To understand the roles of Osr1 and Osr2 in synovial joint development, we focused on development of the elbow joint as that is the earliest joint to form in the developing limb and it has been well described at the morphological and cellular levels. It has been reported that the humerus, ulna and radius elements arise at E11.5 in mice as a continuous Y-shaped mesenchymal condensation with a seemingly uniform population of cells (Hinchliffe and Johnson, 1980). By combining whole mount detection of LacZ and mRNA expression in the Osr1+/− and Osr2+/− embryos, respectively, we found that expression of both Osr1 and Osr2 in the developing E11.5 forelimb overlapped with that of Gdf5 (Fig. 4B, D), one of the earliest markers for joint formation. By performing in situ hybridization using sections through the presumptive elbow joint region, we found that both Osr1 and Osr2 mRNAs are expressed in the presumptive joint cells at E11.5 when the expression of Col2 mRNA is continuous throughout the humerus, ulna and radius elements (Fig. 4E–H). By E12.5, formation of the interzones in the future elbow joint had been initiated, with specific and dramatic downregulation of Col2 mRNA expression (Fig. 4I). At this stage, expression of both Osr1 and Osr2 mRNAs continued to overlap that of Gdf5 mRNA in the interzone cells and are absent in the cartilage anlagen of humerus, ulna, and radius elements (data not shown). These data indicate that Osr1 and Osr2 are expressed in the presumptive joint cells from the onset of joint formation.
Fig. 4.
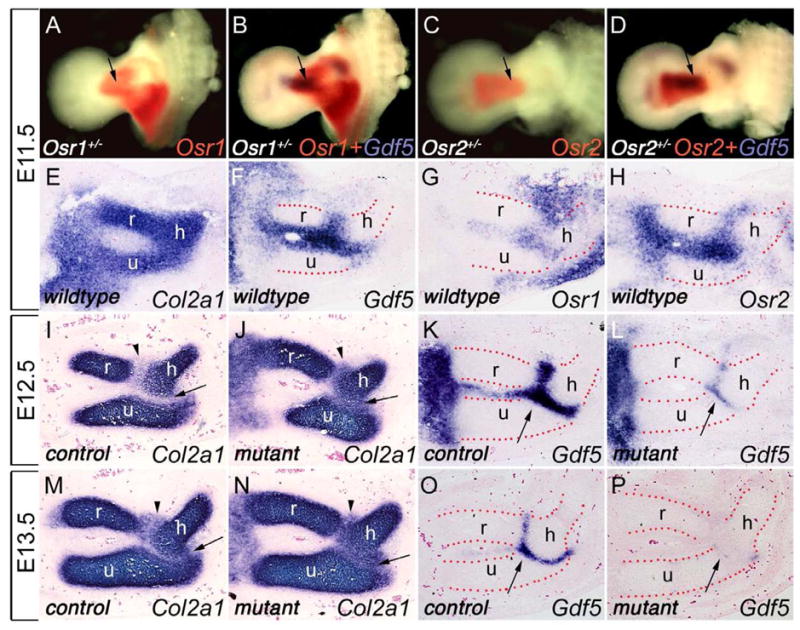
Osr1 and Osr2 are expressed in nascent joint cells and control joint formation. (A–D) Salmon-gal staining and combined Salmon-gal staining with Gdf5 in situ hybridization in limbs of E11.5 Osr1+/− (A and B) and Osr2+/− (C and D) embryos. Osr1 and Osr2 share overlapping expression patterns in the presumptive elbow joint (arrows) with Gdf5. (E) Section in situ hybridization of E11.5 wildtype forelimb showing continuous Col2a1 mRNA expression in the prospective humerus, ulna and radius anlagen. (F–H) Section in situ hybridization of E11.5 wildtype limb detected overlapping expression of Gdf5 (F), Osr1 (G), and Osr2 (H) expression in the presumptive elbow joint cells. (I, J) At E12.5, joint interzone formation in the elbow region of the control embryo (I) was clearly demarcated by dramatically decreased Col2a1 mRNA expression, while the presumptive joint cells in the Osr1f/fOsr2−/−Prrx1-cre mutant littermate showed persistent high levels of Col2a1 mRNA expression (J). Arrows point to the prospective humerus-ulna joint and arrowheads point to the prospective humerus-radius joint. (K, L) At E12.5, Gdf5 was strongly expressed in the interzone cells in the control embryo (arrow in K) but was significantly reduced in the elbow region in the Osr1f/fOsr2−/−Prrx1-cre mutant embryo (arrow in L). (M, N) At E13.5, as the interzone cells in the control embryos starts to differentiate into a three-layered structure with the middle layer remaining negative for Col2a1 expression (M), the Osr1f/fOsr2−/−Prrx1-cre mutant limbs still lack a clear Col2a1-negative interzone in the elbow region (N). (O, P) At E13.5, the joint progenitor cells remain positive for Gdf5 mRNA expression in the control embryo (arrow in O), while the expression of Gdf5 was almost completely downregulated in the joint progenitor cells in Osr1f/fOsr2−/−Prrx1-cre double mutants (arrow in P). h, humerus; r, radius; u, ulna.
We next investigated whether the elbow joint failed to initiate in Osr1f/fOsr2−/−Prrx1-cre mutant embryos. At E11.5, we found that Gdf5 mRNA was expressed in the joint progenitor cells in the elbow region of the double mutant embryos at levels comparable to that of the control embryos (data not shown). At E12.5, the interzones between humerus and radius and between humerus and ulna elements are clearly demarcated by the loss of Col2a1 expression in the control embryos (Fig. 4I). In contrast, Col2a1 expression was only slightly downregulated in these prospective joint regions in the double mutants (Fig. 4J). Moreover, whereas Gdf5 expression was strongly expressed in the interzone cells in the control embryos at this stage, it was dramatically downregulated in the double mutant littermates (Fig. 4K, L). At E13.5, as the interzone cells started to differentiate into a three-layered structure with the middle layer remaining negative for Col2a1 expression and positive for Gdf5 expression in the control embryos (Fig. 4M, O), the Osr1f/fOsr2−/−Prrx1-cre mutant limbs still lacked a clear Col2a1-negative interzone in the elbow region (Fig. 4N). In addition, Gdf5 expression was lost in the joint progenitor cells in these double mutants by this stage (Fig. 4P).
We further investigated whether expression of Wnt9a and Wnt4 were altered in developing limb in Osr1f/fOsr2−/−Prrx1-cre mutant embryos. At E12.5, Wnt9a is only weakly expressed in the elbow joint interzone cells in the control embryos and no significant difference was detected between the control and the Osr1f/fOsr2−/−Prrx1-cre mutant littermates (Fig. 5A, B). In contrast, strong Wnt4 mRNA expression was detected in the cells surrounding the elbow joint interzone in the control embryos at E12.5 but Wnt4 mRNA expression in the corresponding elbow joint cells in the Osr1f/fOsr2−/−Prrx1-cre mutant littermates was significantly reduced (Fig. 5E, F). By E13.5, Wnt9a mRNA expression was clearly detected in the interzone cells between the humerus and radius and between the humerus and ulna in the control embryos (Fig. 5C). However, Wnt9a expression was absent in between the humerus and radius elements but was clearly detected in a subset of cells in between the humerus and ulna elements in the E13.5 double mutant embryos (Fig. 5D). Interestingly, the domain of Wnt9a expression at this stage corresponded with the incomplete fusion of the humerus and ulna at E17.5 (Fig. 3H). Remarkably, while Wnt4 mRNA continued to be expressed in the cells surrounding the elbow interzone in the control embryos at E13.5, its expression in the corresponding cells in the double mutants was almost completely downregulated (Fig. 5G, H). In contrast, Wnt4 expression in the differentiating chondrocytes in the radius and ulna elements, which do not express either Osr1 or Osr2, was similar in the control and Osr1f/fOsr2−/−Prrx1-cre mutant limbs (Fig. 5H). Taken together, these data indicate that joint formation was initiated, as marked by Gdf5 expression at E11.5, but the cellular program of interzone formation was aborted in the absence of both Osr1 and Osr2 function, resulting in lack of separation of the humerus from the radius and ulna elements in the double mutants.
Fig. 5.
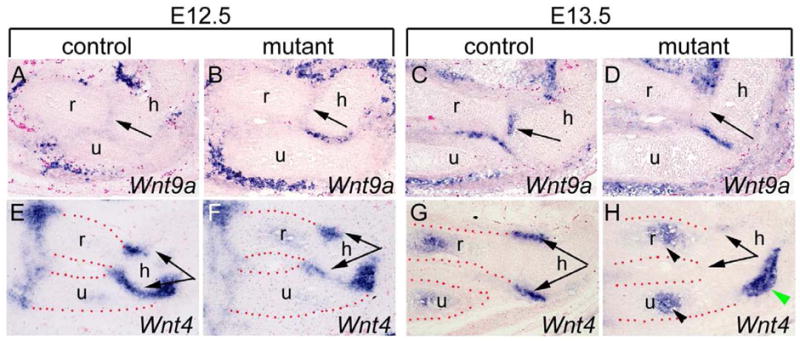
Osr1 and Osr2 acts upstream of Wnt/β-catenin signaling during joint development. (A, B) At E12.5, Wnt9a was weakly expressed in the presumptive elbow joint cells in both the control and the Osr1f/fOsr2−/−Prrx1-cre mutant littermates (arrows). (C, D) At E13.5, Wnt9a mRNA expression was clearly detected in the interzone cells between the humerus and radius and between the humerus and ulna in the control embryos. Wnt9a mRNA expression was absent in between the humerus and radius elements (arrow in D) but was clearly detected in between the humerus and ulna elements in the E13.5 double mutant embryos. (E–H) Strong Wnt4 mRNA expression was detected in mesoderm cells surrounding the elbow joint interzone in E12.5 control embryos (E) while lower levels of Wnt4 expression was detected in the corresponding cells in the E12.5 Osr1f/fOsr2−/−Prrx1-cre mutant littermates (F). (G, H) At E13.5, Wnt4 mRNA was still expressed in the mesoderm cells surrounding the elbow interzone in the control embryo (G), while its expression in the corresponding cells in the Osr1f/fOsr2−/−Prrx1-cre mutant appeared almost completely downregulated (H). Wnt4 expression in the differentiating chondrocytes in the radius and ulna elements (arrowheads in H) was similar in the control and Osr1f/fOsr2−/−Prrx1-cre mutant embryos. Green arrowhead in H points to a Wnt4-expressing tendon tissue that was absent from the control section in G due to differences in the plane of sections. h, humerus; r, radius; u, ulna.
Expression of Osr1 and Osr2 in the developing joint cells do not depend on Wnt/β-catenin signaling
It was previously proposed that Wnt/β-catenin signaling was necessary and sufficient for synovial joint formation (Guo et al., 2004). We investigated whether expression of Osr1 and Osr2 in the joint cells depended on Wnt/beta-catenin signaling. We generated mice homozygous for the loxP-flanked β-catenin allele (Catnbf/f) (Brault et al., 2001) and heterozygous for the Osr2IresCre allele (Lan et al., 2007). Since Cre is active in the Osr2-expressing cells as early as E10.5 in the embryos, we expect the loxP-flanked β-catenin alleles to be inactivated in the joint progenitor cells. Histological analysis of E13.5 embryos showed that the interzone in the elbow joint region of the Catnbf/f;Osr2IresCre/+ mutant embryo was not as clearly organized as in the Osr2IresCre/+ control embryos (Fig. 6A, B), consistent with a requirement for β-catenin in synovial joint formation. However, expression of Osr1 and Osr2 in the developing elbow joint regions was not significantly altered in the Catnbf/f;Osr2IresCre/+ mutant embryos in comparison with the Osr2IresCre/+ control littermates at E13.5 (Fig. 6, C–H), indicating that expression of Osr1 and Osr2 in the developing joint cells do not depend on Wnt/beta-catenin signaling.
Fig. 6.
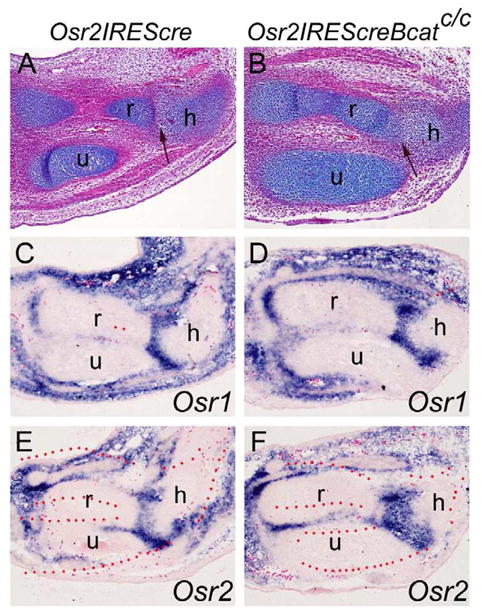
Expression of Osr1 and Osr2 in the developing joint cells was not altered by loss of Wnt/β-catenin signaling. (A, B) HE-stained histological sections through the elbow region of E13.5 Osr2Irescre/+ control (A) and Catnbf/f;Osr2Irescre/+ mutant (B) embryos. Arrow points to the developing joint between humerus and radius, which is disorganized in the mutant (B). (C–F) Section in situ hybridization with Osr1 and Osr2 probes in the elbow region of E13.5 Osr2Irescre/+ (C, E) and Catnbf/f;Osr2Irescre/+ mutant (D, F) embryos. No significant differences in expression of Osr1 or Osr2 were detected in the developing limbs between control and Catnbf/f;Osr2Irescre/+ mutant embryos. h, humerus; r, radius; u, ulna.
Osr1f/fOsr2−/−Prrx1-cre double mutant mice exhibit defects in joint cell differentiation
To maintain the normal function of the synovial joints, articular chondrocytes and synovial fibroblasts synthesize lubricin, which is a water-soluble glycoprotein encoded by Prg4 gene and plays an important role in joint lubrication and synovial homeostasis (Swann and et al., 1981; Schumacher et al., 1994; Flannery et al., 1999; Jay et al., 2000). To investigate whether Osr1 and Osr2 regulate articular cartilage differentiation, we investigated whether expression of Prg4 was altered in developing joints in Osr1f/fOsr2−/−Prrx1-cre mutant embryos. At E15.5, while Prg4 is strongly expressed in the presumptive articular cartilage in both the humerus-radius and the humerus-ulna joints in the elbow region of the control embryos (Fig. 7A, C). In comparison, expression of Prg4 mRNA is dramatically reduced in the Osr1f/fOsr2−/−Prrx1-cre mutant embryos, including in the region where humerus and ulna were not fused (Fig. 7B, D). These results suggest, in addition to an early function in interzone formation, that the function of Osr1 and Osr2 is also required for joint cell differentiation.
Fig. 7.
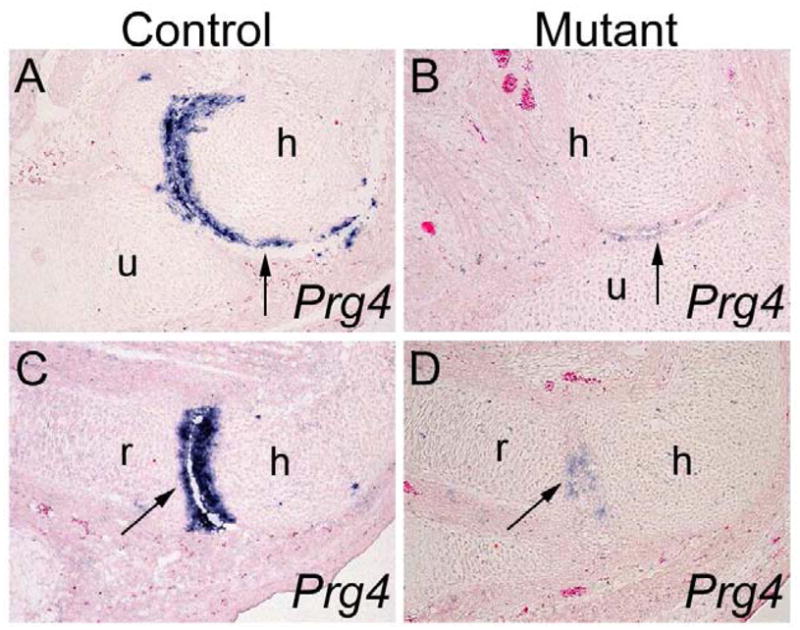
Comparison of Prg4 expression in the developing elbow joints in the Osr1f/fOsr2−/−Prrx1-cre mutant and control embryos. (A–D) While differentiating articular cartilage cells in the elbow joints in the control embryos (A, C) express high levels of Prg4 mRNA at E15.5, only few low Prg4-expressing cells were detected in the elbow joint region in the Osr1f/fOsr2−/−Prrx1-cre littermates (B, D). h, humerus; r, radius; u, ulna.
Discussion
Initiation of joint formation in the developing vertebrate limb involves both specification of the joint cell fate and suppression of the cartilage molecular program in the joint progenitor cells. Whereas recent studies have identified several signaling molecules that are specifically expressed in the presumptive joint cells (reviewed by Archer et al., 2003; Pacifici et al., 2005), little is known about the transcription factors controlling joint formation. In this report, we show that expression of the Osr1 and Osr2 genes are both activated in the presumptive joint cells at the onset of joint initiation and that they play critical roles in regulating synovial joint formation.
Osr1 and Osr2 were initially identified as mammalian homologs of the Drosophila odd-skipped family zinc finger transcription factors (Lan et al, 2001; So and Danielian, 1999). The odd-skipped family in Drosophila consists of four distinct genes, odd, bowl, sob, and drm (Coulter et al., 1990; Green et al., 2002; Hart et al., 1996; Wang and Coulter, 1996). Odd encodes a protein containing four C2H2-type zinc finger motifs, whereas bowl and sob each encode a protein containing five zinc finger motifs highly homologous to ODD. In contrast, drm encodes a small protein containing only two zinc finger motifs, of which only the first finger conforms to the canonical C2H2 sequence and shows high sequence identity to the first zinc finger in the other Odd-skipped family proteins (Green et al., 2002). Mutations in odd, bowl, and drm each resulted in distinct developmental defects (Coulter et al., 1990; Green et al., 2002; Wang and Coulter, 1996). Interestingly, although there is significantly more extensive amino acid sequence identity between ODD and BOWL than that between ODD and DRM proteins, ectopic expression of bowl almost always resulted in different phenotypes from that of expression of odd under the same conditions, whereas ectopic expression of odd and drm in the same tissues often resulted in similar developmental defects (Green et al., 2002; Hao et al., 2003; de Celis Ibeas and Bray, 2003; Hatini et al., 2005; Bras-Pereira et al., 2006). These data suggested that the odd-skipped family transcription factors in Drosophila have largely distinct biochemical functions in vivo that are most likely due to functional domains outside of the zinc finger motifs. In contrast to the Drosophila odd-skipped family, Osr1 and Osr2 are the only members of this family in mammals and they share extensive amino acid sequence identity with each throughout the length of the protein products (Lan et al., 2001). Although targeted null mutations in Osr1 and Osr2 in mice resulted in distinct phenotypes, with heart and urogenital defects in Osr1−/− mice and with cleft palate and open-eyelids in Osr2−/− mice, respectively, (Lan et al., 2004; Wang et al., 2005), we recently demonstrated that expression of Osr1 from the Osr2 locus through targeted gene replacement rescued the craniofacial developmental defects in the Osr2−/− mice (Gao et al., 2009). Thus, we hypothesized that the distinct phenotypes of the Osr1−/− and Osr2−/− mutant mice are most likely due to the distinct developmental expression patterns of these two genes. In this study, we found that expression of both Osr1 and Osr2 are activated in most presumptive joint cells and persist throughout joint development in the developing limbs except that Osr2, but not Osr1, is highly expressed in the joint cells in between several tarsal elements in the ankle region. The corresponding tarsal elements were fused in the Osr2−/− mice whereas all other limb joints developed apparently normally in either the Osr2−/− mice or the Osr1f/f Prrx1-cre mice. However, multiple joints failed to form properly in the Osr1f/f Osr2−/−Prrx1-cre mice. These data confirm that Osr1 and Osr2 function redundantly in the tissues where they are co-expressed during mouse development. The overlapping expression of Osr1 and Osr2 in most developing joints ensures proper development and function of the major synovial joints.
Joint fusion phenotypes partially overlapping with that observed in the Osr1f/f Osr2−/−Prrx1-cre mutant mice have been reported in mice lacking Gdf5, Gdf6, or both Wnt9a and Wnt4 (Settle Jr. et al., 2003; Storm and Kingsley, 1996; 1999; Storm et al., 1994). Gdf5−/−, Gdf6−/−, Gdf5−/−Gdf6−/−, and Wnt9a−/−Wnt4−/− mutant mice all displayed fusion of the carpal and tarsal elements in the wrist and ankle regions, respectively, but they did not have fusion of the long bone elements at the elbow or knee joints as observed in the Osr1f/f Osr2−/−Prrx1-cre mutant mice. Expression of Gdf5, Wnt4, and Wnt9a were all detected in the presumptive joint or peri-joint cells at the onset of joint initiation in the Osr1f/f Osr2−/−Prrx1-cre mutant embryos, indicating that Osr1 and Osr2 are not required for the specification of the joint progenitor cells or for activating expression of these signaling molecules. However, in the absence of Osr1 and Osr2, most of the interzone cells continued to express high levels of Col2 mRNA, suggesting that Osr1/Osr2 function is required for the interzone cells to fully commit to the joint fate. Interestingly, while Gdf5 expression in the joint progenitor cells and Wnt4 expression in the peri-joint cells were downregulated in the Osr1f/f Osr2−/−Prrx1-cre mutant elbow region, a subset of cells in the humerus-ulna joint region maintained Wnt9a mRNA expression (Fig. 5D). Nevertheless, even in the region of the humerus-ulna joint where expression of Wnt9a persisted in the Osr1f/f Osr2−/−Prrx1-cre mutant embryos, those cells failed to upregulate the articular cartilage marker Prg4 (Fig. 7B), suggesting that Osr1/Osr2 plays a critical role in joint cell differentiation. The persistence of Wnt9a expression in some joint progenitor cells in the Osr1f/f Osr2−/−Prrx1-cre mutant embryos, together with our data showing normal expression of Osr1 and Osr2 in the Catnbf/f;Osr2IresCre/+ mutant embryos, suggests that Osr1/2 function in parallel with the Wnt9a signaling pathway during joint interzone formation. On the other hand, the downregulation of Wnt4 in the peri-joint cells in the Osr1f/f Osr2−/−Prrx1-cre mutant embryos suggests that Osr1/2 may be involved in the maintenance of the peri-joint cells and indirectly modulate the Wnt signaling pathway during joint formation. The differences in severity of the phenotype in the different joints may be due to differences in spatiotemporal activation of other molecular pathways, such as the Wnt pathway, during joint development.
Multiple joint fusions, including fusion of the humerus with the ulna and the radius at the elbow region, have also been reported in mice lacking either Noggin or Zeb1 (Brunet et al., 1998; Takagi et al., 1998). Noggin−/− mutant embryos had excessive cartilage formation at the expense of the residual limb mesenchyme, suggesting that the failure of joint formation in this mutant mouse strain resulted from increased chondrogenesis of the limb mesenchyme due to elevated Bmp signaling (Brunet et al., 1998). The Zeb1 protein, containing homeodomain and zinc finger domains, has been shown to interact with Smad proteins and to modulate TGFβ signaling (Postigo, 2003; Nishimura et al., 2006). Interestingly, we recently found that Bmp4 expression was enhanced and expanded in the developing tooth mesenchyme in the Osr2−/− mutant mice (Zhang et al., 2009). One possibility is that Osr1 and Osr2 may function to modulate the activity of the Bmp or TGFβ signaling pathway in the joint progenitor cells. Identification of direct downstream target genes of Osr1 and Osr2 and elucidation of the molecular interactions involving these and other factors in the joint progenitor cells will provide new insights into the molecule mechanisms of joint development.
Acknowledgments
We thank Dr. Christine Hartmann from The Research Institute of Molecular Pathology (IMP) for the instruction on alcian blue staining. We thank Dr. Yingzi Yang from the National Human Genome Research Institute for sharing in situ hybridization probes. This work was supported by NIH/NIDCR grant R01DE013681 and R01DE018401 to RJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archer C, Dowthwaite G, Francis-West P. Development of synovial joints. Birth Defects Res C Embryo Today. 2003;69:144–155. doi: 10.1002/bdrc.10015. [DOI] [PubMed] [Google Scholar]
- Bras-Pereira C, Bessa J, Casares F. Odd-skipped genes specify the signaling center that triggers retinogenesis in Drosophila. Development. 2006;133:4145–4149. doi: 10.1242/dev.02593. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch D, McMahon A, Sommer L, Boussadia O, Kemler R. Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Brunet L, McMahon J, McMahon A, Harland R. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280:1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- Buxton P, Edwards C, Archer C, Francis-West P. Growth/differentiation factor-5 (GDF-5) and skeletal development. J Bone Joint Surg Am. 2001;83A:S23–S30. [PubMed] [Google Scholar]
- Coulter DE, Swaykus EA, Beran-Koehn MA, Goldberg D, Wieschaus E, Schedl P. Molecular analysis of odd-skipped, a zinc finger encoding segmentation gene with a novel pair-rule expression pattern. EMBO J. 1990;9:3795–3804. doi: 10.1002/j.1460-2075.1990.tb07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig F, Bentley G, Archer C. The spatial and temporal pattern of collagens I and II and keratan sulphate in the developing chick metatarsophalangeal joint. Development. 1987;99:383–391. doi: 10.1242/dev.99.3.383. [DOI] [PubMed] [Google Scholar]
- de Celis Ibeas JM, Bray SJ. Bowl is required downstream of Notch for elaboration of distal limb patterning. Development. 2003;130:5943–5952. doi: 10.1242/dev.00833. [DOI] [PubMed] [Google Scholar]
- Dhordain P, Dewitte F, Desbiens X, Stehelin D, Duterque-Coquillaud M. Mesodermal expression of the chicken erg gene associated with precartilaginous condensation and cartilage differentiation. Mech Dev. 1995;50:17–28. doi: 10.1016/0925-4773(94)00322-e. [DOI] [PubMed] [Google Scholar]
- Elders MJ. The increasing impact of arthritis on public health. J Rheumatol Suppl. 2000;60:6–8. [PubMed] [Google Scholar]
- Ellis T, Gambardella L, Horcher M, Tschanz S, Capol J, Bertram P, Jochum W, Barrandon Y, Busslinger M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001;15:2307–2319. doi: 10.1101/gad.200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery C, Hughes C, Schumacher B, Tudor D, Aydelotte M, Kuettner K, Caterson B. Articular cartilage superficial zone protein (SZP) is homologous to megakaryocyte stimulating factor precursor and is a multi-functional proteoglycan with potential growth-promoting, cytoprotective, and lubricating properties in cartilage metabolism. Biochem Biophys Res Commun. 1999;254:535–541. doi: 10.1006/bbrc.1998.0104. [DOI] [PubMed] [Google Scholar]
- Francis-West P, Abdelfattah A, Chen P, Allen C, Parish J, Ladher R, Allen S, Macpherson S, Luyten F, Archer C. Mechanisms of GDF-5 action during skeletal development. Development. 1999;126:1305–1315. doi: 10.1242/dev.126.6.1305. [DOI] [PubMed] [Google Scholar]
- Gao Y, Lan Y, Ovitt CE, Jiang R. Functional equivalence of the zinc finger transcription factors Osr1 and Osr2 in mouse development. Dev Biol. 2009;328:200–209. doi: 10.1016/j.ydbio.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RB, Hatini V, Johansen KA, Liu XJ, Lengyel JA. Drumstick is a zinc finger protein that antagonizes Lines to control patterning and morphogenesis of the Drosophila hindgut. Development. 2002;129:3645–3656. doi: 10.1242/dev.129.15.3645. [DOI] [PubMed] [Google Scholar]
- Guo X, Day T, Jiang X, Garrett-Beal L, Topol L, Yang Y. Wnt/β-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;18:2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao I, Green R, Dunaevsky O, Lengyel J, Rauskolb C. The odd-skipped family of zinc finger genes promotes Drosophila leg segmentation. Dev Biol. 2003;263:282–295. doi: 10.1016/j.ydbio.2003.07.011. [DOI] [PubMed] [Google Scholar]
- Hart MC, Wang L, Coulter DE. Comparison of the structure and expression of odd-skipped and two related genes that encode a new family of zinc finger proteins in Drosophila. Genetics. 1996;144:171–182. doi: 10.1093/genetics/144.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Tabin C. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 2001;104:341–351. doi: 10.1016/s0092-8674(01)00222-7. [DOI] [PubMed] [Google Scholar]
- Hatini V, Green RB, Lengyel JA, Bray SJ, DiNardo S. The Drumstick/Lines/Bowl regulatory pathway links antagonistic Hedgehog and Wingless signaling inputs to epidermal cell differentiation. Genes Dev. 2005;19:709–718. doi: 10.1101/gad.1268005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe J, Johnson D. The development of the vertebrate limb. New York: Oxford University Press; 1980. Limb development in tetrapods; pp. 72–83. [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Holder N. An experimental investigation into the early development of the chick elbow joint. J Embryol Exp Morphol. 1977;39:115–127. [PubMed] [Google Scholar]
- Iwamoto M, Tamamura Y, Koyama E, Komori T, Takeshita N, Williams JA, Nakamura T, Enomoto-Iwamoto M, Pacifici M. Transcription factor ERG and joint and articular cartilage formation during mouse limb and spine skeletogenesis. Dev Biol. 2007;305:40–51. doi: 10.1016/j.ydbio.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James R, Kamei C, Wang Q, Jiang R, Schultheiss T. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- Jay G, Britt D, Cha C. Lubricin is a product of megakaryocyte stimulating factor gene expression by human synovial fibroblasts. J Rheumatol. 2000;27:594–600. [PubMed] [Google Scholar]
- Koyama E, Shibukawa Y, Nagayama M, Sugito H, Young B, Yuasa T, Okabe T, Ochiai T, Kamiya N, Rountree R, Kingsley D, Iwamoto M, Enomoto-Iwamoto M, Pacifici M. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol. 2008;316:62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Kingsley P, Cho E, Jiang R. Osr2, a new mouse gene related to Drosophila odd-skipped, exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mech Dev. 2001;107:175–179. doi: 10.1016/s0925-4773(01)00457-9. [DOI] [PubMed] [Google Scholar]
- Lan Y, Ovitt C, Cho E, Maltby K, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004;131:3207–3216. doi: 10.1242/dev.01175. [DOI] [PubMed] [Google Scholar]
- Lan Y, Wang Q, Ovitt C, Jiang R. A unique mouse strain expressing Cre recombinase for tissue-specific analysis of gene function in palate and kidney development. Genesis. 2007;45:618–624. doi: 10.1002/dvg.20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga G, Lichtler A, Upholt W, Kosher R. Studies on the role of Cux1 in regulation of the onset of joint formation in the developing limb. Dev Biol. 2002;243:44–54. doi: 10.1006/dbio.2001.0559. [DOI] [PubMed] [Google Scholar]
- Logan M, Martin J, Nagy A, Lobe C, Olson E, Tabin C. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Martin J, Bradley A, Olson E. The paired-like homeobox gene Mhox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- Mitrovic D. Development of the diarthrodial joints in the rat embryo. Am J Anat. 1978;151:475–485. doi: 10.1002/aja.1001510403. [DOI] [PubMed] [Google Scholar]
- Nishimura G, Manabe I, Tsushima K, Fujiu K, Oishi Y, Imai Y, Maemura K, Miyagishi M, Higashi Y, Kondoh H, Nagai R. δEF1 mediates TGF-beta signaling in vascular smooth muscle cell differentiation. Dev Cell. 2006;11:93–104. doi: 10.1016/j.devcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Pacifici M, Koyama E, Iwamoto M. Mechanisms of synovial joint and articular cartilage formation: recent advances, but many lingering mysteries. Birth Defects Res C Embryo Today. 2005;75:237–248. doi: 10.1002/bdrc.20050. [DOI] [PubMed] [Google Scholar]
- Pitsillides A, Ashhurst D. A critical evaluation of specific aspects of joint development. Dev Dyn. 2008;273:2284–2294. doi: 10.1002/dvdy.21654. [DOI] [PubMed] [Google Scholar]
- Postigo A. Opposing functions of ZEB proteins in the regulation of the TGFβ/BMP signaling pathway. EMBO J. 2003;22:2443–2452. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Block J, Schmid T, Aydelotte M, Kuettner K. A novel proteoglycan synthesized and secreted by chondrocytes of the superficial zone of articular cartilage. Arch Biochem Biophys. 1994;311:144–152. doi: 10.1006/abbi.1994.1219. [DOI] [PubMed] [Google Scholar]
- Settle S, Rountree R, Sinha A, Thacker A, Higgins K, Kingsley D. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol. 2003;254:116–130. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- So P, Danielian P. Cloning and expression analysis of a mouse gene related to Drosophila odd-skipped. Mech Dev. 1999;84:157–160. doi: 10.1016/s0925-4773(99)00058-1. [DOI] [PubMed] [Google Scholar]
- Später D, Hill T, O’sullivan R, Gruber M, Conner D, Hartmann C. Wnt9a signaling is required for joint integrity and regulation of Ihh during chondrogenesis. Development. 2006;133:3039–3049. doi: 10.1242/dev.02471. [DOI] [PubMed] [Google Scholar]
- Storm E, Huynh T, Copeland N, Jenkins N, Kingsley D, Lee S. Limb alterations in brachypodism mice due to mutations in a new member of the TGF-β superfamily. Nature. 1994;368:639–643. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- Storm E, Kingsley D. Joint patterning defects caused by single and double mutations in members of the bone morphogenetic protein (BMP) family. Development. 1996;122:3969–3979. doi: 10.1242/dev.122.12.3969. [DOI] [PubMed] [Google Scholar]
- Storm E, Kingsley D. GDF5 coordinates bone and joint formation during digit development. Dev Biol. 1999;209:11–27. doi: 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- Stricker S, Brieske N, Haupt J, Mundlos S. Comparative expression pattern of Odd-skipped related genes Osr1 and Osr2 in chick embryonic development. Gene Expr Patterns. 2006;6:826–834. doi: 10.1016/j.modgep.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Swann DA, Slayter HS, Silver FH. The molecular structure of lubricating glycoprotein-I, the boundary lubricant for articular cartilage. J Biol Chem. 1981;256:5921–5925. [PubMed] [Google Scholar]
- Takagi T, Moribe H, Kondoh H, Higashi Y. δEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- Wang L, Coulter DE. bowel, an odd-skipped homolog, functions in the terminal pathway during Drosophila embryogenesis. EMBO J. 1996;15:3182–3196. [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lan Y, Cho E, Maltby K, Jiang R. Odd-skipped related 1 (Odd1) is an essential regulator of heart and urogenital development. Dev Biol. 2005;288:582–594. doi: 10.1016/j.ydbio.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Hu Y, Ramamurthy RTA, Qiu M, Chen Y. Msx1 is required for the induction of Patched by Sonic hedgehog in the mammalian tooth germ. Dev Dyn. 1999;215:45–53. doi: 10.1002/(SICI)1097-0177(199905)215:1<45::AID-DVDY5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lan Y, Chai Y, Jiang R. Antagonistic actions of Msx1 and Osr2 pattern mammalian teeth into a single row. Science. 2009;323:1232–1234. doi: 10.1126/science.1167418. [DOI] [PMC free article] [PubMed] [Google Scholar]