Abstract
Juvenile hormones (JHs) play key roles in regulating metamorphosis and reproduction in insects. The last two steps of JH synthesis diverge depending on the insect order. In Lepidoptera, epoxidation by a P450 monooxygenase precedes esterification by a juvenile hormone acid methyltransferase (JHAMT). In Orthoptera, Dictyoptera, Coleoptera and Diptera epoxidation follows methylation. The aim of our study was to gain insight into the structural basis of JHAMT’s substrate recognition as a means to understand the divergence of these pathways. Homology modeling was used to build the structure of Aedes aegypti JHAMT. The substrate binding site was identified, as well as the residues that interact with the methyl donor (S-adenosylmethionine) and the carboxylic acid of the substrate methyl acceptors, farnesoic acid (FA) and juvenile hormone acid (JHA). To gain further insight we generated the structures of Anopheles gambiae, Bombyx mori, Drosophila melanogaster and Tribolium castaneum JHAMTs. The modeling results were compared with previous experimental studies using recombinant proteins, whole insects, corpora allata or tissue extracts. The computational study helps explain the selectivity towards the (10R)-JHA isomer and the reduced activity for palmitic and lauric acids. The analysis of our results supports the hypothesis that all insect JHAMTs are able to recognize both FA and JHA as substrates. Therefore, the order of the methylation/epoxidation reactions may be primarily imposed by the epoxidase’s substrate specificity. In Lepidoptera, epoxidase might have higher affinity than JHAMT for FA, so epoxidation precedes methylation, while in most other insects there is no epoxidation of FA, but esterification of FA to form MF, followed by epoxidation to JH III.
Keywords: Juvenile hormone synthesis, farnesoic acid, juvenile hormone acid, methyltransferase, P450 epoxidase, Homology modeling
1. INTRODUCTION
Juvenile hormones (JHs) play key roles in regulating metamorphosis and reproduction in insects (Goodman and Granger, 2005). JHs are synthesized and secreted from the corpora allata (CA), a pair of endocrine glands with neural connections to the brain (Tobe and Stay, 1985). To date, seven forms of JH have been fully characterized chemically and physiologically. JH III is the most widespread JH homologue in insects; JH 0, JH I, JH II and 4-methyl-JH I seem exclusive to Lepidoptera (Goodman and Granger, 2005). Diptera produce the 6,7-epoxide of JH III (Goodman and Granger, 2005) and Heteroptera produce skipped bisepoxide JH III (Kotaki et al., 2009).
The biosynthetic pathway of JH III is divided into early and late steps (Belles et al., 2005). The early steps of JH III biosynthesis follow the mevalonate pathway to form farnesyl pyrophosphate (FPP) (Belles et al., 2005). During the late steps, FPP is hydrolyzed by a pyrophosphatase to farnesol (Cao et al., 2009) and then oxidized successively to farnesal and farnesoic acid (FA) (Mayoral et al., 2009a; Baker et al., 1983). The order of the last two steps differs depending on the insect group (Fig. 1). In Lepidoptera, a C-10,11 epoxidation by a P450 monooxygenase converts FA to the epoxy acid (JH acid or JHA) that is subsequently methylated by an O-S-adenosylmethionine-dependent juvenile hormone acid methyltransferase (JHAMT) to form the methyl ester (Reibstein et al., 1976). In Orthoptera, Dictyoptera, Coleoptera and Diptera epoxidation follows methylation (Feyereisen et al., 1981; Tobe and Pratt, 1974; Hammock, 1974; Weaver et al., 1980; Li et al., 2003).
Figure 1.
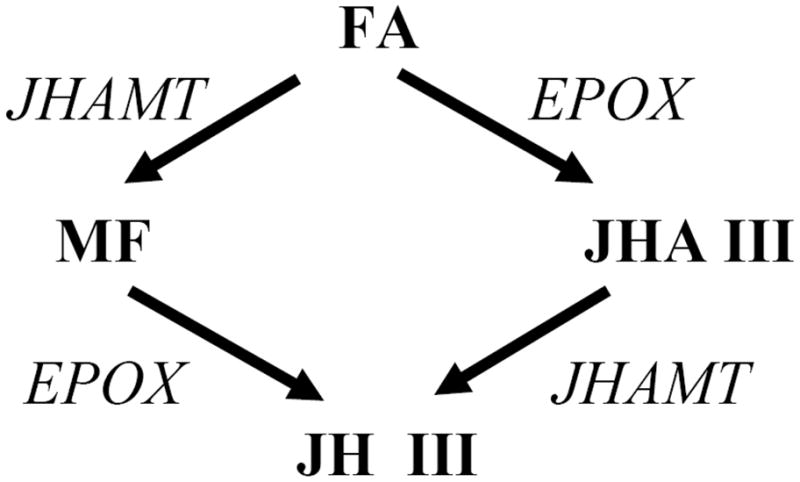
Scheme showing the two alternative sequences of the last two steps of JH synthesis in insects. FA: farnesoic acid; MF: methyl farneosate; JHA III: juvenile hormone acid III; JH III: juvenile hormone III; JHAMT: juvenile hormone acid methyltransferase; EPOX: P450 epoxidase.
The role that the catalytic properties and substrate specificity of JHAMT and epoxidase play in the pathway divergence has been an issue of controversy (Law, 1983; Schooley and Baker, 1985). The minute size of the insect CA has made it difficult to purify and characterize JH biosynthetic enzymes; however the last two enzymes of the pathway were recently molecularly and biochemically described (Shinoda and Itoyama, 2003; Helvig et al., 2004). The recombinant epoxidase from the cockroach Diploptera punctata cannot epoxidize FA, but only convert methyl farneosate (MF) into JH III (Helvig et al., 2004). The recombinant JHAMT enzymes from Bombyx mori (Shinoda and Itoyama, 2003), Drosophila melanogaster (Niwa et al., 2008), Tribolium castaneum (Minakuchi et al., 2008) and Aedes aegypti (Mayoral et al., 2009b) convert FA into MF, as well as JHA into JH III; suggesting that the ability to metabolize both FA and JHA might be an attribute of all insect JHAMTs. To test this hypothesis, homology modeling and docking simulations were performed for A. aegypti JHAMT and four additional insect JHAMTs. Our studies had two aims: 1) to explain the structural basis of substrate specificity for the four JHAMTs that have already been biochemically characterized, and 2) to use these structural analyses to predict the substrate specificity of additional insect JHAMTs that have not been biochemically tested. Every insect JHAMT structure generated by homology modeling was predicted to process FA and JHA since all critical residues interacting with both JH precursors and S-adenosylmethionine (SAM) showed high conservation of identity or function. These results support other analyses suggesting that the order of the methylation or epoxidation reactions in JH synthesis is for the most part imposed by the substrate specificity and affinity of the epoxidase.
2. MATERIALS AND METHODS
2.1 Identification and alignment of sequences
orthologue sequences of A. aegypti JHAMT (AeJHAMT) in other insects were retrieved using blastp (Altschul et al., 1990). The following sequences were aligned using Clustal X (Larkin et al., 2007) and JalView (Clamp et al., 2004): A. aegypti (gi|157112795|), B. mori (gi|112982770|), D. melanogaster (gi|24584607|), T. castaneum (gi|187937187|), Culex pipiens (gi|170049007|), Anopheles gambiae (gi|31210413|), Spodoptera litura (gi|148596804|), Samia cynthia ricini (gi|93115166|), Helicoverpa armigera (gi|148596806|), Nasonia vitripennis (gi|156547661|), Apis mellifera (gi|110756101|) and Acyrthosiphon pisum (gi|193681157|). Ten additional species of Drosophila were also included in the analysis and their accession numbers are provided in the supplementary information.
2.2 Modeling Aedes aegypti JHAMT
the molecular model of AeJHAMT was built by homology modeling using Modeller9v4 (Eswar et al., 2008; Sali and Blundell, 1993). The O-methyltransferases from Anabaena variabilis (PDB ID: 3CCF) and Clarkia breweri (PDB ID: 1M6E) (Zubieta et al., 2003) were chosen as templates on the basis that they had the highest identity to AeJHAMT (20.6% and 16.9%, respectively) among methyl transferases that use a similar catalytic mechanism to transfer the methyl group to a carboxylic acid moiety in their substrates, including the absence of a requirement for a metal cation (e.g. Fe) in the active site for enzymatic activity (Mayoral et al., 2009b). The alignment of the three sequences was done with T-COFFEE (Poirot et al., 2003). Supplementary Figure 1A shows the alignment between the sequences of the two selected templates and the modeled AeJHAMT. We manually corrected the alignment of AeJHAMT to the templates based on the PSIPRED predictions of the template secondary structures (McGuffin et al., 2000). We added constraints to the amino acid residues that interact with SAM when we generated the model, since these key residues in Clarkia are conserved in mosquitoes. In addition, we also checked the secondary structure prediction and FOLD assignments made by the program PHYRE, which is a valuable tool for template selection. In the case of AeJHAMT, the secondary structure predictions were in good agreement with the 3CCF and 1M6E secondary structures. Supplementary Figure 2 displays the structural alignment between the overall folds of the two selected templates and AeJHAMT. The models are colored showing the Root Mean Standard Deviation (RMSD) of the structurally aligned parts (Leach, 2001).
The model with the lowest Discrete Optimized Protein Energy (DOPE) assessment score was subjected to loop refinement using the “loop module” of Modeller9v4. The SCRWL program was then used to regenerate side chains based on a backbone-dependent rotamer library (Bower et al., 1997). The model was then minimized using the AMBER program (Case et al., 2005), with implicit solvent and the ff99sb force field (Hornak et al., 2006) and 10 kcal/molÅ2 restraints on the protein atoms. The final structure was evaluated using WHAT_CHECK (Hooft et al., 1996; the WHAT_CHECK analysis is included as Supplementary Information).
We evaluated an additional Anabaena variabilis SAM-dependent methyltransferase (PDB ID: 3GGD) with 17% sequence identity to AeJHAMT. This protein was not selected as a template because its secondary structure is very different to that predicted for AeJHAMT and critical residues for substrate interaction, such as Q14 and W120, are missing (Supplementary Figure 1B).
2.3 Modeling A. gambiae, B. mori, D. melanogaster and T. castaneum JHAMTs
To build additional insect JHAMT models, we had the choice of using the already optimized AeJHAMT model as template, or start all over using the original Clarkia and Anabaena structures. We chose the former option because the sequence identities between Clarkia and Anabaena and the additional insect JHAMTs to be modeled were often lower than those with AeJHAMT. Using them as template would have added uncertainty to the new models. In contrast, the amino acid identities between AeJHAMT and these four proteins varied between 62.6% (A. gambiae) and 40.6% (T. castaneum); furthermore, the important residues for catalysis and cofactor binding are well conserved.
Using the Modeller9v4 program and the AeJHAMT homology model as template we generated models for A. gambiae, B. mori, D. melanogaster and T. castaneum JHAMTs (Eswar et al., 2008). The alignment of the sequences was performed with ClustalX 2.0.12. (Larkin et al., 2007). The best models were chosen using the DOPE lowest score. Structural alignment and visualization were done with the program VMD (Eargle et al., 2006; Humphrey et al., 1996). Images were rendered with Tachyon (Stone, 1998).
2.4 Docking of substrates on JHAMT models
docking simulations were carried out using the program Autodock4 (Morris et al., 2009). We used the Lamarckian genetic algorithm for the conformational searches. The following parameters were used for all the simulations: a population size of 150 individuals (the population size parameter is related to the number of conformers that are used to start the simulation), 5 million energy evaluations, mutation rates of 0.02, a crossover rate of 0.8 and an elitism value of 1. For each ligand, 100 independent docking runs were performed, and results differing by less than 0.5 Å were clustered together. To validate our docking protocol, SAM was removed from the model and docked again. Afterward, with SAM docked into the protein models, we performed the docking of the substrates: FA, (10R)-JHA, (10S)-JHA, lauric and palmitic acids. Throughout these runs, proteins were kept rigid except for the side chains of Gln-14 and Trp-120, which were allowed to rotate.
3. RESULTS
3.1 A. aegypti JHAMT structure
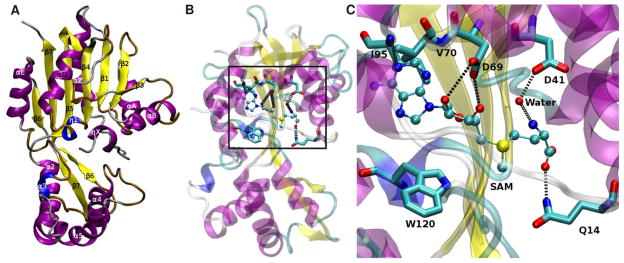
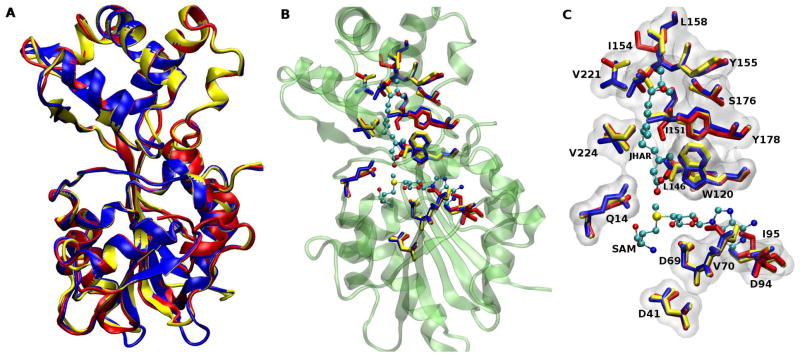
The AeJHAMT structure obtained by homology modeling has the typical S-adenosylmethionine-methyl transferase (SAMT) fold composed of alternating 6-stranded β-sheets with 9 α-helices (Mayoral et al., 2009b) (Fig. 2A). We docked SAM into the apo-structure of AeJHAMT in order to characterize the active site (Fig. 2B). The interaction of AeJHAMT with SAM was very similar to other SAMT-SAM complexes reported in the literature (Zubieta et al., 2003). Asp-69 formed hydrogen bonds with two ribose hydroxyls of SAM, while Asp-41 was able to form a hydrogen bond through a water molecule with the NH3+ moiety of SAM (Fig. 2C). The adenine ring of SAM was located inside a hydrophobic pocket formed by the side chain of Val-70 and Ile-95. Two additional critical conserved residues were identified near SAM, Gln-14 and Trp-120, which bind the carboxyl group of FA or JHA and place them in a suitable conformation for catalysis (Fig. 3). In summary, the complex network of interactions that define the active site of a SAMT was identified in the structure of AeJHAMT, giving support to the validity of our model.
Figure 2. Overall fold of JHAMT.
(A) Homology model of A. aegypti JHAMT. Alpha helices and beta sheets are labeled over their respective structures. Alpha helices are colored in purple, 3_10 helices in blue, extended beta sheets in yellow, turns in ochre and coils in silver. (B) Predicted position of the active site in the overall fold of the protein. Potential hydrogen bonds are shown in black dotted lines. Figure 2B has been rotated ~180 deg relative to Fig. 2A to allow better visualization of the active site. C) Close-up of the active site of the enzyme (Box in B) showing critical residues interacting with SAM. Carbon atoms are colored in cyan, nitrogen atoms in blue, oxygen atoms in red and sulfur atoms in yellow.
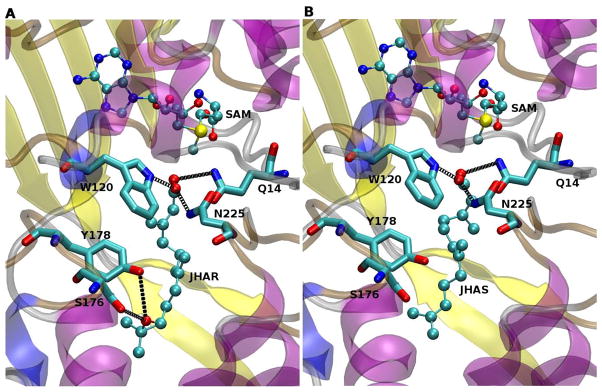
Figure 3. Docking simulations of JHA(10R) and FA.
(A) Predicted docking structure of JHA(10R). (B) Predicted docking structure of FA. Color coding is as described for figure 2. Black dotted lines represent putative hydrogen bonds between the substrates and residues Gln-14 and Trp-120, as well as between the epoxide group of JHA(10R) and Tyr-178 and Ser-176 (Panel A).
3.2 Docking of potential substrates
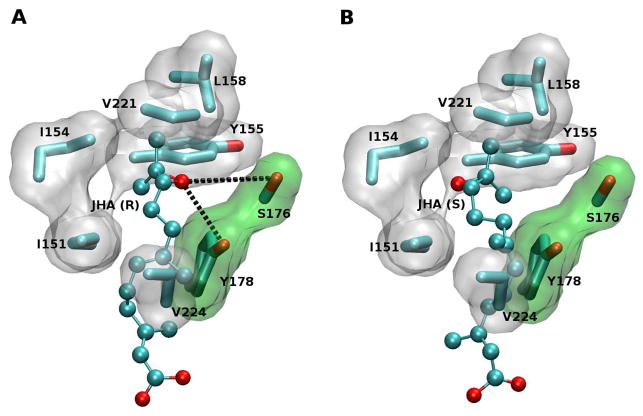
Figures 3A and 3B show the lowest energy conformers for (10R)-JHA and FA, respectively, where the carboxyl group of the substrates are oriented towards the methyl group of SAM, allowing the catalytic transfer. The substrate carboxyl groups of (10R)-JHA and FA fit nicely to form hydrogen bonds with the Trp-120 indole nitrogen and the amide nitrogen of Gln-14, while their carbon chains reside in an hydrophobic pocket formed by Ile-151, Ile-154, Tyr 155, Leu-158, Val-221 and Val-224 (Fig. 4A). Docking results indicated a clear difference in the interaction of (10R)-JHA and (10S)-JHA with the hydrophobic pocket of the binding site of AeJHAMT. Specifically, while the epoxide ring of (10S)-JHA is facing the hydrophobic residues Ile-151 and Ile-154, the (10R)-JHA epoxide ring is facing the opposite side of the pocket, where it could interact more favorably with Ser-176 and Tyr-178 (Fig. 4B). The potential formation of hydrogen bonds between the OH groups of Ser-176 and Tyr-178 and the epoxide group of (10R)-JHA (Fig. 4A, black dotted line) may result in a higher binding affinity of this molecule for the enzyme. This may explain why 10R and not 10S is the substrate of choice for the recombinant AeJHAMT (Mayoral et al., 2009b).
Figure 4. Epoxide recognition pocket.
(A) Close-up showing the docking of JHA(10R). The residues Ser-176 and Tyr-178 are shown with the surface colored in green, as they are both part of the pocket and have an active role in the recognition. Potential hydrogen bonds are shown in black dotted lines. The cover of the binding pocket, with the surface of the residues colored in white, is formed by the residues Ile-151, Ile-154, Leu-158, and Val-221. (B) Close-up showing the docking of JHA(10S) with the epoxide group oriented toward the hydrophobic face of the pocket (Ile-151 and Ile-154) instead of toward Ser-176 and Tyr-178.
Docking simulations of palmitic acid in AeJHAMT failed to find any favorable conformation of the acidic moiety facing SAM and interacting with Gln-14 or Trp-120, probably due to the large tail of palmitic acid that did not fit well into the hydrophobic pocket. On the other hand, we found a conformer for lauric acid that could interact with Gln-14 and Trp-120 but the tail was outside the hydrophobic pocket. Moreover, the energy due to internal constraints of lauric acid was high. Thus, the predicted binding affinity of lauric acid was low compared to those assessed for JHA and FA. These results may explain why AeJHMAT is not able to catalyze the methylation of these two substrates (Mayoral et al., 2009b).
3.3 Comparison with additional insect JHAMT
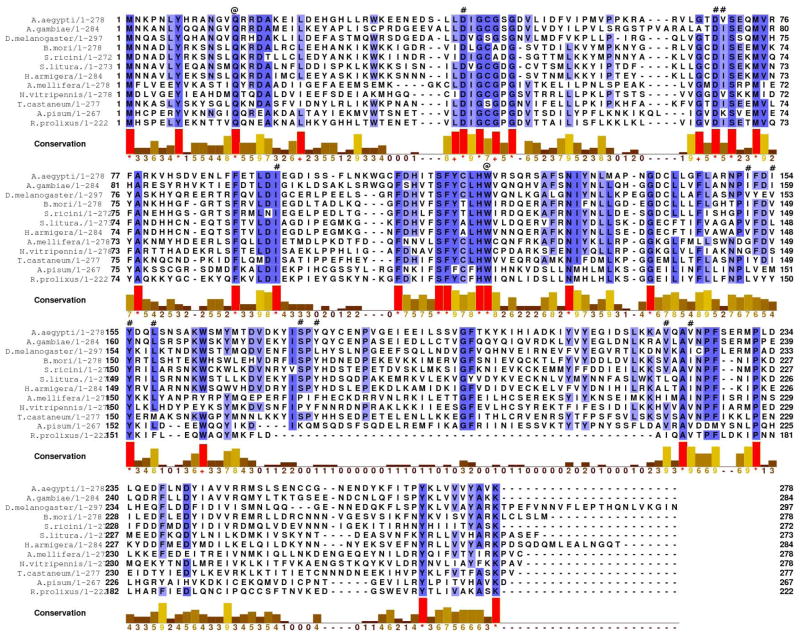
AeJHAMT orthologues were found in other species of insects and a cladogram of the phylogenetic relationship of these sequences was generated (Supplementary Fig. 3). Most of the amino acids identified as important in AeJHAMT for the interactions with the methyl donor (SAM) and the substrates (FA or JHA) are conserved in every JHAMT analyzed (Fig. 5). We generated the structures of D. melanogaster, B. mori, T. castaneum and A. gambiae JHAMTs using homology modeling and AeJHAMT as a template. The moth, fly and beetle species were selected because the biochemical properties of their recombinant proteins have been previously studied. In addition, the malaria mosquito was selected because of its epidemiological relevance. Figure 5 shows most of the amino acids that we have identified as important for the recognition of SAM and the carboxylated substrates (Asp-41, Asp-69, Gln-14, Trp-120 and Ser-176) conserved in the five JHAMTs modeled. For an easier visualization of the results, a structural alignment of only 3 models is displayed in Figure 6 (Aedes, Bombyx and Tribolium). Tyr-178 is conserved in the modeled JHAMTs, excluding that of D. melanogaster, where it is replaced by Leu, a hydrophobic residue that lacks the H-bond donor present in Tyr-178. We believe that this is not a critical substitution because it does not modify the ability of the fruit fly JHAMT to methylate either FA or JHA (Niwa et al., 2008). Nevertheless Ser-176 is conserved in D. melanogaster and provides the H-bond donor residue to interact with the JHA epoxide. Table 1 summarizes the conservation of the amino acids identified as critical for interactions with SAM, FA and JHA in JHAMTs from a set of insect species. Supplementary Table 1 shows that the identified residues are highly conserved in 10 additional species of Drosophila, further emphasizing their importance.
Figure 5. Alignment of several insect JHAMTs.
Residues shading color represents identity, going from blue (100% identity) to white (< 65% identity). Both Gln-14 and Trp-120 in A. aegypti are identical in all sequences analyzed (labeled with @). Other important residues for SAM or ligand interactions are marked with # in the sequence. In the conservation panel: *: Identical residues in all species. +: Conservative substitution in all species (marked in red as well for clarity). 9 to 0 values represent degree of conservation in all species. Conservation is measured as a numerical index reflecting the conservation of physicochemical properties in the alignment (Identity score the highest (*), followed by a numeric score of 9 for the next most conserved group of residues containing substitutions by amino acids included in the same physicochemical class as described by Livingstone, et al., [1993]).
Figure 6. Comparison of overall fold and critical amino acids in three insect JHAMTs.
(A) Overall fold comparison generated by homology modeling of A. aegypti JHAMT (in blue) structurally aligned with models of B. mori JHAMT (in yellow) and T. castaneum JHAMT (in red). (B) Comparison of critical residues present in the JHAMTs of the three insect species: A. aegypti JHAMT residues in blue, B. mori JHAMT residues in yellow and T. castaneum JHAMT residues in red. Residues superimposed on top of the overall fold of AeJHAMT (in green). (C) A close-up showing the comparison of the important amino acids interacting with SAM and JHA(10R) in the three JHAMT species (colors as in B).
Table 1.
Conservation of JHAMT residues critical for interactions with the substrates
| A. aegypti | Q 14 |
D 41 |
D 69 |
V 70 |
I 95 |
W 120 |
L 146 |
I 151 |
I 154 |
Y 155 |
L 158 |
S 176 |
Y 178 |
V 221 |
V 224 |
Activity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. gambiae (*) | = | = | = | I | = | = | = | = | = | = | = | = | = | = | = | Not tested |
| D. melanogaster | = | = | = | I | = | = | = | V | V | = | = | = | L | = | I | Yes for FA and JHA |
| B. mori | = | = | = | I | = | = | = | = | V | = | = | = | = | = | I | Yes for FA and JHA |
| S. ricini | = | = | = | I | = | = | I | = | V | = | = | = | = | = | = | Yes for JHA |
| S. litura | = | = | = | I | = | = | V | V | L | = | = | = | = | L | I | Not tested |
| H. armigera | = | = | = | I | = | = | V | V | V | = | = | = | = | L | I | Not tested |
| T. castaneum | = | = | = | I | = | = | = | = | = | = | M | = | = | = | = | Yes for FA and JHA |
| A. mellifera | = | = | = | I | = | = | = | G | V | = | = | P | F | I | I | Not tested |
| N. vitripennis | = | = | = | = | = | = | I | G | S | = | = | P | F | = | = | Not tested |
| A. pisum | = | = | = | K | = | = | = | L | M | = | = | Q | S | = | = | Not tested |
| R. prolixus | = | = | = | I | = | = | = | L | Y | = | F | ? | ? | I | = | Not tested |
=: Identical amino acid. Letters show conserved substitutions. Underlined bold letters are semi-conserved substitutions. Italic bold letters show non-conserved substitutions. Shaded boxes indicate substitutions not present in any of the three species whose activities have been experimentally tested (these three species are in bold).
? : Residues not known.
C. pipiens has identical substitutions than A. gambiae.
4. DISCUSSION
4.1 JHAMT is a promiscuous enzyme, original evidence from CA studies
the specificity of JHAMT was initially studied using CA extracts. Addition of JH acids stimulated JH synthesis by M. sexta CA but not addition of oleic acid, 9,10-epoxystearic acid, 2-hexadecenoic acid and 9-ket-2-decenoic acid (Reibstein et al., 1976). In the presence of NADPH, JH III was produced from FA with almost no detection of MF; on the other hand, MF was produced from FA-stimulated M. sexta CA extracts in the absence of NADPH (cofactor of the epoxidase), suggesting a lack of specificity of the JHAMT towards FA and JHA III (Reibstein et al., 1976). The JHAMT of the locust Locusta migratoria showed little substrate specificity, esterifying JHA III and FA at similar rates (Pratt et al., 1981). In another locust, Schistocerca gregaria, esterification of the sesquiterpenoid acid precedes epoxidation; and although CA of S. gregaria synthesized epoxy methyl esters from added 3H-JHA, it did so at lower rates than from the 3H-MF (Pratt and Tobe, 1974).
4.2 JHAMT esterifies FA and JHA, confirmation by studies using recombinant enzymes
Shinoda and Itoyama (2003) were the first to describe the enzymatic properties of an insect recombinant JHAMT. In the presence of SAM, this enzyme from B. mori efficiently catalyzed the conversion of FA and JH acids I, II, and III to their cognate methyl esters, although it displayed greater activity towards JHA I and JHA II than JHA III or FA (Shinoda and Itoyama, 2003). Later, studies on the activity of three additional recombinant JHAMTs were published, two from Diptera (Niwa et al., 2008; Mayoral et al., 2009b) and one from Coleoptera (Minakuchi, et al., 2008). All of three enzymes efficiently esterified FA and JHA III (Table 2). In addition, all of them showed enantioselectivity favoring (10R)-JHA III, generating (10S)- and (10R)-JH III isomers at ratios of 2:98 (B. mori), 0.5:99.5 (A. aegypti), 13:87 (T. castaneum) and 20:80 (D. melanogaster), respectively.
Table 2.
Enzymatic activity of purified recombinant JHAMTs on JH III, FA and fatty acids.
| Substrate | A. aegypti (mol /mol/ min) | B. mori (mol /mol/ min) | D. melanogaster kcat (min−1) | T. castaneum (mol /mol/ min) |
|---|---|---|---|---|
| FA | 0.260 ± 0.009 | 0.48 ± 0.07 | 7.7 ± 0.4 | 0.59 ± 0.04 |
| JHA III | 1.033 ± 0.008 | 0.79 ± 0.06 | 10.1 ± 0.5 | 0.56 ± 0.03 |
| Palmitic acid | 0.00089± 0.00012 | NA | 0.1 ± 0.007 | 0.016 ± 0.002 |
| Lauric acid | 0.00010 ± 0.00017 | ND | 0.04 ± 0.008 | 0.002 ± 0.001 |
A. aegypti data are from Mayoral et al., 2009b; B. mori data are from Shinoda and Itoyama, 2003; D. melanogaster data are from Niwa et al., 2008; T. castaneum data are from Minakuchi et al., 2008. FA: farnesoic acid. JHA III: juvenile hormone acid III. NA: not assayed. ND: not detected.
4.3 Structural studies validate that insect’s JHAMTs esterify FA and JHA
the identification of critical active site residues through modeling and docking studies offers a structure-based approach to help both understand the existing JHAMT experimental results and to predict the substrate specificity of untested enzymes of this family. Our aim was to understand the principles of substrate recognition and the transmethylation reaction in insect JHAMTs. Biochemical studies with insect recombinant proteins provided a framework for understanding the catalytic mechanism that underlies substrate recognition and methyl group transfer.
In our modeling and docking studies with AeJHAMT we identified several critical amino acid residues involved in the interactions of the enzyme with the methyl donor (SAM) and with the substrates (FA and JHA). The comparison with the additional three JHAMTs that have been biochemically characterized confirmed that the modeled active sites look very similar among them and showed that all these critical residues are conserved among the four tested proteins, or have substitutions that do not affect the ability of the active site to methylate FA or JHA (Table 2). We predict that A. gambiae, C. pipiens, S. ricini and the nine additional Drosophila species analyzed should be able to utilize both FA and JHA as substrates because all the critical residues are conserved in identity or function with the residues of the four JHAMTs that have been experimentally tested. The same prediction applies to the H. armigera protein, which that has two semi-conservative changes in residues that form the hydrophobic pocket for the carbon chain that should not affect the enzyme specificity.
In contrast, the two hymenopteran JHAMTs present a series of non-conserved substitutions. In A. mellifera and N. vitripennis Ser-176 is replaced by Pro, and Tyr-178 has a conserved substitution by Phe. These two changes suggest that Apis and Nasonia JHAMTs are not able to form a hydrogen bond with (10R)-JHA. In addition, the replacement in these two insects of Ile-151 for a smaller Gly, implies an enlargement of the hydrophobic pocket that could further change the specificity. Longer substrates, such as palmitic or lauric acids, might now be able to fit inside the hydrophobic pocket. Nevertheless, it is important to emphasize that these two hymenopteran proteins should still be able to utilize both FA and JHA as substrates because they conserve the SAM binding pocket, the Trp and Gln residues to recognize the carboxyl moiety of the substrates and a hydrophobic pocket that is able to accommodate FA and JHA.
We also found JHAMT orthologues in two hemimetabola, the hemipterans Acyrthosiphon pisum (pea aphid) and Rhodnius prolixus (kissing bug). Critical residues such as Asp-41, Asp-69, Gln-14 and Trp-120 are conserved. There are also conservative, semi- conservative and non-conservative substitutions (Table 1). The JHAMT sequence in R. prolixus was obtained from the genome project currently in progress; therefore it is a partial sequence that might have some errors (Supplementary Fig. 4). Nevertheless, the two hemimetabola sequences show a high degree of conservation that suggests similar substrate specificity.
We are aware that the structural models done by homology modeling in these studies are based on templates with low homology with AeJHAMT, and consequently, care must be taken in the conclusions derived from them. In the absence of a crystal structure of an insect JHAMT, homology modeling based on proteins with a sequence identity that falls within or below what Chung and Subbiah defined as the twilight zone of 20–25% could be challenging (Chung and Subbiah, 1996). However in this case, the robustness of the AeJHAMT model is supported by the congruence of the docking and biochemical results, both in the substrate specificity and stereospecificity, as well as by the high degree of conservation of the critical amino acids from the five different insect JHAMTs modeled. In summary, even though discrepancy in low conservation zones could be expected, we are confident that the active site region involved in ligand and cofactor binding has been properly characterized.
4.4 JH epoxidase shows narrow substrate specificity
Locusta migratoria JH epoxidase is a microsomal enzyme that specifically converts MF into (10R)-JH III (Feyereisen et al., 1981). Experiments using the cockroach Blaberus giganteus CA and CA homogenates showed that the JH epoxidase is more active on MF than on FA, and it showed high specificity for the 10,11 double bond (Hammock, 1975).
Diploptera punctata recombinant JH epoxidase (CYP15A1) converts methyl (2E,6E)-farnesoate to the natural (10R)-JH III with a very high degree of specificity (98% 10R: 2% 10S). Other terpenoids were not metabolized by CYP15A1, including farnesol, farnesal, FA, JH III, 3,7,11-trimethyl-dodecanol, geranylgeraniol, farnesyl methyl ether and geraniol, as well as the fatty acids lauric acid and arachidonic acid (Helvig et al., 2004). An NADPH-dependent JH epoxidase acting on FA was described from the CA of M. sexta (Reibstein et al, 1976). This enzyme did not efficiently epoxidize MF in the conditions in which the experiments were carried out; the authors argued that FA could be converted to JH III through both branches of the pathway and suggested that Lepidoptera have a different sequence of reactions than cockroaches and locusts (Reibstein et al, 1976). Preliminary experiments using CYP15C1, a recombinant JH epoxidase from B. mori, showed a high stereoselective epoxidation of FA (Shinoda et al., 2007), suggesting that in Lepidoptera substrate specificity of the epoxidase may be the critical factor in determining epoxidation/methylation reaction order. Nevertheless the higher catalytic activity of B. mori JHAMT towards JHA I and JHA II when compared to JHA III or FA might also contribute to the reaction order observed in Lepidoptera (Shinoda and Itoyama, 2003).
4.5 Summary
Results from biochemical studies using corpora allata extracts, kinetic data analysis of recombinant proteins, homology modeling and docking simulations were integrated to identify the potential contribution of specific residues in the recognition of FA and JHA in JHAMTs. Our models support the hypothesis that all JHAMT in insects are able to use FA and JHA as substrates; therefore, the order of the methylation or epoxidation reaction may be primarily imposed by the JH epoxidase’s substrate specificity and affinity. In Lepidoptera, JH epoxidase might have higher affinity than JHAMT for FA, so epoxidation precedes methylation. In most other insects there is no epoxidation of FA, only esterification of FA to form MF, followed by epoxidation to JH III. Further studies on the substrate specificity of recombinant enzymes in mosquitoes and Lepidoptera could experimentally assess the flux of FA and JHA through both pathways and test this hypothesis. In addition, the homology models presented in this study provide valuable information for the identification of critical residues that could be targeted for site-directed mutagenesis aimed at understanding the substrate specificity of insect JHAMTs.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for comments that have significantly improved this manuscript. This work was supported by NIH grant AI 45545 to FGN. The authors acknowledge the NSF Teragrid resource through Grant No. TG-MCA05S010 for providing computational resources and support that have contributed to the results reported in this paper. This work was partially supported by grants from Universidad de Buenos Aires 08-X499 to AGT, ANPCYT 06-5203 to AGT and Conicet PIP 01207 to AGT .
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baker FC, Mauchamp B, Tsai LW, Schooley DA. Farnesol and farnesal dehydrogenase(s) in corpora allata of the tobacco hornworm moth, Manduca sexta. J Lipid Res. 1983;24:1586–94. [PubMed] [Google Scholar]
- Bellés X, Martin D, Piulachs MD. The mevalonate pathway and the synthesis of juvenile hormone in insects. Annu Rev Entomol. 2005;50:181–190. doi: 10.1146/annurev.ento.50.071803.130356. [DOI] [PubMed] [Google Scholar]
- Bower MJ, Cohen FE, Dunbrack RL. Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J Mol Biol. 1997;267:1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- Cao L, Zhang P, Grant DF. An insect farnesyl phosphatase homologous to the N-terminal domain of soluble epoxide hydrolase. Biochem Biophys Res Comm. 2009;380:188–192. doi: 10.1016/j.bbrc.2009.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SY, Subbiah S. A structural explanation for the twilight zone of protein sequence homology. Structure. 1996;4:1123–1127. doi: 10.1016/s0969-2126(96)00119-0. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java Alignment Editor. Bioinformatics. 2004;20:426–7. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Eargle J, Wright D, Luthey-Schulten Z. Multiple Alignment of protein structures and sequences for VMD. Bioinformatics. 2006;22:504–506. doi: 10.1093/bioinformatics/bti825. [DOI] [PubMed] [Google Scholar]
- Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- Feyereisen R, Pratt GE, Hamnett AF. Enzymic synthesis of juvenile hormone in Locusta corpora allata: evidence for a microsomal cytochrome P-450 linked methyl farneosate epoxidase. Eur J Biochem. 1981;118:231–38. doi: 10.1111/j.1432-1033.1981.tb06391.x. [DOI] [PubMed] [Google Scholar]
- Goodman WG, Granger NA. The juvenile Hormones. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 6. Elsevier; 2005. pp. 55–115. [Google Scholar]
- Hammock BD. NADP dependent epoxidation of methyl farneosate to juvenile hormone in the cockroach Blaberus giganteus L. Life Sciences. 1975;17:323–28. doi: 10.1016/0024-3205(75)90479-8. [DOI] [PubMed] [Google Scholar]
- Helvig C, Koener JF, Unnithan GC, Feyereisen R. CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in cockroach corpora allata. Proc Natl Acad Sci USA. 2004;101:4024–4029. doi: 10.1073/pnas.0306980101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Kotaki T, Shinada T, Kaihara K, Ohfune Y, Numata H. Structure determination of a new juvenile hormone from a Heteropteran insect. Organic letters. 2009;11:5234–5237. doi: 10.1021/ol902161x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Law JH. Biosynthesis of juvenile hormones in insects. In: Porter JW, Spurgeon SL, editors. Biosynthesis of isoprenoid compounds. Vol. 2. Wiley and Sons; 1983. pp. 507–534. [Google Scholar]
- Leach AR. Molecular Modeling, principles and applications. 2. Pearson Prentice Hall; Essex, England: 2001. [Google Scholar]
- Li Y, Unnithan C, Hernandez-Martinez S, Feyereisen R, Noriega FG. Activity of the corpora allata of adult female Aedes aegypti: effects of mating and feeding. Insect Biochem Molec Biol. 2003;33:1307–1315. doi: 10.1016/j.ibmb.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- Mayoral JG, Nouzova M, Navare A, Noriega FG. NADP+-dependent farnesol dehydrogenase, a corpora allata enzyme involved in juvenile hormone synthesis. Proc Natl Acad Sci USA. 2009a;106:21091–21096. doi: 10.1073/pnas.0909938106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayoral JG, Nouzova M, Yoshiyama M, Shinoda T, Hernandez-Martinez S, Dolghih E, Turjanski AG, Roitberg AR, Priestap H, Perez M, Mackenzie L, Li Y, Noriega FG. Molecular and functional characterization of a juvenile hormone acid methyltransferase expressed in the corpora allata of mosquitoes. Insect Bioch Mol Biol. 2009b;39:31–37. doi: 10.1016/j.ibmb.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Minakuchi C, Namiki T, Yoshiyama M, Shinoda T. RNAi-mediated knockdown of juvenile hormone acid O-methyltransferase gene causes precocious metamorphosis in the red flour beetle Tribolium castaneum. Febs J. 2008;275:2919–2931. doi: 10.1111/j.1742-4658.2008.06428.x. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Niimi T, Honda N, Yoshiyama M, Itoyama K, Kataoka H, Shinoda T. Juvenile hormone acid O-methyltransferase in Drosophila melanogaster. Insect Biochem Molec Biol. 2008;38:714–720. doi: 10.1016/j.ibmb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Pratt GE, Tobe SS. Juvenile hormone radiobiosynthesised by corpora allata of adult female locust in vitro. Life Sciences. 1974;14:575–86. doi: 10.1016/0024-3205(74)90372-5. [DOI] [PubMed] [Google Scholar]
- Pratt GE, Stott KM, Brooks GT, Jennings RC, Hamnett AF, Alexander BAJ. Utilization of farnesoic acid analogues by the o-methyl transferase from corpora allata of adult female Locusta migratoria. In: Pratt GE, Brooks GT, editors. Juvenile hormone biochemistry. Vol. 15. Elsevier; 1981. pp. 107–121. [Google Scholar]
- Poirot O, O'Toole E, Notredame C. Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reibstein D, Law JH, Bowlus SB, Katzenellenbogen JA. Enzymatic synthesis of juvenile hormone in Manduca sexta. In: Gilbert LI, editor. The Juvenile Hormones. Plenum Press; 1976. pp. 131–146. [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schooley DA, Baker FC. Juvenile hormone biosynthesis. In: Kerkut GA, Gilbert LI, editors. Comprehensive Ins Physiol Biochem Pharm. Vol. 7. Pergamon Press; Oxford: 1985. pp. 363–389. [Google Scholar]
- Shinoda T, Itoyama K. Juvenile hormone acid methyltransferase: a key regulatory enzyme for insect metamorphosis. Proc Natl Acad Sci USA. 2003;100:11986–11991. doi: 10.1073/pnas.2134232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda T, Kozaki T, Namiki T, Mita K. CYP15C1 encodes the cytochrome P450 that catalyzes epoxidation of farnesoic acid to JH acid in the corpora allata of Bombyx mori. Abstracts Ninth International Conference on Juvenile Hormones; York, UK. 2007. p. 34. [Google Scholar]
- Stone J. Master Thesis. Computer Science Department, University of Missouri-Rolla; 1998. An Efficient Library for Parallel Ray Tracing and Animation. [Google Scholar]
- Tobe SS, Pratt GE. The influence of substrate concentrations on the rate of insect juvenile hormone biosynthesis by corpora allata of the desert locust in vitro. Biochem J. 1974;144:107–13. doi: 10.1042/bj1440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe SS, Stay B. Structure and regulation of the corpus allatum. Adv Insect Physiol. 1985;18:305–432. [Google Scholar]
- Zubieta C, Ross JR, Koscheski P, Yang Y, Pichersky E, Noel JP. Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell. 2003;15:1704–1716. doi: 10.1105/tpc.014548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver RJ, Pratt GE, Hamnett AF, Jennings RC. The influence of incubation conditions on the rates of juvenile hormone biosynthesis by corpora allata isolated from adult females of the beetle Tenebrio molitor. Insect Biochem. 1980;10:245–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.