Abstract
The expression of IgG antibodies in Escherichia coli is of increasing interest for analytical and therapeutic applications. In this work, we describe a comprehensive and systematic approach to the development of a dicistronic expression system for enhanced IgG expression in E. coli encompassing: (i) random mutagenesis and high-throughput screening for the isolation of overexpressing strains using flow cytometry, and (ii) optimization of translation initiation via the screening of libraries of synonymous codons in the 5’ region of the second cistron (heavy chain). The effects of different promoters and co-expression of molecular chaperones on full-length IgG production were also investigated. The optimized system resulted in reliable expression of fully assembled IgG at yields between 1 and 4 mg/liter of shake flask culture for different antibodies.
Keywords: Escherichia coli, strain optimization, IgG expression, chemical mutagenesis, fluorescence-activated cell sorting (FACS), translational initiation region (TIR)
1. Introduction
Monoclonal antibodies occupy a prominent position in the drug market and account for over $40 billion in annual sales. There are currently more than 30 commercially available antibody-based therapeutics, including the anti-cancer drugs Herceptin® and Avastin® and the anti-inflammatory drugs Remicade® and Humira®. Currently, more than 280 antibody-based therapeutics are being evaluated in clinical trials (Strohl & Knight, 2009).
Monoclonal antibodies can be expressed either as full-length IgG molecules or as antibody fragments (scFv, Fab or F(ab)2; (Andersen & Reilly, 2004, Humphreys, 2003)). Antibody fragments lack part of or the entire constant region (domains CH1-CH3 and Cκ,λ) and therefore fold more efficiently and can be produced at high yields in microbial hosts (Andersen & Reilly, 2004, Humphreys, 2003). Full-length antibodies, however, are often the preferred therapeutic antibody format due to the properties mediated by the FC domain, namely, significantly longer circulation half-life in mammals and antibody-dependent cell cytotoxicity and complement-dependent cell cytotoxicity (ADCC and CDC, respectively; (Mariani, 1990, Junghans, 1997)). Because the latter two properties depend on glycosylation and because of their relative complex disulfide bond pattern, full-length antibodies are typically expressed in mammalian cell lines, such as CHO or NSO cells (de la Cruz Edmonds et al., 2006, Huang et al., 2007, Liu et al., 2008). However, the selection of stable cell lines appropriate for production is very time-consuming. Additionally, even though IgG yields in CHO cells can reach or exceed 10 g/L of culture, mammalian cell expression requires a long production cycle which in turn translates into much higher capital investment relative to microbial expression systems that benefit from much faster growth rates (Chartrain & Chu, 2008).
Early efforts to express full-length IgGs in bacteria failed, but more recently scientists at Genentech reported the expression of ~1 g/L of correctly folded IgG antibodies in Escherichia coli-fermented cultures (Simmons et al., 2002, Reilly & Yansura, 2010). Bacterially expressed IgGs are aglycosylated and, therefore, do not display ADCC/CDC normally. However, aglycosylated antibody variants that exhibit unique abilities to engage cytotoxic innate immune cells have recently been engineered (Jung et al., 2010). Antibodies lacking ADCC/CDC are in fact preferable in instances where the therapeutic function is receptor agonism or antagonism. Partly because of this reason and partly because of the lower capital investment and production costs, the bacterial expression of IgGs is attracting increasing interest and at least one E. coli expressed antibody is currently in advanced clinical development (http://www.gene.com/gene/pipeline/status/oncology/metmab/)(Martens et al., 2006). Furthermore, full-length IgGs with tailored antigen-binding characteristics have been isolated from combinatorial libraries expressed in E. coli and screened by flow cytometry (Mazor et al., 2007).
While significant yields of fully assembled IgG can be obtained by high-cell-density fermentation (Jung et al., 2010, Simmons et al., 2002), yields in shake flasks rarely exceed 1 mg of assembled full-length antibodies per liter of culture. Increasing the protein yield in shake flasks is particularly important in antibody discovery programs following combinatorial library screening where many variants have to be purified and examined individually for binding and biological function. Unlike large-scale expression which is more efficient when the heavy and light chains are expressed from different promoters on different plasmids (Simmons et al., 2002), in library screening experiments it is preferable that the heavy and light chains be expressed from a single promoter as a dicistronic operon (Mazor et al., 2007).
In this work, we describe a systematic program to enhance IgG yields from a dicistronic expression system in shake flasks by addressing all aspects relevant to protein synthesis and folding in bacteria, namely: (i) effect of promoter strength and signal peptide sequence for the transcription and membrane translocation, respectively, for both the light and heavy chains; (ii) host strain improvement by chemical mutagenesis and high-throughput screening for enhanced expression; (iii) optimization of translation initiation for the second cistron of the dicistronic operon (heavy chain) via mutagenesis of the 5’ region; and (iv) evaluation of the effects of molecular chaperones on IgG yield. In this manner, we were able to obtain up to 6-fold higher yields of full-length IgG compared to wild-type E. coli cells, and the engineered system to produce ~4 mg/L of properly assembled IgG in shake flask culture.
2. Materials and Methods
2.1 Reagents, Strains and Plasmids
All DNA modification enzymes were purchased from New England Biolabs. The E. coli strain JUDE-1 (DH10B (Invitrogen) harboring the 'F' factor derived from XL1-blue (Stratagene)) was utilized for all experiments unless otherwise mentioned. The construction of the IgG expression vectors pMAZ360-M18.1-IgG, pMAZ360-26.10-IgG and pMAZ360-YMF10-IgG has been described previously (Mazor et al., 2007). pMAZ360-9c8-IgG was generated by swapping the VH and VL of M18.1-IgG in pMAZ360-M18.1-IgG with those of 9c8-IgG by using the restriction sites NcoI/NotI and NheI/HindIII, respectively (Sean Carroll and GG, unpublished results). Molecular chaperones and other folding factors were expressed either from pBAD33-based plasmids (Guzman et al., 1995) or from pAR3-based plasmids (Perez-Perez & Gutierrez, 1995). dsbA, dsbC, fkpA, fkpA-dsbA, and tig were expressed from pBAD-DsbA (laboratory stock), pBAD-DsbC (laboratory stock), pBAD-FkpA (Arredondo et al., 2008), pBAD-FkpA-DsbA (Arredondo et al., 2008), and pBAD-Tig (Levy et al., 2001), respectively. dnaK/dnaJ, groEL/groES, HSPA5 (encoding for BiP), secB, and secY/secE were expressed from pAKJ (Perez-Perez et al., 1995), pAG (Perez-Perez et al., 1995), pAR3-BiP (laboratory stock), pAB (Perez-Perez et al., 1995), and pAA4E (Perez-Perez et al., 1995), respectively. The gene encoding for Skp was amplified from E. coli genomic DNA with gene-specific DNA primers and was cloned into pBAD33 by using the restriction sites XbaI-HindIII. The ppiB gene encoding for mouse cyclophilin B (peptidylprolyl isomerase B) was synthesized by gene assembly PCR using codon-optimized oligonucleotides for E. coli expression designed with the assistance of the software DNAWorks (http://helixweb.nih.gov/dnaworks/). Following assembly, the gene was cloned into the available SacI and XbaI site of pBAD33 fused with the pelB leader sequence to its N terminus.
2.2 IgG overexpression
E. coli cells were grown in modified NU medium (Mori, 1979) (4 g/L KH2PO4, 4 g/L K2HPO4, 7 g/L Na2HPO4, 1.2 g/L NH4Cl, 1.2 g (NH4)2SO4, 4 g/L yeast extract, 0.2 % MgSO4, 0.5 % glycerol, 0.05 % glucose). Single bacterial colonies were used to inoculate liquid NU overnight cultures containing the appropriate antibiotics (100 µg/mL ampicillin with and without 40 µg/mL chloramphenicol). The following day, cultures were diluted 1:50 in 4 ml of fresh NU media and grown at 37 °C to an optical density at 600 nm (OD600) of 0.7 with shaking. At that point, the temperature was decreased to 25 °C and after a temperature equilibration period of 5–20 min, protein expression was induced by the addition of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for approximately 16 h. When molecular chaperones and other folding factors were co-expressed, 0.01% of L-arabinose was added to liquid medium prior to inoculation of the cells from the overnight culture.
2.3 Fluorescent Labeling and fluorescence-activated cell sorting (FACS)
Digoxigenin-dipyrromethene boron difluoride (digoxigenin-BODIPY) was purchased from Invitrogen. For labeling, cells were harvested by centrifugation and resuspended in cold 5X Tris-KCl buffer (250 mM Tris-HCl, pH 7.4, and 750 mM KCl) (Sarkar et al., 2008) and 200 nM digoxigenin-BODIPY followed by 1 h incubation at room temperature with shaking. Flow cytometry and sorting were carried out essentially as described previously (Skretas & Georgiou, 2009). Briefly, the cell fluorescence (530/30 nm) exhibited upon fluorescent antigen binding to IgG was monitored using a Becton-Dickinson FACSAria™ flow cytometer and analyzed with FACSDiva software. For FACS screening, cells were initially gated on a side-scatter (SSC-H) versus forward-scatter (FSC-H) plot. 2×105 clones corresponding to the top 1–3% fluorescent events were isolated, grown on LB agar plates with the appropriate antibiotics, harvested, and then grown in liquid NU media as described above, and subjected to repeated rounds of PECS screening using FACS.
2.4 SDS-PAGE and Western blotting
Cells were lysed by brief sonication cycles over ice and the lysate was clarified by centrifugation (12,000 X g for 20 min) and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Samples were normalized so that each lane contained an amount of total protein corresponding to the same number of cells as judged by OD600 measurements. SDS–PAGE was performed on 4–20 or 12% gels under reducing (2.5% (v/v) 2-mercaptoethanol) and non-reducing conditions and gels were stained with GelCode Blue® (Pierce). For Western blot analyses, proteins were transferred to polyvinylidene fluoride (PVDF) membranes for 16 h at 110 mA or 2 h at 350 mA. Membranes were blocked with 5% non-fat dry milk in phosphate-buffered saline containing 0.1% Tween-20 (PBST) for approximately 1 h at room temperature. Following a triple washing step with PBST, membranes were incubated with a 1:2,000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories) in PBST containing 0.5% non-fan dry milk at room temperature for approximately 1 h. After triple washing with PBST again, the probed proteins were visualized on X-ray film with SuperSignal West Pico chemiluminescent substrate (Pierce). The intensity of the band corresponding to fully assembled IgG was compared with serially diluted IgG1 standard (clinical grade trastuzumab antibody (anti-p185HER2, 4D5; kindly provided by Michael Rosenblum). Densitometry was carried out using a Quantity One™ (Biorad) densitometer.
2.5 Enzyme-linked immunosorbent assay (ELISA)
50 µl/well of 4 µg/ml solutions of digoxigenin-bovine serum albumin (BSA) (Invitrogen) were used to coat 96-well ELISA plates (Corning Incorporated) for 16 h at 4 °C with shaking followed by blocking with 50 µl/well of a 2% nonfat milk solution in PBS (PBSM) and shaking at room temperature for 2 h. Clarified cell lysates were diluted in PBSM so that each sample contained total protein corresponding to an equal number of cells as judged by OD600. 50 µl of protein samples were added to the wells and allowed to bind for 1 h at room temperature followed by triple washing step with PBS + 0.5% Tween-20 (PBST). 50 µl of HRP-conjugated goat anti-human IgG (heavy and light chain) antibodies (Jackson ImmunoResearch Laboratories) diluted 1:4,000 into PBSM were then added and allowed to bind for 1 hour with shaking. After discarding the content of the wells, ELISA plates were visualized by the addition of 50 µl/well of Onestep™ Ultra TMB-ELISA (Thermo Scientific, Waltham, MA) and quenched with the addition of 50 µl/well of 2 N H2SO4.
2.6 Chemical Mutagenesis
Chemical mutagenesis was carried out essentially as described in previous work (Zhan et al., 2004). Briefly, JUDE-1 cells harboring pMAZ360-26.10-IgG were grown in LB medium containing 100 µg/ml of ampicilin to an OD600 of ~0.3. The cells contained in 5 ml of culture were collected by centrifugation at 2,800 × g for 10 min, washed twice with cold citrate buffer (0.1M Sodium Citrate, pH=5.5) and resuspended in 1.9 ml of the same buffer, and treated with 50 µg/ml of N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) (Sigma) at 37°C for 15, 30 and 45 min. After MNNG treatment, the cells were mixed, washed twice with phosphate buffer (0.1 M KH2PO4, pH=7.0) and resuspended in 2 ml of the same buffer.
2.7 Construction of translational initiation region (TIR) libraries
A QuickChange® sire-directed mutagenesis kit (Stratagene) was used to introduce SpeI and StuI restriction sites into the pMAZ360-26.10-IgG vector that flanks the pelB leader sequence of the VL-Ck gene to generate pMAZ360-26.10-TIR. The double stranded DNA libraries of the heavy and light chain genes containing the randomized TIRs were prepared by annealing the forward and reverse oligonucleotides shown in Supplementary Materials and Methods. The theoretical diversity of each library was ~3.7×104. For the construction of the light chain library, pMAZ360-26.10-TIR was digested with SpeI and StuI and ligated with similarly digested light chain dsDNA fragments using T4 ligase. For the construction of the heavy chain library, pMAZ360-26.10-TIR was digested with NdeI and NheI and ligated with similarly digested heavy chain dsDNA fragments using T4 ligase. The generated plasmid libraries were transformed into JUDE-1 competent cells together by electroporation yielding more than 106 clones. The quality of each library was assessed by sequencing 40–60 library members and it was found that all contain distinct nucleic acid sequences without insertions or deletions.
2.8 IgG purification
IgG antibodies were overexpressed in 500 ml cultures as described above and were purified from the growth medium supernatant and the soluble fraction of cell lysates using protein A sepharose (Thermo scientific) affinity chromatography.
3. Results
3.1 Development of a FACS-based assay for monitoring IgG expression at the single-cell level
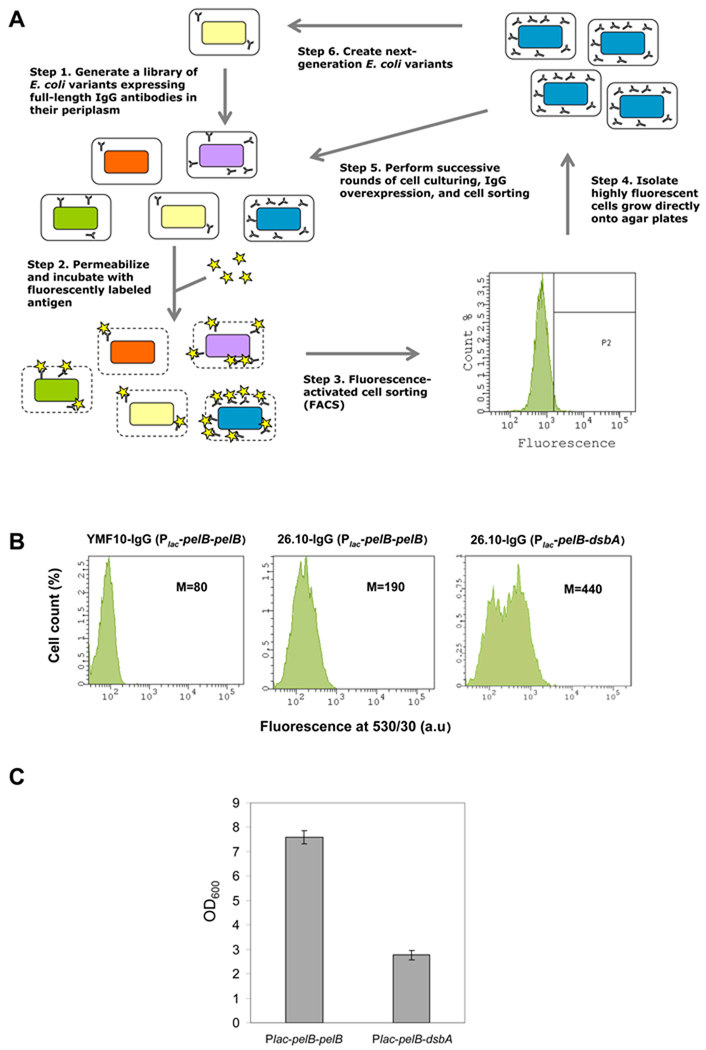
We developed an assay for monitoring antibody expression at the single-cell level via flow cytometry by modifying a technique developed earlier by our group termed periplasmic expression followed by cytometric sorting (PECS) (Chen et al., 2001). Briefly, E. coli cells expressing full-length IgGs in the periplasm are incubated in a high-osmolarity buffer that renders their outer membrane permeable to antigens (Fig. 1A). Addition of an excess of a fluorescently labeled small-molecule antigen allows the antigen to equilibrate into the periplasmic space and bind to the properly assembled IgG molecules. Thus, cell fluorescence is proportional to the number of functional, antigen-binding molecules in the periplasm. Clones containing mutations that increase IgG expression, display higher fluorescence and can be isolated by FACS. An important characteristic of PECS is that despite outer membrane permeabilization, the bacterial cells retain their viability and sorted cells can thus be immediately subjected to additional rounds of screening (Figure 1A).
Figure 1. Single-cell Fluorescent Detection of IgG Expression.
(A) General scheme of E. coli mutagenesis and isolation of clones displaying increased IgG expression using periplasmic expression with cytometric screening (PECS). (B) Fluorescence histograms of E. coli JUDE-1 cells over-expressing either 26.10 IgG or YMF10 IgG (anti-PA antibody (Mazor et al., 2007)); negative control) with different combinations of leader peptides for heavy and light chain periplasmic export. Statistics correspond to a population of 10,000 cells. M: mean fluorescence intensity. (C) Growth of E. coli JUDE-1 cells over-expressing 26.10 IgG antibody with different leader peptides for heavy and light chain periplasmic export. OD600: optical density at 600 nm, Plac-pelB-pelB: lac promoter with pelB leader peptide for light and heavy chain periplasmic export, Plac-pelB-dsbA: lac promoter with pelB leader peptide for periplasmic export of light chain and dsbA leader peptide for periplasmic export of heavy chain.
Full-length and properly assembled IgGs have been expressed in E. coli from a dicistronic operon consisting of the light (VL-CL) and heavy (VH-CH1-CH2-CH3) chain genes fused with leader peptides that target both chains for periplasmic export under the control of an inducible promoter (Mazor et al., 2007). For expression optimization studies, we used as a model the IgG1 variant of the 26.10 scFv antibody which binds to the heart condition drug digoxin and its aglycone digoxigenin with high affinity (Chen et al., 1999, Mazor et al., 2007). This antibody was selected as a model system because both digoxin and digoxigenin are small-molecule antigens appropriate for use as ligands in PECS (molecular weights 781 and 391 Da, respectively).
Three different E. coli strains were evaluated for IgG expression: JUDE-1 (DH10B (Invitrogen) harboring the 'F' factor derived from XL1-blue (Stratagene)), MC4100A (Santini et al., 2001), and KS303 (MC1000 lpp 5508; (Strauch & Beckwith, 1988)). The latter strain contains an lpp deletion and therefore lacks the major outer membrane lipoprotein, a defect that confers a significant increase in the permeability of the outer membrane without eliciting cell lysis (Ni et al., 2007). Each of these strains was transformed with the vector pMAZ360-26.10-IgG, which expresses the 26.10 IgG heavy and light chains as fusions with a pelB export leader under the control of the lac promoter as part of a dicistronic operon (Mazor et al., 2007). Cells were grown at 25 °C, IgG expression was induced with IPTG, cells were labeled with 200 nM digoxigenin-BODIPY in 5X Tris-KCl (a buffer that results in optimized outer membrane permeabilization (Sarkar et al., 2008)), and bacterial fluorescence was analyzed using flow cytometry. Highest levels of fluorescence compared to a negative control were observed in JUDE-1 (Figure 1B and data not shown) and therefore this strain was selected for further studies.
The effect of different promoters (lac, trc, tet, araBAD, T7) and leader peptides (the Sec pathway-dependent leader sspelB (Manting & Driessen, 2000) or the signal recognition particle (SRP) pathway-dependent signal sequence ssdsbA (Schierle et al., 2003)) on IgG yield were evaluated by flow cytometry and Western blotting (Figure 1B and data not shown). These experiments showed that a lac promoter with sspelB and ssdsbA export of light and heavy chains respectively (lac-pelB-dsbA plasmid), gave the highest level of assembled IgG (Figure 1B). However, this combination of leader peptides and promoters had a significant effect on cell growth and resulted in low final cell yields (Figure 1C). These effects were alleviated when the ssdsbA was replaced with sspelB (Figure 1C). Therefore, we elected to use sspelB for the export of both the heavy and the light chains.
3.2 Random mutagenesis and screening for chromosomal lesions that result in enhanced IgG yield
JUDE-1 cells carrying the pMAZ360-26.10-IgG vector were subjected to random mutagenesis using the chemical mutagen N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) as described in section 2.6. The resulting mutants were pooled to form the pre-sort library, propagated in liquid NU medium, and IgG expression was induced by the addition of IPTG as described in section 2.2. After labeling with 200 nM digoxingenin-BODIPY, ~107 cells were subjected to PECS screening using FACS. The cells exhibiting the top 1–3% fluorescence were collected in liquid medium, plated, propagated in liquid medium again, and subjected to additional rounds of PECS. The percent viability, (number of colony forming units divided by the number of sorted events measured FACS) of the chemically mutated cells in the utilized PECS buffer (5x Tris-KCl) was around 10%, whereas the parental strain JUDE-1 exhibited a viability of 30–40% under the same conditions. An increase in the average BODIPY fluorescence intensity was observed in every round until the fluorescent signal became saturated after the fifth round. At that point the mean fluorescence of the population was approximately 2.5-fold higher than the BODIPY fluorescence exhibited by the initial library (Figure 2A).
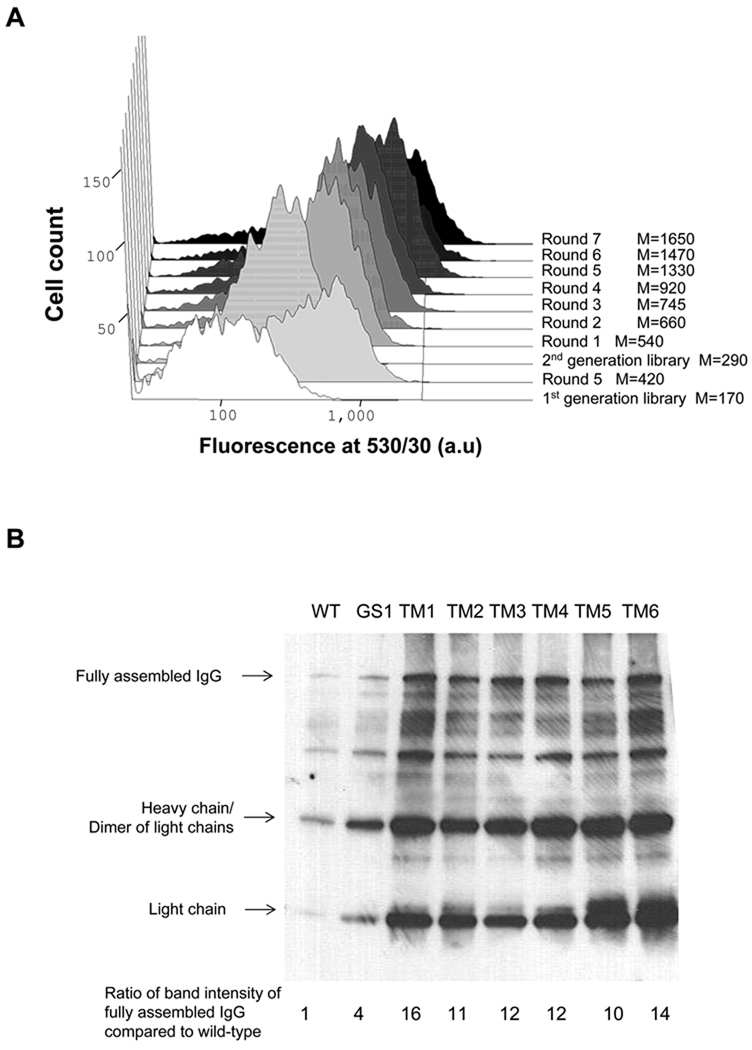
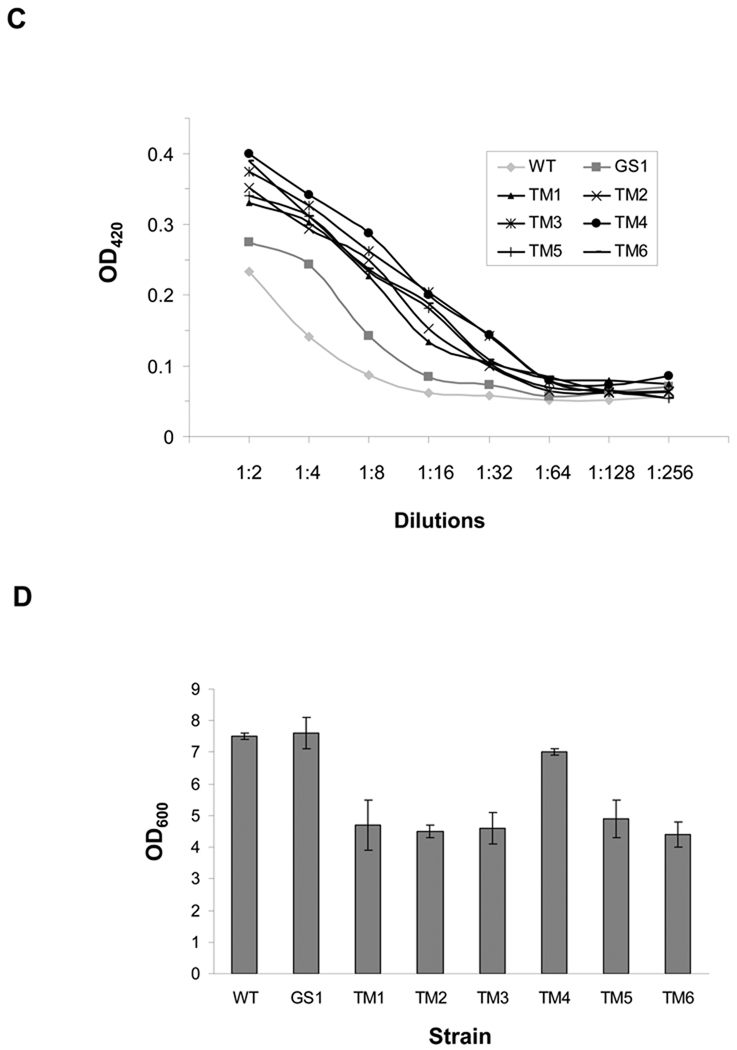
Figure 2. Screening of Chemically Mutated E. coli for IgG Expression.
(A) FACS screening of mutated JUDE-1 cells expressing 26.10 IgG labeled with digoxigenin-BODIPY. M: mean fluorescence intensity. (B) Western blotting of 26.10 IgG expressed in isolated clones after two rounds of mutagenesis and screening. The increase in the yield of fully assembled IgG relative to wild-type (WT) JUDE-1 (lower panel) was determined by scanning densitometry. (C) ELISA assays of IgG level in cell lysates. OD420: optical density at 420 nm. (D) Growth of isolated mutant clones after 16 h of 26.10-IgG overexpression. Experiments were carried out in triplicate and the error bars correspond to one standard deviation from the mean values. In (B) and (C) protein samples were normalized by OD600 so as to contain total protein that corresponds to the same number of bacterial cells. OD600: optical density at 600 nm.
Three hundred single clones were selected at random from the fifth round population for further analysis. 26.10 IgG expression was evaluated first by ELISA of whole cell lysates using immobilized digoxigenin and then the seven clones with the highest ELISA signals, termed GS1–GS7, were further analyzed for expression of fully assembled IgG by Western blotting. All seven clones were found to yield enhanced production of properly assembled antibody compared to the parental E. coli strain JUDE-1 (Figure 2B and data not shown). As expected, curing of the pMAZ360-26.10-IgG plasmid resulted in loss of the ELISA signals, while retransformation with fresh pMAZ360-26.10-IgG vector restored the ELISA signal in 5/7 clones to the level of the original isolates (Figure 2C and data not shown).
A second round of chemical mutagenesis and screening was performed by pooling GS1–GS5 clones carrying the pMAZ360-26.10-IgG plasmid, followed by exposure to MNNG. After seven rounds of FACS screening as above, the fluorescent signal saturated at a level nearly 10-fold higher than that of the parental JUDE-1 cells (Figure 2A). Seven clones, termed TM1–TM7, from the seventh round of sorting were randomly selected for further study and shown to exhibit approximately 5-fold higher cell fluorescence, which was dependent on the presence of the pMAZ360-26.10-IgG plasmid. Among TM1–TM7, six clones regained fluorescence upon retransformation with fresh vector (TM1–TM6). Western blot analysis revealed that TM1–TM6 produce 10–20 times more fully assembled IgG compared to the parental JUDE-1 cells (Figure 2B) and this was accompanied by markedly higher ELISA signals (Figure 2C). For comparison, bacterial clones isolated after the first round of mutagenesis, resulted in approximately 3-fold enhancement in the yield of 26.10 IgG (Figure 2B). However, with the exception of TM4, the other E. coli mutant strains isolated after the second round of mutagenesis grew more slowly and reached a lower final cell culture density after overnight growth compared to wild-type JUDE-1 (Figure 2D). For this reason, TM4 was selected for further studies.
It is interesting to note that the increased yield of fully assembled IgG in the TM mutants was accompanied by an appreciable increase in the production of the heavy and light chains (Figure 2B) indicating that the selected chromosomal lesions in these mutants affected protein synthesis and/or export and stability in the periplasm.
3.3 Isolation of heavy chain TIR variants
The IgH, IgL dicistronic operon in pMAZ360-26.10-IgG results in accumulation of an excess of light chain (first cistron gene product) over heavy chain (Figure 2B and data not shown). Swapping the order of the light and heavy chain ORFs of the IgG operon of the pMAZ360 vector did not improve IgG assembly (data not shown). We reasoned that a more balanced ratio of heavy:light chain synthesis may facilitate proper assembly. In E. coli, the protein synthesis rate is often critically dependent on the rate of translation initiation which in turn is influenced by the propensity of the 5’ end of the mRNA around the Shine-Dalgarno sequence to form stable secondary structure that occludes ribosome binding. Even single nucleotide changes in the translation initiation region, which extends from the 5’ end of the RBS to about 20 nucleotides downstream of the start codon, can affect the efficiency of translational initiation (Kudla et al., 2009, Simmons & Yansura, 1996). Several previous studies have demonstrated that the TIR region can be engineered to enhance heterologous protein expression and secretion (Simmons & Yansura, 1996, Bandmann & Nygren, 2007, Ahn et al., 2008). Similarly, optimized TIR sequences have been shown to be important for the expression of full-length IgGs in a system where the IgH and IgL were expressed from monocistronic operons (Simmons et al., 2002).
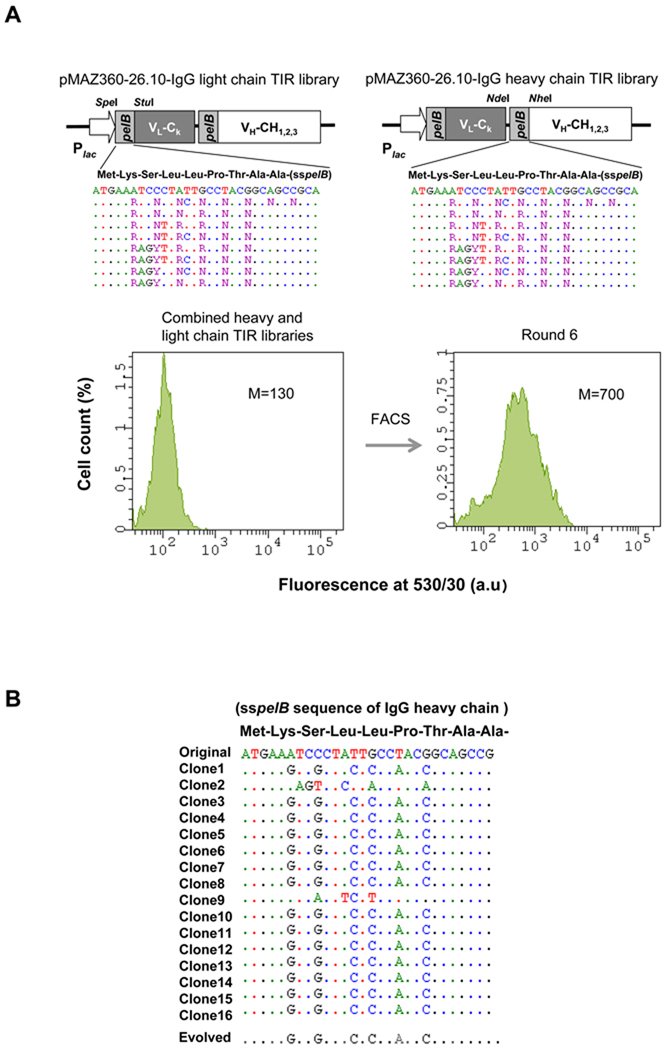
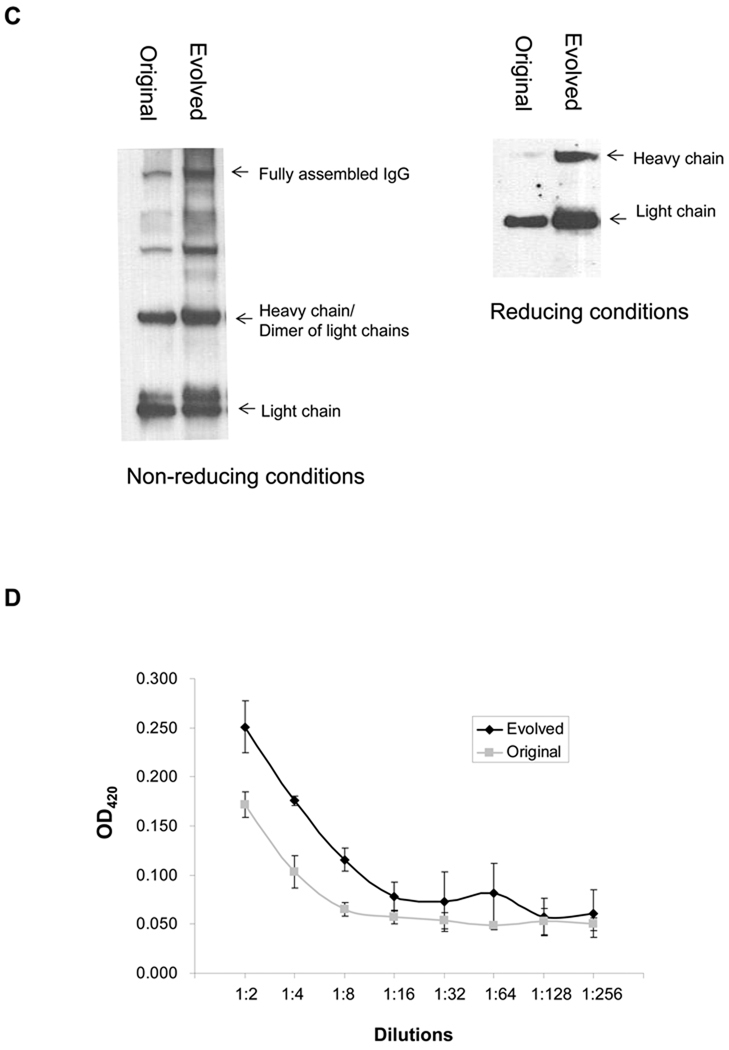
In order to select for a TIR sequence that confers a more favorable ratio of IgH:IgL, degenerate oligonucleotides were used to generate two libraries of pMAZ360-26.10-IgG plasmids carrying silent mutations in the TIR of both the heavy chain or light chain genes. The theoretical diversity for each of the heavy and light chain libraries was 3.7 × 104 (Figure 3A). First, we attempted to transform the constructed library into the evolved TM4 strain. The acquired mutations in TM4, however, had a dramatically decreasing effect on the transformation efficiency of the strain. Thus, we focused on wild-type JUDE-1 cells. Approximately 106 colonies of E. coli JUDE-1 cells transformed with the TIR libraries in pMAZ360-26.10-IgG were pooled and subjected to successive rounds of PECS screening for enhanced binding to digoxigenin-BODIPY using FACS. The fluorescence signal was saturated after the sixth round of sorting (Figure 3A), at which point 90 colonies were picked randomly, grown in microtiter-well plates and the expression of functional antibody protein in whole-cell lysates was determined by ELISA. Interestingly, the TIR region in 15 clones showing the highest ELISA signals were found to contain mutations only in the heavy chain TIR. 13/15 clones had a common nucleotide sequence (Figure 3B). Western blotting and ELISA analysis of IgG expressed in JUDE-1 cells using the consensus TIR sequence revealed a 2- to 3-fold improvement in IgG assembly (Figure 3C) leading to a proportional increase in antigen binding as detected by ELISA (Figure 3D).
Figure 3. Screening of a Library for Optimized Translational Initiation Region (TIR) Sequences.
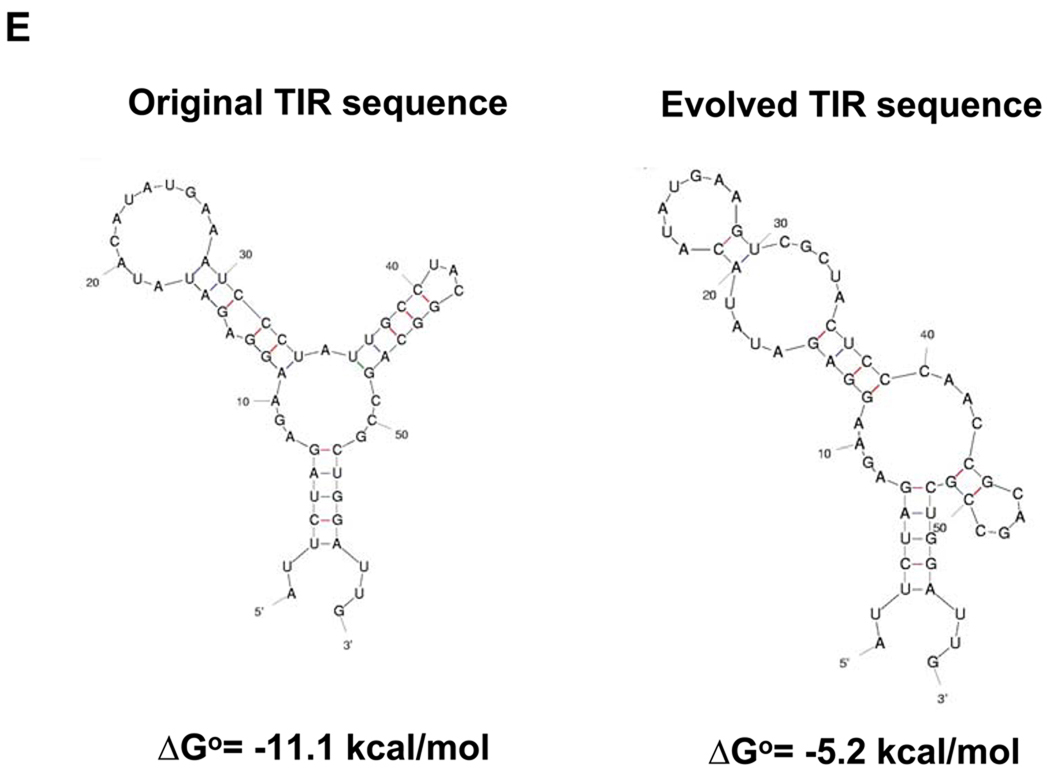
(A) Construction of TIR libraries for light and heavy chain genes and screening by FACS. M: mean fluorescence intensity, sspelB: pelB signaling sequence (B) Nucleotide sequences of the heavy chain TIRs of the isolated clones. (C) Western blotting of JUDE-1 cells carrying the pMAZ360-26.10-IgG vector either with the original or the selected heavy chain TIR sequences under non-reducing (left) and reducing (right) conditions. All lanes were loaded with total protein corresponding to the same number of cells as judged by OD600. (D) ELISA determination of antigen-binding activity in cell lysates from mutant clones. Experiments were carried out in triplicate and the error bars correspond to one standard deviation from the mean values. (E) Prediction of mRNA secondary structure for the original and the evolved heavy chain TIR sequences using the Mfold software (Zuker, 2003). ΔGo: Gibbs free energy of unfolding.
The thermodynamic stability of the RNA encompassing the TIR sequence, i.e. a 60 nucleotide-long region that included the ribosome-binding site and 30 nucleotides downstream of the ATG initiation codon was calculated using two different algorithms, Mfold and RBS calculator. Mfold (Zuker, 2003), predicts mRNA secondary structure, whereas the RBS calculator (Salis et al., 2009) predicts relative translational initiation rates by using a free energy model to calculate the total change in Gibbs free energy upon binding of the 30S ribosome complex to a ribosome binding site on the mRNA. Consistent with the experimental results, Mfold predicted that the selected mutant sequences had a lower ΔG of folding relative to the parental gene (Figure 3E). Similarly, the RBS calculator program predicted that the selected TIR sequence should result in 10-fold higher translational initiation rate relative to that of the TIR sequence contained in the original vector (35370 vs. 3730 of arbitrary units, respectively).
3.4 Effect of Molecular Chaperone and Cysteine Oxidoreductase Co-expression
Co-expression of molecular chaperones has been shown in a number of studies to assist the production and folding of several recombinant proteins, including antibody fragments (Levy et al., 2001, Lin et al., 2008, Mavrangelos et al., 2001, Zhang et al., 2002, Jurado et al., 2002). The yield of the 26.10 IgG expressed in the TM4 mutant strain in cells with and without co-expression of various chaperones and enzymes known to influence heterologous protein expression, was evaluated. Specifically, the effect of co-expression of the periplasmic chaperones Skp and FkpA, the cytoplasmic molecular chaperones/co-chaperones DnaK/DnaJ, GroEL/GroES and the peptidyl-prolyl cis/trans isomerase trigger factor, the human endoplasmic reticulum molecular chaperone BiP, the mouse ER peptidyl-prolyl cis-trans isomerase Cyclophilin B (Feige et al., 2009), the periplasmic disulfide oxidoreductases and isomerases DsbA and DsbC (Zhang et al., 2002), and an engineered FkpA-DsbA chimera (Arredondo et al., 2008) was investigated. Furthermore, we tested the effect of co-expressing the components of the Sec secretory pathway SecB and SecE/SecY which mediate export into the periplasm. We found that co-expression of DsbA, DsbC, BiP, and SecE/SecY individually resulted in 2- to 3-fold increased yield of fully assembled 26.10 IgG (Table 1). The most significant effect was observed in cells co-expressing DsbA, which increased the yield of assembled, antigen-binding antibody by more than 3-fold (Table 1). This result is consistent with very recent findings that DsbA overexpression enhances the yield of properly folded IgG in the periplasmic space of E. coli cells (Reilly & Yansura, 2010).
Table 1.
Effect of co-expressing molecular chaperones and other folding factors on the expression and of fully assembled 26.10-IgG in TM4 cells.
| Co-expressed folding factor |
Expression of full-length 26.10-IgG (in arbitrary units) |
|---|---|
| No factor | 100 |
| Trigger factor | 57 ± 5 |
| DnaK/DnaJ | 136 ± 21 |
| GroEL/GroES | 144 ± 12 |
| Skp | 120 ± 14 |
| BiP | 198 ± 10 |
| SecB | 68 ± 7 |
| SecY/SecE | 206 ± 3 |
| DsbA | 319 ± 27 |
| DsbC | 236 ± 13 |
| Cyclophilin B | 73 ± 2 |
| FkpA | 73 ± 2 |
| FkpA-DsbC | 83 ± 5 |
3.5 Optimized expression of various IgG antibodies
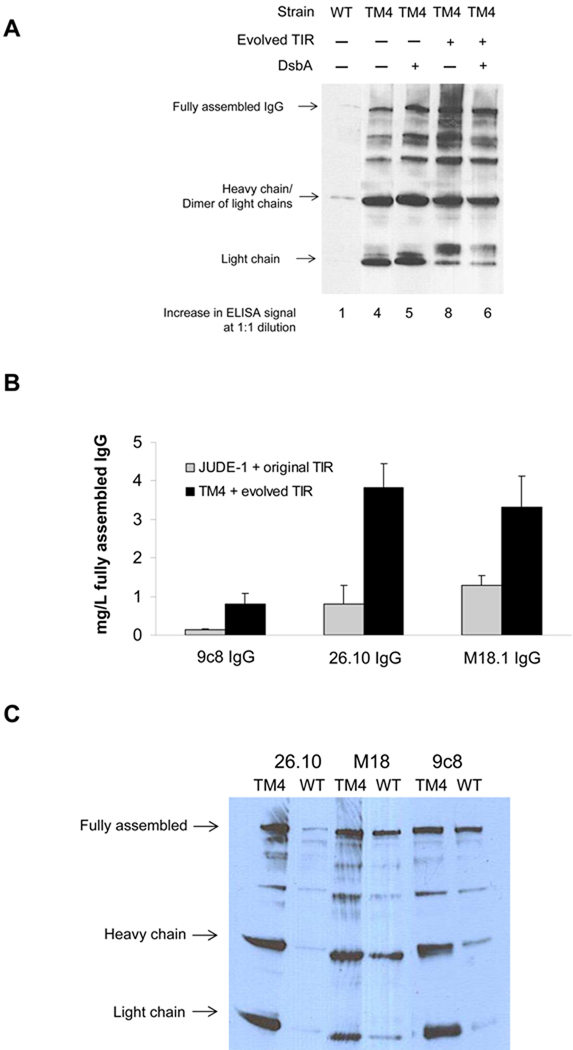
The 26.10 IgG was expressed in the TM4 mutant strain carrying the pMAZ360-26.10-IgG vector with and without the selected, optimized IgH TIR sequence and in the presence or absence of DsbA co-expression (Figure 4A). Unfortunately, the effects of the optimized TIR sequence and DsbA co-expression in the TM4 mutant were not additive (Figure 4A). Thus, for subsequent studies we elected to employ TM4 with the engineered TIR to avoid the need for having a second plasmid for DsbA expression.
Figure 4. Yield of Various Fully Assembled IgGs in Shake Flask Cultures.
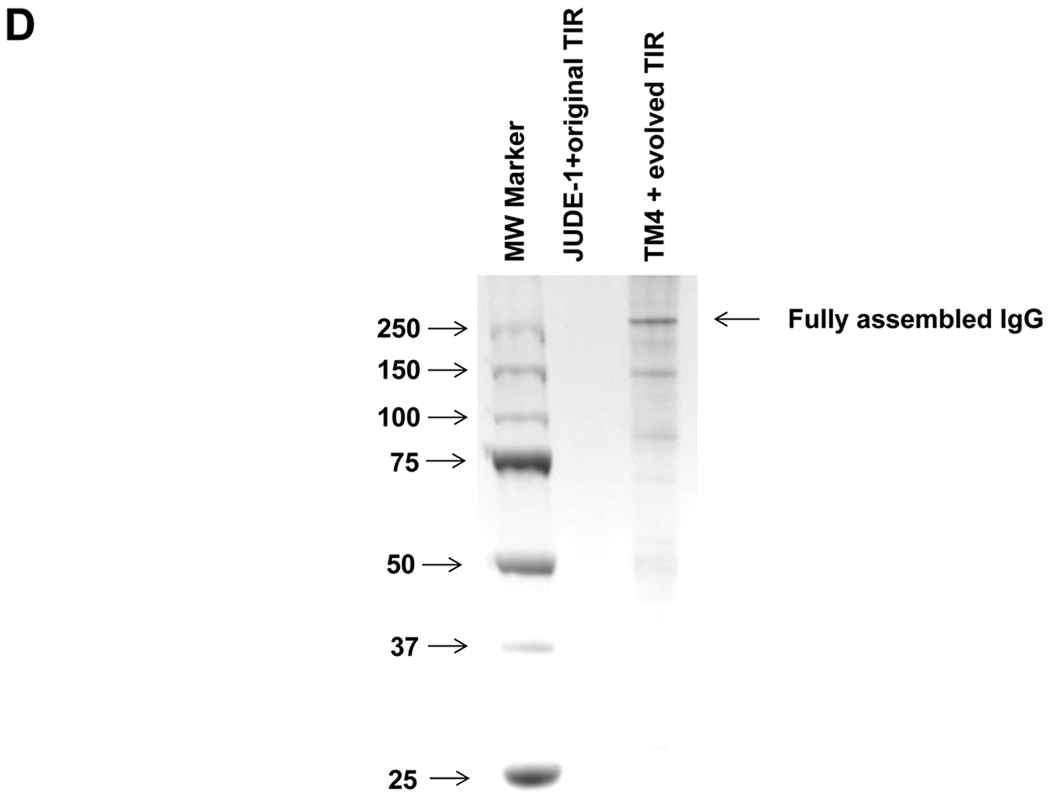
(A) Yield of assembled 26.10 IgG in different combinations of strain, heavy chain TIR sequence, and DsbA co-expression determined by Western blotting and ELISA (bottom). WT: E. coli JUDE-1 cells. (B) Production of full-length antibody for different IgG variants expressed either in JUDE-1 cells carrying the original pMAZ360 vector or in TM4 carrying the pMAZ360 vector containing the engineered heavy chain TIR sequence. Experiments were carried out in triplicate and the error bars correspond to one standard deviation from the mean values. (C) Western blot analysis of the production of full-length antibody for different IgG variants expressed either in JUDE-1 or TM4 cells carrying pMAZ360 vector with the original heavy chain TIR sequence. All lanes were loaded with total protein corresponding to the same number of cells as judged by OD600. (D) SDS-PAGE analysis of 26.10 IgG following protein A affinity chromatography of cell lysates from either JUDE-1 cells carrying the pMAZ360-26.10-IgG vector with the original heavy chain TIR sequence or TM4 cells carrying the pMAZ360-26.10-IgG vector with the engineered heavy chain TIR sequence. MW: molecular mass in kDa.
We compared the yield of fully assembled IgG in E. coli TM4 from plasmids expressing heavy and light chains as a dicistronic operon transcribed from Plac and containing the engineered TIR, with that obtained using the parental strain JUDE-1 and unmodified TIR. Three different IgG antibodies were used: the anti-digoxin 26.10 antibody, the 9c8 nanomolar affinity anti-complement factor C1S antibody (Sai Reddy and GG, unpublished results), and M18.1, a humanized antibody that binds to the Bacillus anthracis Protective Antigen (PA) with high affinity; (Mazor et al., 2007). All three antibodies are human IgG1 variants with a C kappa light chain and variable regions that originated from different murein germ lines. IgG overexpression was carried out in 100 mL shake flask cultures. The accumulation of fully assembled IgG in cell lysates and in the extracellular fluid was determined by Western blotting and the yield was estimated by densitometric analysis of the band intensity and comparison to serial dilutions of a commercially available IgG1 (Herceptin®) as standard (Figure 4B). As observed also in previous work where full-length IgG antibodies expressed using a dicistronic system had been purified to near homogeneity (Mazor et al., 2007), wild-type E. coli cells accumulated 0.1–1.3 mg of fully assembled IgGs per liter of shake flask culture. Use of the optimized expression system, however, resulted in a 6-fold increase for 9c8-IgG, 4-fold increase in the yield of 26.10 IgG and a 2.5-fold increase for M18.1 IgG, yielding up to 4 mg/L of full-length antibody. This generally observed expression-enhancing effect occurs partly due to the beneficial mutations acquired in TM4 and partly due to the use of the evolved TIR, as the expression of different IgG antibodies in TM4 cells but without the use of the optimized TIR sequence resulted in smaller increases in the production of fully assembled IgG (Figure 4C). Highly purified 26.10-IgG antibody expressed with the the optimized expression system could be obtained from cell lysates by protein A affinity chromatography (Figure 4D).
4. Discussion
In eukaryotic cells, antibody biogenesis is critically dependent on interactions of the newly secreted polypeptides in the ER with a network of molecular chaperones and with protein disulfide bond isomerases (Melnick & Argon, 1995, Vanhove et al., 2001, Elkabetz et al., 2005). Recapitulating IgG assembly in vitro has been challenging. Evidence from biophysical studies suggests that the rate limiting step in the assembly of the FAB domain of IgGs is the folding of the CH chain which is normally disordered and folds upon association with the CL (Feige et al., 2009). Very recently, protocols for the efficient refolding of IgG from E. coli inclusion bodies have been developed (Hakim & Benhar, 2009). However, the expression of soluble and correctly assembled IgG antibodies is usually preferable for routine studies in antibody discovery and for large-scale preparative purposes. Achieving high-level IgG biogenesis in bacteria which lack the mammalian molecular chaperone network requires a fine balance between protein synthesis, secretion and folding. The formation of disulfide bonds within each of the immunoglobulin domains in IgG and in the hinge region joining the two heavy chains are essential for the assembly of the native tertiary structure. Although a variety of engineered E. coli strains that enable disulfide bond formation in the cytoplasm are now available (Faulkner et al., 2008), expression of correctly assembled full-length IgG in bacteria having an oxidizing cytoplasm has not been demonstrated. Therefore, full-length antibody production in bacteria requires export into the oxidative environment of the periplasmic space, as well as slow growth, slow protein synthesis rates and balanced ratio of heavy/light chain production that favor proper chain association (Levy et al., 2001, Mazor et al., 2007, Simmons et al., 2002).
In this work, we report the systematic optimization of IgG antibody expression in bacteria from a dicistronic operon by examining transcription, translation, in vivo protein folding and in parallel with genetic approaches. The light and heavy chains were placed in a dicistronic operon transcribed from an inducible promoter, because: (i) a dicistronic operon is desirable for the recovery of antibody DNA during the screening of IgGs from libraries (Mazor et al. 2007, 2010); (ii) monocistronic expression of heavy and light chain genes fused the pelB signal peptide and transcribed from the lac promoter resulted in lower IgG yields relative to the dicistronic system used here (data not shown). It should be noted, however, that a monocistronic expression system that capitalized on a proprietary PhoA promoter system and the StII signal peptide has been used for preparative production (Simmons et al., 2002). The dicistronic operon for the expression of the heavy and light chains was cloned into the previously described pMAZ360 vector (Mazor et al., 2007), but it could alternatively be also incorporated into the E. coli chromosome. A modification of a high-throughput technique (PECS) developed earlier for the isolation of antibodies with desired antigen specificity (Chen et al., 2001), was employed for the monitoring of antibody expression in E. coli at the single-cell level and for the screening of mutant libraries. Two rounds of chemical mutagenesis, each followed by several rounds of FACS sorting, led to the isolation of mutant bacteria that displayed a marked increase in the production of fully assembled IgG, as monitored both by ELISA and Western blotting. Mutant strains were shown to accumulate higher levels of heavy and light polypeptides indicating that the chromosomal lesions affect protein synthesis in a manner that favorably affects the formation of fully assembled antibodies. It should be noted that increased protein synthesis does not necessarily translate into higher IgG yields, as evident by the fact that expression from stronger promoters (T7, trc) had a detrimental effect on the IgG yield (data not shown). Consequently, in the mutant strain TM4, mutations in the chromosome must have had a subtle effect on the rate of transcription or translation that in turn was favorable for protein folding in the periplasm. However, we can not rule out the possibility that the acquired mutations affect the degradation or translocation of the antibody heavy and light chain in a manner that favors their accumulation. Because this strain was obtained following two rounds of chemical mutagenesis we anticipate that it will contain multiple mutations whose individual effects on protein expression will require extensive additional analyses. The genetic lesions that result in enhanced full-length IgG production in TM4 can be identified by standard mutation mapping techniques (Zhan et al., 2004) coupled with the flow cytometric screening assay described here, or by whole genome sequencing (MacLean et al., 2009, Srivatsan et al., 2008). Combinatorial library screening was also used to isolate optimized TIR sequences that enhanced the translation of the second cistron (heavy chain) and thus resulted in a more favorable heavy:light chain ratio in the periplasm. The requirement for a balanced expression of heavy and light chains for the production of correctly assembled antibodies or FAB fragments has also been observed in earlier studies (Humphreys, 2003, Simmons et al., 2002). Interestingly, combinatorial screening yielded TIR sequences that were predicted to display less stable RNA secondary structure and enhance translation initiation. This genetic verification of model predictions suggests that computational tools, namely Mfold, and more importantly, the RBS calculator are reliable tools for improving translation. To our knowledge, this work is the first attempt to engineer full-length IgG expression by generating E. coli mutant libraries and employing high-throughput screening to isolate the desired overexpression strains.
Finally, as part of this study we also examined the effects of co-expression of several molecular chaperones and enzymes that are known to assist protein folding. It was hypothesized that co-expression of cytoplasmic chaperones may assist in the secretion of the heavy and light chains into the periplasm by preventing the cytoplasmic aggregation of the pre-protein, containing the signal peptide (Berges et al., 1996). However, no significant effect on IgG yield was observed in this case. At the same time, co-expression of the translocon components SecY/E resulted in a reproducible 2-fold enhancement in the amount of fully assembled IgG, suggesting that the capacity of the secretory apparatus may be limiting. Similar to these results, Perez-Perez et al. in earlier studies showed that co-expression of SecY/E enhanced the secretion of human interleukin-6 in E. coli by 1.2 to 10.8 fold (Perez-Perez et al., 1996, Perez-Perez et al., 1994). A similar enhancement in the production of full-length IgG was observed upon overexpression of the eukaryotic chaperone BiP. BiP has been found previously to promote antibody folding together with protein disulfide isomerase (PDI) in vitro (Mayer et al., 2000). In a cell-free IgG production system utilizing E. coli lysate, however, BiP was found ineffective in increasing the yields of fully assembled IgG (Frey et al., 2008). A more significant increase in IgG yield was obtained by co-expressing the periplasmic oxidoreductase DsbA or the disulfide bond isomerase DsbC. The beneficial effect of co-expression of cysteine oxidoreductases on the folding of multi-disulfide-bond proteins including FAB antibody fragments is well established (Georgiou & Segatori, 2005, Levy et al., 2001). Consistent with these observations, full-length IgG could be produced with an in vitro transcription-translation system containing E. coli lysate only in the presence of a disulfide bond isomerase/oxidoreductase, PDI or DsbC, while the redox potential of the bacterial lysate was found to be critical for the yields of intact IgG antibody (Frey et al., 2008).
While the co-expression of molecular chaperones and enzymes that assist folding was found to enhance IgG yields in TM4, the introduction of the engineered TIR did not show a further synergistic effect. A likely explanation for this result is that the simultaneous expression of three secreted polypeptides, i.e. the heavy and light chains as well as the chaperone/foldase saturates the export machinery. However other explanations, such as a decrease in the level of transcription from the araBAD promoter in TM4, for example, cannot be ruled out. Regardless of the mechanism, this finding underscores the need to balance a diverse array of physiological processes in the course of engineering cells for enhanced heterologous protein expression.
In summary, the above studies: (i) highlight the utility of high-throughput screening for strain and vector optimization for protein expression; (ii) demonstrate a systematic approach for addressing various biological processes that can impact transcription, protein synthesis, secretion and folding, and importantly (iii) led to the development of an expression system that resulted in reproducibly 3- to 6-fold higher expression of fully assembled IgG for three antibodies comprising different germline V genes and complementarity determining regions (CDRs). The engineered bacterial expression system is thus likely to be useful for the rapid expression of diverse antibodies generated in antibody discovery programs. Such evolved systems could be analyzed and optimized further, e.g. by metabolic analysis and engineering (Boghigian et al., 2010), to generate specialized antibody-production strains. Finally, we note that the systematic approach applied to the optimization of antibody expression described here could serve as a model for enhancing the expression of other secreted heterologous proteins in bacteria.
Supplementary Material
Acknowledgements
We would like to thank Yariv Mazor, Thomas J. Van Blarcom, Xin Ge and Sang Teak Jung for useful discussions and William Kelton for critical reading and comments on the manuscript. We also thank Michael Rosenblum for kindly providing clinical grade Trastuzumab (Herceptin®) antibody. This work was supported by the National Institute of Health grant GM 55090 and by the Clayton Foundation.
Abbreviations
- IgG
immunoglobulin G
- TIR
translational initiation region
- PECS
periplasmic expression followed by cytometric sorting
- FACS
fluorescence-activated cell sorting
- MNNG
N-methyl-N'-nitro-N-nitrosoguanidine
- IPTG
isopropyl-β-D-thiogalactopyranoside
- PA
Bacillus anthracis Protective Antigen
- ELISA
enzyme-linked immunosorbent assay
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- BODIPY
dipyrromethene boron difluoride
- BSA
bovine serum albumin
- PVDF
polyvinylidene fluoride
- SRP
signal recognition particle
- OD600
optical density at 600 nm
- LB
Luria-Bertani
- PBS
phosphate-buffered saline
- HRP
horseradish peroxidase
- ORF
open reading frame
- PDI
protein disulfide isomerase
- CDR
complementarity determining regions
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn JH, Keum JW, Kim DM. High-throughput, combinatorial engineering of initial codons for tunable expression of recombinant proteins. J Proteome Res. 2008;7:2107–2113. doi: 10.1021/pr700856s. [DOI] [PubMed] [Google Scholar]
- Andersen DC, Reilly DE. Production technologies for monoclonal antibodies and their fragments. Curr Opin Biotechnol. 2004;15:456–462. doi: 10.1016/j.copbio.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Arredondo S, Segatori L, Gilbert HF, Georgiou G. De novo design and evolution of artificial disulfide isomerase enzymes analogous to the bacterial DsbC. J Biol Chem. 2008;283:31469–31476. doi: 10.1074/jbc.M803346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandmann N, Nygren PA. Combinatorial expression vector engineering for tuning of recombinant protein production in Escherichia coli. Nucleic Acids Res. 2007;35:e32. doi: 10.1093/nar/gkl1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges H, Joseph-Liauzun E, Fayet O. Combined effects of the signal sequence and the major chaperone proteins on the export of human cytokines in Escherichia coli. Appl Environ Microbiol. 1996;62:55–60. doi: 10.1128/aem.62.1.55-60.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boghigian BA, Seth G, Kiss R, Pfeifer BA. Metabolic flux analysis and pharmaceutical production. Metabolic Engineering. 2010;12:81–95. doi: 10.1016/j.ymben.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Chartrain M, Chu L. Development and production of commercial therapeutic monoclonal antibodies in Mammalian cell expression systems: an overview of the current upstream technologies. Curr Pharm Biotechnol. 2008;9:447–467. doi: 10.2174/138920108786786367. [DOI] [PubMed] [Google Scholar]
- Chen G, Dubrawsky I, Mendez P, Georgiou G, Iverson BL. In vitro scanning saturation mutagenesis of all the specificity determining residues in an antibody binding site. Protein Eng. 1999;12:349–356. doi: 10.1093/protein/12.4.349. [DOI] [PubMed] [Google Scholar]
- Chen G, Hayhurst A, Thomas JG, Harvey BR, Iverson BL, Georgiou G. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS) Nat Biotechnol. 2001;19:537–542. doi: 10.1038/89281. [DOI] [PubMed] [Google Scholar]
- de la Cruz Edmonds MC, Tellers M, Chan C, Salmon P, Robinson DK, Markusen J. Development of transfection and high-producer screening protocols for the CHOK1SV cell system. Mol Biotechnol. 2006;34:179–190. doi: 10.1385/mb:34:2:179. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y, Argon Y, Bar-Nun S. Cysteines in CH1 underlie retention of unassembled Ig heavy chains. J Biol Chem. 2005;280:14402–14412. doi: 10.1074/jbc.M500161200. [DOI] [PubMed] [Google Scholar]
- Faulkner MJ, Veeravalli K, Gon S, Georgiou G, Beckwith J. Functional plasticity of a peroxidase allows evolution of diverse disulfide-reducing pathways. Proc Natl Acad Sci U S A. 2008;105:6735–6740. doi: 10.1073/pnas.0801986105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, Buchner J. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell. 2009;34:569–579. doi: 10.1016/j.molcel.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Haslbeck M, Hainzl O, Buchner J. Synthesis and characterization of a functional intact IgG in a prokaryotic cell-free expression system. Biol Chem. 2008;389:37–45. doi: 10.1515/BC.2008.007. [DOI] [PubMed] [Google Scholar]
- Georgiou G, Segatori L. Preparative expression of secreted proteins in bacteria: status report and future prospects. Curr Opin Biotechnol. 2005;16:538–545. doi: 10.1016/j.copbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim R, Benhar I. "Inclonals": IgGs and IgG-enzyme fusion proteins produced in an E. coli expression-refolding system. MAbs. 2009;1:281–287. doi: 10.4161/mabs.1.3.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li Y, Wang YG, Gu X, Wang Y, Shen BF. An efficient and targeted gene integration system for high-level antibody expression. J Immunol Methods. 2007;322:28–39. doi: 10.1016/j.jim.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Humphreys DP. Production of antibodies and antibody fragments in Escherichia coli and a comparison of their functions, uses and modification. Curr Opin Drug Discov Devel. 2003;6:188–196. [PubMed] [Google Scholar]
- Jung ST, Reddy ST, Kang TH, Borrok MJ, Sandlie I, Tucker PW, Georgiou G. Aglycosylated IgG variants expressed in bacteria that selectively bind FcgammaRI potentiate tumor cell killing by monocyte-dendritic cells. Proc Natl Acad Sci U S A. 2010;107:604–609. doi: 10.1073/pnas.0908590107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans RP. Finally! The Brambell receptor (FcRB). Mediator of transmission of immunity and protection from catabolism for IgG. Immunol Res. 1997;16:29–57. doi: 10.1007/BF02786322. [DOI] [PubMed] [Google Scholar]
- Jurado P, Ritz D, Beckwith J, de Lorenzo V, Fernandez LA. Production of functional single-chain Fv antibodies in the cytoplasm of Escherichia coli. Journal of Molecular Biology. 2002;320:1–10. doi: 10.1016/S0022-2836(02)00405-9. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R, Weiss R, Chen G, Iverson BL, Georgiou G. Production of correctly folded Fab antibody fragment in the cytoplasm of Escherichia coli trxB gor mutants via the coexpression of molecular chaperones. Protein Expr Purif. 2001;23:338–347. doi: 10.1006/prep.2001.1520. [DOI] [PubMed] [Google Scholar]
- Lin B, Renshaw MW, Autote K, Smith LM, Calveley P, Bowdish KS, Frederickson S. A step-wise approach significantly enhances protein yield of a rationally-designed agonist antibody fragment in E. coli. Protein Expr Purif. 2008;59:55–63. doi: 10.1016/j.pep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Liu C, Dalby B, Chen W, Kilzer JM, Chiou HC. Transient transfection factors for high-level recombinant protein production in suspension cultured mammalian cells. Mol Biotechnol. 2008;39:141–153. doi: 10.1007/s12033-008-9051-x. [DOI] [PubMed] [Google Scholar]
- MacLean D, Jones JDG, Studholme DJ. Application of 'next-generation' sequencing technologies to microbial genetics. Nature Reviews Microbiology. 2009;7:287–296. doi: 10.1038/nrmicro2122. [DOI] [PubMed] [Google Scholar]
- Manting EH, Driessen AJ. Escherichia coli translocase: the unravelling of a molecular machine. Mol Microbiol. 2000;37:226–238. doi: 10.1046/j.1365-2958.2000.01980.x. [DOI] [PubMed] [Google Scholar]
- Mariani G, Strober W. Immunoglobulin metabolism. In: Metzger H, editor. Fc Receptors and the Action of Antibodies. American Society of Microbiology. 1990. pp. 94–177. [Google Scholar]
- Martens T, Schmidt NO, Eckerich C, Fillbrandt R, Merchant M, Schwall R, Westphal M, Lamszus K. A novel one-armed anti-c-Met antibody inhibits glioblastoma growth in vivo. Clin Cancer Res. 2006;12:6144–6152. doi: 10.1158/1078-0432.CCR-05-1418. [DOI] [PubMed] [Google Scholar]
- Mavrangelos C, Thiel M, Adamson PJ, Millard DJ, Nobbs S, Zola H, Nicholson IC. Increased yield and activity of soluble single-chain antibody fragments by combining high-level expression and the Skp periplasmic chaperonin. Protein Expr Purif. 2001;23:289–295. doi: 10.1006/prep.2001.1506. [DOI] [PubMed] [Google Scholar]
- Mayer M, Kies U, Kammermeier R, Buchner J. BiP and PDI cooperate in the oxidative folding of antibodies in vitro. J Biol Chem. 2000;275:29421–29425. doi: 10.1074/jbc.M002655200. [DOI] [PubMed] [Google Scholar]
- Mazor Y, Van Blarcom T, Mabry R, Iverson BL, Georgiou G. Isolation of engineered, full-length antibodies from libraries expressed in Escherichia coli. Nat Biotechnol. 2007;25:563–565. doi: 10.1038/nbt1296. [DOI] [PubMed] [Google Scholar]
- Melnick J, Argon Y. Molecular Chaperones and the Biosynthesis of Antigen Receptors. Immunology Today. 1995;16:243–250. doi: 10.1016/0167-5699(95)80167-7. [DOI] [PubMed] [Google Scholar]
- Mori H, Yano T, Kobayashi T, Shimizu S. High density cultivation of biomass in fed-batch system with DO-Stat. J Chem Engi Japan. 1979;12:313–319. [Google Scholar]
- Ni Y, Reye J, Chen RR. lpp deletion as a permeabilization method. Biotechnol Bioeng. 2007;97:1347–1356. doi: 10.1002/bit.21375. [DOI] [PubMed] [Google Scholar]
- Perez-Perez J, Barbero JL, Marquez G, Gutierrez J. Different PrlA proteins increase the efficiency of periplasmic production of human interleukin-6 in Escherichia coli. J Biotechnol. 1996;49:245–247. doi: 10.1016/0168-1656(96)83990-3. [DOI] [PubMed] [Google Scholar]
- Perez-Perez J, Gutierrez J. An arabinose-inducible expression vector, pAR3, compatible with ColE1-derived plasmids. Gene. 1995;158:141–142. doi: 10.1016/0378-1119(95)00127-r. [DOI] [PubMed] [Google Scholar]
- Perez-Perez J, Marquez G, Barbero JL, Gutierrez J. Increasing the efficiency of protein export in Escherichia coli. Biotechnology (N Y) 1994;12:178–180. doi: 10.1038/nbt0294-178. [DOI] [PubMed] [Google Scholar]
- Perez-Perez J, Martinez-Caja C, Barbero JL, Gutierrez J. DnaK/DnaJ supplementation improves the periplasmic production of human granulocyte-colony stimulating factor in Escherichia coli. Biochem Biophys Res Commun. 1995;210:524–529. doi: 10.1006/bbrc.1995.1691. [DOI] [PubMed] [Google Scholar]
- Reilly DE, Yansura DG. Production of Monoclonal Antibodies in E. coli. In: Shire SJ, Gombotz W, Bechtold-Peters K, Andya J, editors. Current Trends in Monoclonal Antibody Development and Manufacturing. New York: Springer; 2010. pp. 295–308. [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nature Biotechnology. 2009;27 doi: 10.1038/nbt.1568. 946-U112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini CL, Bernadac A, Zhang M, Chanal A, Ize B, Blanco C, Wu LF. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 2001;276:8159–8164. doi: 10.1074/jbc.C000833200. [DOI] [PubMed] [Google Scholar]
- Sarkar CA, Dodevski I, Kenig M, Dudli S, Mohr A, Hermans E, Pluckthun A. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci U S A. 2008;105:14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierle CF, Berkmen M, Huber D, Kumamoto C, Boyd D, Beckwith J. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J Bacteriol. 2003;185:5706–5713. doi: 10.1128/JB.185.19.5706-5713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LC, Reilly D, Klimowski L, Raju TS, Meng G, Sims P, Hong K, Shields RL, Damico LA, Rancatore P, Yansura DG. Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. J Immunol Methods. 2002;263:133–147. doi: 10.1016/s0022-1759(02)00036-4. [DOI] [PubMed] [Google Scholar]
- Simmons LC, Yansura DG. Translational level is a critical factor for the secretion of heterologous proteins in Escherichia coli. Nat Biotechnol. 1996;14:629–634. doi: 10.1038/nbt0596-629. [DOI] [PubMed] [Google Scholar]
- Skretas G, Georgiou G. Genetic analysis of G protein-coupled receptor expression in Escherichia coli: inhibitory role of DnaJ on the membrane integration of the human central cannabinoid receptor. Biotechnol Bioeng. 2009;102:357–367. doi: 10.1002/bit.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Han Y, Peng JL, Tehranchi AK, Gibbs R, Wang JD, Chen R. High-Precision, Whole-Genome Sequencing of Laboratory Strains Facilitates Genetic Studies. Plos Genetics. 2008;4 doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch KL, Beckwith J. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci U S A. 1988;85:1576–1580. doi: 10.1073/pnas.85.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohl WR, Knight DM. Discovery and development of biopharmaceuticals: current issues. Curr Opin Biotechnol. 2009;20:668–672. doi: 10.1016/j.copbio.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Vanhove M, Usherwood YK, Hendershot LM. Unassembled Ig heavy chains do not cycle from BiP in vivo but require light chains to trigger their release. Immunity. 2001;15:105–114. doi: 10.1016/s1074-7613(01)00163-7. [DOI] [PubMed] [Google Scholar]
- Zhan X, Gao J, Jain C, Cieslewicz MJ, Swartz JR, Georgiou G. Genetic analysis of disulfide isomerization in Escherichia coli: expression of DsbC is modulated by RNase E-dependent mRNA processing. J Bacteriol. 2004;186:654–660. doi: 10.1128/JB.186.3.654-660.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li ZH, Wang F, Fang M, Yin CC, Zhou ZY, Lin Q, Huang HL. Overexpression of DsbC and DsbG markedly improves soluble and functional expression of single-chain Fv antibodies in Escherichia coli. Protein Expr Purif. 2002;26:218–228. doi: 10.1016/s1046-5928(02)00502-8. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.