Abstract
A panel of biomarkers may improve predictive performance over individual markers. Although many biomarker panels have been described for ovarian cancer, few studies used pre-diagnostic samples to assess the potential of the panels for early detection. We conducted a multi-site systematic evaluation of biomarker panels using pre-diagnostic serum samples from the Prostate, Lung, Colorectal, and Ovarian Cancer (PLCO) screening trial.
Using a nested case-control design, levels of 28 biomarkers were measured laboratory-blinded in 118 serum samples obtained before cancer diagnosis and 951 serum samples from matched controls. Five predictive models, each containing 6–8 biomarkers, were evaluated according to a pre-determined analysis plan. Three sequential analyses were conducted: blinded validation of previously established models (Step 1); simultaneous split-sample discovery and validation of models (Step 2); and exploratory discovery of new models (Step 3). Sensitivity, specificity, sensitivity at 98% specificity, and AUC were computed for the models and CA125 alone among 67 cases diagnosed within one year of blood draw and 476 matched controls. In Step 1, one model showed comparable performance to CA125, with sensitivity, specificity and AUC at 69.2%, 96.6% and 0.892, respectively. Remaining models had poorer performance than CA125 alone. In Step 2, we observed a similar pattern. In Step 3, a model derived from all 28 markers failed to show improvement over CA125.
Thus, biomarker panels discovered in diagnostic samples may not validate in pre-diagnostic samples; utilizing pre-diagnostic samples for discovery may be helpful in developing validated early detection panels.
Keywords: Early Detection, Screening, Biomarkers, Validation, Study Design
Introduction
Ovarian cancer is the fifth leading cause of cancer death among women in the US. While early detection might reduce ovarian cancer mortality, there is currently no proven effective early detection tool for the disease.
In the last decade, many serum biomarkers or panels of biomarkers have been reported to detect ovarian cancer with higher sensitivity and specificity than the best marker currently available, CA125 (1–4). With one exception (5), such studies utilized serum samples collected at the time of diagnosis, and generally included a high proportion of cases with advanced stage disease. Further, few of these biomarkers or panels have been evaluated in a rigorous validation study. Thus, their utility for screening, which requires detection at an asymptomatic phase, cannot be determined. This general scenario is not limited to ovarian cancer – for virtually all of the major cancers, many promising predictive biomarkers have been identified, but few have been tested rigorously in pre-diagnostic specimens (specimens collected before clinical manifestation of the disease from asymptomatic subjects).
This report is the second of two companion reports describing a multi-site, simultaneous, coordinated effort to systematically evaluate the performance of biomarkers for early detection of ovarian cancer using a nested case-control design and stored, pre-diagnostic serum samples obtained from the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. The first report details the developmental process for selecting the best biomarkers from phase II (diagnostic) and phase III (pre-diagnostic) specimens to be included in a final panel of biomarkers from a larger pool of candidate markers (6). This report proposes a novel, systematic approach for un-biased evaluation of classification models combining multiple biomarkers and presents the performance results in pre-diagnostic samples of five predictive models derived from the first report.
Materials and Methods
PLCO Biorepository
The pre-diagnostic serum samples used in the current study were from the PLCO biorepository. PLCO is a randomized controlled cancer screening trial evaluating various screening tests for the four PLCO cancers. Over 150,000 healthy subjects ages 55–74 from across the U.S. were randomized to a screening or usual care arm at ten screening sites from 1993 to 2001. The primary outcome of the trial is to assess whether routine screening can reduce cancer-specific mortality (7, 8). The overall screening protocol has been described elsewhere (8). For ovarian cancer screening, women with at least one ovary at baseline received a CA125 blood test at each of six annual screenings, and a transvaginal ultrasound (TVU) at the first four screenings (9). Subjects who tested positive for either CA125 or TVU were referred to their local physicians who determined the diagnostic workup procedures. Any diagnosis of cancer and its stage, grade, and initial treatment, were obtained. Subjects with positive tests but no cancer diagnosis continued to undergo annual screenings. Cancers diagnosed in between screens, or after the screening period ended, were identified through annual surveys of cancer and vital status. Data on demographics, risk factors and dietary information were collected through multiple questionnaires administered at baseline and during the follow up period.
Blood samples were collected from intervention arm subjects at each of the six annual screens (10). Therefore, up to six serial bloods may be available for a given subject. The collection of biospecimens was approved by the NCI Special Studies Institutional Review Board (OH97-C-N041) and by the local Institutional Review Board for each of the screening sites. Informed consent was obtained from all subjects who provided blood samples to be stored for future research. Blood samples were processed in several different ways to obtain serum, plasma, buffy coat, red blood cells, or cryo-preserved whole blood.
Study Coordination
Six investigator groups participated in this study; each group’s proposal was approved by the PLCO Etiologic and Early Marker Studies (EEMS) Review Panel1, on the basis of scientific merits, to use PLCO pre-diagnostic specimens to evaluate a panel of biomarkers for early detection of ovarian cancer. The specific markers included in each panel are shown in Table 1. The rationale for selecting these markers is detailed in the companion report (6). Most of these markers had been previously shown to differentiate clinical cases from control subjects with high sensitivity and specificity (2–5, 11–13).
Table 1.
Summary of the biomarker panels
| Panel A | Panel B | Panel C | Panel D | Panel E | |
|---|---|---|---|---|---|
| PI Institution | Yale University | Partners | Fred Hutchinson Cancer Research Center |
MD Anderson Cancer Center/NCI DCEG |
University of Pittsburgh |
| Sample Allocation | 1X200ul | 2x300ul | 1X300ul | 1X100ul | 1X200ul |
| Marker Name (Gene Symbol) | Assayed, used in Step 1, Step 2 or Step 3 | ||||
| Apolipoprotein A-I (APOA1) | 1, 2 | ||||
| Beta-2-microglobulin (B2M) | 3 | ||||
| B7-H4 (VTCN1) | 1, 2 | ||||
| CA125 (MUC16) | 1, 2 | 1, 2, 3 | 1, 2 | 1, 2* | 1, 2 |
| CA15-3 (MUC1) | 1, 2 | ||||
| CA19-9 | X | ||||
| CA72-4 | 1, 3 | 1 | |||
| CTAPIII (PPBP) | 1, 2 | ||||
| EGFR (EGFR) | 1, 2 | ||||
| Eotaxin (CCL11) | 1 | ||||
| HE4 (WFDC2) | 1, 2, 3 | 1, 2 | 1, 2 | ||
| Hepcidin-25 (HAMP) | X | ||||
| IGFBPII (IGFBP2) | 1 | ||||
| IGF-II (IGF2) | 1, 2 | ||||
| ITIH4 (ITIH4) | X | ||||
| Kallikrein-6 (KLK6) | X | ||||
| Leptin (LEP) | 1, 2 | ||||
| Mesothelin (MSLN) | 1 | ||||
| MIF (MIF) | 1, 2 | ||||
| MMP-3 (MMP3) | 1 | ||||
| MMP-7 (MMP7) | 1, 2 | ||||
| OPN (SPP1) | 1, 2 | ||||
| Prolactin (PRL) | 1, 2 | 1, 2 | |||
| Secretory Leukocyte Protease Inhibitor (SLPI) | 3 | ||||
| Spondin 2 (SPON2) | 1 | ||||
| sVCAM-1 (VCAM1) | 1 | ||||
| Transferrin (TF) | X | ||||
| Transthyretin (TTR) | 1, 2 | ||||
1, 2, 3 denotes the steps in which the marker was used in the model. X denotes that the marker was assayed in sera but not used in any step model.
The MD Anderson/NCI group used CA125 values obtained from the PLCO Trial.
The NCI PLCO leadership assumed overall coordination of these studies, with the investigators’ consent, input and collaboration, in order to standardize sampling, statistical methods, and data interpretation across the studies.
Common sampling plan
Figure 1 shows the subject selection criteria. Among 24,650 eligible subjects, 118 cases of pathologically confirmed (through May 2006) invasive ovarian, primary peritoneal, and fallopian tube cancers with appropriate consents and available samples were identified. Both screen-detected cases (identified from diagnostic workup subsequent to a positive CA125 or TVU test), and clinically diagnosed cases were included. For each case, eight controls were randomly selected from 24,473 healthy subjects without cancer: four general population controls, 2 controls with a family history of breast or ovarian cancers, and two controls with elevated CA125. These special controls were included to assess the performance of the models in high risk populations but were not included in primary analyses. Controls were frequency-matched by age and calendar year of blood draw. For each study subject, a single serum sample closest and prior to diagnosis (proximate sample) was selected for laboratory analysis.
Figure 1.
Sampling scheme. Note: 119 cases and 952 controls were originally selected following the sampling scheme. One case was excluded due to a coding error, and one control was excluded due to a consent issue, leaving 118 cases and 951 controls in the final sample set. LMP denotes tumors of low malignant potential.
Common data analysis plan
The common data analysis plan was formulated to clearly distinguish between validation and discovery, both of which were to be accommodated in the overall analysis strategy. In this study, validation refers to “hypothesis testing”, i.e. the assessment of a “locked-in” marker panel and classification model conducted on blinded samples. A stepwise analytic plan, as described below, was developed to accommodate the multiple study goals (i.e. validation and discovery), while ensuring uniformity in study conduct and data interpretation.
Step 1: Validation of previously established models
Step 1 was designed to determine the performance of each marker panel and associated classification model that was pre-established in diagnostic samples. Each investigator team was required to predict case/control status on each sample, using the classification model. The predictions, along with individual marker values and description of the model, were submitted to PLCO so that the PLCO statistician could calculate performance indicators for each marker panel on behalf of the investigators, who would remain blinded to the samples throughout the entire process.
Step 2: Simultaneous discovery and validation of models
Step 2 was designed to determine whether models tested in Step 1 could be improved if investigators were given the opportunity to train (discover) on a subset of the PLCO pre-diagnostic samples and subsequently validate on the remaining blinded samples. Upon completion of Step 1, the data set was thus randomly split 50–50 (done by a third party programmer who was not involved in the data analysis) into a training set (60 cases and 476 controls), which was subsequently unblinded, and a validation set (58 cases and 475 controls), which remained blinded. Covariates associated with the samples, including demographic, lifestyle, medical history and other common risk factors such as hormone use status were made available at this point for model building. As in Step 1, investigators were required to predict the case/control status of each sample in the validation set. Prediction models were based on 60 cases and 239 general population controls in the training set. The predictions, along with the modified classification model, were submitted to the PLCO statistician for computation of performance measures as in Step 1. Note that investigators were not provided their Step 1 results until after Step 2 had been completed to ensure total blinding.
Step 3: Exploratory discovery of new models
Step 3 was for discovery of new panels and models. At this time, PLCO released (un-blinded) the full dataset to all investigator teams so that they could further refine panels and models by training on the entire sample set2. This approach increased the sample size and study power, but it did not provide an opportunity for validation in an independent sample set within PLCO. We report here the results of a single family of logistic models developed on the pooled set of markers across all sites (“pan-site model”). The fixed subset of markers used across this family was determined based on performance over all 118 cases and 476 general population controls; distinct individual models in the family corresponded to different intervals from blood draw to diagnosis (e.g., ≤ 1 year, 1–2 years) and were fit on these cases and all general population controls.
Statistical methods
The primary performance measures were agreed upon a priori; these included sensitivity, specificity, area under the Receiver Operating Characteristic (ROC) curve (AUC), and sensitivity at 98% specificity (SE98). Note the latter two measures could be computed only for those models that produced a propensity score (i.e., a one dimensional quantity in which increasing values indicate a greater likelihood of disease). For comparison, all of these measures were also computed for CA125 alone (using the PLCO trial cutoff of ≥35 U/ml).
Performance measures for the Step 1 models (and the Step 3 model) were computed for both the training and validation data sets, whereas performance measures for the Step 2 models were computed for the validation set only. However, to facilitate direct comparison of Step 1 and Step 2 models, we also computed the Step 1 performance measures limited to what became the validation set in Step 2. For three of the panels (A, C, D), both the Step 1 and 2 models incorporated propensity scores; thus each allowed for computation of AUC and SE98. For these, we directly compared AUC and SE98 across each step and tested the null hypothesis of no difference between the steps; for AUC, the statistical test accounted for the fact that the AUCs at the two steps were correlated (14). The remaining two models (B, E) incorporated a propensity score at only Step 1 or Step 2. For these we could not compare AUC or SE98 across steps; however, we could compare the sensitivity at the fixed level of specificity (e.g. X%) achieved by the model without the propensity score. Specifically, we adjusted the propensity score of the model with such a score to find a cutoff that gave a specificity of X and then computed the corresponding sensitivity of the model at that cutoff.
Results
Table 1 shows the markers assayed and their inclusion in the models for Step 1 through Step 3. Note that two investigator groups (MDACC and NCI DCEG) had independently developed the same panel, resulting in five distinct marker panels3. A total of 28 individual biomarkers were assayed. CA125 was included in all panels, and HE4 was included in 3 of the 5 panels. The pan-site (Step 3) model included CA125, HE4, CA72.4, SLPI, and B2M. The Step 1 and Step 2 models ranged from linear combinations of log marker values to highly non-linear expressions incorporating the maximum of a set of normalized marker values. One model (B2) incorporated family history of breast and ovarian cancer and smoking status in addition to marker values. Detailed description of each model is provided in Supplemental Materials.
Table 2 shows the demographic features of all cases and controls originally selected for this study. All subjects were 55 or older (by design) and most were white.
Table 2.
Population characteristics of the samples
| Race/Ethnic Group | Case (N=118) |
Control (N=951) |
|---|---|---|
| White, Non-Hispanic | 106 (89.8%) | 872 (91.7%) |
| Black, Non-Hispanic | 6 (5.1%) | 47 (4.9%) |
| Hispanic | 2 (1.7) | 9 (0.9%) |
| Asian | 3 (2.5%) | 21 (2.2%) |
| American Indian | 1 (0.8%) | 2 (0.2%) |
| Age at Serum Draw | ||
| 55–59 | 14 (11.9%) | 120 (12.6%) |
| 60–64 | 37 (31.4%) | 295 (31%) |
| 65–69 | 38 (32.2%) | 304 (32%) |
| 70–74 | 24 (20.3%) | 192 (20.2%) |
| 75–79 | 5 (4.2%) | 40 (4.2%) |
| All | 118 (100%) | 951 (100%) |
Although 118 cases were originally included in the study, the interval from the date of the sample (blood draw) to diagnosis varied widely, ranging from 12 to 2898 days, with a majority (N=67, 57%) being less than one year. For the purpose of comparing the performance of the models to each other and to CA125 alone, we restricted our analysis to the 67 cases within one year and 26 cases 1–2 years of diagnosis. Table 3 shows the distribution of histologic type, stage and grade of these cases. Additionally, only the general population controls were used for the main analyses.
Table 3.
Characteristics of cases diagnosed within 2 years from blood draw (N=93)
| Cancer type | ≤12 months N(%) |
13–24 months N(%) |
|---|---|---|
| Ovarian | 48 (71.6) | 23 (88.5) |
| Primary peritoneal | 10 (14.9) | 3 (11.5) |
| Fallopian tube | 9 (13.4) | 0 (0.0) |
| Histologic type | ||
| Serous cystadenocarcinoma | 40 (59.7) | 14 (53.8) |
| Mucinous cystadenocarcinoma | 1 (1.5) | 0 (0.0) |
| Endometrioid adenocarcinoma | 7 (10.4) | 5 (19.2) |
| Clear Cell Cystadenocarcinoma | 3 (4.5) | 1 (3.8) |
| Undifferentiated Carcinoma | 1 (1.5) | 0 (0.0) |
| Other | 2 (3.0) | 0 (0.0) |
| Adenocarcinoma, NOS/ Carcinoma, NOS | 11 (16.4) | 5 (19.2) |
| Malignant granulosa | 2 (3.0) | 1 (3.8) |
| Stage | ||
| Stage I/II | 17 (25.4) | 11 (42.3) |
| Stage III | 42 (62.7) | 9 (34.6) |
| Stage IV | 8 (11.9) | 5 (19.2) |
| Unknown | 0 (0.0) | 1 (3.8) |
| Grade | ||
| Well differentiated | 2 (3.0) | 3 (11.5) |
| Moderately differentiated | 13 (19.4) | 5 (19.2) |
| Poorly differentiated | 41 (61.2) | 14 (53.8) |
| Unknown | 11 (16.4) | 4 (15.4) |
| Mode of Detection | ||
| Screen Detected | 49 (73.1) | 1 (3.8) |
| CA125+; TVU− | 17 | 0 |
| CA125+; TVU+ | 15 | 0 |
| CA125+; TVU not done | 8 | 0 |
| CA125− TVU+ | 9 | 1 |
| Not Screen Detected | 18 (26.9) | 25 (96.2) |
| CA125−; TVU− | 9 | 16 |
| CA125−; TVU not done | 7 | 8 |
| ߓCA125 not done; TVU not done | 2 | 1 |
| Total | 67 | 26 |
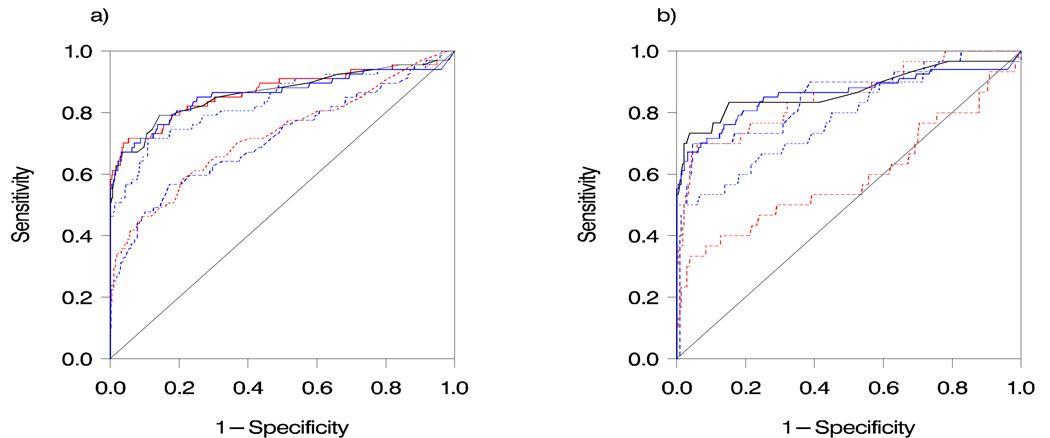
Table 4 displays the results for the five models for Steps 1 and 2. For the within-one-year cases, one model (B1) had comparable performance to CA125 alone in Step 1, with sensitivity, SE98, specificity, and AUC of 69.2%, 64.6%, 96.6% and 0.892, respectively, compared to 63.1%, 64.6%, 98.5% and 0.890 for CA125. Three of the models (A1, C1, E1) showed substantially lower sensitivity (34.3% to 37.9%), SE98 (25.4% to 32.8%) and AUC (0.712 to 0.721) than CA125. Finally, one model (D1) employed a low cutoff, resulting in very low specificity (32.2%) but high sensitivity (95.4%); AUC (0.858) and SE98 (52.3%) were slightly lower than that of CA125. In Step 2, we observed the same general pattern as in Step 1 in the within-one-year cases, with one model (B2) performing comparably to CA125 alone and the remaining models showing overall lower performance. Figure 2 compares the receiver operating characteristic (ROC) curves for the models, CA125 and the Step 3 pan-site model. For the 1–2 year cases, sensitivity ranged from 0% to 21.4% for the models and 0% for CA125 alone.
Table 4.
Results of models from Steps 1 to 3
| Model | Sensitivity* ≤ 12 mo % (95% CI) |
Sensitivity* 13–24 mo % (95% CI) |
Specificity* % (95% CI) |
ROC*♦ Area (95% CI) |
Sensitivity at 98% Specificity♦ % (95% CI) |
|---|---|---|---|---|---|
| Step 1 | N=67 | N=26 | N=476 | N=67 | |
| A1 | 34.3 (23–46) | 7.7 (1–25) | 96.8 (95.2–98.4) | 0.721 (0.64–0.80) | 32.8 (22–44) |
| B1 | 69.2 (58–80) | 12.5 (3–31) | 96.6 (94.9–98.3) | 0.892 (0.84–0.95) | 64.6 (53–76) |
| C1 | 34.3 (23–46) | 11.5 (2–30) | 95.1 (93.1–97.1) | 0.712 (0.63–0.79) | 25.4 (15–36) |
| D1 | 95.4 (90–99) | 76.0 (59–93) | 32.2 (27.4–36.5) | 0.858 (0.80–0.92) | 52.3 (40–64) |
| E1 | 37.9 (26–50) | 3.9 (0–20) | 89.8 (87.0–92.6) | N/A† | N/A† |
| CA125‡ | 63.1 (51–75) | 0.0 (0–13) | 98.5 (97.4–99.6) | 0.890 (0.84–0.94) | 64.6 (53–76) |
| Step 2 †† | N=30 | N=15 | N=237 | N=30 | |
| A2 | 53.3 (35–71) | 6.7 (0–32) | 96.6(94.3–98.8) | 0.852 (0.77–0.94) | 36.7 (20–54) |
| B2 | 80.0 (66–94) | 21.4 (5–50) | 92.2(88.7–95.7) | N/A† | N/A† |
| C2 | 70.0 (54–86) | 6.7 (0–32) | 91.9 (88.4–95.4) | 0.848 (0.76–0.94) | 46.7 (29–64) |
| D2 | 55.2 (37–73) | 0.0 (0–22) | 86.9 (82.5–91.3) | 0.810 (0.72–0.90) | 51.7 (34–69) |
| E2 | 30.0 (14–46) | 13.3(2–40) | 96.2 (93.7–98.7) | 0.590 (0.46–0.72) | 23.3 (8–38) |
| CA125‡ | 72.4 (56–89) | 0.0 (0–22) | 97.9 (96.0–99.8) | 0.898 (0.82–0.98) | 72.4 (56–89) |
| Step3 (Pan-site) | N/A§ | N/A§ | N/A§ | 0.911 (0.86–0.96) | 68.2 (57–80) |
Calculated based on cut-off specified by the model. Note that we included all general population controls in the calculation of specificity and ROC; these as a whole were comparable to the one year cases with respect to the matching variables of age and calendar year of blood draw.
Calculated based on the 67 cases diagnosed ≤12 months from blood draw.
This model did not produce a propensity score, thus the measure cannot be calculated.
Using data previously obtained in PLCO for the same subject and study year as the samples in the current study, and a cutoff of ≥35 U/ml.
For Step 2 model, data from the validation set is shown.
The pan-site model did not have a cutoff, therefore no sensitivity or specificity can be calculated.
Figure 2.
Receiver operating characteristic (ROC) curves for Step 1 models (a) and Step 2 models (b), compared to that of CA125 and the Step 3 (pan-site) model in the within-one-year cases. Black solid: CA125 alone; blue solid: Step 3 model; blue dotted: Panel D; blue dashed: Panel C; red solid: Panel B; red dotted: Panel A; red dashed: Panel E. Note: Figure 2b curves are based on validation set only.
Table 5 displays direct comparisons of the performance of the Step 1 and Step 2 models for each panel for the within-one-year cases. For the three panels (A, C & D) where the AUC could be computed at both steps, AUC was statistically significantly improved in two of the panels (with an increase of about 0.10) but was significantly worsened in the other panel (decrease of about 0.10). For the other two panels (B & E), sensitivity at fixed specificity was slightly lower at Step 2 than Step 1.
Table 5.
Direct comparison of Step 1 and Step 2 model results* for the ≤12 month cases
| Model | Sensitivity at Fixed Specificity§ | ROC AUC | ||
|---|---|---|---|---|
| Step 1 | Step 2 | Step 1 | Step 2 | |
| A | 36.7 | 36.7 | 0.753 | 0.852† |
| B | 86.7 | 80.0 | ||
| C | 33.3 | 46.7 | 0.754 | 0.848† |
| D | 55.2 | 51.7 | 0.899† | 0.810 |
| E | 37.9 | 36.7 | ||
Both Step 1 and Step 2 results are restricted to validation set only to facilitate direct comparison.
Specificity fixed at 98% (in both steps) for panels A, C, and D. For panel B, specificity was fixed (in both steps) at 92.2%, the level of the B2 model, while for panel E specificity was fixed (in both steps) at 91.1%, the level of the E1 model for the validation set.
Significantly elevated compared to model in other Step (p < 0.05)
The Step 3 model showed an AUC of 0.911 and a SE98 of 68.2% in the within-one-year cases (Table 4). Its performance in these cases was only slightly, and not statistically significantly, better than both that of CA125 alone and that of the best performing Step 1 or 2 model, namely B1. For the 1–2 year cases, the AUC of the model was 0.740, compared to 0.642 for CA125 (p=0.14) (not shown in Table 4).
We also examined the performance of each prediction model in high risk women using the special controls (data not shown). None of the models (at Step 1 or 2) displayed a significant difference in specificity (or sensitivity) between women who reported a family history of breast or ovarian cancer and those who did not. We also observed that, all models except E1 and E2 showed significantly decreased specificity (and significantly increased sensitivity) in women with elevated CA125 compared to those with normal CA125.
Discussion
The importance of using appropriate specimens for biomarker research at all stages has gained much attention in recent years, largely due to the fact that many enthusiastic reports of promising biomarkers have turned out not to be reproducible. This phenomenon has been examined by numerous investigators (15–20). One of the major problems is bias introduced by systematic differences between the case and control specimens used for biomarker discovery that inflates the performance of the markers for cancer diagnosis (21–24).
In 2008, Pepe et al described the PRoBE (prospective-specimen collection, retrospective-blinded-evaluation) design to “eliminate common biases that pervade the biomarker research literature" (25). In the PRoBE approach, a "case-control" analysis is conducted among subjects followed in a cohort study with prospectively collected specimens. Many potential biases are prevented when specimens are collected and handled in a "blinded" manner, before the diagnosis is known. The current PLCO analysis, designed before the PRoBE approach was described, utilized essentially the same approach.
The idea that a panel of biomarkers could perform significantly better than individual biomarkers alone has gained popularity in recent years. A number of studies have shown improved performance of a panel of ovarian cancer biomarkers over CA125 alone (26–28) when used in diagnostic samples. Contrary to these reports, the current study found that the inclusion of additional biomarkers appeared to add little to CA125 when used in pre-diagnostic samples.
The current study is significant in several ways. First, it provides the first example of a coordinated, systematic approach to biomarker validation using pre-diagnostic samples. Second, the findings raise a question about the current paradigm for biomarker development, namely, using diagnostic samples for discovery and validating them in pre-diagnostic samples. It is possible that markers discovered in diagnostic samples are significantly differentially expressed only when the tumor becomes large, or clinically apparent. Such markers may have little value for early detection.
As mentioned in the companion paper (6), the fact that screening with CA125 was ongoing at the time of the blood draws may have affected the resulting estimates of the sensitivity (and AUC) of CA125, especially with regard to the more-than-one-year cases. Specifically, subjects with elevated CA125 tended to be diagnosed within one year of blood draw due to follow up of the PLCO screening; thus such cases were selectively excluded from the pool of more-than-one-year cases. Indeed, the sensitivity of CA125 in the 13–24 month cases here was exactly zero (using the PLCO cutoff of 35 U/ml). Since each of the models described here incorporates the CA125 level, the ongoing screening may have also affected the estimates of their performance, although probably not to as great an extent. For the purpose of comparing the performance of the models to that of CA125 alone, we believe that the results restricted to the within-one-year cases provide for a reasonably unbiased assessment. For cases further removed from the blood draw, including the 13–24 month cases, however, the comparison may be somewhat biased in favor of the models
As a next step, it is possible that incorporating longitudinal marker values obtained from serial samples may improve the performance of a marker or a panel of markers. It has been shown that utilizing serial CA125 values improved its performance (29). To this end, two of the current investigator teams have already been approved to use the PLCO serial samples to further evaluate their marker panels and models.
A lesson learned from this exercise is that more attention needs to be directed toward biomarker discovery. A critical aspect of study design should be choosing the appropriate specimens according to the intended use of the biomarker. For biomarkers of early disease, it may be necessary to use pre-diagnostic samples for discovery. We hope this study serves as a catalyst for further discussions on how to move the biomarker field forward toward clinical applications.
Supplementary Material
Acknowledgements
The authors thank Dr. Philip Prorok, Division of Cancer Prevention, National Cancer Institute, the Screening Center investigators and staff of the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, Mr. Tom Riley and staff, Information Management Services, Inc., Ms. Barbara O’Brien and staff, Westat, Inc. Most importantly, we acknowledge the study participants for their contributions to making this study possible.
Financial support: This work was supported in part by contracts from the National Cancer Institute to 10 PLCO screening centers, a coordinating center and a central analytic laboratory; grants from the National Cancer Institute (P50 CA083636 to NU, P50 CA083639 to RCB, 5U01 CA86381 to DWC, R01 CA127913 and U01 CA084986 to GM); and grants from Golfers Against Cancer and the Wiley Mossy Foundation to RCB.
Footnotes
More information about access to PLCO biorepository resource is available on the website www.plcostars.com or plco.cancer.gov (note that this is a resource for studying many cancers, not just prostate, lung, colorectal, and ovarian cancers).
Some investigator teams wished to remain blinded to the validation set for future studies. In this case, the full dataset was released to a statistician designated by the team.
This overlap was not identified until the marker data had been submitted to PLCO, due to the complexity of the project, and the fact that the markers eventually measured were somewhat different from those originally proposed. Both panels were measured using the same ProteinChip platform developed by Vermillion. Correlation coefficients between the MDACC and NCI DCEG data for the same marker ranged from 0.22 (APOA1) to 0.94 (ITIH4). The PIs agreed to jointly represent the panel (based on MDACC dataset) in this report.
References
- 1.Petricoin EF, Ardekani AM, Hitt BA, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z, Bast RC, Jr, Yu Y, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 3.Gorelik E, Landsittel DP, Marrangoni AM, et al. Multiplexed Immunobead-Based Cytokine Profiling for Early Detection of Ovarian Cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:981–987. doi: 10.1158/1055-9965.EPI-04-0404. [DOI] [PubMed] [Google Scholar]
- 4.Visintin I, Feng Z, Longton G, et al. Diagnostic markers for early detection of ovarian cancer. Clin Cancer Res. 2008;14:1065–1072. doi: 10.1158/1078-0432.CCR-07-1569. [DOI] [PubMed] [Google Scholar]
- 5.Anderson GL, McIntosh M, Wu L, et al. Assessing Lead Time of Selected Ovarian Cancer Biomarkers: A Nested Case-Control Study. J Natl Cancer Inst. 2010;102:26–38. doi: 10.1093/jnci/djp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cramer DW, Bast RC, Berg CD, et al. Ovarian Cancer Biomarker Performance in Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial Specimens. doi: 10.1158/1940-6207.CAPR-10-0195. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gohagan JK, Prorok PC, Hayes RB, Kramer BS. The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: history, organization, and status. Control Clin Trials. 2000;21 doi: 10.1016/s0197-2456(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 8.Prorok PC, Andriole GL, Bresalier RS, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 9.Buys SS, Partridge E, Greene MH, et al. Ovarian cancer screening in the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial: Findings from the initial screen of a randomized trial. Am J Obstet Gynecol. 2005;193:1630–1639. doi: 10.1016/j.ajog.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Hayes RB, Reding D, Kopp W, et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin Trials. 2000;21 doi: 10.1016/s0197-2456(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 11.Mor G, Visintin I, Lai Y, et al. Serum protein markers for early detection of ovarian cancer. Proc Natl Acad Sci U S A. 2005;102:7677–7682. doi: 10.1073/pnas.0502178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer C, Duan X, Hawley S, et al. Systematic Evaluation of Candidate Blood Markers for Detecting Ovarian Cancer. PLoS ONE. 2008;3:e2633. doi: 10.1371/journal.pone.0002633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore LE, Fung ET, McGuire M, et al. Evaluation of apolipoprotein A1 and posttranslationally modified forms of transthyretin as biomarkers for ovarian cancer detection in an independent study population. Cancer Epidemiol Biomarkers Prev. 2006;15:1641–1646. doi: 10.1158/1055-9965.EPI-05-0980. [DOI] [PubMed] [Google Scholar]
- 14.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 15.Sturgeon CM, Hoffman BR, Chan DW, et al. National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines for Use of Tumor Markers in Clinical Practice: Quality Requirements. Clin Chem. 2008;54:e1–e10. doi: 10.1373/clinchem.2007.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potter JD. Epidemiology informing clinical practice: from bills of mortality to population laboratories. Nat Clin Prac Oncol. 2005;2:625–634. doi: 10.1038/ncponc0359. [DOI] [PubMed] [Google Scholar]
- 17.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 18.Diamandis EP. Proteomic patterns to identify ovarian cancer: 3 years on. Expert Rev Mol Diagn. 2004;4:575–577. doi: 10.1586/14737159.4.5.575. [DOI] [PubMed] [Google Scholar]
- 19.Contopoulos-Ioannidis DG, Ntzani EE, Ioannidis JPA. Translation of highly promising basic science research into clinical applications. Am J Med. 2003;114:477–484. doi: 10.1016/s0002-9343(03)00013-5. [DOI] [PubMed] [Google Scholar]
- 20.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 21.Ransohoff DF, Gourlay ML. Sources of Bias in Specimens for Research About Molecular Markers for Cancer. J Clin Oncol. 2009;28:698–704. doi: 10.1200/JCO.2009.25.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorpe JD, Duan X, Forrest R, et al. Effects of Blood Collection Conditions on Ovarian Cancer Serum Markers. PLoS ONE. 2007;2:e1281. doi: 10.1371/journal.pone.0001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker SG, Kramer BS, McIntosh M, Patterson BH, Shyr Y, Skates S. Evaluating markers for the early detection of cancer: overview of study designs and methods. Clin Trials. 2006;3:43–56. doi: 10.1191/1740774506cn130oa. [DOI] [PubMed] [Google Scholar]
- 24.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–149. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 25.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–1438. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore RG, McMeekin DS, Brown AK, et al. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol Oncol. 2009;112:40–46. doi: 10.1016/j.ygyno.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das PM, Bast RC., Jr Early detection of ovarian cancer. Biomark Med. 2008;2:291–303. doi: 10.2217/17520363.2.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Yu Y, Xu F, et al. Combining multiple serum tumor markers improves detection of stage I epithelial ovarian cancer. Gynecol Oncol. 2007;107:526–531. doi: 10.1016/j.ygyno.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skates SJ, Menon U, MacDonald N, et al. Calculation of the risk of ovarian cancer from serial CA-125 values for preclinical detection in postmenopausal women. J Clin Oncol. 2003;21:206s–210s. doi: 10.1200/JCO.2003.02.955. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.