Abstract
Imagine a pathogen that is spreading radially as a circular wave through a population of susceptible hosts. In the interior of this circular region, the infection dies out due to a subcritical density of susceptibles. If a mutant pathogen, having some advantage over wild-type pathogens, arises in this region it is likely to die out without leaving a noticeable trace. Mutants that arise closer to the infection wavefront have access to more susceptible hosts and thus are more likely to become established and perhaps (locally) outcompete the original pathogen. Among the factors (position, transmission rate, pathogen-induced death rate) that influence the fate of a mutant, which are most important? What does this tell us about the types of mutants that are likely to invade and become established? How do such tendencies serve to steer the evolution of pathogens in a spatial setting? Do different types of models of the same phenomena lead to similar conclusions?
We address these issues from the point of view of an individual-based stochastic spatial model of host–pathogen interactions. We consider the probability of a successful invasion by a single mutant as a function of the transmissibility and virulence strengths and the mutant position in the wavefront. Next, for a version of the model in which mutations arise spontaneously, we obtain analytical and simulation results on the mean time to a successful invasion. We also use our model predictions to gain insight into experimental data on bacteriophage plaques. Finally, we compare our results to those based on ordinary and partial differential equations to better understand how different models might influence our predictions on the fate of a mutant pathogen.
Keywords: Pathogen, SIR, Mutant invasion, Spatial structure, Interacting particle system, CA, Bacteriophage
1. Introduction
The spread of a pathogen through a spatially distributed population of susceptible hosts typically occurs along traveling wavefronts. The shape, size, smoothness, etc., of such a wavefront depends on host distribution, transmission and mortality rates. When the pathogen arises from a localized outbreak, the wavefront is roughly circular in shape, at least when environmental conditions and host density are essentially spatially homogeneous. Examples include the spread of a viral plaque in a lawn of bacteria on a Petri dish, and the transmission of a plant pathogen through a forest. Even when pathogens are randomly dispersed at a density that is not too high, they will initially form isolated waves of infection wherever host density is sufficiently high to support them.
This spatial structure in the spread of a pathogen plays an essential role in determining when and where mutant pathogens arise, as well as their probability of successfully invading and contributing to the evolution of pathogen virulence in a spatially extended population of hosts. The goal of this paper is to provide a theoretical understanding of these issues based on a spatially explicit stochastic model. We compare the resulting predictions to empirical observations and to the behavior predicted by differential equation models.
In the case where the pathogen dynamics (infection and pathogen-associated host death) operate on a faster time-scale than host dynamics (births and natural deaths), there is little or no regrowth of susceptible hosts in the wake of the pathogen wave. In this situation, the success probability of a mutant pathogen is greatly enhanced if it happens to arise in the vicinity of the wavefront where it will have access to a sufficient number of susceptible hosts. Mutants that arise behind the wavefront are subject to the rigors of spreading among host populations that are at low density or are spatially fragmented. Of course, the outcomes depend on things like the rate of pathogen spread (transmissibility) and the death rate due to infection (virulence).
Unlike their spatial counterparts, the key to homogeneously mixed host–pathogen systems and the ordinary differential equations (ODE)) that model them is that each pathogen has equal access to all susceptible hosts. In this simplified setting, the outcomes of competition between different pathogens are determined by the basic reproductive ratio (cf. Murray, 1989), which in turn is governed by the ratio of the transmission and removal rates of the pathogens. The result is that, if two pathogens are competing for a single host species, the pathogen with the largest ratio dominates and eventually drives the competing pathogen to extinction. If pathogens are allowed to mutate in this setting, the direction of evolution is typically guided by a combination of trade-offs between transmissibility and virulence and the afore-mentioned optimization of the basic reproductive ratio (May and Anderson, 1983). We will see that predictions based on these classical mass-action differential equations—modeling, for example, competition between different strains of a bacteriophage in a chemostat—are quite different from predictions based on spatial models.
There have been numerous studies relating the importance of spatial structure in models of host–pathogen systems; these include Sato et al. (1994), Rand et al. (1995), Haraguchi and Sasaki (2000), and Boots et al. (2004). A point we wish to emphasize, and which has been emphasized elsewhere, is that the fate of a mutant pathogen is determined by stochastic events that are influenced largely by interactions among a relatively small number of individuals, not bulk densities. We will see that, in addition to the values of transmissibility and virulence, the spatial location of a mutant pathogen plays a key role in determining the probability that it becomes established in the population.
The model we study is an individual-based stochastic spatial version of an SIR model (S = susceptible, I = infective, R = removed). Such models belong to the family of interacting particle systems (IPS) and are sometimes referred to as (asynchronously updated) probabilistic cellular automata (CA). We note that these models approximate the stochastic, spatial, individual-based nature of real host–pathogen systems. To illustrate the effects of these features of the dynamics, we will compare our results with those predicted by (mass-action) ODE and reaction–diffusion equations (RDE). The former effectively average out all spatial structure and randomness, relating information that is present only in bulk averages. The latter are also based on an averaging out of randomness, but this time on a local scale, leading to some preservation of spatial information such as the appearance of constant-speed traveling wavefronts.
1.1. Related studies
Bacteriophages (of the lytic sort) are viruses that infect, reproduce within, and lyse bacterial cells. Phagebacteria systems are ideal for experimental evolution studies in which short generation times and high mutation rates allow one to see evolution in “real time.” These microbial host–pathogen systems can be studied in spatially structured environments to gain important insight into the role of space in their evolution (Koch, 1964; Yin, 1993; Yin and McCaskill, 1992). When inoculated in isolated spots on a lawn of bacteria growing on an agar plate, the phage form circular clearings (or plaques) that are visible with the unaided eye. These radially expanding plaques often exhibit characteristic bulges that arise from mutant strains that propagate more quickly than the wild-type phage.
Yin (1993) investigated the appearance of such mutants in radially propagating plaques of the bacteriophage T7 grown on Escherichia coli host bacteria on agar plates. By sampling the phage plaque along equally spaced radii, several mutant colonies were detected. These mutants, arising from deletions in the phage genome, produced phage strains that spread more quickly than the wild-type phage. In Section 2.3, we will further discuss Yin's empirical work and use our model to provide a theoretical framework for understanding these results.
Haraguchi and Sasaki (2000) studied the evolution of virulence in a host–pathogen IPS quite similar to the one studied in our paper, except that their model takes a more long-term point of view and host reproduction plays a key role. They found that the natural spatial clustering of the host population leads to evolutionarily stable strategies with intermediate transmission rates. To some extent, our study can be thought of as revealing the detailed spatial dynamics of pathogen spread and mutation within a cluster of susceptibles. Evolution, as directed by ecological factors and selective pressures, occurs on several hierarchical levels. The afore-mentioned paper, as well as others such as Rand et al. (1995), view the dynamics from a more global point of view in which susceptible and infected hosts are essentially in equilibrium. Within that broader context, there are battles being fought on smaller length scales that drive the system. Our study, as compared to these earlier ones, is thus geared more toward small-scale transient dynamics in which we seek to predict which mutations are likely to gain a foothold in the larger scale equilibrium dynamics. We find, for example, that mutant pathogens invade by claiming an ever-widening swath of susceptibles in such a way that the resulting carnage due to mutant pathogens displays a “clonal wedge” pattern.
In Edmonds et al. (2004) a simulation study was undertaken for a model of population growth (not a host–pathogen system). They considered the fate of a neutral mutant in an expanding population. Their model consists of a 25 × 100 grid of demes, each harboring up to N individuals and updated in discrete time steps by reproduction, migration, and random culling of individuals from demes exceeding a certain size. Each simulation was started with a wild type individual seeded in a deme on the left side of the grid. After the wave of population expansion reached some predetermined distance, a single mutant with the same growth and migration characteristics was placed randomly at a spot along the wave front. They considered the probability that the mutant colony had at least one descendent colonizing a deme on the right side when the entire grid is saturated, as well as how the place of origin of the mutation relates to the location of the centroid of the mutant population. Among other things, they observed that the success probability for a given invasion attempt is enhanced when the mutant originates in a sparse region of the non-mutant wave.
1.2. The model
The model studied here is an individual-based lattice model known as an IPS model. Our simulations take place on a 300 × 300 grid of sites, each in one of the states: S = susceptible, I1 = infected by (wild-type) pathogen 1, I2 = infected by (mutant) pathogen 2, R = removed or dead. Site x will change its state at a rate that depends on that state and the states of all sites in some neighborhood of x:
| (1) |
where fi(x) denotes the fraction of neighbors of site x that are in state Ii. (The neighborhood chosen for the simulations consists of the eight nearest neighbors of x, sometimes referred to as the Moore neighborhood.) Thus, susceptibles become infected by a pathogen at a rate that is proportional to the number of neighboring individuals infected by that pathogen, infected individuals die or are otherwise removed at constant rates, and a wild-type pathogen mutates to a mutant pathogen at a constant rate. In keeping with our emphasis on the dynamics of rapid localized spread of pathogens in an environment of susceptibles, the model does not allow for reproduction by susceptibles and the only deaths are those incurred as a result of infection. We refer to βi as the transmission rate for pathogen i, δi as the mortality rate (or virulence) associated with pathogen i, and μ the mutation rate.
To understand the spatial dynamics that drive the invasions, we mention a few standard results for these spatial SIR models. The first thing to note is that if a single infective is placed amid a field of susceptibles, the infection will spread along an essentially circular ring whose radius increases linearly with time. (Actually, the rate of radial spread is constant only in the limiting sense, as the radius goes to infinity and the curvature goes to zero, but for practical purposes the difference is not noticeable.) Behind the infective wavefront sit the removed individuals and perhaps a few lucky susceptibles that escaped infection. This is all characterized mathematically by the “shape theorem” in the IPS literature (Durrett, 1988). This behavior is reflected in partial differential equation models of SIR dynamics via traveling waves and is readily observed experimentally in bacteriophage plaques (Koch, 1964; Yin, 1993; Yin and McCaskill, 1992) and other spatially extended host–pathogen systems.
In the lattice-based model, this circular shape is easy to understand. Since the infection spreads by direct contact between susceptibles and infectives, the rate at which a susceptible on the boundary of the wave becomes infected is proportional to the number of neighboring lattice sites that contain an infective. Alternatively, one can think of the infectives on the wavefront as racing to claim susceptible sites. Susceptible sites that are lagging behind the wave (corresponding to dents in the wavefront) will typically be in contact with more infectives than a typical susceptible at a smooth part of the wave, and hence it has a higher infection rate. This acts to fill in the dents along the wavefront. Similarly, protrusions in the wave lead to neighboring susceptibles that are in contact with very few infectives. Thus the average speed of such a part of the wavefront will be smaller than the smooth part of the wave. In other words, although stochasticity results in the wavefront being irregularly shaped, the aforementioned differences in local transmission rates result in the wavefront being self-correcting, thus maintaining the global circular shape of the wave.
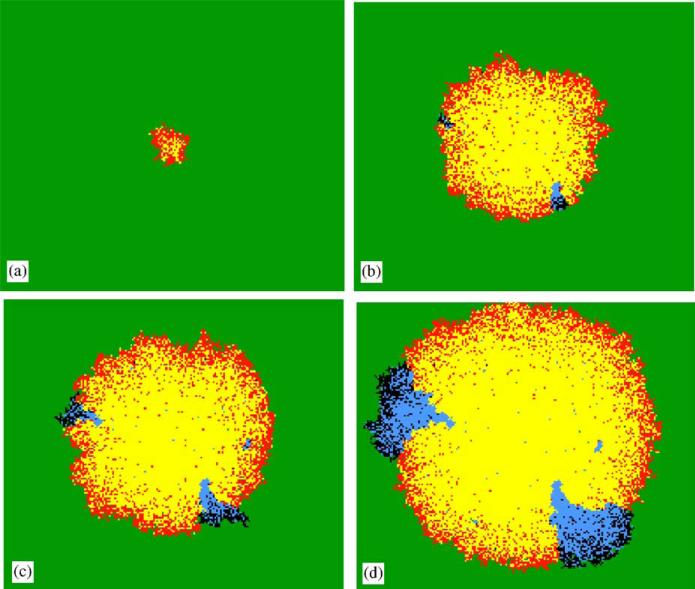
In Fig. 1, we see stages in a typical run of the simulation, with β1 = 0.002, δ1 = 0.0002, β2 = 0.0024, δ2 = 0.00022, and positive mutation rate μ = 10–7. The wild-type pathogen starts spreading radially through the susceptibles as prescribed by the shape theorem. There is a thin, roughly circular, wavefront of infectives and the interior of the circle is mostly occupied by dead susceptibles, possibly with a few remaining susceptibles that escaped infection. As this wild-type pathogen epidemic spreads, the “active” ring of infection (i.e. the set of hosts currently infected by a wild-type pathogen) builds up more and more infectives simply because the radius is increasing. If each such infective has a small probability μ of being infected by a mutant pathogen (with parameters β2 and δ2 that allow it to spread faster than the wild-type pathogen), then eventually such a mutant will appear somewhere along the wavefront. Based on factors such as the spatial position of the mutant among susceptibles and wild-type infectives, and the values of infectivity and virulence for both pathogen strains, this mutant might either die out completely or start to spread successfully, eventually claiming a larger and larger share of the available susceptibles. In the latter case, the sites that were ever infected by the mutant pathogen trace out a sector that is roughly wedge shaped and can end up bulging past the reaches of the wild-type wave. Such sectors have been observed in a number of settings, including the aforementioned bacteriophage plaques. In the next section, we examine this behavior more closely with an emphasis on trying to understand the factors that influence whether or not a mutant pathogen can successfully invade.
Fig. 1.
A typical simulation of the spontaneous mutation IPS model (green = susceptibles, red wild-type infectives, yellow = dead wild-type infectives, black = mutant infectives, blue = dead mutant infectives). (a) wild-type infection wave begins; (b) two mutant infective invasions getting started; (c) mutant infective colonies starting to emerge, with one unsuccessful invasion also evident; (d) clonal wedge pattern of two fully developed mutant pathogen colonies. Note that the “live” infectives are mostly on the edge. The clonal wedge pattern, evident only when we distinguish between different types of dead infectives, provides a historical perspective on the development of the spatial invasions. The parameter settings are β1 = 0.002, δ1 = 0.0002, β2 = 0.0024, δ2 = 0.00022, and μ = 0.0000001.
2. Behavior of the IPS model
We consider two sets of simulations for the IPS model. In the first (with mutation rate μ = 0), we explore the effect of the position of the mutant pathogen (as well as its transmission and mortality rates) on the probability of successful invasion. We initiate a wave of pathogen spread by placing a small number of wild-type pathogens in the middle of a sea of susceptibles. After this wave has spread for a predetermined amount of time, we place a single mutant pathogen near the wavefront. We consider mutant locations that are on the leading edge of the wave as well as various distances ahead of the wave. The reason for also considering mutant locations in advance of the wild-type wavefront was to be better able to compare our invasion results to those predicted by partial differential equation models, in which densities correspond to local averages. These “jumping” mutants can also lend some insight to situations for which the mutants have a larger dispersal range than the wild-type pathogen. In bacteriophages, for example, one evolutionary strategy that a mutant might employ is to delay lysing of the host cell with a resultant increase in “burst size.” This could lead to mutant phage being sent ahead of the wild-type infective wave.
In the second set of simulations, the wave of pathogen spread is initiated as before, but now mutant pathogens appear spontaneously (i.e. we have mutation rate μ>0). To implement this, we assume that each newly formed infective has a small probability of arising from the mutant type pathogen. In this model, we consider only one mutant type so as to focus on how the parameters for that particular mutant affect the dynamics of invasion. In addition, a host can be infected by only one pathogen. We are particularly interested in the mean time for the first successful invasion and how it depends on the parameters. Since this is intended to model actual mutants, they always appear at the site of a new infection (i.e. a nearest neighbor of an I1) and hence there is no discussion of the distance from the wave front.
An important point to keep in mind is that there is a critical value, rc, for the ratio of the transmission rate and the death rate (cf., Durrett, 1988). More specifically, in the spatial epidemic model with a single pathogen type having parameters β and δ, the epidemic can spread with positive probability if β/δ>rc, and dies out with probability one if β/δ<rc. With the neighborhoods we are using, simulations show that this critical ratio is approximately 3.5 (actually, somewhere between 3.4 and 3.5). Thus, all the pathogens we simulate must satisfy such an inequality to have any chance of persisting.
2.1. The fate of a single mutant
Here, we consider the model with μ = 0 and manually introduce a single mutant pathogen to a spreading wave of wild-type pathogens. Each simulation is initialized on a 300 × 300 lattice with a small (circular) cluster of 41 sites in state I1 at the center of lattice, and all other sites susceptible. We run the simulation for a total of 250 units of time, where one “unit of time” corresponds to an average of 90, 000 (= 300 × 300) sites updating in our continuous-time model. Running the process for this amount of time allows the relevant dynamics to unfold, but prevents the wave of infection from reaching the boundary. Thus, we need not be concerned with behavior at the boundary of the lattice.
After 30 units of time, we place a single mutant (I2) along a radius in a randomly chosen direction at a predetermined distance in advance of the wave. A random angle is chosen and the pathogen is placed along a radial line with that angle and a fixed distance from the edge of the wavefront. More specifically, a point along the radius at distance d from the wavefront is actually obtained by moving along the radius Euclidean distance d and then picking the nearest lattice site to that point; the distances chosen in the simulations correspond to d = 0, 2, 4, 6, 8, and 10. Distance 0, of course, means that the mutant arises at a point on the edge of the infective wave, in contact with both hosts and pathogens.
We say that a successful invasion occurs if at time 250 there are at least 90 I2 individuals present on the lattice; otherwise, the invasion is said to fail. In the simulation results that follow, we fix the wild-type parameters β1 = 0.002, δ1 = 0.0005 (β1/δ1 = 4). The probability of a successful invasion, as presented in Figs. 2–4, was computed as the proportion of runs that led to such behavior; each data point is based on 300 runs for a given setting of β2, δ2.
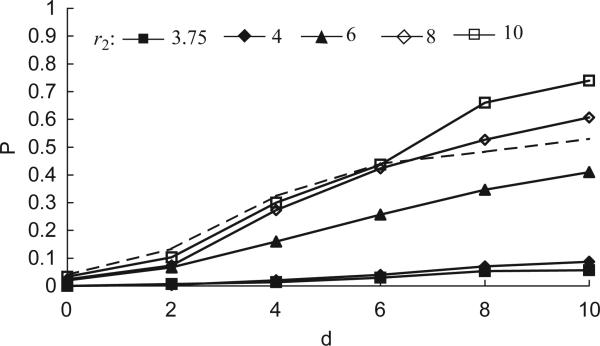
Fig. 2.
Invasion probability as a function of mutant position d and transmission rate β2, for a fixed ratio β2/δ2 = 10 that is significantly larger than the wild-type ratio. (Wild-type parameters β1 = 0.002, δ1 = 0.0005, β1/δ1 = 4 are held constant.) The dashed curve represents the success probability for a pathogen that has exactly the same transmission and mortality rates as the wild type. For β2<β1, the mutant pathogen has a difficult time, with invasion probability essentially tracking that of a neutral mutant until the distance from the wavefront is so large that there is not much chance for competition. When the mutant has the same transmission rate as the wild type, the effect of the larger ratio is seen to be significant. As we increase the mutant transmission rate past that of the wild type, the invasion probability increases somewhat but most of the gains are made at d = 0. This indicates the importance of transmission rate in determining whether the mutant is able to escape the initial congestion of the wavefront.
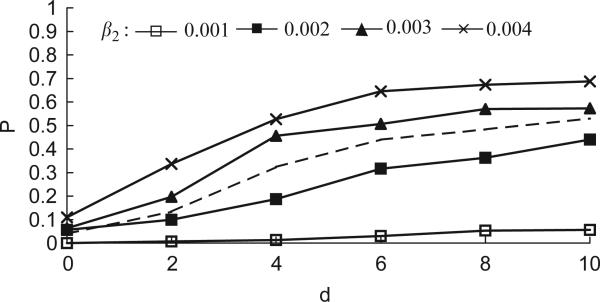
Fig. 4.
Invasion probability as a function of mutant position d and ratio r2 = β2/δ2, for a fixed value of transmission rate β2 = 0.001 that is smaller than that of the wild-type. Other settings are as in Fig. 2.
The simulation results shown below indicate that the probability of a successful invasion depends on the relative values (for mutant and wild type) of both the transmission rates (βi) and the ratio (βi/δi) of transmission rate and virulence. Contrast this to the situation for mass-action models (see Section 3) in which the ratio alone determines the outcome of an attempted mutant invasion.
In Fig. 2, we have a fixed mutant ratio β2/δ2 = 10, which is significantly greater than the wild-type ratio β1/δ1 = 4. We plot the invasion probability as a function of distance from the edge of the wild-type wavefront for each of four different values of the mutant transmission rate β2. As a baseline for comparison, we also plot the invasion probability for a neutral mutant; i.e. a mutant pathogen with the same transmission and mortality rates as the wild type (and hence the wild-type ratio, too). Perhaps the most interesting parts of this graph (and the two that follow) are the data points at distance d = 0. These correspond to mutants that arise most naturally from a wild-type pathogen at the wavefront. As d increases, we put the mutant in a better position to escape the initial congestion of wild-type infectives and enter into territory that has a high density of susceptibles. This gives us an idea of the relative importance of this initial phase of the invasion process. As was mentioned earlier, consideration of positive distances is also useful for comparison with models based on reaction–diffusion equations (Section 3) and for dealing with pathogens that can have different dispersal characteristics.
From the graph we can see that, if β2<β1, the mutant pathogen has a very hard time invading, even though it has a much higher ratio. Once the pathogen has higher transmission rate than the wild type, the invasion probability increases significantly. Large values of d should not be accorded too much significance since such a mutant, if it survives initially, has a chance to spread substantially before encountering and having to compete for susceptibles with any wild-type infectives, and hence be counted as a successful invader. However, the levelling off of the invasion probability for large d does point out something important. Namely, there is a maximum invasion probability (less than one) that can be attained. This is due to the fact that, even if the mutant pathogen is not competing with the wild type, the invading colony is initiated by a single mutant and hence can be snuffed out by chance early on. For example, in the case with β2 = 0.004 (and hence δ2 = 0.0004 to ensure a ratio of 10), the probability of the single mutant infective dying before infecting any susceptibles is δ2/(δ2 + β2) = 1/11. Thus, this demographic stochasticity leads to a maximum invasion probability of less than 0.91.
In Fig. 3, we fixed the ratio of the mutant pathogen at 3.75, which is a bit less than that of the wild type, but still larger than the critical ratio. Clearly, the invasion probability is significantly reduced from that in the case of ratio 10, but if the transmission rate for the mutant pathogen is higher than that of the wild type, it can invade. Arguing as above, we see that a significant fraction of the failed invasions can be accounted for by demographic stochasticity at the beginning of the invasion. Also notice that at d = 0 the data point for the neutral mutant is slightly below the one for smaller β2. This seeming discrepancy is accounted for by the stochastic nature of the data points and is within a 95% confidence interval (error bars not shown). Not surprisingly, d = 0 is where the outcomes are most variable since a successful invasion from the edge of the wavefront requires that the mutant pathogen first escape the congested area near wild-type infectives.
Fig. 3.
Invasion probability as a function of mutant position d and transmission rate β2, for a fixed ratio β2/δ2 = 3.75 that is smaller than the wild-type ratio (but still larger than the critical ratio). Other settings are as in Fig. 2.
To emphasize the importance of the transmission rate, we consider in Fig. 4 a fixed value of β2 = 0.001 (lower than wild-type transmission rate) and plot the invasion probability as a function of distance and ratio. For mutants arising at the edge of the infective wave (d = 0), invasion is almost impossible, regardless of the ratio. Even when the mutant has the advantage of starting ahead of the wild-type wavefront, it outperforms neutral mutants only when d>6 and the ratio is much higher than that of the wild type. This is another strong illustration of the role transmissibility plays in invasion.
2.1.1. Predicting invadability
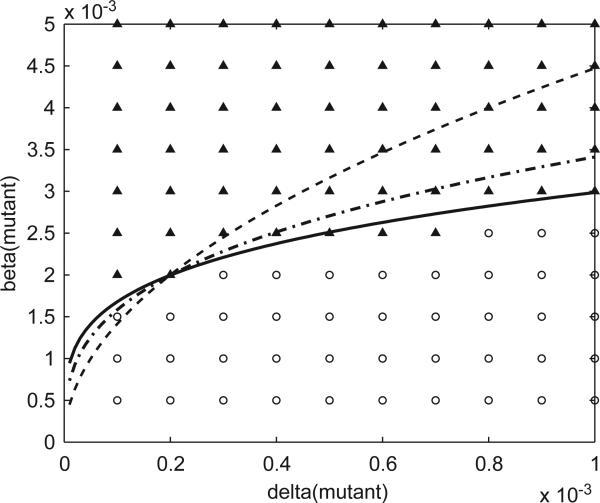
The above data indicate that the invasion probability for a mutant pathogen arising in a wave of wild-type infectives depends on both the transmission rates βi and the ratios ri = βi/δi, with the βi's having a greater effect. In Fig. 5, we fix the values of the wild-type parameters, β1 and δ1 (and hence r1), and record the parameter values for the mutant that lead to a successful invasion (from distance d = 0) in a fraction of at least 0.05 of 300 runs for each such setting. The value of 0.05 for the cutoff probability was chosen to match the d = 0 invasion probability for a neutral mutant, as seen in Figs. 2–4. We propose the following
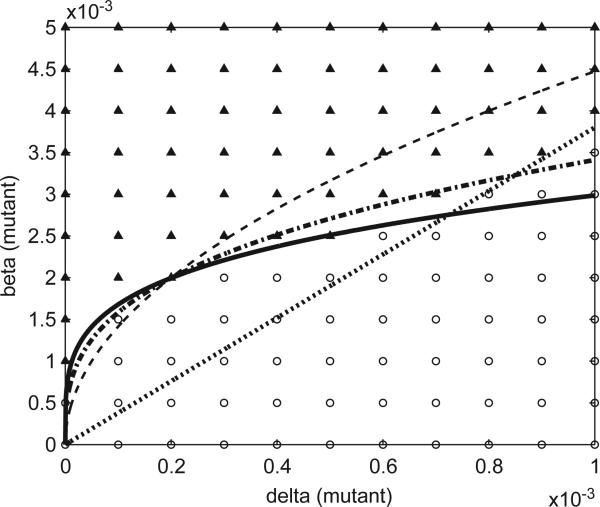
Fig. 5.
Invadability threshold for a single mutant as a function of δ2, β2 (with δ1 = 0.0002, β1 = 0.002, β1/δ1 = 10 held constant). The dark triangles represent parameter values for which a fraction of at least 0.05 of the 300 runs resulted in a successful mutant invasion; the open circles correspond to invasion frequencies of less than 0.05. The dotted straight line gives the critical ratio r2 = β2/δ2 = 3.5. The solid curve gives values of δ2, β2 satisfying the relation ; dash-dot curve ; dashed curve β2r2 = β1r1. The data indicate that for invasion to occur the parameters must correspond to points above both the critical ratio line and the solid curve, as specified in the invasion condition.
In other words, parameter values for a successful mutant should lie above the curve given by β2(β2/δ2)1/3 = K1, where K1 = β1(β1/δ1)1/3 is constant; they should also lie above the line given by β2/δ2 = 3.5 that determines the critical ratio for spread in the absence of competition. The data in Fig. 5 show an excellent fit to the proposed invasion condition. We performed several other simulations for different settings of the wild-type parameters, and in each case found excellent agreement with the invasion condition.
The proposed invasion condition for the spatially explicit IPS model should be compared to that for a mass-action model (see Section 3) which requires r2>r1. Such conditions play an important role in the evolution of virulence and transmissibility.
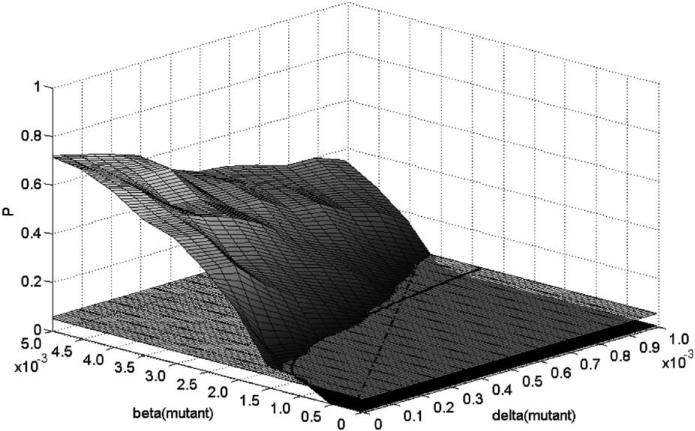
The above invasion condition and figure refer to parameter values for a single mutant pathogen (relative to the wild type) that give it “a chance” to invade. The actual probability of invasion is another matter. In Fig. 6, we plot the invasion probability surface and the cutoff plane as a function of the mutant parameters. The two threshold curves specified in the invasion condition are also plotted. As the parameters move into the invadability region further from these boundary curves, the frequency of successful invasion rises rapidly, again with more sensitivity to changes in the transmission rate than to the ratio.
Fig. 6.
Invasion probability surface corresponding to the parameters in Fig. 5. Also illustrated are the cutoff probability plane and the threshold curves. The straight line gives the critical ratio r2 = 3.5 and the solid curve gives the parameters satisfying the relation .
2.2. Spontaneous mutations in an expanding wave of infection
In these simulations, we used the same starting configuration as above, but instead of manually introducing a single mutant at a prescribed time, mutations appear spontaneously at rate μ>0; i.e. we add a transition I1 → I2 at rate μ.
We define the first invasion time T to be the time it takes until the total number of mutants (I2's) on the whole 300 × 300 lattice is at least 90. This simple criterion provides a good match with what we saw in the simulations. It is important not to set the threshold too low since there can be a number of mutants that arise but fail to invade. Also, once a successful invasion is well under way, the number of mutant infectives increases very rapidly.
We use these spontaneous-mutation simulations to investigate two important features of spatial host–pathogen dynamics. First, we derive an equation for the mean of the first invasion time based on the probability that a single mutant will invade (as in Section 2.1). We then compare our predictions to the spontaneous-mutation simulations. Second, we use our simulations to provide some insight into the behavior of the bacteriophage plaques described by Yin (1993).
2.2.1. Mean invasion time for mutant pathogens
If one mutant pathogen has, for example, twice the invasion probability as another, how much faster does it invade on average? Here, we derive an approximation to the mean invasion time for a mutant pathogen (characterized by its invasion probabilities and mutation rate) arising along the circular wavefront of a wild-type pathogen spreading in a uniform field of susceptible hosts. Thus, we provide an analytic expression that relates the single mutant invasion probabilities to the simulations of spontaneous mutations. Such expressions can be quite valuable in that they allow one to estimate parameters from observations. This is especially true for spatial models of microbial systems, where obtaining accurate spatial information on small-scales can be a challenge.
So, suppose our mutant pathogen has infectivity and virulence parameters β2, δ2, resp., and let p = p(β2, δ2) denote the probability that a single mutant can invade. This is the quantity that was estimated from simulations earlier and, of course, also depends on the parameters, β1, δ1 of the wild-type pathogen. (Note that we are ignoring the effects of the curvature of the wavefront, and hence the time of such an invasion, on the invasion probability. This effect is negligible when μ is small since, with very high probability, mutants arise only after the radius of the wave is fairly large.)
Consider, as in the spontaneous mutation simulations, that we start with a small number of I1's initiating a (circular) wave of infection in a sea of S's. Assume that the mutant type I2 arises at mutation rate μ. Since the radius of the I1 wave increases linearly with time, we can write n(t) = 2πct for the approximate number of infected edge sites in the I1 wave, for some constant c that encompasses the speed and mean thickness of the traveling wave of infectives. These are the sites that are subject to mutation. For example, I2 mutants will arise at rate M(t) = μn(t) = μ2πct at time t. Obviously, as the ring of infected sites gets larger, there are more sites available to mutate. Now, each new I2 mutant that arises has, on average, probability p to actually invade, so the I2 invasion rate at time t is given by pM(t). Thus, the cumulative invasion intensity in time interval [0, t] is given by
| (2) |
Now, since mutants are rare and the number of sites subject to mutation is large, interference between newly arisen mutants can be ignored and the appearance of invading mutants along the wavefront can be approximated by a Poisson process. Thus, if T denotes the time of the first successful I2 invasion, then
| (3) |
and hence the expected time for the first successful invasion by an I2 mutant is
| (4) |
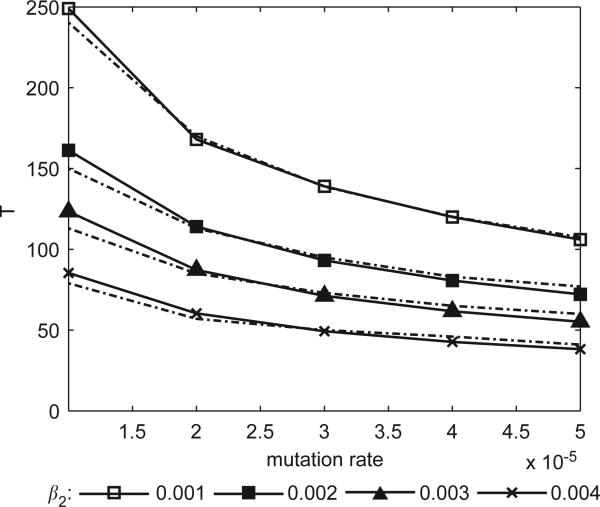
In Fig. 7, we get an idea of the “trend” in the mean invasion time for the spontaneous-mutation IPS model as compared to the trend predicted by Eq. (4), both as functions of mutation rate. The goal here was to see how well our prediction of a dependence on mutation rate matched the actual values generated by the spatial model; after all, in a non-spatial model, one would expect the mean invasion time to decay much more quickly with μ. With this in mind, we skirted the problem of estimating the constants p and c by simply fitting a curve of the form to the data curve. If p and c were known, the fitted curve would be shifted up or down but the shape would be the same. As can be seen, the dependence agrees strongly with the results of the IPS simulations. (Note that, in Figs. 7 and 8, quantities of the form represent numerical average times based on 300 runs for a specific set of parameters, while E(T) denotes the mathematical expectation. These quantities should, of course, be approximately equal.)
Fig. 7.
Dark curves show mean invasion time, , based on 300 runs as a function of mutation rate in the spontaneous mutation IPS model. The lighter companion curve for each of these gives the best fit of the form ; cf. Eq. (4). The value of the constant a is simply chosen to overlay the two curves as much as possible so as to determine how well the dependence on mutation rate is matched. In all simulations, we fix the values β1 = 0.002, r1 = 4, and r2 = 5.
Fig. 8.
Comparison of values of (solid squares), based on spontaneous mutation simulations, and p/p′ (open squares), based on single mutant simulations; cf. Eq. (6). The numbers on the horizontal axis correspond to different parameter settings for the two mutant pathogens. If we write T1, T2, T3, T4 for the transmission rates 0.004, 0.003, 0.002, 0.001, then the six points on the horizontal axis correspond to the parameter settings of (T1, T2), (T1, T3), (T1, T4), (T2, T3), (T2, T4), (T3, T4). Held constant are r1 = r2 = 10, β1 = 0.002, and δ1 = 0.0002.
The exponentially decaying tail probability in Eq. (3) indicates that, as long as p is not too close to zero, we will see a successful invasion within a reasonable amount of time with very high probability. This is, in fact, what we saw in the simulations—little chance for successful invasions early on, but their appearance is almost guaranteed within some intermediate time window. This same strong predictability for the appearance of mutant pathogens in a radially expanding wave has also been observed experimentally; see, for example, the “convergent diversity” in bacteriophage plaques in Yin (1993).
This effect can also be explained by the fact that the invasion intensity given in Eq. (2) is not constant in time for a given radially expanding pathogen. Clearly, as the radius increases there are more pathogens to mutate. This is partly a consequence of our local viewpoint. Of course, if one were to consider a more global view with hosts and pathogens at equilibrium, then one would have a constant rate of mutant invasion as long as the spatial window was large enough to maintain constant densities of hosts and infectives in the window.
Now consider two different mutant pathogens, with the corresponding infected states denoted by I2 and , resp. Write β2, δ2, resp., , for the infectivity and virulence parameters of I2, resp., , and let p = p(β2, δ2) and denote the corresponding single mutant invasion probabilities. Assume further that mutant type I2 arises at mutation rate μ and type at rate μ′.
Then the ratio of mean invasion times for these two mutants is
| (5) |
For example, to decrease E(T) by a factor 1/10, one would need the mutation rate (or the success probability) to increase by a factor 100.
Note that the above formula provides a method for estimating parameters. For example, suppose two different mutants have the same mutation rate (or, in an experimental evolution setting, perhaps the mutants are artificially added at the same time to different wild-type pathogen waves). If one can observe the average times to invasion, and , for the two different mutant pathogens then the relative invasion probabilities for single mutants can be estimated by
| (6) |
thereby obtaining what can be considered local information from global information. We checked to see how well this approximation works by computing invasion probabilities via single mutant simulations, as in the previous section, and then using spontaneous mutation simulations to calculate the mean invasion times. A comparison of the resulting quantities from the formula is given in Fig. 8.
2.3. Comparison to Yin's bacteriophage experiments
Yin (1993) investigated the emergence of mutant phage in radially spreading phage plaques. Some spatial resolution was obtained by analyzing stab samples along several radial lines. Phage mutants that arose by deleting parts of the wild-type phage genome were able to spread into the bacterial host population faster than the wild-type phage and consequently could sometimes become established in the plaque at high enough densities to be detectable.
We compare Yin's data to predictions based on the above IPS simulations with spontaneous mutations. Although the model in this paper does not explicitly account for all the complexities of phage dynamics (e.g. latent period, burst size), it does seem to be close enough in certain essential aspects to help explain the observations made by Yin. (We have also developed a more complex spatial model that incorporates many of the specific features of phages. As pertaining to the issues of this paper, the outcomes of simulations with the phage model were very similar, at least qualitatively, to the ones presented here for the generic host–pathogen model. In other respects, the phage model is quite different. That model will be explored in another paper.)
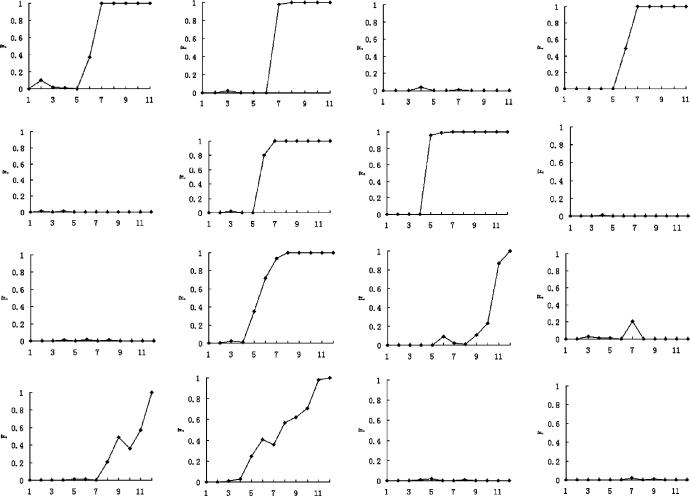
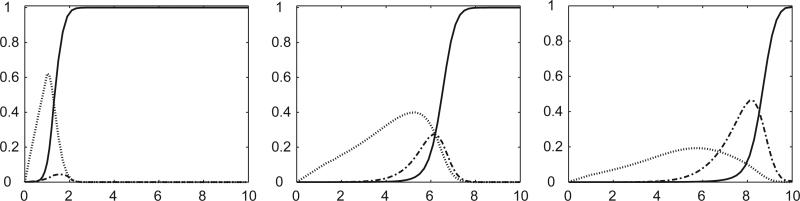
In Yin's experiments, the accumulation of mutant phage density along radial stabs followed patterns similar to those in our simulations (Fig. 9). Yin only reported three such curves. In Fig. 10, we show several examples from simulations giving the variety of behavior that can arise for a given mutant and wild type. Much of the stochasticity results from the roughness of the traveling wavefront and the random positions of the mutants. It is clear that the shape of the radial mutant density curves can depend on the time of the invasion that impinges on a given radius, together with the position of the radius relative to the clonal wedge (e.g. through the center or near the edge). In addition, we see that certain parameter values lead to more variability in the invasion curves.
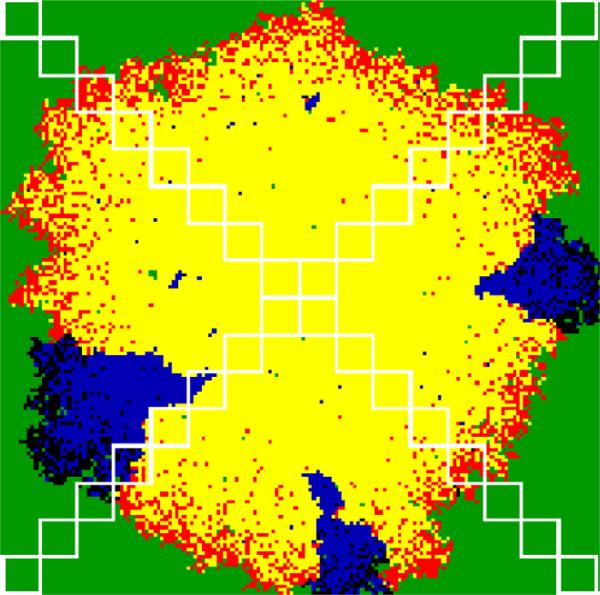
Fig. 9.
Sampling regions along four equally spaced radii used to generate the invasion curves in Fig. 10. Each square contains 100 sites (10 × 10) and is intended to correspond to a “stab sample” in Yin's experiments. The colors in the simulation are as in Fig. 1.
Fig. 10.
Invasion curves corresponding to the radial sampling regions in Fig. 9. Each row corresponds to one (completed) run of the simulation, with the four graphs in each row giving the fraction of mutant pathogens in sampling squares 1,2, etc., along each of the four different radii. The top two rows were obtained with parameter settings β1 = 0.002, δ1 = 0.00016, β2 = 0.004, δ2 = 0.00032, and μ = 10–7; the bottom two rows were obtained with parameter settings β1 = 0.002, δ1 = 0.00016, β2 = 0.0023, δ2 = 0.00018, and μ = 10–7. Note that larger values of β2 lead to less variability in the invasion curves.
In addition to these curves, Yin sampled plaque-derived mutants and wild-type phage and then grew them in shaker cultures. These were diluted, plated, and counted to obtain short-term growth curves for each. These curves suggested similar, though not identical, rates of spread for the mutants found along two different radii. (They were actually genetically distinct variants.) Yin noted, however, that the similarity of these curves did not seem to coincide with the very different rates of emergence along the radial samples. To explain this we note that, as discussed above, even identical mutants can lead to very different radial sample curves due to the shape and stochasticity of the clonal wedges. Of course, another factor could simply be that the mutants arose on the plates due to an advantage realized in the spatial plaque. As we have seen from our model, advantages in plate cultures do not necessarily translate to advantages in liquid cultures; even when they do, the effects can be very different. (Recall that in liquids, the ratio of transmission rate to mortality rate is the key to competition between phages, while on plates the transmission rate plays the dominant role; cf. Fig. 5.) Although this explanation has not been shown to be the only one consistent with Yin's observations, it at least suggests that what were thought to be conflicting results need not be at odds at all.
Yin also estimated the probability of mutation (deletion) to be 10–8 based on the fact that approximately 108 phage had been produced by the time the first invasion was detected. Although this estimate is consistent with values obtained in liquid culture (Scearce et al., 1991), it is probably misleading for the spatial cultures. It should never be assumed a priori that rates in spatial models are similar to those in well-mixed systems. For example, events that require contact or physical proximity (such as that between infected and susceptible cells to produce a new infective) can be quite different in liquid and plate cultures. A phage particle in a well-mixed liquid has access, in theory, to all susceptible cells during a short time interval, while a phage particle in a plate culture only has access to nearby cells. In addition, the contacts in liquid are fleeting, while those in plates are more stable. At any rate, as we have seen, not all mutants are presented with the same opportunity to invade. Those that arise behind the wavefront have little chance of invading (and hence of being detectable). Even those that arise at the forefront of the wave are not guaranteed to successfully invade. This suggests that the mutation rate in Yin's experiments is actually significantly higher than the estimated value of 10–8.
3. Comparison with differential equation models
There are several types of models used to study host–pathogen systems. In addition to individual-based lattice models, such as the one discussed above, there are also models based on differential equations. These sacrifice detail and stochasticity in favor of tractability. In some cases the simpler models do a good job of predicting behavior captured by the particle system models, but this is not always the case. In this section, we briefly explore two types of differential equation models that have been used often to study host–pathogen systems. We ask whether predictions about the fate of a mutant pathogen agree for the different models or if, on the contrary, different models produce different predictions. If the latter is true, it is important to understand how the choice of model can bias the results being sought. For example, different models might suggest different ‘strategies’ for the evolution of virulence.
Our interest is in determining how the parameters affect the ability of the mutant pathogen to invade when it arises at low density. We will see that, although there are some similarities, the three models can lead to different predictions. Since our main interest is in the behavior of the IPS model, we will keep this section rather brief.
3.1. Ordinary differential equations
A mean-field (or mass-action) ODE that is related to the IPS is
| (7) |
Here, λ represents a constant immigration rate for new susceptibles. Many similar models have been studied (cf. May and Anderson, 1983; Bremermann and Thieme, 1989; Nowak and May, 1994), including systems modeling chemostat growth and incorporating additional mechanisms of host growth and death. It has been established that in the above system, as well as others for which the pathogens are horizontally transmitted (no vertical transmission) and neither coinfection nor superinfection is allowed, the pathogen with the highest basic reproductive ratio, R0—defined as the number of secondary infections produced by a single infective introduced into a population consisting of all susceptibles—will outcompete and eventually replace the other pathogen. This outcome is independent of the initial densities of the two pathogens. For the above system, the basic reproductive ratio for pathogen i is βiS/δi. Since each pathogen encounters the same density of susceptibles in these ODE models, the pathogen with the highest ratio βi/δi is the one that wins (i.e. persists forever and drives the other pathogen density to zero). Thus, for example, in a well-mixed liquid containing susceptibles and a pathogen with transmissibility β1 and virulence δ1, a mutant (or externally introduced) pathogen with transmissibility β2 and virulence δ2 will invade and eventually replace the original pathogen provided β2/δ2>β1/δ1. (It is assumed here that each pathogen is viable in the sense that it satisfies an inequality of the form βi/δi>rc(λ), where the critical value rc(λ)≥1 is a non-increasing function of λ.)
We remark that, while our IPS model has no regrowth of susceptibles, some sort of replenishment of susceptibles is necessary in the ODE model since there is no refuge from the pathogens. In the IPS model, and in the RDE model below, we imagine a localized infection spreading into a limitless field of susceptibles. There is no notion of spatial localization in an ODE, so we are restricted to more of an equilibrium analysis. On the contrary, in the spatial models our focus is more on the non-equilibrium invasion dynamics that take place along a traveling wavefront of infectives. Despite this difference in the points of view of the two models, we feel it is important to compare them since many studies deal exclusively with ODE models and much of the existing folklore is based on such models. As we have seen, the ratio of transmissibility to virulence, which completely determines the outcome in a well-mixed liquid environment, is not the only factor deciding the fate of a mutant in the IPS model. We will also see a difference with the deterministic spatial model that we treat next.
3.2. Reaction– diffusion equations
Consider the one-dimensional RDE
| (8) |
where the parameters have similar interpretations as before, and D is the diffusion coefficient. This is a partial differential equation that is related to the particle system in a natural way via a ‘fast-stirring’ limit (Durrett and Neuhauser, 1994; Krone, 2004). Roughly speaking, this says that if, in our original IPS model, between state changes at different sites we were to allow the ‘particles’ to jiggle around a bit by exchanging the states at some nearby sites, then the above RDE (with the same rates as in the IPS) would serve as a good approximation to the medium-scale spatial dynamics of the particle system. By this last part, we mean that the density at a spatial point for the solution to the RDE is approximately equal to the average density for a suitably chosen group of sites in some spatial window in the particle system. Since we are more interested in the original particle system than the one with fast stirring, one must accept the fact that, while the above RDE will still behave roughly the same as the particle system in certain respects, the connection between the corresponding parameters in the two models is no longer as tight. Thus, the RDE that most closely matches the behavior of the IPS model may have somewhat different parameter values. This being said, it is often the case that one can learn a lot about the behavior of one model from the behavior of the other. Using a one-dimensional RDE to compare to our two-dimensional IPS model is also reasonable since the circular traveling wave solutions for the analogous two-dimensional RDE—the solutions that reflect the behavior of the radially expanding pathogen wavefronts in the IPS—approach those of the one-dimensional RDE above as the radius of the circular wavefront increases. In other words, locally, the circular wavefront eventually looks like a planar wave.
It is standard (cf. Murray, 1989) that if one of the pathogens is absent, the resulting two-type system gives rise to traveling wave solutions under appropriate initial conditions. In the system containing only types S and Ii the (asymptotic) wave speed is , where i = 1 or 2, provided βiS>δi. To compare to the mutant invasion setting for the particle system model, the idea is to consider such a traveling wave solution for I1 and introduce a tiny pulse of the mutant pathogen I2 at some position in the wave front. This perturbation spoils, at least temporarily, the traveling wave solution involving S and I1. Mathematically, this is difficult to treat since at least one of the pathogens is trying to spread into a region with non-constant susceptible density. We solved these equations numerically and found that, depending on the parameters, one of the pathogens will die out and the other will develop a traveling wave solution. See Fig. 11 for such a solution at three different time points.
Fig. 11.
Invasion in a traveling wave for RDE model. Parameters are β1 = 0.002, δ1 = 0.0005, β2 = 0.0023, δ2 = 0.0005, and D = 1. Solid curve gives susceptible density, dark dashed curve gives density of mutant infectives, and light dashed curve gives density of wild-type infectives. Three snapshots show the mutant arising at low density in the wavefront of the wild-type pathogen and then steadily taking over while the wild type dies out.
While the comparison between the particle system and the ODE was somewhat forced due to the lack of any spatial structure in the latter, comparing the particle system to the reaction–diffusion model is quite natural. Not only do both account for spatial structure, but they also capture the wavelike nature of pathogen spread into a population of susceptibles—the shape theorem for the particle system and the traveling wave solution for the RDE. The traveling wave can be thought of as a macroscopic manifestation of the more fine-grained, stochastic details afforded by the shape theorem.
To investigate the conditions under which a mutant pathogen will successfully invade in the one-dimensional reaction–diffusion model, we began with a traveling wave of the wild-type infectives moving to the right into a constant density of susceptibles (Murray, 1989). At one of six possible positions in the wavefront, we then added a very small pulse of the mutant pathogen and followed the resulting waves of pathogens traveling to the right. The chosen positions for the mutant pulse were equally spaced with the one furthest to the left aligning with the left edge of the wild-type wave, the next one in the center of the wild-type wave, the third position aligning with the front edge of the wild-type wave (as in the first frame of Fig. 11), and so on, with three more positions at equally spaced increments in advance of the wave. Keeping in mind that a density modeled by a partial differential equation represents an average of values in some neighborhood of a point, this variable placement of mutants is similar to the particle system simulations in Section 2.1 with a single mutant introduced at several distances from the edge of the wild-type wave. Of course, the RDE is deterministic, so there is no randomness in the outcome; either the mutant invades or it does not.
When interpreting the outcomes of such mutant invasions in the RDE, we considered two points of view. Firstly, we looked to see how invadability was affected by the spatial location of the mutant relative to the wild-type wavefront, the transmission rates, and the wave speeds. Secondly, to compare more directly with the single mutant simulations for the IPS, we placed the mutant at the third of our six positions within the wild-type wavefront and asked how the parameters βi and δi affected the ability of the mutant to invade. Roughly speaking, the results showed that the RDE model predictions for invadability were quite similar to those from the IPS model. Recall that the ODE model was not very helpful in determining the fate of a mutant in the IPS model.
We found that the fate of the mutant was influenced by the relative values of the speeds, the transmission rates, and the position of the mutant. First of all, the speed of the mutant had to be greater than that of the wild type to invade. (Here, speed of pathogen i refers to the speed of a single pathogen wave with full access to susceptibles.) When c2<c1, the mutant did not invade. If c2>c1, then the mutant was able to invade regardless of initial position when β2>β1. When c2>c1 and β2<β1, the mutant could invade only when it started out not too far behind the wavefront. Thus, the wave speeds and transmission rates were key features in determining invadability in the RDE.
Next, we compared the invadability behavior of the RDE to that for the IPS. In Fig. 12 we identify the parameter values, β2 and δ2, that led to invasion in the RDE; of course, there are no probabilities to consider in this deterministic model. These invasion points were found to lie in a region that was fairly well approximated by the invasion region for the IPS simulations, indicating that the invadability conditions for a single mutant were roughly similar in these two models. The two invasion data points in Fig. 12 that fell below the curve β2 · (β2/δ2)1/3 = β1 · (β1/δ1)1/3 satisfied the conditions c2>c1 and β2>β1, and so were to be expected. We conclude that, as far as predicting the possibility of a successful invasion by a single mutant pathogen near the leading edge of an expanding wave of wild-type infectives, the RDE model is very similar in behavior to the IPS model and certainly a better predictor than the ODE model. Of course, the finer details such as the actual probability of an invasion do not even make sense in the RDE model.
Fig. 12.
Invadability for a single mutant in the RDE model as a function of δ2, β2 (with δ1 = 0.0002, β1 = 0.002, β1/δ1 = 10 held constant). The dark triangles represent parameter values for which the RDE resulted in a successful mutant invasion; the open circles correspond to failed invasion. (There are no probabilities in this deterministic model.) The straight line giving the critical ratio in Fig. 5 is not applicable here. The solid curve gives values of δ2, β2 satisfying the relation ; dash-dot curve ; dashed curve β2r2 = β1r1.
4. Discussion
When a pathogen encounters a sufficiently high density of susceptible hosts in a spatially extended setting, an infectious wave results. Such traveling waves of pathogens have been observed in the spread of epidemics (Murray, 1989) and in spatially structured experimental systems (Yin, 1993; Yin and McCaskill, 1992). The fact that such waves are common is not surprising mathematically; they are predicted by traveling wave solutions to RDE and by the shape theorem for IPS models.
During the spread of an infectious wavefront into a population of susceptible hosts, mutant pathogens can arise. It is the goal of this paper to understand the factors that determine the fate of such mutants. In order to account for spatial structure and the stochastic effects that arise due to the small numbers of individuals involved in local interactions and the initial phases of a mutant invasion, we used a stochastic individual-based lattice model (IPS). We compared the predictions of our model to those of several differential equation models to see how model assumptions can influence results. It was seen, for example, that conditions leading to the invasion of a mutant pathogen in the IPS model are quite different from those in an ODE model and rather close to those in a reaction–diffusion model.
We studied the IPS model via two sets of simulations. In the first, we introduced a single mutant infective to the infectious wavefront of a wild-type pathogen. The probability that such a mutant could successfully invade was considered as a function of its position relative to the wild-type wavefront and the relative values of transmission and mortality rates for mutant and wild-type pathogens. This led us to formulate an invasion condition that showed good agreement with the simulations. It also illustrated a striking difference with invasion conditions for mass-action ODE models, which are based on the basic reproductive ratios of the two pathogens.
In the second set of simulations, we allowed mutant pathogens to arise spontaneously from wild-type infectives. This permitted us to estimate the mean time to a mutant invasion. We derived an analytical formula for the mean invasion time based on the invasion probability of a single mutant (from the first set of simulations) and the mutation rate. We were thus able to link the two sets of simulations in a way that allows one to estimate microscopic parameters from macroscopic observations. We also used these simulations and our understanding of the dynamics of the model to gain considerable insight into the appearance of mutants in bacteriophage plaques, as studied by Yin (1993).
Acknowledgements
We thank Jim Bull for suggesting this problem and for pointing us to Yin's work. We also thank Holly Wichman, Caitlin Coberly, and Darin Rokyta for discussions about bacteriophage. Yongtao Guan generously provided advice on coding the simulations. WW and SMK were supported in part by the National Institutes of Health award number P20 RR 16448 from the COBRE program of the National Center for Research Resources. Finally, we thank the reviewers for comments that led to improvements in the presentation.
References
- Boots M, Hudson PJ, Sasaki A. Large shifts in pathogen virulence relate to host population structure. Science. 2004;303:842–844. doi: 10.1126/science.1088542. [DOI] [PubMed] [Google Scholar]
- Bremermann HJ, Thieme HR. A competitive-exclusion principle for pathogen virulence. J. Math. Biol. 1989;27:179–190. doi: 10.1007/BF00276102. [DOI] [PubMed] [Google Scholar]
- Durrett R. Lecture Notes on Particle Systems and Percolation. Wadsworth; Belmont, CA: 1988. [Google Scholar]
- Durrett R, Neuhauser C. Particle systems and reaction diffusion equations. Ann. Probab. 1994;22:289–333. [Google Scholar]
- Edmonds CA, Lillie AS, Cavalli-Sforza LL. Mutations arising in the wave front of an expanding population. Proc. Natl Acad. Sci. USA. 2004;101:975–979. doi: 10.1073/pnas.0308064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi Y, Sasaki A. The evolution of parasite virulence and transmission rate in a spatially structured population. J. Theoret. Biol. 2000;203:85–96. doi: 10.1006/jtbi.1999.1065. doi:10.1006/jtbi.1999.1065. [DOI] [PubMed] [Google Scholar]
- Koch AL. The growth of viral plaques during the enlargement phase. J. Theoret. Biol. 1964;6:413–431. doi: 10.1016/0022-5193(64)90056-6. [DOI] [PubMed] [Google Scholar]
- Krone SM. Spatial models: stochastic and deterministic. Math. Comput. Modelling. 2004;40:393–409. [Google Scholar]
- May RM, Anderson RM. Epidemiology and genetics in the coevolution of parasites and hosts. Proc. R. Soc. London B. 1983;219:281–313. doi: 10.1098/rspb.1983.0075. [DOI] [PubMed] [Google Scholar]
- Murray JD. Mathematical Biology. Springer; Berlin: 1989. [Google Scholar]
- Nowak MA, May RM. Superinfection and the evolution of parasite virulence. Proc. R. Soc. London B. 1994;255:81–89. doi: 10.1098/rspb.1994.0012. [DOI] [PubMed] [Google Scholar]
- Rand DA, Keeling M, Wilson HB. Invasion, stability and evolution to criticality in spatially extended, artificial host–pathogen ecologies. Proc. R. Soc. London B 259. 1995:55–63. [Google Scholar]
- Sato K, Matsuda H, Sasaki A. Pathogen invasion and host extinction in lattice structured populations. J. Math. Biol. 1994;39:251–268. doi: 10.1007/BF00163881. [DOI] [PubMed] [Google Scholar]
- Scearce LM, Pierce JC, McInroy B, Masker W. Deletion mutagenesis independent of recombination in bacteriophage T7. J. Bacteriol. 1991;173:869–878. doi: 10.1128/jb.173.2.869-878.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J. Evolution of bacteriophge T7 in a growing plaque. J. Bacteriol. 1993;175:1272–1277. doi: 10.1128/jb.175.5.1272-1277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, McCaskill JS. Replication of viruses in a growing plaque: a reaction–diffusion model. Biophys. J. 61. 1992:1540–1549. doi: 10.1016/S0006-3495(92)81958-6. [DOI] [PMC free article] [PubMed] [Google Scholar]