Abstract
Proteases play central roles in cell invasion by Toxoplasma gondii and other apicomplexan parasites. Herein we report the cloning and characterization of a novel secretory putative metalloproteinase, Toxolysin 4 (TLN4). T. gondii tachyzoites store TLN4 in the micronemes and secrete it in response to elevated calcium, suggesting a possible role in cell invasion. TLN4 is initially synthesized as a large (~260 kDa) precursor, which is extensively processed into multiple proteolytic fragments within the parasite secretory system. At least some of these proteolytic fragments remain associated in a large molecular complex. Whereas precomplementation with the TLN4 cDNA allowed disruption of the TLN4 gene, multiple attempts to directly knockout TLN4 without precomplementation failed. TLN4 knockout parasites were detected by PCR in transfected populations but were lost from the cultures during drug selection and growth suggesting that TLN4 contributes to parasite fitness.
Keywords: maturation, secretion, protease, protein complex
1. Introduction
Toxoplasma gondii is an obligate intracellular parasite of the phylum Apicomplexa, which includes other significant human pathogens such as Plasmodium falciparum, the most lethal agent of malaria. T. gondii infection, toxoplasmosis, can be particularly severe in immunocompromised individuals or the developing fetus [1]. Cell invasion by T. gondii is characterized by the sequential secretion of distinct organelles including micronemes and rhoptries [2]. Micronemes are the first to release their contents, which includes proteins (MICs) that confer apical attachment of the parasite to the host cell surface and penetration into the target cell. Subsequently, rhoptry neck proteins (RONs) and rhoptry bulb proteins (ROPs) are secreted. RONs assemble together with the MIC protein TgAMA1 to form the moving junction (MJ) [3,4], which is a complex arrangement of proteins promoting the juxtaposition of the host and parasite plasma membranes during the formation of the parasitophorous vacuole membrane (PVM). Posterior translocation of the MJ, which is driven by the parasite’s actin-myosin motor system, establishes the parasitophorous vacuole (PV) derived from the host plasma membrane [5]. It is also at this moment that ROPs are introduced into the host cell via small vesicles (evacuoles) that thereafter fuse with the PV [6]. MICs, RONs, and ROPs are proteolytically processed at several steps before and during cell invasion, but to date only a few T. gondii secretory or surface proteases have been identified.
Insulin-degrading enzymes (IDEs) are proteases found throughout the tree of life including in bacteria, fungi, plants and animals [7]. Human IDE was first characterized for its role in degrading insulin after uptake into cells [8]. However, subsequent studies showed that it has broad substrate specificity, cleaving and inactivating several small proteins and peptides including insulin [8], β-amyloid [9], glucagon [10], insulin-like growth factors I and II [11], transforming growth factor α [12], β-endorphin [13] and others. With this broad specificity, recognition of the substrates by human IDE more likely depends on the tertiary structure of the peptide rather than its amino acid sequence. Also, IDEs have been localized in the cytosol, peroxisomes, endosomes and even at the surface of some cells depending on the cell type, suggesting several roles for these enzymes [14–17]. IDEs are members of the M16A zinc metalloproteinase subfamily within the ME clan. They are characterized by an inverted zinc metalloproteinase core motif (HXXEH), which is located in the first 200 amino acid residues of the N-terminus of the protein. Zinc acts as an essential cofactor in the catalytic reaction.
The T. gondii genome encodes four members of the M16A family, which are termed toxolysins (TLN). TLN4, which was initially identified in a proteomic screen of T. gondii tachyzoite secretory products [18], displays a signature motif (HXXEHX69EX6E) for M16A metalloproteinase subfamily. Here we show that TLN4 is extensively processed within the parasite, but these products appear to remain associated in a large molecular complex. TLN4 is principally localized within the micronemes and is secreted in a calcium dependent manner, consistent with discharge from the micronemes. Conventional disruption of TLN4 locus was repeatedly unsuccessful, suggesting TLN4 contributes to parasite fitness.
2. Materials and methods
2.1 Plasmids
The TLN4 cDNA was amplified by PCR from a T. gondii RH-strain cDNA library (V.B. Carruthers, unpublished) in five different PCR reactions to cover its 7308 base pairs (bp). The primers were designed according to the sequence established in ToxoDB (www.toxodb.com), accession number TGME49_006510. We took advantage of four endogenous restriction sites found in the TLN4 cDNA to sequentially assemble the cDNA fragments in pBluescriptSK+. The first PCR covers the ATG start codon (bold) and an AvrII restriction site at the position -5 (5’- ggactagt cctaggcaagatgaggaggccctgtc-3’) to a unique BstZ171 restriction site at position 1410 (5’-ccttcggagtatacacggagcg-3’) of the TLN4 cDNA. SpeI was added upstream of an AvrII restriction site in the forward primer. Subsequent PCR reactions covered positions 1410 (BstZ171) (5’-cgctccgtgtatactccgaagg-3’) to 2916 (MfeI) (5’-ccggacaattggcttgttg-3’), positions 2916 (MfeI) (5’-caacaagccaattgtccg-3’) to 4739 (StuI) (5’-cgtctctgaggcctcgccag-3’), positions 4739 (StuI) (5’-ctggcgaggcctcagagacg-3’) to 6104 (XcmI) (5’-cgactccaggaagttgctggtggatc -3’) and position 6104 (XcmI) (5’-gatccaccagcaacttcctggagtcg -3’) to the stop codon at position 7308 (bold) (5’-gtcgatatctcaggcatgcaccactggaac-3’). An EcoRV site was added downstream of the stop codon. Subsequently, the 5’ untranslated region beginning −938 to the start codon of TLN4 and 3’ untranslated region +1227 to the stop codon were amplified from RH strain genomic DNA. These DNA fragments were fused to TLN4 cDNA using KpnI and AvrII restriction sites and EcoRV and XbaI restriction sites, respectively. We took advantage of the endogenous restriction site AvrII to fuse the 5’ untranslated region without affecting the consensus ribosome binding sequence adjacent to the ATG start codon.
To generate the TLN4-A-HA2 construct, a 12-bp ApaI-HpaI linker was inserted in frame at DNA position 1089 by fusion PCR [19]. This dual restriction site insertion added 4 amino acids between the leucine at amino acid position 363 and alanine at position 364 (Leu363- Gly- Pro-Val-Asn-Ala364) of TLN4. We used the ApaI-HpaI restriction sites to insert two copies of the Haemophilus influenzae hemagglutinin (HA) epitope [20]. A similar procedure was used to generate the TLN4-IA1-HA2 alleles, except in this case a 12-bp NsiI-NcoI linker was inserted in frame at DNA position 2547 by fusion PCR. This insertion added 4 amino acids between the lysine at position 849 and the glutamic acid at position 850 (Lys849-Met-His-Pro-Trp-Glu850). We used the newly introduced restriction sites to insert two copies of HA. Vectors containing the tagged TLN4 constructs were linearized using KpnI (NEB) and purified by phenol/chloroform extraction. Freshly egressed RH tachyzoites were co-transfected with 50 ug of the TLN4 vectors and 5 ug of the pDHFR-TS containing the selectable marker dihydrofolate reductase-thymidylate synthase by electroporation. Drug selection was performed as described below.
2.2 TLN4 Antibody
TLN4 antibody was generated against a recombinant fragment of the enzyme as follows. DNA encoding TLN4 amino acids 209–733 was PCR amplified, gel purified and cloned in frame with a hexa-histidine tag at the C-terminus in the vector pET21a(+) (Novagen) using NdeI and NotI. The resulting plasmid, pET21a(+)-TLN4 286-733, was transformed into E. coli BL21 DE3 and a single clone was grown in LB ampicillin (100 mgml−1) to A600 0.5 and induced with 1mM isopropyl-β-D-thiogalactopyranoside at 37°C, for 4 h. Bacteria were harvested and resuspended in CelLytic™ solution (Sigma) containing 0.5 mgml−1 benzonase (Sigma) and protease inhibitor cocktail Complete mini (Roche Diagnostics). The mixture was vortexed for 10 min and sonicated to ensure complete lysis. Insoluble material was discarded by centrifugation at 13,000 rpm, 4°C, for 10 min. The supernatant fraction was incubated with nickel chelation resin (Novagen) pre-charged with 400 mM NiSO4 and washed with 4 M NaCl, 480 mM imidazole, 160 mM Tris-HCl, pH 7.9). Recombinant TLN4 was eluted with 4 M imidazole, 2 M NaCl, 80 mM Tris-HCl, pH 7.9 before concentrating the relevant fractions and exchanging the buffer to PBS using an Amicon Ultra 15 Centrifugal filter device (Millipore). Recombinant TLN4 was emulsified with Freund’s complete adjuvant (Sigma) and injected in the peritoneal cavity of 5 Swiss Webster mice (25 ug/injection), with subsequent boosting in Freund’s incomplete adjuvant every 2 weeks for 6 weeks. Serum was collected five days following the last immunization.
2.3 Parasite culture and manipulation
DNA transfection by electroporation and routine culture of human foreskin fibroblasts (HFF) and RH-strain based T. gondii parasites have been described previously [21]. Selection for DHFR-TS was performed by including 1 µM pyrimethamine in the normal media. Selection for hypoxanthine-xanthine-guanine phosphoribosyltransferase (HXGPRT) was performed using 25 µgml−1 mycophenolic acid and 50 µgml−1 xanthine.
2.4 Immunoblotting
Electrophoresis and immunoblotting were performed using standard protocols [22]. Briefly, T. gondii cell lysates were separated by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes using a Transblot Semi-Dry Transfer Cell (Bio-Rad). Membranes were blotted with mouse anti-TLN4 or rat-HA mAb 3F10 (Roche Applied Science). Secondary antibodies used were peroxidase-conjugated goat anti-rat, anti-mouse or anti-rabbit (Jackson Immuno Research).
2.5 Pulse-Chase Immunoprecipitation
Freshly egressed tachyzoites (2×108 ml−1 in 400 ul labeling medium: Delbecco’s Modified Eagle’s Medium containing 1% FBS and 10 mM HEPES, with or without addition of 50 ugml−1 brefeldin A) were “pulse” metabolically labeled for 10 min at 37°C with 540 µCi 35S methionine and cysteine (Express Protein labeling mix, Perkin Elmer). Samples were “chased” by the addition of 3.6 ml of warm labeling medium containing 5 mM each of unlabeled methionine and cysteine. Samples (800 µl) were collected at various times and placed on ice before the parasites were pelleted, washed and lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.5, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl; supplemented with DNAse and RNAse). Similar banding patterns were seen regardless of whether protease inhibitors were included in the lysis buffer. TLN4 was immunoprecipitated by sequential addition of mouse anti-TLN4 (2 ul) and protein G sepharose beads (30 µl of 25% slurry; GE Healthcare), solubilized in reducing SDS-PAGE sample buffer, resolved by 7.5% SDS-PAGE, stained with Coomassie blue, soaked in fluorogenic solution (Enhance™, GE Healthcare) and revealed by phosphorimaging using a Typhoon trio imaging system (GE Healthcare).
2.6 Coimmunoprecipitation
For coimmunoprecipitation, 100 µl of 50% slurry protein G sepharose beads were washed 4 times with RIPA buffer and incubated with rat anti-HA or mouse anti-TLN4 for 1 h in RIPA buffer. Beads were washed twice with 1 ml of 0.2 M sodium borate pH 9.0 and rotated in a solution of 25 mM dimethyl pimelimidate (Sigma) in sodium borate for 45 min. Dimethyl pimelimidate was added for a second time, rotated 45 min and washed with 0.2 M sodium borate. The reaction was quenched by adding 1 ml 0.2 M ethanolamine pH 8.0, rotated 1 h and washed again with RIPA buffer. Freshly egressed parasites from two T25 cell culture flasks were harvested, resuspended in RIPA buffer and rotated for 30 min. Insoluble material was removed by centrifugation, 13,000 rpm, 4°C, 10 min. Supernatant was tumbled with the cross-linked beads for 2 h. Beads were washed four times with 1 ml RIPA buffer. Immunoprecipitated proteins were resuspended in 100 µl of sample buffer, boiled 5 min, resolved by 10% SDS-PAGE and immunoblotted as described above.
2.7 In vitro cross-linking
Freshly egressed tachyzoites were harvested and resuspended in 1% Triton X-100 in PBS. Cell extracts were divided in two tubes. The first tube was untreated and the second one was treated for 30 min with 2 mM ethylene glycolbis(succinimidylsuccinate) (EGS) (Pierce). After quenching the reaction with 50 mM Tris-HCl, pH 7.5 for 15 min., cross-linked products were analyzed by 7.5% SDS-PAGE and immunoblotted as described above.
2.8 Native blue gel electrophoresis (NBGE)
Freshly egressed tachyzoites were harvested and resuspended in 200 µl of 150 mM NaCl, 50 mM Tris-HCl pH 7.0. Triton X-100 was added at a final concentration of 1% and the solution was subjected to bath sonication. DNA was digested by the addition of 1 unit µl−1 benzonase with 2 mM MgCl2 for 30 min at RT. After centrifugation at 13,000 rpm at 4°C for 30 min, the supernatant was collected in new tubes and the samples were resolved using a NativePAGE Novex Bis-Tris gel system according to the manufacture’s instructions (Invitrogen). Briefly, 40 µl of each sample was mixed with 4X sample buffer (50 mM Bis Tris pH 7.2, 50 mM NaCl, 10% Glycerol, 0.0001% Ponceau) and 5% G-250 Coomassie. Samples were loaded and electrophoresed at 4°C with dark blue cathode buffer for the first third of the gel and light blue cathode buffer for the next two thirds. Immunoblotting was performed as described above.
2.9 Immunofluorescence assays (IFA)
HFF cells grown in eight well chamber slides (Lab-Tek, Fisher) were infected with parasites for 24 h. Slides were washed with PBS, fixed with 4% paraformaldehyde for 20 min, permeabilized in 0.1% Triton X-100 in PBS for 10 min, washed with PBS and blocked for 30 min with PBS containing 10% FBS. Fixed parasites were incubated for 1 h with mouse anti-TLN4, rabbit anti-TgMIC2 [23] or rat anti-HA mAb 3F10 (Roche Applied Science) and washed 3 times in PBS containing 1% FBS and 1% NGS (wash buffer). Slides were subsequently incubated with wash buffer containing 1 ugml−1 4,6-diamidino-2-phenylindol (DAPI) and Alexa-594 anti-rabbit IgG, Alexa-488 anti-mouse IgG or Alexa 488 anti-rat IgG (Invitrogen). Images were collected using a Zeiss Axio Observer Z1 inverted microscope equipped with an Axiocam MRM CCD camera.
2.10 Excretory/secretory antigens (ESA)
ESA was collected as described previously [24]. Briefly, freshly egressed parasites from two T25 cell culture flasks were syringed, filtered and chased with DMEM, 20 mM HEPES and pellet. Parasites were resuspended at 4×108 ml−1 in DMEM, 20 mM HEPES, 1% FBS. One 100 µl sample of parasites was left untreated whereas a parallel sample was pre-treated with 100 µM 1,2-bis(2-aminophenoxy)ethane-N,N,N,N-tetraacetic acid-acetoxymethyl ester (BAPTA-AM; Calbiochem), and secretion was induced with 1% (v/v) ethanol for 2 min at 37°C. Constitutive secretion was also analyzed by incubating the parasites for 20 min at 37°C without ethanol or BAPTA-AM. Secretion was arrested by incubation on ice. Samples were centrifuged twice at 3,000 rpm, for 10 min at 4°C and the resulting parasite fraction (pellet) and ESA fraction (supernatant) were analyzed by 10% SDS-PAGE and immunoblotting.
2.11 Attempted targeted disruption of TLN4
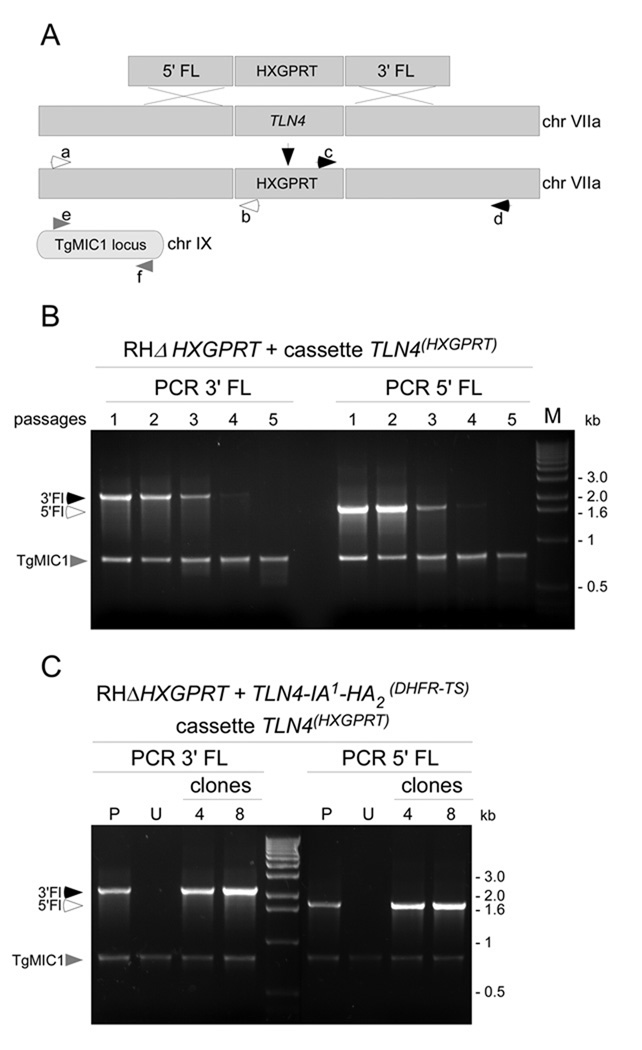
TLN4 genomic flanking sequences were obtained from the ToxoDB website (www.toxodb.com). The 5’UTR sequence corresponding to -3079 to -1 upstream the TLN4 start codon was PCR amplified with primers 5’-cggggtaccgtggaagccgatacgagag-3’ and 5’- agggggccccttgcctaggaggggagaa-3’. The resulting product was cloned into pMiniHXG [25], which contains an HXGPRT selectable marker flanked by the DHFR-TS promoter and terminator, using KpnI and ApaI. Subsequently, the TLN4 3’UTR sequence corresponding to positions 1 to 3361 downstream of the stop codon was PCR amplified with primers 5’- ctagtctagaaagtgggttgtgcgcattaaac-3’ and 5’-ttccttttttcgccggcgggcagaagacgttgagcttc-3’ and cloned into the XbaI and NotI of pMiniHXG. This plasmid, named pMini-5Fl-TLN4-HXGPRT-3FL-TLN4, was used as template to PCR amplify a segment starting from −1500 upstream of the start codon and +1500 downstream of the stop codon with primers 5’-gatgtttgtcgaagctatctatctgcg-3’ and 5’-gcgtcgcacacgaaagagctc-3’. This PCR product was purified and transfected by electroporation in RHΔHXGPRT. Drug selection was initiated 24 hr after transfection as described above. Genomic DNA was extracted after each passage. Analytical PCR was performed to detect correct homologous recombination of the cassette at the TLN4 locus using the primers illustrated in Figure 4A: a, 5’-agttgcagccagaggcagaagcaagtcc-3’; b, 5’- cagtcagataacaggtgtagcg-3’; c, 5’-gcgggtgacgcagatgtgcgtgtatcc-3’; d, 5’- gaaaagtgtctgcgtgttagcagc-3’; e, 5’-tagtaagcagccagcgtcac-3’; and f, 5’-ctttgcaaatccgttctaggtc-3’.
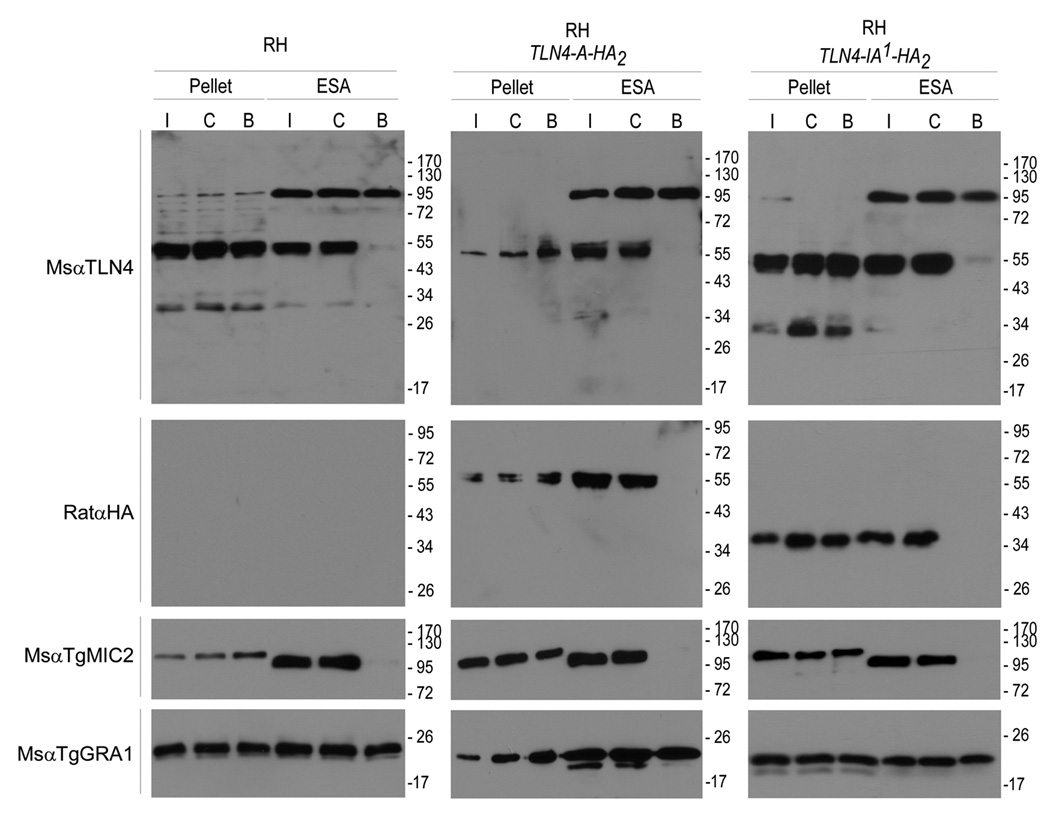
Figure 4.
Analysis of TLN4 secretion. Tachyzoites of WT RH or tagged strains were purified and treated before isolating parasite pellet and ESA fractions. Treatments included induced (I) secretion with ethanol for 2 min, constitutive (C) secretion for 20 min, or BAPTA-AM (B) inhibition of secretion. Samples were probed with the antibodies indicated on the left.
3. Results and Discussion
3.1 TLN4 is extensively processed within the parasite
TLN4, which consists of thirteen exons on chromosome VIIa, is predicted to encode a 256 kDa protein with a signal peptide at the N-terminus (Fig. 1A). Functional domains were identified using Interpro (http://www.ebi.ac.uk/interpro/). The first domain of TLN4 exhibits the characteristics of an active core of the M16A metalloproteinase subfamily. Also present are three so-called inactive domains (IA), which are common in metalloproteinases of the M16 subfamilies. Inactive domains, which are structurally related to the active domain, contribute to the proteolytic event by forming a clamp that accommodates the substrate. The first inactive domain is distinct whereas the second and third inactive domains overlap with one another. Metalloproteinases of the M16A subfamily are often large proteins with predicted sizes in excess of 100 kDa. However, TLN4 is especially large because of its long C-terminal extension, which lacks homology to other proteins and terminates in a repeat domain (RD) consisting of 8 nearly perfect 28 aa repeats (Fig. 1A). The repeat domain contains a relatively high density of single nucleotide polymorphisms between T. gondii strains (www.toxodb.org), suggesting that it is under selective pressure or is otherwise prone to accumulating mutations.
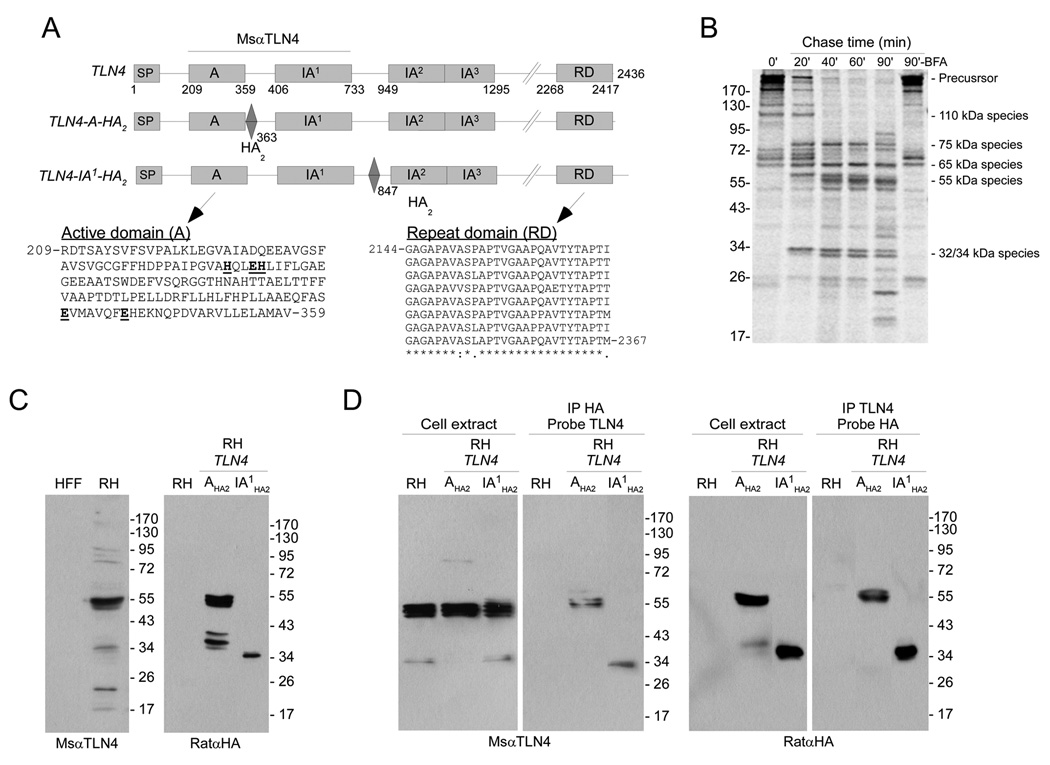
Figure 1.
TLN4 is extensively processed. (A) Schematic illustrations of TLN4 including domain features and placement of epitope tags. Amino acid positions of each domain are indicated underneath the TLN4 illustration and the positions of the HA2 epitope tag are indicated below the TLN4-A-HA2 and TLN4-IA1-HA2 illustrations. As demarked by a horizontal line, MsαTLN4 recognizes the active (A) and first inactive (IA1) domain of TLN4. Also shown are the sequences of the A domain and repeat domain (RD). The A domain active site residues in the HXXEHX69EX6E are emboldened and underlined. Note the 8 nearly perfect 28 aa repeats in the RD domain. Perfectly conserved positions (8 identical residues) are indicated with an asterisk, mostly conserved positions (6 or 7 identical residues) are denoted with a colon and partially conserved positions (5 or fewer identical residues) are marked with a period. (B) Pulse-chase immunoprecipitation analysis of TLN4 proteolytic processing. Tachyzoites were pulse labeled with 35S-Met and 35S-Cys for 15 min and chased with unlabeled amino acids for the indicated times. TLN4 fragments were immunoprecipitated with MsαTLN4 and analyzed by 7.5% SDS-PAGE and phosphorimaging. Positions of molecular weight standards (in kilodaltons, left) and the major TLN4 species (right) are marked. (C) Immunoblotting analysis of TLN4 products. The A and IA1 domains exist in a 55 kDa product based on recognition of untagged TLN4 by MsαTLN4 (left panel) and recognition of TLN4-A-HA2 by RatαHA (right panel, center lane). The linker region between IA1 and IA2 is a 34 kDa product based on recognition of TLN4-IA1-HA2 by RatαHA (right panel, right lane). (D) Reciprocal immunoprecipitation and immunoblotting analysis of TLN4 products. Shown are immunoblots of parasite cell extracts or immunoprecipitates with the indicated antibodies used for immunoprecipitation or probing.
Although proteases are often initially synthesized as inactive zymogens that undergo proteolytic processing (maturation) to become active enzymes, M16A proteases typically do not undergo such processing. To determine if TLN4 is proteolytically processed, we pulse labeled tachyzoites with 35S-Met and 35S-Cys for 15 min and chased with nonradioactive amino acids for 20, 40, 60 or 90 min. TLN4 was immunoprecipitated with a mouse antibody directed against recombinant TLN4 active domain (A) and the first inactive domain (IA1)(Fig. 1A), and the products were analyzed by SDS-PAGE and phosphorimaging (Fig. 1B). Several intensely labeled large precursor species (>200 kDa) were seen immediately following the labeling period (0 min chase) along with additional less intense smaller species. In the 20 min chase sample, we observed several processed products of 75 kDa, 67–72 kDa, 65 kDa, 60 kDa and 34 kDa. The 75 kDa, 65 kDa and 34 kDa species remained stable in the 40, 60, and 90 min chase samples whereas the 67–72 kDa and 60 kDa species diminished coincident with the emergence of new species at 55 kDa (consisting of a series of closely migrating products), 50 kDa, and 32 kDa. At 90 min, we also noticed two small new products of 17 kDa and 26 kDa. The complexity of the immunoprecipitates (at least 18 species in the 90 min chase sample) suggests that TLN4 is multiply processed, and that the A and IA1 domains to which the antibody was raised probably remain associated with other parts of the protein and possibly other proteins. TLN4 maturation was nearly completely arrested by pretreating the parasites with brefeldin A (BFA), a fungal metabolite that inhibits protein transport to the trans-Golgi, indicating that most of the processing occurs within or beyond the trans-Golgi. All T. gondii MICs and ROPs that undergo maturation are processed within or beyond the trans-Golgi except TgSUB1, which is initially processed in the parasite endoplasmic reticulum [26]. Some of the TLN4 precursor species might also be processed in the ER. To our knowledge this is the first example of an M16A metalloproteinase that undergoes proteolytic maturation.
Next we performed immunoblotting to identify fragments containing the A and IA1 domains. As shown in Figure 1C, the TLN4 antibody principally recognized the 55 kDa closely migrating species that were noted above and several minor species (including 34 kDa and 22 kDa) in an RH tachyzoite lysate. That these reactive species are derived from the parasite was confirmed by the absence of reactivity to HFF host cell proteins. The high molecular weight precursor species were not observed, consistent with the extensive maturation of TLN4. The heterogeneity of the 55 kDa product suggests that regions immediately flanking this species of TLN4 are particularly susceptible to processing. The intensity of the 34 kDa species varies between experiments, possibly because of instability and degradation.
To investigate in more detail the processing of TLN4, we engineered two different epitope tagged expression constructs driven by the TLN4 promoter. The constructs contain two copies of the HA epitope tag after the A domain (TLN4-A-HA2) or the first inactive domain (TLN4-IA1-HA2)(Fig. 1A). After confirming the DNA sequences of these constructs, we stably introduced them into RH strain parasites and isolated single clones. In lysates of parasites expressing TLN4-A-HA2, anti-HA recognized the 55 kDa species, indicating that this product encompasses the A and IA1 domains (Fig. 1C). Smaller, less abundant species were also observed. Parasites expressing TLN4-IA1-HA2 show a distinct 34 kDa band, possibly corresponding to the ~32 kDa (untagged) product seen in the pulse-chase analysis.
Although we attempted to analyze processing of the C-terminal region by generating a construct tagged with HA at the extreme C-terminus of TLN4, we were unable to detect a signal by immunoblotting lysates of stably transformed parasites. We also transfected parasites with expression constructs of TLN4 containing various domain truncations, but none of them gave a signal on immunoblots. We additionally tried different fusion tags (TLN4-IA1-myc2 and TLN4-IA1-GFP) and different promoters (SAG1P-TLN4-IA1-HA2, DHFRP-TLN4-IA1-HA2), but again no expression was detected. These observations suggest that TLN4 is intolerant of certain modifications and expression from heterologous promoters.
That the 55 kDa species encompasses the A and IA1 domains was further confirmed by reciprocal immunoprecipitation of TLN4-A-HA2 with anti-HA or MsαTLN4 and immunoblotting with anti-HA or MsαTLN4 (Fig. 1D). The 34 kDa species from TLN4-IA1-HA2 was similarly recognized by both anti-HA and MsαTLN4 in reciprocal immunoprecipitation and immunoblotting indicating shared epitopes or association within a complex of processed products.
3.2 TLN4 exists in a high molecular weight complex
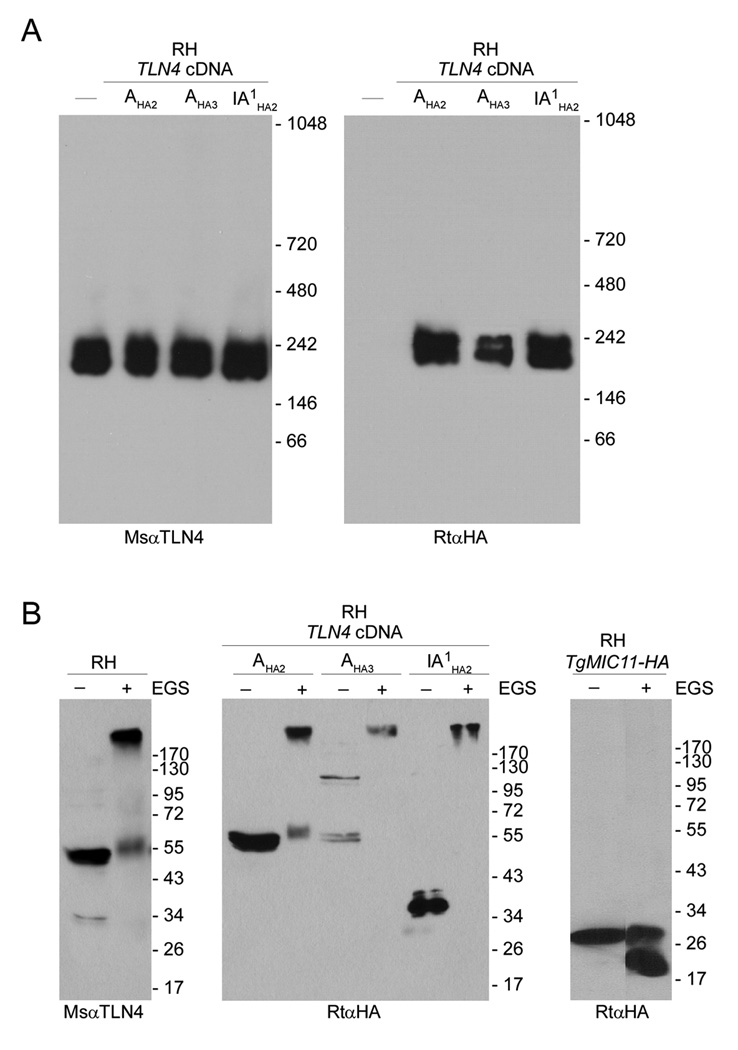
Two types of experiments were performed to further test if the processed products of TLN4 remain associated together. First, we examined the migration of TLN4 by native blue gel electrophoresis (NBGE) whereby protein extracts from RH parasites and parasites expressing TLN4-A-HA2 or TLN4-IA1-HA2 were electrophoresed under non-denaturing conditions, transferred to a PVDF membrane and immunoblotted with antibodies to TLN4 or HA. As shown in Figure 2A, we observed a closely spaced doublet of ~200 kDa and ~230 kDa in all of the strains probed with anti-TLN4 (left panel). The same two species were observed in TLN4-A-HA2 and TLN4-IA1-HA2 lysates probed with anti-HA (Fig. 2A, right panel). Second, we performed a cross-linking experiment in which parasite protein extracts from the three strains were treated with ethylene glycolbis(succinimidylsuccinate) (EGS) and analyzed by SDS-PAGE and immunoblotting. Figure 2B, left panel shows that the 55 kDa and 34 kDa species detected by anti-TLN4 in RH shift up to a high molecular weight species after cross-linking. Similarly, when probed with anti-HA the 55 kDa species TLN4-A-HA2 and the 34 kDa species in TLN4-IA1-HA2 cross-link into a high molecular weight species (Fig. 2B, middle panel). This cross-linked species, which is estimated to be 285 kDa, is of a similar size to that of the native complex observed by NBGE. In contrast, TgMIC11-HA [27], which is not known to exist in a complex, failed to cross-link into a high molecular weight species (Fig. 2B, right panel), confirming the specificity of the cross-linking reaction. Collectively, these findings demonstrate that at least some of the TLN4 processed products remain associated after maturation. Since the size of the cross-linked product is similar to the precursor species seen in the pulse-chase experiment, it is possible that all of the TLN4 products remain together in a complex. Alternatively, a subset of the processed products might associate with one or more additional parasite protein(s) to form the high molecular weight species. While the stoichiometry of the TLN4 complex is unknown, at least one IDE is known to be active as a homodimer [28]; hence, a third possibility is that fragments encompassing the active and inactive domains of TLN4 assemble into a homodimeric complex with two copies of each domain.
Figure 2.
TLN4 exists in a high molecular weight protein complex. (A) Native blue gel electrophoresis (4–16% polyacrylamide gradient) immunoblots of extracts from RH parasites or RH parasites stably transfected with the tagged cDNA of TLN4 (TLN4-A-HA2 or TLN4-IA1-HA2). Probing with the MsαTLN4 or with the rat anti-HA shows a doublet corresponding to TLN4. (B) SDS-PAGE immunoblotting analysis of parasite extracts cross-linked with EGS. Proteins were extracted from the aforementioned strains treated with solvent (DMSO) or EGS and separated by 7.5% SDS-PAGE. Probing with MsαTLN4 or with the RatαHA shows the shift of TLN4 in a high molecular weight complex. TgMIC11-HA [27] failed to shift after cross-linking, indicating the specificity of the reaction.
3.3 TLN4 principally localizes to micronemes and is secreted in a calcium-dependent manner
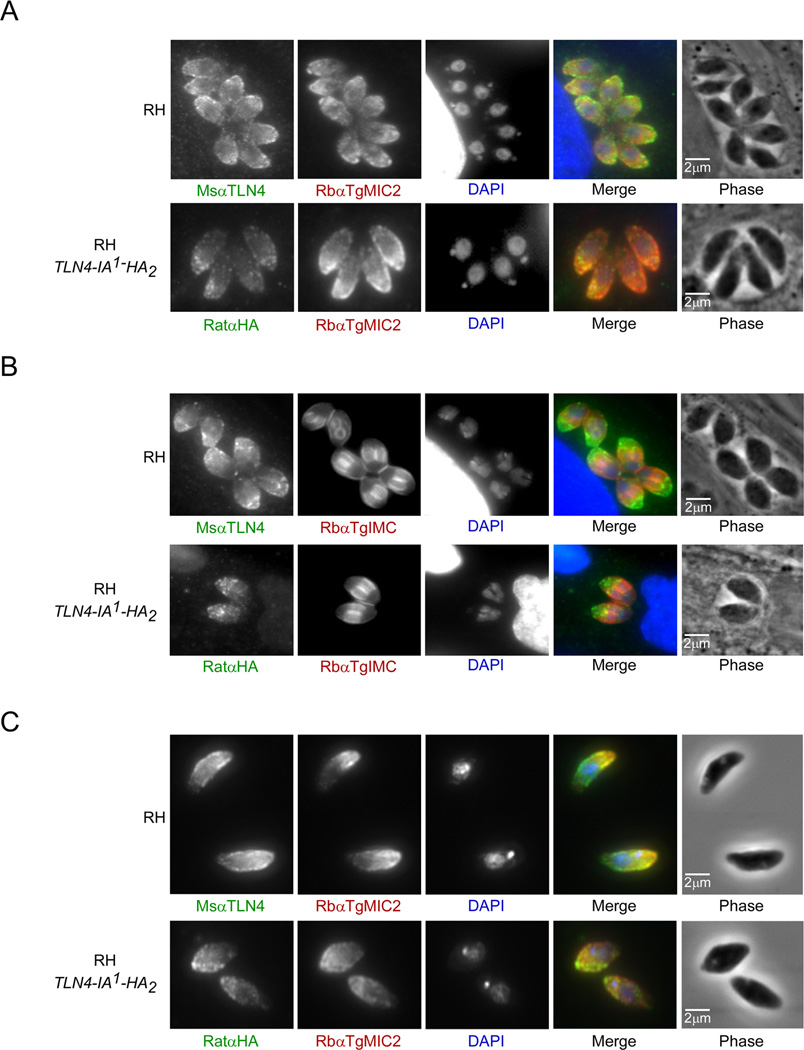
Next, to determine the subcellular localization of TLN4, we examined intracellular and extracellular tachyzoites by immunofluorescence microscopy. TLN4 localized to the apical periphery of the parasite, which substantially overlapped with the well-characterized microneme protein TgMIC2 [23] in both intracellular and extracellular parasites (Fig. 3A–C, upper panels). TLN4 staining appeared to extend further to the basal extremity of the parasite than that of MIC2 in extracellular parasites, indicating that a subpopulation of TLN4 might reside in a distinct location (Fig. 3C, upper panel). We also used the antibody directed against HA to locate the tagged versions of TLN4 in strains expressing TLN4-A-HA2 and TLN4-IA1-HA2. While TLN4-A-HA2 staining was too faint to reliably determine its subcellular distribution, TLN4-IA1-HA2 showed an apical staining pattern that also largely colocalized with TgMIC2 (Fig 3A–C, lower panels). The localization of TLN4 in the micronemes suggests a possible role in parasite egress, gliding motility, and/or cell invasion. Also, we noted that intracellular parasites undergoing cell division showed a particularly strong signal for TLN4 in the extreme apical end of the parasite (Fig. 3A and B), indicating a potential role of TLN4 in the biogenesis of micronemes during replication.
Figure 3.
TLN4 resides principally in the micronemes. Immunofluorescence microscopy of intracellular (A and B (dividing parasites)) or extracellular (C) parasites of RH or RH expressing TLN4-IA1-HA2 (upper and lower panels, respectively). Parasites were fixed with 4% paraformaldehyde and stained with MsαTLN4 (green) and RbαTgMIC2 (red) antibodies or RatαHA (green) and RbαTgMIC2 (red) antibodies.
To determine if TLN4 is secreted from the parasites in a calcium-dependent manner similar to other microneme proteins, we probed immunoblots of ESA with anti-TLN4 and anti-HA. The 55 kDa and 34 kDa species of TLN4 were secreted from all three strains tested (RH, TLN4-A-HA2, and TLN4-IA1-HA2) under conditions of induced (1% ethanol, 37°C, 2 min) or constitutive (no ethanol, 37°C, 20 min) secretion (Fig. 4). Pretreatment with the calcium antagonist BAPTA-AM blocked secretion of TLN4 in a manner similar to TgMIC2 but had no effect on secretion of the dense granule protein TgGRA1. Anti-TLN4 detected a 95 kDa product that is enriched in the ESA and unaffected by BAPTA-AM. This product is likely a cross-reactive protein since it is not recognized by anti-HA in the TLN4-A-HA2, and TLN4-IA1-HA2 strains.
3.4 TLN4 is probably required for parasite fitness
To further investigate the function of TLN4, we attempted to genetically ablate TLN4. Tachyzoites of strain RHΔHXGPRT [25] or RHΔku80Δhxg, which has increased homologous targeting efficiency [29], were transfected with a knockout construct consisting of 1.5 kb of TLN4 flanking regions on either side of an HXGPRT selectable marker cassette. At each passage under drug selection, parasite genomic DNA was isolated for PCR analysis of correct integration using primers outside of the flanks with primers that anneal within the HXGPRT cassette (Fig. 5A). Also, as a control for the genomic DNA we amplified a fragment of the TgMIC1 gene in separate, but parallel, reactions that were combined for gel analysis. TLN4 PCR products diagnostic of correct integration of the knockout construct were detected in the early passages, but these products diminished rapidly between passages 3 and 4, and were absent in passage 5. In contrast, the intensity of the TgMIC1 product remained constant. Attempts to isolate TLN4 knockout clones in the early passes were repeatedly unsuccessful. We also failed to obtain a TLN4 knockout strain using constructs bearing chloramphenicol acetyl transferase (CAT) or pyrimethamine-resistant dihydrofolate reductase-thymidylate synthetase (DHFR-TS). Detection of knockout parasites in the early passes suggests that the TLN4 locus is receptive to homologous recombination, but the loss of such parasites indicates that TLN4 deficient parasites have a selective disadvantage in the population.
Figure 5.
TLN4 is probably required for parasite fitness. (A) Schematic of the gene replacement strategy involving double crossover replacement of the TLN4 gene with the selectable marker gene HXGPRT. PCR primers used for detection of knockout parasites are indicated. (B) PCR analysis of transfected populations under drug selection shows progressive loss of TLN4 recombinants with increasing passes based on amplification of the 3’ flank or 5’ flank. PCR amplification of an unrelated gene, TgMIC1, was included as a control for equal amounts of genomic DNA in the reactions. (C) Precomplementation with TLN4-IA1-HA2 allows disruption of TLN4. RHΔHXGPRT parasites stably transfected with TLN4-IA1-HA2 were transfected with the TLN4 knockout construct and drug selected as a population (P) and as two independent clones (4 and 8). Untransfected (U) parasites were included as a negative control.
To determine if the endogenous copy of TLN4 could be disrupted when an extra copy of TLN4 is provided we precomplemented RHΔHXGPRT parasites with the TLN4-IA1-HA2 expression plasmid. After transfecting the TLN4 knockout construct, the resulting drug resistant population of RHΔhxgTLN4-IA1-HA2 parasites remained positive for correct integration at the TLN4 locus after 5 passages (Fig. 5C, lane “Pop”). After isolating clones from the population, two (clones 4 and 8) out of twenty-five were positive for correct homologous recombination and replacement of TLN4, confirming that the TLN4 locus is genetically receptive. This experiment further demonstrates that the TLN4-IA1-HA2 cDNA is functional and that addition of two copies of the HA epitope after the inactive domain doesn’t interfere with TLN4 function.
Multiple attempts to directly disrupt TLN4 in Ku80-deficient parasites using knockout constructs with DHFR or CAT were also unsuccessful despite isolating parasite clones soon after transfection. We were also unsuccessful in repeated attempts to generate a tetracycline regulated (tet-off) conditional knockout mutant of TLN4 either by promoter replacement or by disrupting TLN4 after precomplementing with a regulatable copy of the TLN4 cDNA. In the latter case, expression of the regulatable copy of TLN4 may have been too low to allow disruption of the TLN4 gene.
Although the function of TLN4 remains unknown, it is reasonable to think that it contributes to microneme dependent processes such as egress, gliding motility, and parasite invasion of host cells. If TLN4 functions in parasite egress, it could contribute to the degradation of the host cytoskeletal structures that encase intracellular parasites. With respect to gliding motility, it is intriguing to note that TLN4 is located near the middle of a genetic locus on the chromosome VIIa that was identified by linkage analysis to be associated with the cell migration and transmigration activities of the parasite [30]. For cell invasion, TLN4 could act as a maturase for other microneme proteins or serve as a regulator of such proteins during entry.
Acknowledgements
We thank Tracey Schultz for technical expertise and Peter Bradley, My-Hang Huynh, and Jessica Beauchamp for critically reading the manuscript. This work was supported by a fellowship from the National Science and Engineering Research Council of Canada (J.L.) and National Institutes of Health grant AI046675 (V.B.C.).
Abbreviations
- A
active domain
- BAPTA-AM
1,2-bis(2-aminophenoxy)ethane-N,N,N,N-tetraacetic acid-acetoxymethyl ester
- BFA
brefeldin A
- CAT
chloramphenicol acetyl transferase
- DHFR-TS
dihydrofolate reductase-thymidylate synthase
- EGS
ethylene glycolbis(succinimidylsuccinate)
- ESA
excretory/secretory antigens
- HA
hemagglutinin epitope
- HFF
human foreskin fibroblast
- HXGPRT
hypoxanthine-xanthine-guanine phosphoribosyltransferase
- IFA
immunofluorescence assay
- IA
inactive domain
- IDE
insulin degrading enzyme
- NBGE
native blue gel electrophoresis
- MIC
microneme protein
- RIPA
radioimmunoprecipitation buffer
- ROP
rhoptry protein
- RON
rhoptry neck protein
- PV
parasitophorous vacuole
- PVM
parasitophorous vacuole membrane
- PVDF
polyvinylidene fluoride
- TLN4
toxolysin 4
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
GenBank Accession number: in process, submission # 1424514
REFERENCES
- 1.Carter AO, Frank JW. Congenital toxoplasmosis: epidemiologic features and control. CMAJ. 1986;135:618–623. [PMC free article] [PubMed] [Google Scholar]
- 2.Carruthers VB, Sibley LD. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol. 1997;73:114–123. [PubMed] [Google Scholar]
- 3.Alexander DL, Mital J, Ward GE, Bradley P, Boothroyd JC. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 2005;1:17. doi: 10.1371/journal.ppat.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lebrun M, Michelin A, El Hajj H, Poncet J, Bradley PJ, Vial H, et al. The rhoptry neck protein RON4 re-localizes at the moving junction during Toxoplasma gondii invasion. Cell Microbiol. 2005;7:1823–1833. doi: 10.1111/j.1462-5822.2005.00646.x. [DOI] [PubMed] [Google Scholar]
- 5.Sibley LD. Toxoplasma gondii: perfecting an intracellular life style. Traffic. 2003;4:581–586. doi: 10.1034/j.1600-0854.2003.00117.x. [DOI] [PubMed] [Google Scholar]
- 6.Hakansson S, Charron AJ, Sibley LD. Toxoplasma evacuoles: a two-step process of secretion and fusion forms the parasitophorous vacuole. EMBO J. 2001;20:3132–3144. doi: 10.1093/emboj/20.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirsky IA, Broh-Kahn RH. The inactivation of insulin by tissue extracts; the distribution and properties of insulin inactivating extracts. Arch Biochem. 1949;20:1–9. [PubMed] [Google Scholar]
- 8.Mirsky IA. Insulinase, insulinase-inhibitors, and diabetes mellitus. Recent Prog Horm Res. 1957;13:465–471. [PubMed] [Google Scholar]
- 9.Farris W, Leissring MA, Hemming ML, Chang AY, Selkoe DJ. Alternative splicing of human insulin-degrading enzyme yields a novel isoform with a decreased ability to degrade insulin and amyloid beta-protein. Biochem. 2005;44:6513–6525. doi: 10.1021/bi0476578. [DOI] [PubMed] [Google Scholar]
- 10.Kirschner RJ, Goldberg AL. A high molecular weight metalloendoprotease from the cytosol of mammalian cells. J Biol Chem. 1983;258:967–976. [PubMed] [Google Scholar]
- 11.Roth RA, Mesirow ML, Yokono K, Baba S. Degradation of insulin-like growth factors I and II by a human insulin degrading enzyme. Endocr Res. 1984;10:101–112. doi: 10.3109/07435808409035411. [DOI] [PubMed] [Google Scholar]
- 12.Garcia JV, Gehm BD, Rosner MR. An evolutionarily conserved enzyme degrades transforming growth factor-alpha as well as insulin. J Cell Biol. 1989;109:1301–1307. doi: 10.1083/jcb.109.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Safavi A, Miller BC, Cottam L, Hersh LB. Identification of gamma-endorphin-generating enzyme as insulin-degrading enzyme. Biochem. 1996;35:14318–14325. doi: 10.1021/bi960582q. [DOI] [PubMed] [Google Scholar]
- 14.Qiu WQ, Folstein MF. Insulin, insulin-degrading enzyme and amyloid-beta peptide in Alzheimer's disease: review and hypothesis. Neurobiol Aging. 2006;27:190–198. doi: 10.1016/j.neurobiolaging.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Lynch JA, George AM, Eisenhauer PB, Conn K, Gao W, Carreras I, et al. Insulin degrading enzyme is localized predominantly at the cell surface of polarized and unpolarized human cerebrovascular endothelial cell cultures. J Neurosci Res. 2006;83:1262–1270. doi: 10.1002/jnr.20809. [DOI] [PubMed] [Google Scholar]
- 16.Bondy CA, Zhou J, Chin E, Reinhardt RR, Ding L, Roth RA. Cellular distribution of insulin-degrading enzyme gene expression. Comparison with insulin and insulin-like growth factor receptors. J Clin Invest. 1994;93:966–973. doi: 10.1172/JCI117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, et al. Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- 18.Zhou XW, Kafsack BF, Cole RN, Beckett P, Shen RF, Carruthers VB. The opportunistic pathogen Toxoplasma gondii deploys a diverse legion of invasion and survival proteins. J Biol Chem. 2005;280:34233–34244. doi: 10.1074/jbc.M504160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 20.Kolodziej PA, Young RA. Epitope tagging and protein surveillance. Methods Enzymol. 194:508–519. doi: 10.1016/0076-6879(91)94038-e. 199. [DOI] [PubMed] [Google Scholar]
- 21.Roos DS, Donald RG, Morrissette NS, Moulton AL. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 1994;45:27–63. doi: 10.1016/s0091-679x(08)61845-2. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch EF, Maniatis T, editors. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Wan KL, Carruthers VB, Sibley LD, Ajioka JW. Molecular characterisation of an expressed sequence tag locus of Toxoplasma gondii encoding the micronemal protein MIC2. Mol Biochem Parasitol. 1997;84:203–214. doi: 10.1016/s0166-6851(96)02796-x. [DOI] [PubMed] [Google Scholar]
- 24.Carruthers VB, Blackman MJ. A new release on life: emerging concepts in proteolysis and parasite invasion. Mol Microbiol. 2005;55:1617–1630. doi: 10.1111/j.1365-2958.2005.04483.x. [DOI] [PubMed] [Google Scholar]
- 25.Donald RG, Roos DS. Gene knock-outs and allelic replacements in Toxoplasma gondii: HXGPRT as a selectable marker for hit-and-run mutagenesis. Mol Biochem Parasitol. 1998;91:295–305. doi: 10.1016/s0166-6851(97)00210-7. [DOI] [PubMed] [Google Scholar]
- 26.Miller SA, Binder EM, Blackman MJ, Carruthers VB, Kim K. A conserved subtilisin-like protein TgSUB1 in microneme organelles of Toxoplasma gondii. J Biol Chem. 2001;276:45341–45348. doi: 10.1074/jbc.M106665200. [DOI] [PubMed] [Google Scholar]
- 27.Harper JM, Zhou XW, Pszenny V, Kafsack BF, Carruthers VB. The novel coccidian micronemal protein MIC11 undergoes proteolytic maturation by sequential cleavage to remove an internal propeptide. Int J Parasitol. 2004;34:1047–1058. doi: 10.1016/j.ijpara.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Song ES, Juliano MA, Juliano L, Hersh LB. Substrate activation of insulin-degrading enzyme (insulysin). A potential target for drug development. J Biol Chem. 2003;278:49789–49794. doi: 10.1074/jbc.M308983200. [DOI] [PubMed] [Google Scholar]
- 29.Huynh MH, Carruthers VB. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot Cell. 2009;8:530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, et al. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science. 2006;314:1776–1780. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]