Abstract
Abro1 (also known as KIAA0157) is a scaffold protein that recruits polypeptides to assemble the BRISC (BRCC36-containing isopeptidase complex) deubiquitinating (DUB) enzyme. The four subunits of BRISC enzyme include Abro1, NBA1, BRE, and BRCC36 proteins. The DUB activity of the BRISC enzyme is exclusively directed against Lys63-linked polyubiquitin that does not have a proteolytic role but regulates protein function. In this report, we identified Abro1 as a specific interactor of THAP5, a zinc finger transcription factor that is involved in G2/M control and apoptosis. Abro1 was predominantly expressed in the heart and its protein level was regulated following experimentally induced myocardial ischemia/reperfusion (MI/R) injury. Furthermore, in patients with coronary artery disease (CAD), there was a dramatic increase in Abro1 protein level in the myocardial infarction (MI) area. Increase in Abro1 lead to a significant reduction in Lys63-linked ubiquitination of specific protein targets. Reducing the Abro1 protein level exacerbated cellular damage and cell death of cardiomyocytes due to MI/R injury. Additionally, overexpression of Abro1 in a heterologous system provided significant protection against oxidative stress-induced apoptosis. In conclusion, our results demonstrate that Abro1 protein level substantially increases in myocardial injury and coronary artery disease and this up-regulation is part of a novel cardioprotective mechanism. In addition, our data suggest a potential new link between Lys63-specific ubiquitination, its modulation by the BRISC DUB enzyme, and the development and progression of heart disease.
1. Introduction
Protein ubiquitination is a posttranslational modification that affects many cellular processes including protein degradation, transcription, DNA repair, cell cycle, and apoptosis (for a recent review see Chen et al. [1]). Ubiquitin is a small polypeptide that can be covalently attached to target proteins via its carboxyl-terminus [2]. Ubiquitination can occur as a single molecule attached to the target protein via a lysine (monoubiquitination) or as a ubiquitin chain (polyubiquitination) [1]. Ubiquitin chains are formed by the conjugation of monomers employing one of the seven lysine residues present in ubiquitin [2]. The most common and understood forms of polyubiquitination involve Lys48 (K48) or Lys63 (K63)-linked chains. K48-linked polyubiquitin chains usually target a protein for degradation by the proteasome [3,4]. K63-linked polyubiquitin has a non-proteolytic role and regulates protein function, subcellular localization, and protein-protein interactions [5]. Ubiquitin modifications on a protein can be reversed by a process that involves numerous enzymes known as deubiquitinating enzymes (DUBs) [6]. There are over 100 DUBs in mammalian cells, belonging to five distinct families [4]. As is the case with ubiquitination, deubiquitination is a highly regulated process and has been implicated in many cellular functions including gene expression [7], DNA repair [8], cell cycle control [9], kinase activation [10], and apoptosis [11].
An important role for protein polyubiquitination in the normal function of the heart as well as in the development of human heart disease has begun to emerge [12-14]. Recent reports suggest that the ubiquitinproteasome system (UPS) can be involved in myocardial ischemia/reperfusion (MI/R) injury and cardiac hypertrophy [4,13,15-17]. In other studies, hyperubiquitination of proteins was found in the heart of patients with dilated cardiomyopathy (DCM) [18]. Limited information exists on the role of deubiquitination enzymes (DUBs) in cardiomyocytes. Most DUBs consist of multi-protein complexes, and in many cases the constituent subunits or the physiological substrates of these enzymes remain unknown [4]. The normal function of DUBs is very important because mutations in DUB genes have been implicated in the development of several human diseases including cancer and neurodegeneration [19-21].
We have recently reported the isolation and characterization of THAP5, a human zinc finger nuclear protein that is hyper-expressed in the human heart [22]. The normal function of THAP5 in cardiomyocytes is unclear but when it was overexpressed in a heterologous system, it induced cell cycle arrest [22]. To further investigate the molecular mechanism of THAP5 function, we used the yeast two-hybrid system to isolate THAP5 interactors. One such interactor isolated in this screen was the Abro1 protein. Abro1 has recently been shown to be a component of the BRISC (BRCC36-containing isopeptidase complex) enzyme [23]. This complex has DUB activity that is specifically directed towards K63-linked polyubiquitin chains. Abro1 functions as a scaffold protein that recruits the rest of the proteins found in the BRISC enzyme [24]. These proteins include NBA1 [25], BRE, and BRCC36 [23]. The same polypeptides are also components of yet another BRCC36 containing complex, the BRCA1-A complex, which includes BRCA1, RAP80, Abraxas, and BARD1 [26]. Abraxas is a homolog of Abro1 and these proteins share 39% sequence homology at their amino-terminus. This homologous sequence represents the interaction domain for the common subunits NBA1, BRE, and BRCC36 [23] that are present in both BRISC and BRCA1-A complexes [27,28]. Abraxas and Abro1 have different carboxyl-terminal sequences, and in Abraxas this domain binds the BRCA1 protein that targets the BRCA1-A complex to specific DNA damage foci [26].
In the present work, we show that THAP5 interacts with the carboxyl-terminal domain of Abro1 to become part of the BRISC enzyme. Since Abro1 is expressed predominantly in the heart, we investigated the normal function of this protein and its potential involvement in heart disease. Abro1 protein levels show a dramatic increase in the myocardial infarction area in patients with CAD. A similar increase in Abro1 protein levels was also observed in the hearts of mice following MI/R injury. Inhibiting the Abro1 protein induction exacerbated the cellular damage and apoptosis of cardiomyocytes following MI/R. Overexpression of Abro1 provided substantial protection against oxidative stress-induced apoptosis. Abro1 induction was associated with an increase in Lys63-linked deubiquitination of specific target proteins. Our results clearly demonstrate that Abro1 protein is regulated in experimentally induced MI/R and in the hearts of patients with CAD. Furthermore, we show that the upregulation of Abro1 is part of a cardioprotective mechanism that may play a significant role in the development and/or progression of heart disease.
2. Materials and Methods
2.1. Yeast Two-Hybrid Screen
We used the yeast two-hybrid system to screen a HeLa as well as a melanocyte cDNA library as previously described [22,29,30]. The bait used was THAP5 protein (aa 1-395) expressed from the pGilda vector (Clontech) as a LexA fusion protein. Several interacting proteins were identified in this screen. One of these interactors isolated from the melanocyte cDNA library was a partial cDNA clone of a previously described protein called Abro1, also known as KIAA0157 [26]. The full-length cDNA for Abro1 encodes 415 amino acids and was isolated from a Marathon Ready human heart cDNA library (Clontech). The presence and stability of the recombinant proteins in yeast cells were monitored by Western blot analysis using LexA-antibodies (for bait) or HA-antibodies (for preys).
2.2. Cell Culture
Rat embryonic ventricular myocardial cell line (H9c2) was obtained from ATCC and grown in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal calf serum, 2mM L-glutamine, 100units/ml penicillin, and 100μg/ml streptomycin (Invitrogen) [31].
2.3. Interaction Between THAP5 and Abro1 Proteins in Human Cardiomyocytes
Human heart tissue (100mg) was ground in liquid nitrogen and homogenized using an Ultra-Turrax T8 (IKA-werke) in ice-cold lysis buffer (150mM NaCl, 20mM Tris-HCl pH 7.6, 1mM CaCl, 1mM MgCl, 10% Glycerol, 1% NP40) that included a protease and a phosphatase inhibitor cocktail (Roche). Homogenate was cleared by centrifugation for 10min at 14,000g and the protein concentration was determined using the BioRad assay (BioRad). Approximately 400μg of total lysate was pre-cleared by mixing it with protein G-Agarose beads (Roche) for 1 hour followed by incubation with THAP5 polyclonal antibody [22] for 2 hours at 4°C. Protein G-Agarose beads were then added and allowed to bind overnight at 4°C. Immunoprecipitates were collected by brief centrifugation, washed extensively with lysis buffer, and resolved by SDS-PAGE. They were then electro-transferred onto a PVDF membrane and probed with a rabbit polyclonal Abro1 antibody (Bethyl Laboratories) followed by a secondary anti-rabbit-IgG trueBlot HRP-conjugated (eBioscience). The immunocomplex was visualized using the ECL substrate (Pierce).
2.4. Northern Blot Analysis of Abro1 mRNA Expression in Human Tissues
A human mRNA tissue blot (Clontech) representing twelve human tissues was probed with a radiolabeled Abro1 cDNA. The blot was hybridized with the radiolabeled probe at 42°C, washed at 65°C, and subjected to autoradiography [32]. To verify that an equal amount of mRNA was present on each lane, the blot was stripped and re-probed for β-actin mRNA expression.
2.5. Localization of Abro1 Protein in human cardiomyocytes
To investigate the subcellular localization of Abro1 protein in cardiomyocytes, we used adult human myocytes isolated from donor hearts as previously described [33-35]. Fixed cells were permeabilized, and double-stained with rabbit anti-Abro1 (Bethyl Laboratories) and mouse anti-α actinin (sarcomeric) (Sigma) followed by secondary anti-Rabbit-CY3 and anti-mouse-FITC antibodies (Jackson). Auto-fluorescence due to lipofuscins was blocked using Sudan black and the slides were mounted using Fluoromount-G solution containing DAPI to stain nuclei. The slides were observed using a Leica SP5II confocal microscope system (Leica).
2.6. Abro1 mRNA expression (RT-PCR) in cardiomyocytes
Primary neonatal rat and mouse cardiomyocytes were isolated from the hearts of 2-day-old Sprague-Dawley rats or 3-day-old C57BL6 mouse pups respectively, as previously described [36-38]. Total RNA was isolated from adult human cardiomyocytes, and neonatal mouse or rat cardiomyocytes cultured for 48 hours, using RNeasy mini kit according to the manufacturer's instructions (Qiagen). Equal amounts of RNA (1μg) were treated with DNase and reverse transcribed using a cDNA synthesis kit for RT-PCR (Roche). A semi-quantitative RT-PCR was performed with the following primers: Abro1 mouse and rat specific primers: Fw 5′-CCCAATCTAGGCAATACT AGCC-3′, Rw 5′-GGTCCTCGTCAGGATGT; Abro1 human specific: Fw 5′-GAATCTTGTCAGGCAGAAG-3′; Rw 5′- TTAAATCTGGGAGGTCTGAGTG-3′. The following GAPDH primers were used as an internal control: Fw 5′-CATCACCATCTTC CAGGAGCGAG-3′ and Rw 5′-CACCACCTTCTTGATGTCATCA-3′. PCR products were analyzed on a 1.5% agarose gel. The relative levels of Abro1 mRNA expression were normalized to GAPDH levels.
2.7. Expression of Abro1 Protein in the Heart of Patients With Coronary Artery Disease (CAD)
Human cardiac tissues were prepared as described [39]. Briefly, human cardiac tissue samples were taken from the left ventricles of failing human hearts that were explanted in the course of heart transplantation. The study's protocol was approved by the local ethics committee, and written informed consent given by patients, according to the National Disease Research Interchange (NDRI). Hearts from patients with end-stage heart failure who were undergoing cardiac transplantation because of CAD (Coronary Artery Disease) or Dilated Cardiomyopathy (DCM) were investigated. Healthy donor hearts that were ultimately rejected for transplantation because of technical reasons were also included in this study as healthy tissue control. Human heart tissue lysates were prepared as described in Method section 2.3. Approximately 20μg of heart extract was resuspended in SDS-sample buffer and boiled for 3 minutes. Samples were resolved by SDS-PAGE and electro-transferred onto PVDF membranes (Pall Life Sciences) using a Semi-Dry cell Transfer Blot (Bio-Rad). 4% nonfat dry milk in TBST buffer was used to block any non-specific binding. The membrane was incubated with Abro1 (Bethyl Laboratories) polyclonal antibody (1:500), followed by a secondary HRPconjugated goat anti-rabbit antibody (Jackson ImmunoResearch) (1:15000), and visualized by ECL (Pierce).
2.8. Myocardial Ischemia/Reperfusion
Mice were anesthetized with 2% isoflurane. Myocardial I/R was induced by temporarily exteriorizing the heart, via a left thoracic incision, and placing a 6-0 silk suture slipknot around the left anterior descending coronary artery. After 30 minutes of MI, the slipknot was released and the myocardium was reperfused for 3 hours. At the end of the reperfusion, the heart was quickly removed and the heart ventricles were harvested for protein analysis. Sham operated control mice (Sham, MI/R) underwent the same surgical procedure except that the suture placed under the left coronary artery was not tied.
2.9. In vivo siRNA-mediated Abro1 Knock-down (Abro1-KD)
We utilized siRNA gene silencing technique to knock-down Abro1 expression in mouse hearts as described [40]. Two mouse-specific Abro1 siRNAs with the following nucleotide sequences GGCAGUAUGUGCAGAUGUA and GGACAUCAGGGCAAUUUAU were used together. For control, siRNA oligos of the same size having scrambled nucleotide sequences were used. All siRNAs were obtained from Dharmacon. The siRNAs were diluted in 5% glucose and mixed with in vivojet PEI (Genesee Scientific). Adult wild type mice C57BL/6 were anesthetized with 2% isoflurane, and their hearts were exposed via left thoracotomy at the fifth intercostals space. Abro1 specific siRNA, 20μl (0.8μg/μl), or negative control was delivered via three separate intramyocardial injections (32G needle) to temporarily blanch the left ventricular free wall [40,41]. Forty-eight hours after siRNA injection, the mice were subjected to MI/R.
2.10. Measurement of Caspase-3 Activity
Apoptotic cell death was determined by caspase-3 activation using a fluorometric kit (R&D System). Briefly, myocardial tissue was homogenized in ice-cold caspase lysis buffer (50mM Hepes pH 7.4, 0.1% Chaps, 5mM DTT, 0.1mM EDTA, 0.1% Triton-X100). 50μg of tissue lysates were used to perform the fluorometric assay according to the manufacturer's instructions. The fluorescence emission of the 7-amino-4-trifluoromethylcoumarin (AFC), released on proteolytic cleavage of the fluorogenic substrate DEVDAFC by active caspase-3, was measured using the Biotek FL600 microplate fluorescence reader (400 nm excitation; 505 emission wavelength). Caspase-3 activity was directly proportional to the fluorescence signal and was expressed as nmol AFC per hour per milligram protein.
2.11. Over-expression of Abro1 and Apoptosis Assay
H9c2 cells were plated in 6-well plates and transfected with either EGFP-C1 empty vector or EGFP-Abro1 plasmid using Fugene HD transfection reagent (Promega). Eighteen hours after transfection, cells were treated with two concentrations of H2O2 (100 or 200μM) for 18hours [42,43]. The percentage of apoptotic cells in the transfected population was estimated by staining with phycoerythrinconjugated Annexin V at room temperature for 15 min in 1X binding buffer followed by analysis on a FACSCalibur flow cytometer (BD Biosciences).
2.12. Western Blot Analysis
Sham or MI/R tissues were prepared as described above. Lysates (50μg for Abro1 analysis or 100μg for Ub-K63 detection) were separated by SDS-PAGE, transferred to PVDF membranes and blocked using 4% skim milk in TBST. Membranes were then probed with the following primary antibodies: rabbit anti-Abro1 (Bethyl Laboratories) or mouse anti-Ub-K63 (eBioscience). The membranes were washed and incubated with either goat anti-rabbit or goat anti-mouse horseradish peroxidase-conjugate antibodies (Jackson Immunoresearch) for 1 hour. The immunoblot was visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). The blot densities were analyzed with GeneTools analysis software.
2.13. Statistical Analysis
All quantitative data were expressed as mean ± standard deviation. Differences among groups were analyzed by one-way analysis of variance followed by Tukey's post hoc t-test. A value of p < 0.05 was considered significant.
3. Results
3.1. Isolation of Abro1 and its Interaction with THAP5
We used the yeast two-hybrid system to isolate THAP5 interacting proteins. Two different cDNA libraries prepared from either HeLa cells or from human primary melanocytes were used in order to screen as many diverse proteins as possible. The bait used was the full length THAP5 protein fused to LexA (LexA-THAP51-395). The screen was performed as previously described [29] and several THAP5 interactors were identified. One such specific interactor was isolated from both cDNA libraries. Further analysis identified this cDNA to encode a partial polypeptide of Abro1. Using specific primers and rapid amplification of cDNA ends we were able to isolate the full-length cDNA for the human Abro1 protein. A cDNA fragment encoding the full length Abro11-415 protein was cloned back into the pJG4-5 vector. The interaction of Abro11-415 with the different domains of THAP5 was monitored. Figure 1A shows that Abro11-415 interacts with the full-length THAP5 protein. This interaction involves two specific areas of the THAP5 protein: one that is present at the amino-terminus (aa 1-162) and another at the carboxyl-terminus (aa 162-395). THAP5 is predominantly expressed in the human heart [22]. Therefore, we used human heart extracts to verify that THAP5 and Abro1 proteins can interact in vivo. A specific antibody [22] was used to precipitate endogenous THAP5 protein, and the presence of Abro1 protein in the complex was monitored by Western blot analysis. Figure 1B shows that THAP5 protein interacts with Abro1 in human heart to form a stable complex that is easily precipitated. Pre-immune serum was used as a control to verify the specificity of THAP5-Abro1 interaction.
Fig. 1.

Interaction of Abro1 with THAP5 in yeast and in human cardiomyocytes. (A) Yeast colonies were transformed with plasmids encoding the indicated baits. Full-length Abro1 was used as prey. Blue color results from a positive protein-protein interaction. (B) THAP5 antibody was used to precipitate the endogenous protein as well as any THAP5 associated proteins in human heart lysate. The precipitated complex was resolved by SDS-PAGE, and the presence of Abro1 in the complex was monitored using a specific antibody. Lane 3 shows that Abro1 co-precipitated with THAP5, and there was no interaction when pre-immune serum was used (lane 2). Lane 1 shows total heart lysate. The “*” indicates a non-specific protein band seen in all our co-IP experiments.
3.2. Expression of Abro1 mRNA in Various Human Tissues
Abro1 protein can be divided into three functional domains. There is an ABR domain at the amino-terminus, a coiled-coil domain (c-c) in the middle of the protein, and a targeting domain in the carboxyl-terminus (Figure 2A). Both the ABR domain and the coiled-coil regions have been previously shown to mediate specific interactions with NBA1, BRE, and BRCC36 [23]. These two domains show 39% homology to another scaffold protein, Abraxas (Abra1) [26]. The carboxyl-terminal sequence (aa 260-415) is unique and shows no similarity to any protein currently present in the GenBank. The targeting domain mediates the specific interaction of Abro1 with THAP5 (results not shown). To investigate the expression of Abro1 in human tissues, a Northern blot was probed with a radioactive cDNA fragment that corresponds to the full-length protein (aa 1-415). Figure 2B shows Abro1 mRNA is expressed at high level in the human heart but some expression is also present in the muscle, brain, kidney, and small intestine. The high expression of Abro1 mRNA in the human heart suggests this protein might have a significant role in this tissue.
Fig. 2.
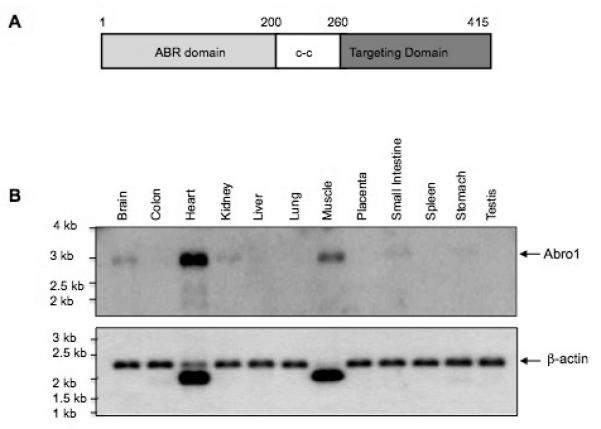
Functional domains and expression of Abro1 mRNA in human tissues. (A) Schematic representation of the different domains of Abro1 protein. The protein can be divided into three functionally and structurally distinct domains. The amino-terminal ABR domain has been previously shown to mediate interaction with BRE and NBA1 proteins. The coiled-coil (c-c) domain has also been shown to interact with BRCC36. The carboxyl-terminal domain of Abro1 (targeting domain) is unique in its amino acid sequence and interacts with THAP5. (B) A commercially available Northern blot (Clontech) containing 2μg/lane poly mRNA from various adult human tissues was probed with 32P-Abro1 cDNA. A single transcript of about 3.0 kb long was detected. Abro1 is predominantly expressed in the heart although some expression is also seen in muscle, brain, kidney, and small intestine. β-actin probe was used to verify that equal amounts of mRNA were present in each lane.
3.3. Subcellular Localization of Abro1 Protein
Human adult cardiomyocytes were stained with Abro1 antibody and the subcellular localization of the protein was monitored using confocal microscopy. Figure 3A shows Abro1 is predominantly found in the cytoplasm of cardiomyocytes. These results are also supported by a recent study that has also found the BRISC enzyme to be present in the cytoplasm [24,27]. We monitored Abro1 mRNA levels in human, mouse and rat cardiomyocytes to verify the expression and relative levels. Adult human cardiomyocytes, and neonatal mouse as well as rat cardiomyocytes were used for these experiments. Figure 3B shows that Abro1 mRNA is clearly expressed in all isolated cardiomyocytes and the highest level was found in human cells.
Fig. 3.
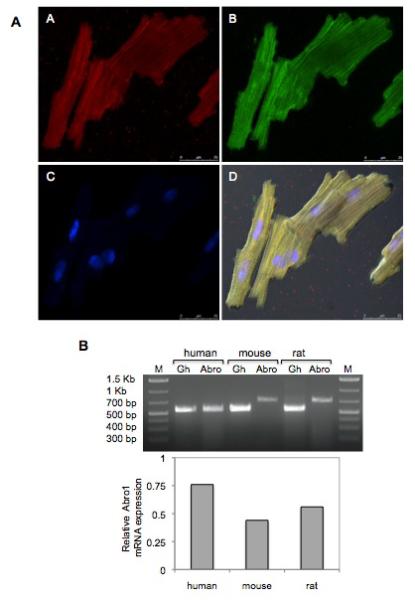
Subcellular localization of Abro1 protein and mRNA expression in human, mouse, and rat cardiomyocytes. (A) Adult human cardiomyocytes were stained with Abro1 antibody (panel A) as well as the sarcomeric specific marker anti-α-actinin (panel B). Panel C shows DAPI staining of the same cells and panel D shows the merged images including differential interference contrast (DIC). Abro1 is predominantly found in the cytoplasm of the cardiomyocytes. (B) Abro1 mRNA expression in cardiomyocytes was detected by RT-PCR. PCR products were obtained using species-specific PCR primers and analyzed on a 1.5% agarose gel. The first and last lanes represent DNA size markers. “Abro” indicates the PCR product obtained with human Abro1-specific primers (560bp), or mouse and rat specific primers (730bp). Abro1 gene expression was normalized to GAPDH (Gh) (570bp).
3.4. Expression of Abro1 Protein in the Human Heart and Deregulation in Patients With Coronary Artery Disease
Abro1 mRNA is predominantly expressed in the human heart suggesting this protein may play an important role in the normal function of cardiomyocytes and/or the development of heart disease. Therefore, we monitored Abro1 protein levels in the hearts from six patients with coronary artery disease (CAD). Using Western blot analysis, we compared Abro1 protein levels at the remote zone (RZ) and the myocardial infarction area (MI) on each heart. Figure 4A shows that there was a significant increase in Abro1 protein level in the MI area compared to the RZ on the same heart. The MI area is the part of the heart that sustains the maximum damage in CAD. Figure 4B shows the expression of Abro1 protein in healthy human heart tissues. The Abro1 protein level in these normal heart tissues is low compared to the protein level seen in the MI area. There was also no significant difference in Abro1 protein levels between the hearts of patients with Dilated Cardiomyopathy (DCM) and control (donor) hearts (Supplemental Figure 1S).
Fig. 4.
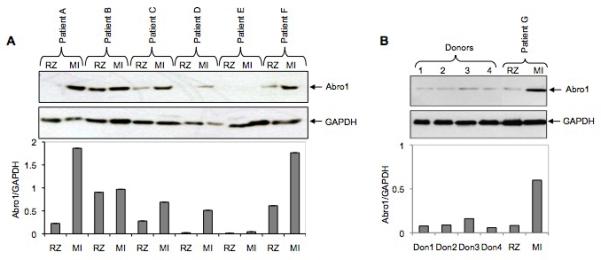
Abro1 protein levels in heart tissues from patients with coronary artery disease (CAD) and healthy donors. (A) Heart tissue extracts from six patients (A to F) with CAD were used in a Western blot to monitor the expression of Abro1 protein. Extracts were prepared from two areas on each heart that corresponded to the remote zone (RZ) and the myocardial infarction area (MI). In most patients, there was a substantial increase in Abro1 protein level in the MI area as compared to the RZ area of the same heart. The same blot was also probed with GAPDH antibody to verify equal loading. The bottom panel shows Abro1/GAPDH ratio calculated after densitometry analysis. (B) Heart tissue lysates from four healthy donors (donors 1 to 4) and one CAD patient (patient G) were used in a Western blot analysis using Abro1 antibodies. The same blot was also probed with GAPDH antibody to verify equal loading. Bottom panel shows Abro1/GAPDH ratio calculated after densitometry analysis.
3.5. Abro1 Protein Level is Regulated in Mice Following MI/R Injury
We investigated the potential regulation of Abro1 protein levels in mouse hearts following ischemia/reperfusion (MI/R) injury. For this experiment, four sham operated animals and four MI/R animals were used. In the Western blot, only two animals from each group are shown since the rest had similar levels of Abro1 expression. The expression of Abro1 protein in the heart extracts of these mice was monitored as described in the methods. Figure 5 shows that the expression of Abro1 protein substantially increased (at least three-fold) in the hearts of mice that underwent MI/R compared to sham operated controls.
Fig. 5.
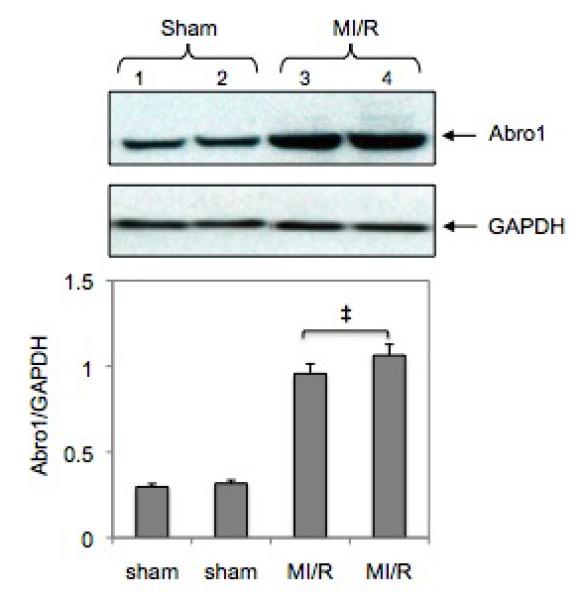
Regulation of Abro1 protein in the mouse heart following MI/R injury. Myocardial ischemia was induced for 30 minutes followed by 3 hours of reperfusion. Heart tissue extracts from sham-operated mice (n=4, two representative samples were used) or mice that underwent MI/R (n=4, two representative samples were used) were analyzed by a Western blot. There was an increase in the Abro1 protein level in the ischemic hearts (lanes 3 and 4) compared to the sham operated (lane 1 and 2). GAPDH antibody was used to verify the amount of protein present in each lane. Bottom panel shows Abro1/GAPDH ratio after densitometry analysis. Data are means ± standard deviation in animal group (n=4 hearts/group). ‡ P<0.05 vs. sham.
3.6. Downregulation of Abro1 Protein During MI/R Exacerbates Cell Injury and Apoptosis
We utilized an in vivo siRNA gene silencing technique to reduce Abro1 protein levels in mouse hearts. We used a mixture of two different siRNAs that were specific for Abro1 mRNA. As a control, we used a siRNA of the same size but with scrambled nucleotide sequence. Following siRNA injections, MI/R was induced and the expression of Abro1 was monitored by Western blot analysis (Figure 6A). We also monitored the levels of the K63-linked ubiquitinated proteins under the same conditions. Figure 6B shows MI/R leads to a reduction of the K63-linked ubiquitination, but the ubiquitination is partially restored when specific siRNA was used to inhibit the Abro1 protein level. These results clearly show an inverse correlation between the level of Abro1 protein and the specific K63-ubiquitination observed in the heart extracts from animals that underwent MI/R.
Fig. 6.
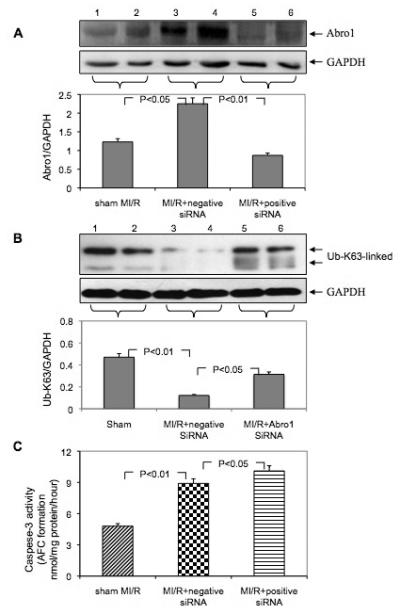
Downregulation of Abro1 protein followed by MI/R exacerbates cell injury and apoptosis. Following siRNA injection, the mice underwent MI/R, and the expression of Abro1 was monitored by Western blot analysis. (A) Western blot analysis of Abro1 protein expression from heart lysates (n=7-13 hearts/group, two representative samples of each group are presented). Lanes 1 and 2 show Abro1 protein expression in sham-operated mice. Lanes 3 and 4 show the expression of Abro1 in heart lysates from mice treated with a non-specific siRNA (negative siRNA) followed by MI/R. Lanes 5 and 6 show Abro1 expression from heart lysates in mice treated with Abro1 a specific siRNA (positive siRNA). The same blot was also probed for GAPDH expression to verify equal protein loading on each lane. The Abro1/GAPDH ratio was calculated after densitometry analysis. (B) The same heart extracts were also analyzed for K63-linked ubiquitination in MI/R+non-specific siRNA and MI/R+Abro1 specific siRNA. GAPDH antibody was used to verify the amount of protein present in each lane and the ratio of UbK63/GAPDH is shown in the bottom panel. (C) The degree of cellular damage and apoptosis was estimated by monitoring caspase-3 activity. MI/R led to a significant increase in caspase-3 activity (MI/R+non-specific siRNA) compared to control sham-operated animals. In mice that were treated with Abro1 specific siRNAs, caspase-3 activity showed a further, significant increase (MI/R+Abro1 specific siRNA). Data are means ± standard deviation in animal group (n=7-13 hearts/group).
The degree of cellular damage and apoptosis in the heart was estimated by measuring caspase-3 activity. Figure 6C shows the substantial induction in Abro1 protein level after MI/R caused a significant increase in caspase-3 activity. In the mice that were treated with Abro1 siRNAs prior to MI/R, caspase-3 activity showed a further increase over the control, suggesting that Abro1 inhibition led to more cellular damage and apoptosis.
3.7. Abro1 Overexpression has a Protective Role Against H2O2-Induced Apoptosis
H9c2 cells were transfected with GFPAbro1 or GFP vector alone and H2O2 was used to induce apoptosis. The percentage of apoptotic cells in the population of transfected cells was monitored by Annexin V staining and flow cytometry. Figure 7 shows that when cells were treated with 100μM H2O2, approximately 32% of the GFP transfected cells stained positive for Annexin V. In contrast, cells that were transfected with GFPAbro1 under the same conditions showed substantial resistance to apoptosis and only 15% stained positive for Annexin V. A similar result was obtained when a higher concentration (200μM) of H2O2 was used. In this case, the GFP vector-alone transfected cells sustained over 50% apoptosis compared to 30% for the GFP-Abro1 expressing cells.
Fig. 7.
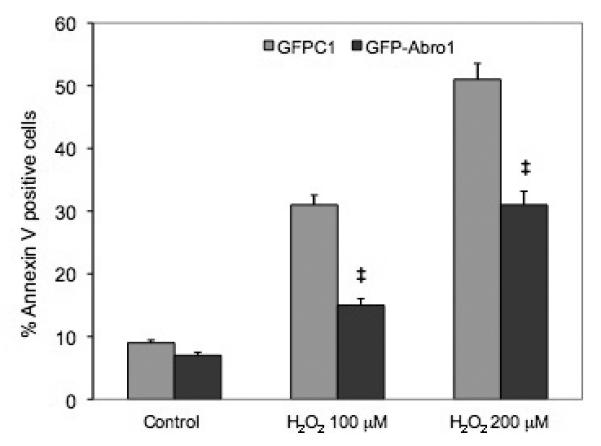
Abro1 overexpression protects H9c2 cells from H2O2 induced apoptosis. H9c2 cells were transfected with empty GFP vector or GFP-Abro1 and apoptosis was induced using 100μM or 200μM of H2O2 for 18 hours. Cells were stained with Annexin V and analyzed using a FACScalibur Flow Cytometer (BD Biosciences). Data are mean ± standard deviation of four independent experiments. ‡ P<0.05 vs. GFP-vector.
3.8. Abro1 Upregulation Following MI/R Leads to a Significant Decrease in K63-linked Ubiquitination of Specific Proteins
Since Abro1 is part of the BRISC enzyme, we investigated if its upregulation during MI/R would be associated with an increase in K63-linked deubiquitination. We used specific antibodies that recognize K63-linked ubiquitinated proteins on a Western blot to probe mouse heart lysates. This antibody recognized three major K63-ubiquitinated polypeptides in the heart lysates. These proteins had an approximate molecular weight of 62, 55, and 48 kDa (Figure 8, lanes 1 and 2) respectively. These polypeptides could represent distinct proteins or could be one protein with different ubiquitin ratio. Alternatively, they could be proteolytic products. The intensity of these K63-linked ubiquitinated protein bands showed a dramatic decrease in the two mouse hearts that underwent MI/R (Figure 8, lanes 3 and 4). These results suggest that Abro1 induction leads to K63-specific deubiquitination of specific target protein(s) in the mouse heart.
Fig. 8.
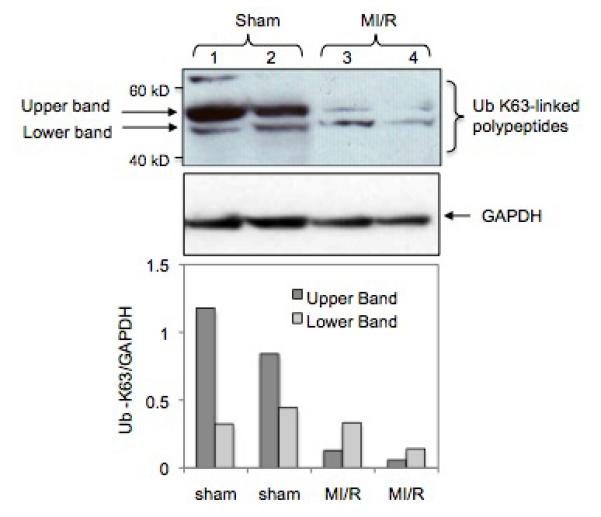
MI/R leads to a decrease in K63-linked polyubiquitination of specific target proteins. Western blot analysis of heart extracts prepared from mouse hearts that underwent MI/R (lanes 3 and 4) or sham operated (lanes 1 and 2). A specific antibody that recognizes K63-linked ubiquitinated proteins was used as described in the experimental procedures. The bottom panel shows the Ub-K63/GAPDH ratio of the two major K63-linked ubiquitinated polypeptides found in the heart extracts. The K63-linked ubiquitination on these proteins was significantly reduced in the heart extracts from mice that underwent MI/R. GAPDH antibody was used to verify equal loading per lane. The bottom panel shows Ub-K63/GAPDH ratio that was calculated after densitometry analysis of the respective upper and lower bands.
4. Discussion
We have previously reported the characterization of THAP5, a zinc finger protein that is hyper-expressed in the human heart [22]. The normal function of THAP5 remains unknown, but studies using HEK293 cells suggested a potential involvement of this protein in the regulation of cell cycle as well as cell death [22]. To further investigate the normal function of THAP5, we used a yeast two-hybrid system to isolate potential interactors of this protein. In this screen, we isolated Abro1 as a specific THAP5 interactor. Abro1 is a homolog of Abraxas, a scaffold protein used to assemble the various polypeptides that compose the BRCA1-A complex [26]. This complex includes Abraxas, BRCA1, BARD1, RAP80, BRCC36, BRE and NBA1 [25,26]. The BRCA1-A complex is required for DNA damage response, and Abraxas has been shown to be necessary for G2/M checkpoint control, resistance to DNA damage, and DNA repair [44,45]. The BRCA1-A complex has both E3 ligase as well as deubiquitination activity and operates exclusively in the nucleus whereas the BRISC complex is found in the cytoplasm [27,46]. There is a 39% amino acid sequence similarity between Abro1 and Abraxas at the amino-terminal part of the proteins that encompasses the ABR (aa 1-200) and the coiled-coil domains (aa 201-260) [26]. The carboxyl-terminus is unique in both proteins and for Abraxas, this domain interacts with BRCA1. Abro1 does not interact with BRCA1 and its carboxyl-terminus shows no homology to any protein currently present in the GenBank. This unique carboxyl-terminus of Abro1 is involved in the specific interaction with the THAP5 protein that we observed in both the yeast and in mammalian cells. A recent study identified Abro1 to be part of a K63-linked DUB enzyme called BRISC (BRCC36-containing isopeptidase complex) [23]. BRISC is a four-subunit DUB enzyme that includes Abro1, BRCC36 (a zinc metalloprotease, member of the JAMM/MPN+ family) [23], BRE and NBA1 [25]. In this BRISC enzyme, Abro1 acts as a scaffold protein that brings together the other three polypeptides to form the complex. Furthermore, Abro1 binding to BRCC36 promotes the catalytic activity of the enzyme, although this heterodimer has minimal activity when compared to the complete four-subunit BRISC complex [23,24,27].
We monitored the expression of Abro1 in various human tissues as well as in isolated cardiomyocytes derived from human, mouse and rat tissues. The isolated cardiomyocytes clearly expressed Abro1. Abro1 mRNA was predominantly expressed in the human heart, but lower expression was also detected in the skeletal muscle, brain, kidney and small intestine. In addition, using coimmunoprecipitation experiments we were able to detect an Abro1-THAP5 complex in human heart. These results suggested that Abro1 like its interactor THAP5, might have an important function in normal cardiomyocytes and/or heart disease. We have previously reported that THAP5 is regulated in the MI area, in the heart of patients with CAD [22]. In this report, we show that Abro1 protein is also regulated in CAD. There was a significant increase in Abro1 protein level in the MI area, when compared to the RZ of the same heart. The MI area is the part of the heart that sustains maximum damage and cell death in CAD patients. To expand on these studies, we performed MI/R injury in mice using standard procedures. These experiments showed that Abro1 protein level significantly increased in the heart of mice that underwent MI/R.
The induction of Abro1 protein in CAD that was also replicated in MI/R could have a proapoptotic function or could be part of a cardioprotective response. To further investigate the role of Abro1 in MI/R injury, we used specific siRNAs to inhibit the level of this protein in the hearts of mice prior to MI/R. In these experiments, inhibition of Abro1 led to an increase in K63-linked ubiquitination and significantly elevated caspase-3 activity in cardiomyocytes when compared to the control mice. These results suggested that Abro1 has a cardioprotective function in this animal model of cardiac injury. Furthermore, we expanded on these studies using the rat myocardial cell line, H9c2. H9c2 cells were transfected with GFP-Abro1 and apoptosis was induced using two different concentrations of H2O2 as previously described [29]. The overexpression of Abro1 provided significant protection of these cells against H2O2 induced apoptosis suggesting Abro1 induction is part of a protective mechanism against cell death.
Since Abro1 is part of BRISC, a K63-linked DUB enzyme, we investigated if induction of Abro1 protein during MI/R coincides with a decrease in K63-linked ubiquitination in heart lysates. We used a specific antibody that recognizes K63-linked ubiquitinated proteins and detected several K63-linked ubiquitinated polypeptides in the heart extracts. These K63-ubiquitinated proteins could represent a single polypeptide with a different ubiquitin ratio or they could be proteolytic fragments of a larger precursor. The degree of the ubiquitination of these polypeptides, as indicated by the intensity of the bands, was dramatically reduced in the heart of mice following MI/R.
These results suggest that Abro1 induction is associated with an increase in K63-linked deubiquitination of specific cardiac protein(s) and that this activity has a cardioprotective function. The identity of the K63-linked polyubiquitinated protein(s) that are substrates of the BRISC enzyme in the heart and how their function is regulated via K63-linked deubiquitination are currently unknown.
The identification of this novel cardioprotective mechanism involving K63-linked DUB can provide new therapeutic targets for the development of drugs that could be used in patients with CAD. We expect drugs that are able to increase the protein level of Abro1, or the K63-linked DUB activity of the BRISC enzyme would protect cardiomyocytes from apoptosis.
Lys63-specific deubiquitination by the BRISC complex is regulated following heart injury and CAD
Lys63-specific deubiquitination protects cardiomyocytes from oxidative stress and MI/R injury
Abro1/KIAA0157, the scaffold protein of the BRISC complex is expressed predominantly in human heart
Abro1/KIAA0157 protein level are regulated in the heart of patients with CAD
Supplementary Material
Acknowledgements
We thank members of the Zervos' lab for comments and suggestions and Mark Yerkes for critical reading of the manuscript. This work was supported by the National Institutes of Health Grant 5R01DK55734 (to A.S.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: none declared
References
- 1.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33(3):275–86. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Kaiser P, Huang L. Global approaches to understanding ubiquitination. Genome Biol. 2005;6(10):233. doi: 10.1186/gb-2005-6-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyes-Turcu FE, Wilkinson KD. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem Rev. 2009;109(4):1495–508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–97. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9(6):536–42. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventii KH, Wilkinson KD. Protein partners of deubiquitinating enzymes. Biochem J. 2008;414(2):161–75. doi: 10.1042/BJ20080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel JA, Grant PA. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res. 2007;618(1-2):135–48. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19(24):2925–40. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 9.Song L, Rape M. Reverse the curse--the role of deubiquitination in cell cycle control. Curr Opin Cell Biol. 2008;20(2):156–63. doi: 10.1016/j.ceb.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komada M. Controlling receptor downregulation by ubiquitination and deubiquitination. Curr Drug Discov Technol. 2008;5(1):78–84. doi: 10.2174/157016308783769469. [DOI] [PubMed] [Google Scholar]
- 11.Shan J, Brooks C, Kon N, Li M, Gu W. Dissecting roles of ubiquitination in the p53 pathway. Ernst Schering Found Symp Proc. 2008;(1):127–36. doi: 10.1007/2789_2008_105. [DOI] [PubMed] [Google Scholar]
- 12.Li HH, Willis MS, Lockyer P, Miller N, McDonough H, et al. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J Clin Invest. 2007;117(11):3211–23. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson C, Ike C, Willis PWt, Stouffer GA, Willis MS. The bitter end: the ubiquitinproteasome system and cardiac dysfunction. Circulation. 2007;115(11):1456–63. doi: 10.1161/CIRCULATIONAHA.106.649863. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez JE, Schisler JC, Patterson C, Willis MS. Seek and destroy: the ubiquitinproteasome system in cardiac disease. Curr Hypertens Rep. 2009;11(6):396–405. doi: 10.1007/s11906-009-0069-7. [DOI] [PubMed] [Google Scholar]
- 15.Mearini G, Schlossarek S, Willis MS, Carrier L. The ubiquitin-proteasome system in cardiac dysfunction. Biochim Biophys Acta. 2008;1782(12):749–63. doi: 10.1016/j.bbadis.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Willis MS, Patterson C. Into the heart: the emerging role of the ubiquitin-proteasome system. J Mol Cell Cardiol. 2006;41(4):567–79. doi: 10.1016/j.yjmcc.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Carrier L, Schlossarek S, Willis MS, Eschenhagen T. The ubiquitin-proteasome system and nonsense-mediated mRNA decay in hypertrophic cardiomyopathy. Cardiovasc Res. 85(2):330–8. doi: 10.1093/cvr/cvp247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weekes J, Morrison K, Mullen A, Wait R, Barton P, et al. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3(2):208–16. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 19.Gong B, Leznik E. The role of ubiquitin C-terminal hydrolase L1 in neurodegenerative disorders. Drug News Perspect. 2007;20(6):365–70. doi: 10.1358/dnp.2007.20.6.1138160. [DOI] [PubMed] [Google Scholar]
- 20.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16(9):1 097–112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 21.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17(3):331–9. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Balakrishnan MP, Cilenti L, Mashak Z, Popat P, Alnemri ES, et al. THAP5 is a human cardiac-specific inhibitor of cell cycle that is cleaved by the proapoptotic Omi/HtrA2 protease during cell death. Am J Physiol Heart Circ Physiol. 2009;297(2):H643–53. doi: 10.1152/ajpheart.00234.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, et al. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. Embo J. 2009;28(6):621–31. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper EM, Boeke JD, Cohen RE. Specificity of the BRISC deubiquitinating enzyme is not due to selective binding to K63-linked polyubiquitin. J Biol Chem. 2009;285(14):10344–52. doi: 10.1074/jbc.M109.059667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23(6):729–39. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316(5828):1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson-Fortin J, Shao G, Bretscher H, Messick TE, Greenberg RA. Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J Biol Chem. 285(40):30971–81. doi: 10.1074/jbc.M110.135319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng L, Wang J, Chen J. The Lys63-specific deubiquitinating enzyme BRCC36 is regulated by two scaffold proteins localizing in different subcellular compartments. J Biol Chem. 285(40):30982–8. doi: 10.1074/jbc.M110.135392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cilenti L, Soundarapandian MM, Kyriazis GA, Stratico V, Singh S, et al. Regulation of HAX-1 anti-apoptotic protein by Omi/HtrA2 protease during cell death. J Biol Chem. 2004;279(48):50295–301. doi: 10.1074/jbc.M406006200. [DOI] [PubMed] [Google Scholar]
- 30.Faccio L, Fusco C, Viel A, Zervos AS. Tissue-specific splicing of Omi stress-regulated endoprotease leads to an inactive protease with a modified PDZ motif. Genomics. 2000;68(3):343–7. doi: 10.1006/geno.2000.6263. [DOI] [PubMed] [Google Scholar]
- 31.Hescheler J, Meyer R, Plant S, Krautwurst D, Rosenthal W, et al. Morphological, biochemical, and electrophysiological characterization of a clonal cell (H9c2) line from rat heart. Circ Res. 1991;69(6):1476–86. doi: 10.1161/01.res.69.6.1476. [DOI] [PubMed] [Google Scholar]
- 32.Faccio L, Fusco C, Chen A, Martinotti S, Bonventre JV, et al. Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J Biol Chem. 2000;275(4):2581–8. doi: 10.1074/jbc.275.4.2581. [DOI] [PubMed] [Google Scholar]
- 33.del Monte F, Kaumann AJ, Poole-Wilson PA, Wynne DG, Pepper J, et al. Coexistence of functioning beta 1- and beta 2-adrenoceptors in single myocytes from human ventricle. Circulation. 1993;88(3):854–63. doi: 10.1161/01.cir.88.3.854. [DOI] [PubMed] [Google Scholar]
- 34.del Monte F, O'Gara P, Poole-Wilson PA, Yacoub M, Harding SE. Cell geometry and contractile abnormalities of myocytes from failing human left ventricle. Cardiovasc Res. 1995;30(2):281–90. [PubMed] [Google Scholar]
- 35.del Monte F, Harding SE, Dec GW, Gwathmey JK, Hajjar RJ. Targeting phospholamban by gene transfer in human heart failure. Circulation. 2002;105(8):904–7. doi: 10.1161/hc0802.105564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natarajan AR, Rong Q, Katchman AN, Ebert SN. Intrinsic cardiac catecholamines help maintain beating activity in neonatal rat cardiomyocyte cultures. Pediatr Res. 2004;56(3):411–7. doi: 10.1203/01.PDR.0000136279.80897.4C. [DOI] [PubMed] [Google Scholar]
- 37.De Windt LJ, Lim HW, Taigen T, Wencker D, Condorelli G, et al. Calcineurin-mediated hypertrophy protects cardiomyocytes from apoptosis in vitro and in vivo: An apoptosis-independent model of dilated heart failure. Circ Res. 2000;86(3):255–63. doi: 10.1161/01.res.86.3.255. [DOI] [PubMed] [Google Scholar]
- 38.Sreejit P, Kumar S, Verma RS. An improved protocol for primary culture of cardiomyocyte from neonatal mice. In Vitro Cell Dev Biol Anim. 2008;44(3-4):45–50. doi: 10.1007/s11626-007-9079-4. [DOI] [PubMed] [Google Scholar]
- 39.Song G, Campos B, Wagoner LE, Dedman JR, Walsh RA. Altered cardiac annexin mRNA and protein levels in the left ventricle of patients with end-stage heart failure. J Mol Cell Cardiol. 1998;30(3):443–51. doi: 10.1006/jmcc.1997.0608. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Lau WB, Gao E, Tao L, Yuan Y, et al. Cardiomyocyte-Derived Adiponectin is Biologically Active in Protecting Against Myocardial Ischemia/Reperfusion Injury. Am J Physiol Endocrinol Metab. 2010;298(3):E663–70. doi: 10.1152/ajpendo.00663.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao L, Wang Y, Gao E, Zhang H, Yuan Y, et al. Adiponectin. An Indispensable Molecule in Rosiglitazone Cardioprotection Following Myocardial Infarction. Circ Res. 2010;106(2):409–1. doi: 10.1161/CIRCRESAHA.109.211797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eguchi M, Liu Y, Shin EJ, Sweeney G. Leptin protects H9c2 rat cardiomyocytes from H2O2-induced apoptosis. Febs J. 2008;275(12):3136–44. doi: 10.1111/j.1742-4658.2008.06465.x. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Cui M, Sun L, Jia Z, Bai Y, et al. Angiopoietin-1 protects H9c2 cells from H2O2-induced apoptosis through AKT signaling. Biochem Biophys Res Commun. 2007;359(3):685–90. doi: 10.1016/j.bbrc.2007.05.172. [DOI] [PubMed] [Google Scholar]
- 44.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316(5828):1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316(5828):1202–5. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 46.Baer R, Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev. 2002;12(1):86–91. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


