Abstract
The heparan sulfate (HS) chains of heparan sulfate proteoglycans (HSPG) are “ubiquitous” components of the cell surface and the extracellular matrix (EC) and play important roles in the physiopathology of developmental and homeostatic processes. Most biological properties of HS are mediated by interactions with “heparin-binding proteins” and can be modulated by exogenous heparin species (unmodified heparin, low molecular weight heparins, shorter heparin oligosaccharides and various non-anticoagulant derivatives of different sizes). Heparin species can promote or inhibit HS activities to different extents depending, among other factors, on how closely their structure mimics the biologically active HS sequences. Heparin shares structural similarities with HS, but is richer in “fully sulfated” sequences (S domains) that are usually the strongest binders to heparin/HS-binding proteins. On the other hand, HS is usually richer in less sulfated, N-acetylated sequences (NA domains). Some of the functions of HS chains, such as that of activating proteins by favoring their dimerization, often require short S sequences separated by rather long NA sequences. The biological activities of these species cannot be simulated by heparin, unless this polysaccharide is appropriately chemically/enzymatically modified or biotechnologically engineered. This mini review covers some information and concepts concerning the interactions of HS chains with heparin-binding proteins and some of the approaches for modulating HS interactions relevant to inflammation and cancer. This is approached through a few illustrative examples, including the interaction of HS and heparin-derived species with the chemokine IL-8, the growth factors FGF1 and FGF2, and the modulation of the activity of the enzyme heparanase by these species. Progresses in sequencing HS chains and reproducing them either by chemical synthesis or semi-synthesis, and in the elucidation of the 3D structure of oligosaccharide–protein complexes, are paving the way for rational approaches to the development of HS-inspired drugs in the field of inflammation and cancer, as well in other therapeutic fields.
Keywords: Heparins, Heparan sulfates, Inflammation, Chemokines, Cytokines, Cancer, Glycotherapeutics
1. Introduction
Heparan sulfate (HS) is a glycosaminoglycan (GAG) often referred to as a “ubiquitous” component of cell surfaces and extracellular matrix (ECM) in the form of HS proteoglycans (HSPGs) in which its polysaccharide chains are covalently attached to peptide/protein cores (Bernfield et al., 1999; Whitelock and Iozzo, 2005; Iozzo, 2005; Bishop et al., 2007; Dreyfuss et al., 2009; Iozzo et al., 2009). Owing to the difficulties in unraveling their complex and diversified structures and functions, HSPGs and their HS chains have been examined only relatively recently. With continuous refinement of methods for their detection and structural analysis and perception of their implication in important pathophysiological processes, HS is arousing increasing interest and is being investigated also with the aim of generating HS-inspired glycotherapeutics (Coombe and Kett, 2005; Bishop et al., 2007; Lindahl, 2007; Laremore et al., 2009; Lindahl and Li, 2009). The well established anticoagulant/antithrombotic drug heparin, which is extracted from animal organs where it is largely stored in the granules of mast cells (Lindahl et al., 1998; Dreyfuss et al., 2009), shares several structural features with HS (Lyon and Gallagher, 1998; Casu and Lindahl, 2001). Heparin species (comprising unfractionated and low-molecular weight heparins, their fractions, fragments, and their variously modified forms) are regarded as agonists or antagonists of HS functions. Most of the biological activities of HS are exerted through interaction with “heparin binding proteins” (Conrad, 1998; Capila and Linhardt, 2002; Coombe, 2008; Lindahl and Li, 2009). Although some of these interactions involve peptide moieties of the HSPG (Bishop et al., 2007; Iozzo et al., 2009), the vast majority are associated with sequences within the GAG chains. This mini review deals with actual or potential modulation of these interactions and the corresponding pathophysiological functions using exogenous heparin species, and focuses on applications in inflammation and cancer. Applications of these species in other therapeutic areas, such as the major field of anticoagulation and thrombosis (Bourin and Lindahl, 1993; Linhardt, 2003) and the prospective ones in viral, Alzheimer's, and malaria diseases (Lindahl, 2007; Lindahl and Li, 2009) are outside the scope of this article. For HS and heparin relevant to inflammation and cancer, we choose to focus on paradigmatic examples rather than attempting to cover the expanding information relating to specific roles of HS, which are covered in other reviews (Linhardt, 2003; Götte, 2003; Iozzo, 2005; Whitelock and Iozzo, 2005; Liu and Sasisekharan, 2005; Gallagher, 2006; Fears and Woods, 2006; Taylor and Gallo, 2006; Bishop et al., 2007; Coombe, 2008; Rek et al., 2009; Fux et al., 2009; Iozzo et al., 2009; Lindahl and Li, 2009).
Among the functions of the HS chains of HSPGs in inflammation and cancer, highlighted in numerous papers (reviewed, i.a., by Parish, 2006 and Yip et al., 2006), a major one is their ability to act as co-receptors for growth factor receptor signaling, whereby they promote cell proliferation and angiogenesis. Other inflammation and cancer-related functions are exerted in cell invasion and metastasis, either directly or following cleavage of HS chains by heparanase (Parish, 2006; Fux et al., 2009). HSPGs, such as syndecan-1 on cell surface (Sanderson and Yang, 2008) and perlecan in basement membranes (Iozzo et al., 2009) can also modulate cell adhesion and migration (Yip et al., 2006). In this mini review, the interactions of HS chains of HSPGs are briefly described from a structural point of view, with the aim of estimating to what extent they can be simulated (agonized or antagonized) by exogenous heparin species. Among open issues of structure–activity relationships, the role of natural or induced local flexibility along heparin/HS chains in affecting protein binding and biological properties will be discussed and, in this context, present and potential pharmaceutical applications of heparin fragments and derivatives will also be presented.
2. Similarities and differences between the structure of heparan sulfate and heparin
Although HS and heparin are localized in different cellular compartments, their structures are qualitatively similar, since they share some common sequences, although represented in quite different proportions depending on the extent of post-polymer biosynthetic transformation of the common precursor N-acetyl-heparosan (Lindahl et al., 1998). In heparin, the N-acetyl groups of d-glucosamine (GlcNAc) in the precursor are largely replaced by N-sulfate groups, and the d-glucuronic acid (GlcA) residues are predominantly converted to l-iduronic acid (IdoA) with subsequent 2-O-sulfation of most of these latter residues, as well as 6-O-sulfation of most of the glucosamine residues. On the other hand, HS still contains large amounts of GlcNAc and GlcA residues and is less O-sulfated than heparin (Lindahl et al., 1998; Lyon and Gallagher, 1998; Casu and Lindahl, 2001). Major sequences in HS are usually represented by N-acetylated domains (NA) constituted by repeating disaccharide units of (GlcA–GlcNAc)n and those of heparin by sulfated domains (S) constituted by extended repeating units of the trisulfated disaccharide IdoA2SO3–GlcNSO36SO3. However, both GAGs, and especially HS, display high degrees of structural heterogeneity and variability, with some aminosugar residues in NA domains being also 6-O-sulfated. In minor proportions, S domains contain some GlcA, IdoA and 2-O-sulfated GlcA residues. A few aminosugar residues bear unsubstituted amino groups and others a 3-O-sulfate group, this latter especially in the pentasaccharide sequence of the active site for antithrombin. The relative content of various units in HS varies significantly depending on animal species and, more markedly, on organs and tissues (reviewed in Laremore et al., 2009). The relative content is also distinct for different heparins (reviewed in Casu, 2005). However, it should be recalled that heparins are purified products and some of their chains could conceivably be lost and some residues modified during “purification” steps. Statistically, HS and heparin can be differentiated in terms of their N-acetyl/N-sulfated GlcN, GlcA/IdoA uronic acids and in their O-sulfate group content (reviewed in Lyon and Gallagher, 1998; Casu and Lindahl, 2001).
An important difference between the structures of HS and heparin is in the arrangement of their major units along the polysaccharide chains. Heparin is characterized by relatively long sequences of repeating trisulfated disaccharide units separated by relatively short, undersulfated sequences containing both IdoA and GlcA residues (NA/NS domain). The NA and S domains of HS are arranged in blocks, separated by “mixed” NA/NS domains containing both N-acetylated and N-sulfated GlcN, as well as GlcA and IdoA (or IdoA2SO3) residues (Gallagher, 2006). A model for such an arrangement of the rat skin fibroblast HS chain (Murphy et al., 2004) is depicted in the upper part of Fig. 1. It should be noted that HS from different sources are characterized by varied domain lengths and spacing between them. As an example, rat liver HS is much richer in higher sulfated (S) regions, and its chains are shorter than in rat skin fibroblast HS (Lyon and Gallagher, 1998). HS and heparin of different origins also differ in their degree of 6-O-sulfation and the location of 6-O-sulfated GlcN residues along HS chains, which seems to be controlled not only by the action of 6-O-sulfotransferases during biosynthetic transformations, but also by post-biosynthetic “editing” exerted on cell surfaces by extracellular 6-O-endosulfatases (Ai et al., 2006). Notably, 6-O-sulfate groups were found at precise, central positions in rat skin fibroblast HS (hexasaccharide) sequences which had been excised from S-regions (Merry et al., 1999).
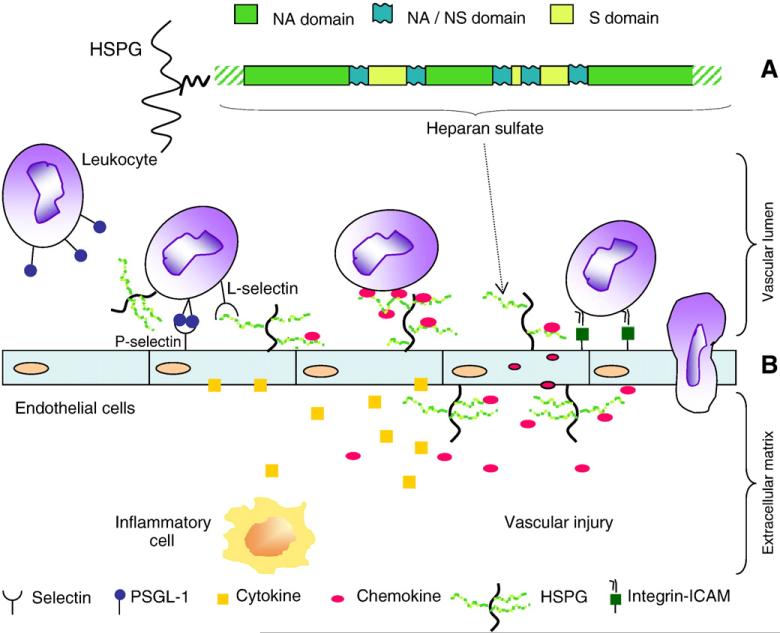
Fig. 1.
(A) Model of a section of a HS chain (adapted from Murphy et al., 2004). (B) Multiple functions for HS during inflammatory reactions. HS participates in each of the major steps of leukocyte extravasation, such as facilitating L-selectin-dependent cell rolling, chemokine transport across the endothelium, and the chemokine presentation to leukocytes, which results in integrin activation (redrawn from Wang et al., 2005; Fig. 1 of Suppl. Inform.). ICAM: Intercellular adhesion molecule.
3. Heparin- and HS-binding proteins
3.1. General aspects
Most of the biological activities of heparin and HS are associated with complexation with proteins (Conrad, 1998; Capila and Linhardt, 2002; Dreyfuss et al., 2009; Lindahl and Li, 2009). Although heparin (or HS)/protein molar ratios in these complexes are usually 1:1, sufficiently long saccharide chains can also form 1:2 complexes either with the same protein or two different proteins (i.e., a growth factor and its receptor). Different topologies of these complexes are depicted in Lindahl and Li (2009).
The interaction of heparin with antithrombin (AT) was the first to be investigated in depth (reviewed in Lindahl and Li, 2009) and led to the rational design of potent synthetic antithrombotic drugs (van Boekel and Petitou, 1993). Such an interaction is considered as the paradigmatic example of “specific” binding of a heparin/HS sequence to a protein, involving conformational changes in the protein and a dramatic increase in its activity. Notably, the minimal active sequence has been identified in the pentasaccharide GlcNAc(or NSO3)6SO3–GlcA–GlcNSO3,3,6SO3–IdoA2SO3–GlcNSO3,6SO3, containing the central 3-O-sulfated glucosamine residue which is taken as a marker of the active site for AT. The pentasaccharide sequence is contained in only about one third of the chains constituting heparin, and much less in those of HS (reviewed in Casu and Lindahl, 2001; Lindahl and Li, 2009). The heparin/HS interactions with AT and other coagulation proteins are covered in numerous articles and reviews. It should be noted that the large majority of the biologically relevant, non-AT-mediated interactions of heparin/HS do not require the above mentioned pentasaccharide sequence and that a detailed coverage of interactions of heparin/HS species with other proteins would need considerably more space than is available here. In this mini review these interactions will be summarized and discussed for a few examples only, which are considered to be relevant to the potential development of antiinflammatory and anticancer drugs. These examples include interaction with some chemokines (as exemplified by interleukin-8) and growth factors (exemplified by FGF1 and FGF2), as well as some structural aspects of interactions with the HS-cleaving enzyme heparanase involved in inflammation and cancer.
Among HSPGs relevant to inflammation and cancer, we chose syndecan-1 (CD138) as a paradigmatic example. Functions of HS chains of syndecan-1 as coreceptor for insoluble ligands (e.g. ECM fibronectin and laminin) affecting cell adhesion, as coreceptor for soluble ligands (e.g. growth factors, chemokines, and cytokines) that form part of a signaling complex with their signal transducing receptors, and as effector in other functions, are reviewed in Bernfield et al. (1999). Recent studies unraveled the role of syndecan-1 in stimulating endothelial invasion and angiogenesis in myeloma and other cancers (Purushothaman et al., 2010). Other HSPGs, such as other members of the syndecan family (Bernfield et al., 1999; Götte, 2003; Bishop et al., 2007) and basement membrane proteoglycans such as perlecan, agrin and collagen XVIII (Iozzo and San Antonio, 2001; Iozzo, 2005; Iozzo et al., 2009) have emerged as protagonists of angiogenesis and other biological functions. Besides sharing structural and functional domains with extracellular matrix proteins, growth factors and surface receptors, the basement membrane proteoglycans contribute to the storage of (HS-binding)-cytokines and growth factors, but also in presenting them in a more active configuration to their receptors (Iozzo et al., 2009). Some of these interactions, which modulate cancer growth and angiogenesis and share similarities with those of syndecans at the cell surface, were recently reviewed (Whitelock and Iozzo, 2005; Iozzo, 2005; Whitelock et al., 2008; Iozzo et al., 2009). Structural aspects of HS-protein interactions are discussed in more detail in recent reviews (Imberty et al., 2007; Gandhi and Mancera, 2008; Skidmore et al., 2008; Rudd et al., 2009).
HS–protein interactions play several roles in inflammation (Taylor and Gallo, 2006; Parish, 2006) and cancer (Yip et al., 2006; Bishop et al., 2007; Lindahl and Li, 2009). An essential feature of any inflammatory response is the rapid recruitment of leukocytes from the blood to the site of inflammation. This recruitment process requires the leukocytes to migrate through the blood-vessel wall and enter tissues via a multistep mechanism known as extravasation. Key steps during extravasation are as follows: the initial attachment and rolling of leukocytes on the inflammed endothelium; activation of leukocytes by endothelial-bound chemokines; stable adherence of the activated leukocytes to endothelium; and migration of leukocytes along chemokine gradients, into the target tissue (Parish, 2006). As shown schematically in the lower part of Fig. 1, HS chains of HSPGs play three major roles at different stages during the entry of leukocytes into sites of inflammation: they facilitate l-selectin-dependent cell rolling, promote chemokine transport across the endothelium, and assist chemokine presentation to leukocytes, which results in integrin activation (Wang et al., 2005). Inhibition of leukocyte rolling promoted by selectins plays a role also in early events of tumor metastasis (reviewed in Borsig, 2004). Though prevented by heparin (reviewed in Borsig, 2004), the P-selectin-dependent cell rolling in vascular lumen seems not mediated by HS, but mostly by sulfatides (Borsig et al., 2002). On the other hand, L-selectin effects are both mediated by HS (Wang et al., 2005) and inhibited by heparins (Koenig et al., 1998; Hostetter et al., 2007).
3.2. Heparin/HS chains as stabilizers of dimeric proteins
The interaction with HS chains is thought to provide a mechanism for retaining chemokines on cell surfaces, facilitating the formation of chemokine gradients (Johnson et al., 2005). Immobilization of chemokines to endothelial cells by HSPGs has been demonstrated both in vitro and in vivo. This interaction is thought to enable chemokines to concentrate at functional sites. Both HS binding and the ability to form higher-order oligomers are essential for the activity of chemokines in vivo (Johnson et al., 2005). Aspects of the molecular basis and functional implications of chemokine interactions with HS have been discussed (Wang et al., 2005; Parish, 2006; Imberty et al., 2007; Rek et al., 2009; Lortat-Jacob, 2009).
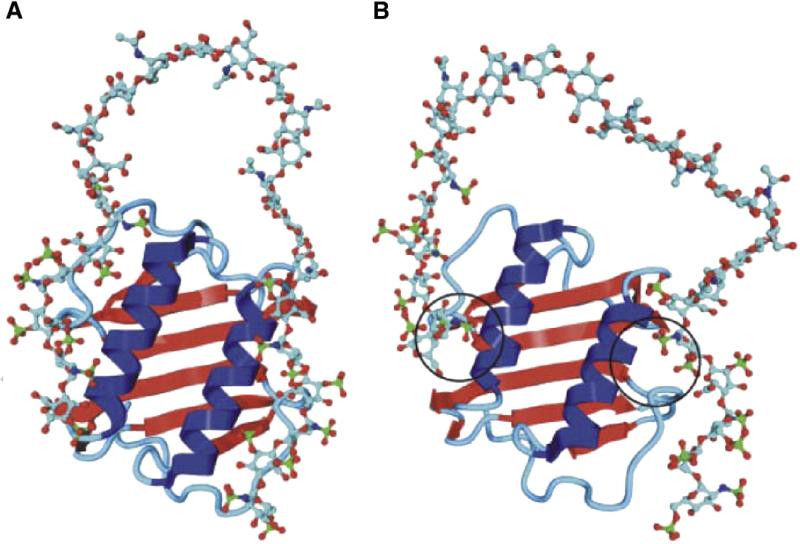
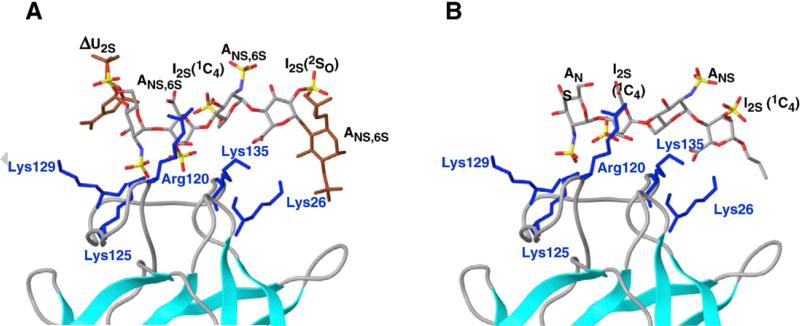
Interleukin-8 (IL-8; CXCL8 according to the new nomenclature) provides a paradigmatic example of a HS-dependent process and of the function of HS chains in assembling two protein molecules. In the case of IL-8, such a complex consists of a flat array of β-pleated sheet with the α-helices of the two monomers arranged as antiparallel rods on top. Biochemical studies suggested that the dimeric form of the protein mediates the binding of 22–24 mer sequences of HS consisting of two ~6 monosaccharide long sulfated (S) blocks separated by an N-acetylated (NA) domain consisting of ≤14 monosaccharide residues. According to the proposed model, the HS chain binds in a horseshoe fashion over the helical region of the dimeric protein (Spillmann et al., 1998). Heparin and heparin oligosaccharides are unable to promote IL-8 dimerization (Goger et al., 2002), most probably because they do not possess sufficiently long NA-domains flanking their S-domains. An NMR and molecular modeling docking study essentially confirmed the horseshoe model for IL-8 (Fig. 2A; Krieger et al., 2004). As illustrated in Fig. 2B (Krieger et al., 2004), molecular dynamics simulation also indicated that contacts with only one disaccharide portion for each sulfated hexasaccharide sequence was sufficient for binding to IL-8 (Krieger et al., 2004), and was confirmed in experiments using synthetic oligosaccharides on microarrays (de Paz et al., 2007). In another context, a trisulfated disaccharide sequence has been also shown to be sufficient for high affinity binding to the chemokine RANTES (CCL5) (Johnson et al., 2005; Rek et al., 2009). The topology of complexes of heparin/HS oligosaccharides with a number of other chemokines widely differ from each other (Imberty et al., 2007; Lortat-Jacob, 2009). An electrospray ionization mass spectrometric study of complexes formed by individual components of a HS octasaccharide library with the chemokines MCP-1(CCL2), MCP-2(CCL8), and MCP-3(CCL7) indicated that the HS sulfation is the foremost requirement for chemokine binding. However, within octasaccharides of constant charge density, N-acetylation was also observed to augment binding, suggesting that there may be as yet undiscovered specificity in the chemokine–HS interaction (Schenauer et al., 2007). Another example of HS chains that stabilize dimeric heparin-binding proteins is provided by their interaction with the growth factor VEGF165, which controls the formation and growth of blood vessels under physiological and pathological conditions (Robinson et al., 2006). Aspects of the molecular basis for the relative flexibility of HS domains are discussed in Section 3.
Fig. 2.
Modeled complex of dimeric IL-8 and a heparin 24-mer composed of two hexasaccharides constituted of “fully sulfated” sequences connected by a nonsulfated N-acetylated, GlcA-containing dodecamer, before (A) and after (B) molecular dynamics simulation. In both binding sites, the interaction between protein and oligosaccharide focused on a disaccharide (Krieger et al., 2004).
3.3. Heparin/HS chains and fragments as promoters of tertiary complexes and signaling
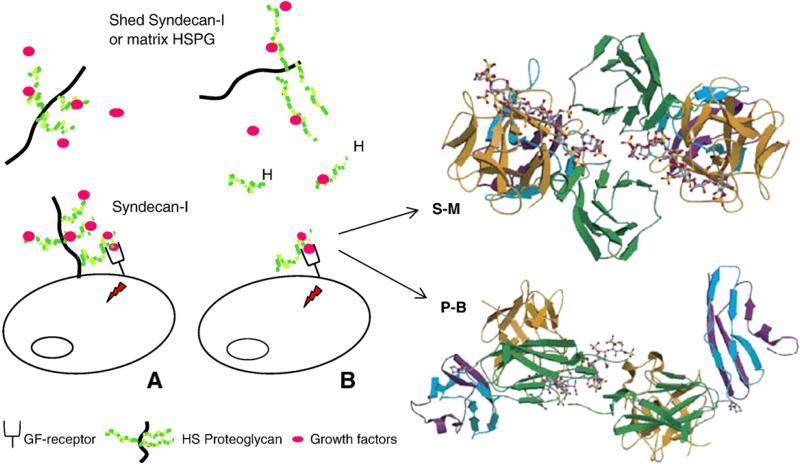
HS interactions regulate the activity of the growth factors activity through a number of distinct mechanisms, including extending the half-life, controlling diffusion, and differentially modulating interactions with the receptor tyrosine kinases (Bernfield et al., 1999; Mohammadi et al., 2005). The interaction of the HS chains of a cell surface HSPG with growth factors is exemplified in Fig. 3A for syndecan-1, which is expressed in various human cancers and may promote tumorigenesis by regulating tumor cell spreading and adhesion, proliferation and angiogenesis (Bernfield et al., 1999; Fears and Woods, 2006). Syndecan-1 binds to factors that promote angiogenesis and tumor cell proliferation (such as FGF2, VEGF, and IL-8) and its expression is regulated by some of these factors (Sanderson and Yang, 2008). FGF2 is stored in a dormant form by soluble syndecan-1 and is released, upon cleavage of its HS chains by heparanase, as a mitogenic complex with still relatively large heparin-like oligosaccharides (Bernfield et al., 1999). Biological functions similar to those exhibited by the HS chains of syndecans are also performed by the HS chains of perlecan, an important basement membrane HSPG. The major functions of perlecan, such as storing and stabilizing chemokines and growth factors, and presenting them in more active configurations to their receptors, have been covered in recent reviews, which also highlighted the dual role of the proteoglycan as a proangiogenic and antiangiogenic molecule (Iozzo, 2005; Whitelock et al., 2008; Iozzo et al., 2009).
Fig. 3.
(A) Function of HS in growth factor (GF)-mediated signaling. (B) Activation of signaling by exogenous heparin (H) or a heparin-like HS fragment released by heparanase. The figure also shows two competing crystallographic models for the formation of ternary complexes involving FGF, FGFR, and a heparin dodecasaccharide. S–M: Schlessinger–Mohammadi symmetrical 2:2:2 complex for FGF2–FGFR1c; P–B: Pellegrini–Blundell asymmetrical 2:2:1 complex for FGF1–FGFR2c (structures redrawn from Mohammadi et al., 2005).
FGF1 and FGF2 — the prototypical members of the fibroblast growth factor (FGF) family — are the most extensively studied growth factors, mainly because of their potent angiogenic activity, exerted by triggering mitogenic signals through formation of ternary complexes with HS and FGF receptors (FGFR) (reviewed in Presta et al., 2005; Mohammadi et al., 2005). In these complexes, HS chains keep together the growth factor and its receptor allowing functional contacts that promote active signaling. As illustrated in Fig. 3B, exogenous heparin can displace HS from these complexes and support signaling (Presta et al., 2005). Most of the 23 known FGFs have high affinity for heparin (Asada et al., 2009). Competitive binding and activity studies using variously sulfated heparin/HS oligosaccharides have defined the minimal size and structure of sequences able to bind individual FGFs (selected examples in Pye et al., 2000; Kreuger et al., 2001; Delehedde et al., 2002; Jemth et al., 2002). The role of 6-O-sulfation in binding FGFs has been the most extensively studied (see Ashikari-Hada et al., 2004). 6-O-sulfation is required for binding FGF1, but not for FGF2; however, at least one 6-O-sulfate group is needed to allow formation of ternary complexes involving FGFRs and for signaling (reviewed in Lyon and Gallagher, 1998; Casu and Lindahl, 2001). High-resolution gel chromatography studies permitted to evaluate the molecular size of FGF–FGFR–heparin complexes. Theses studies suggest that the initial event in FGF signaling, at least in the case of FGF1, may be driven by the co-operative binding of two FGF1 monomers to a single heparin fragment (Robinson et al., 2005). Rationalization of the minimum length of heparin/HS chains and the requirements for 6-O-sulfation have been sought and discussed in support of one or the other X-ray structures of ternary complexes of heparin decasaccharides with FGF1 and FGF2 and their corresponding receptors (Fig. 3B). These crystal structures opened a debate on the spatial arrangement of components of ternary complexes (the Pellegrini–Blundell (P–B) asymmetrical vs the Schlessinger–Muhammadi (S–M) symmetrical model) in actual biological systems (Mohammadi et al., 2005; Harmer et al., 2006). It was proposed that the two arrangements are equally stable in solution for ternary complexes of both FGF1 and FGF2, and that the FGF2/FGFR/heparin decasaccharide may co-exist in both the asymmetrical and symmetrical forms, however, always with the 2:2:1 stoichiometry of the asymmetric model (Goodger et al., 2008). The length of the oligosaccharide chain appears to influence the preference for one or the other arrangement of FGF and receptor molecules, with a 2-O-sulfate iduronate residue at the non-reducing end of heparin tetra- and hexasaccharides being essential for the growth factor-mediated mitogenic activity of these saccharides (Goodger et al., 2008).
Sequence and sulfation pattern requirements for binding and signaling are somewhat different for different growth factors (Ashikari-Hada et al., 2004; Asada et al., 2009) and growth factor–receptor complexes (Zhang et al., 2009), indicating some degree of specificity in several systems. Using libraries of HS-related oligosaccharides generated by chemo-enzymatic methods, common binding motifs required for binding to FGF1 and FGF2 have been shown to be IdoA2SO3–GlcNSO36SO3–IdoA2SO3 and IdoA2SO3–GlcNSO3–IdoA2SO3, respectively (Kreuger et al., 2001; Jemth et al., 2002). Apparent discrepancies among different groups and experimental systems have been found as regards the structure and minimal size required for binding the growth factors and stimulating cell proliferation (reviewed in Mohammadi et al., 2005). A tetrasaccharide size was found to be sufficient for binding FGF1 and FGF2, but heparin/HS octa- or longer oligosaccharides are required for the biological activity (Delehedde et al., 2002; Mohammadi et al., 2005). Interestingly, a synthetic sulfated hexasaccharide, which does not contain the above mentioned trisaccharide motif required for FGF1 binding of HS fragments, stimulates the growth factor more efficiently than natural oligosaccharides (Angulo et al., 2004). Since this hexasaccharide displays sulfate groups only on one side of its 3D chain structure and is unable to promote FGF1 dimerization, these findings suggest that FGF1 dimerization is not a general and absolute requirement for biological activity (Angulo et al., 2004). However, though important in drug design, this atypical hexasaccharide may not reproduce a normal physiological mechanism.
A particular level of complexity has been revealed for binding and signaling requirements of hepatocyte growth factor/scattered factor (HGF/SF), for which a minimum growth factor binding sequence is a disulfated trisaccharide comprising an internal iduronate flanked by monosulfated hexosamine residues. However, these sequences also recognize quite different carbohydrate backbones and sulfation patterns such as those of dermatan sulfate, indicating in this case a lack of any apparent positional requirement for sulfation (Deakin et al., 2009).
3.4. Heparin/HS species as substrates for heparanase
The ECM provides a scaffold for cell growth, migration, differentiation and survival. ECM-remodeling enzymes are therefore expected to profoundly affect cell and tissue function. Heparanase is an endo-β-d-glucuronidase capable of cleaving the HS side chains of HSPGs at a limited number of sites. Enhanced heparanase expression in human tumors correlates with metastatic potential, tumor vascularity, and reduced postoperative survival of cancer patients (reviewed in Vlodavsky et al., 2008; Fux et al., 2009). Among actions relevant to inflammation and cancer, heparanase enhances shedding of soluble syndecan-1, stimulates tumor angiogenesis and endothelial invasion (Yang et al., 2007a; Sanderson and Yang, 2008; Purushothaman et al., 2010) and increases sulfation of its HSPG HS chains, thereby facilitating their interaction with chemokines (reviewed in Li and Vlodavsky, 2009). Thus, heparanase is an attractive target for anticancer drug discovery and for new treatment of inflammation, including delayed type hypersensitivity (DTH) of skin reactions, inflammatory bowel disease (IBD), Crohn's disease, ulcerative colitis, and rheumatoid arthritis (RA), where the enzyme is overexpressed (reviewed in Parish, 2006; Li and Vlodavsky, 2009). Whether heparanase is overexpressed as a defence mechanism, or acts in concert with pro-inflammatory stimuli to aggravate pathological conditions, is an open question. Heparanase may influence several stages of the inflammatory response depicted in Fig. 1, especially by altering the chemokine-orienting role of intact HSPGs. However, the net effect of heparanase on extravasation of leukocytes is still unknown. The use of transgenic mice deficient of heparanase as recently described to study the impact of the protein on airway leukocytes (Waern et al., 2010) will certainly provide information on these and other important aspects.
Heparanase cleaves HS chains at the level of GlcA residues between sulfated GlcN (or GlcNAc) residues (Sandbäck-Pikas et al., 1998), with preference for certain structural environments (Okada et al., 2002). Heparin acts both as a substrate and an inhibitor of the enzyme (Sandbäck-Pikas et al., 1998; Naggi et al., 2005). Heparin oligosaccharides lacking GlcA residues are not cleaved by heparanase. The 3-O-sulfated GlcA-containing pentasaccharide corresponding to the active site for AT is cleaved by heparanase, providing the means of determining the activity of the enzyme; AT-binding octasaccharides compete with the homologous pentasaccharide as substrates and also show some inhibitory activity (Bisio et al., 2007).
The structure of heparanase has been defined to some detail and of the active site and heparin/HS binding sites have been identified (reviewed in Fux et al., 2009). Besides its enzymatic activity, which is exerted predominantly on HS chains of overexpressed syndecan-1 (Yang et al., 2007a) and perlecan (Reiland et al., 2004), heparanase also elicits nonenzymatic, HS-independent activities, such as enhanced Akt phosphorylation, that may have an impact on cancer- and inflammation related pathologies (reviewed in Fux et al., 2009).
4. Issues of specificity. Influence of local flexibility
It has been speculated that HS chains of HSPG in different organs are biosynthetically engineered in order to perform specific protein binding and biological functions. However, similar protein binding properties and corresponding biological functions have been observed for oligosaccharides of the same size, but with somewhat different degrees of sulfation and sulfation patterns (reviewed in Lindahl and Li, 2009). In most cases, the strongest protein binding was observed for the “fully sulfated” (i.e., N-, 6-O-, and 2-O-sulfated) natural heparin/HS sequences, and binding can be further potentiated by their chemical oversulfation. This suggests that extra-sulfate groups may be compatible, but not essential for high-affinity binding (Lindahl and Li, 2009). It has become increasingly evident that more than one sulfation pattern is compatible with high-affinity binding of an oligosaccharide to a given protein. As an extreme example, HGF/SF recognizes different sulfation patterns not only in HS, but also in dermatan sulfate, a GAG with a different carbohydrate backbone (Deakin et al., 2009). The attractive concept of a “HS code” is being gradually replaced by the concept that “specificity” of protein binding by HS is governed more by tissue-dependent arrangement of component blocks (i.e., length and spacing of S, NA, and mixed sequences) than by precise location of sulfate groups along these sequences (Kreuger et al., 2006; Lindahl and Li, 2009). A scheme was proposed that explains graded affinity of sulfate groups of an oligosaccharide and the basic residues of a protein as a function of the “complementarity” of charges (Lindahl and Li, 2009). The basic concept behind the scheme is that, whereas interaction between a HS domain and a target protein may require a precise minimal pattern of sulfate groups, there is no biosynthetic mechanism to ensure generation of such precise sequences devoid of any other sulfate groups. Biosynthesis seems indeed regulated to affect levels of N-, 2-O-, and 6-O-sulfation, and thus the probability of generating a specific binding sequence that may be hidden among redundant residues. On the other hand, the proposed scheme does not rule out the essential participation of HS segments containing rare residues (such as GlcN with unsubstituted amino groups or bearing a 3-O-sulfate group).
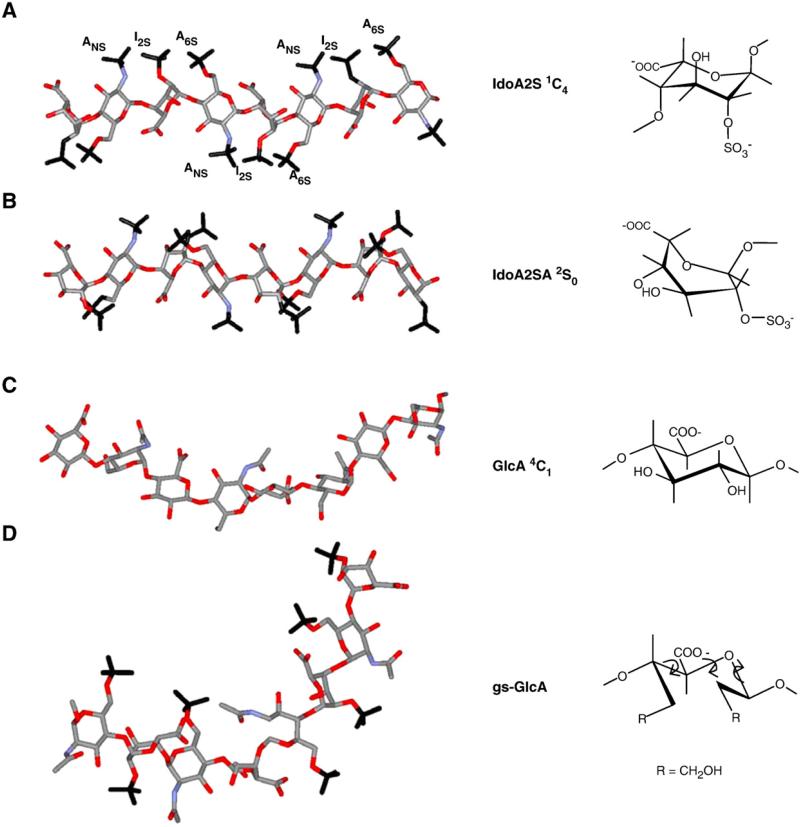
The relative flexibility of different structural HS domains has been often discussed, especially in connection with its role in providing the GAG chain segments with appropriate orientation of substituents to establish favorable “contacts” with the protein binding sites. As discussed in Section 3.2 for IL-8 and VEGF165, the N-acetylated and/or mixed (transition) domains of HS are thought to constitute the flexible segments of HS chains (Spillmann et al., 1998). Protein-induced changes in the local conformation of heparin oligosaccharides have been identified in a number of crystal structures, especially involving FGFs (Raman et al., 2003). These changes, which result in local deviations (“kinks”) in the helical axis of the oligosaccharide to provide optimal ionic and van der Waals contacts with the protein, are generated by slight changes in the backbone torsion angles in addition to changes in the shape of the iduronate rings (Raman et al., 2003). Conformational flexibility is conferred on iduronic acid-containing GAG sequences by the co-existence of equienergetic conformations of iduronate (IdoA and/or IdoA2SO3) residues, which is regarded as a molecular device to facilitate binding to proteins (Casu et al., 1998; Rudd et al., 2009). As illustrated in Fig. 4A and B for a segment of heparin/HS chain, a change of an IdoA2SO3 residue from the 1C4 to the 2S0 conformation, both existing in equilibrium in internal sequences of the polysaccharides, drastically changes the shape of this residue and the orientation of its appended sulfate group. Fig. 4A and B also show that an iduronate residue in the 1C4 conformation (with two axial glycosidic bonds) is more contracted than the one in the 2S0 conformation (with two semi-equatorial glycosidic bonds) (Raman et al., 2003). Comparison with a similar model of N-acetyl heparosan (Fig. 4C) shows that the GlcA residue of the heparin/HS precursor, and of their corresponding NA regions in the mature polysaccharides as well, span a somewhat larger space because of their diequatorial glycosidic bonds. Co-crystal structures in which a protein selects one of the two equienergetic conformations in a heparin oligosaccharide have been reported, a notable example being a FGF2-bound hexasaccharide, in which one sulfated iduronate residue is in the chair 1C4 conformation and the other in the skew-boat 2S0 conformation (Fig. 5A). Protein binding also drives the conformational equilibrium toward one of the two conformations of iduronate residues in solution, as demonstrated in AT-bound oligosaccharides for IdoA2SO3 within the pentasaccharide sequence of the active site for AT and for an IdoA outside this sequence (Guerrini et al., 2008). However, examples of solution conformations of heparin oligosaccharides with iduronate residues driven toward the 2S0 form as a consequence of interaction with a growth factor have not been yet reported. A NMR and molecular modeling study of a non-6-O-sulfated tetrasaccharide in the absence and presence of FGF2 indicates that its IdoA2SO3 residue retains the 1C4 conformation prevalent in free solution (Fig. 5B; Guglieri et al., 2008). The finding that the conformer populations of IdoA (or IdoA2SO3) are affected by O-sulfation of the neighboring glucosamine residues (for 6-O-sulfation, see Murphy et al., 2008) is most probably relevant to the protein-binding properties of heparin/HS sequences.
Fig. 4.
Energy-minimized structures of heparin/HS S chains (A,B) and NA chains, these latter represented by the biosynthetic common precursor N-acetyl heparosan, C), and of one low-energy conformer of glycol-split N-acetyl heparin (D) compatible with experimental Nuclear Overhauser Effect values. Structures A and B (redrawn from Mulloy et al., 1993) illustrate the dramatic influence of changes in the conformation (from 1C4 to 2S0) of IdoA2OSO3 residues on spacing of sulfate groups along the chains (ANS and A6S = N-sulfate and 6-O-sulfate groups of GlcN residues; I2S = 2-O-sulfate group of IdoA residues). The rigid (4C1) conformation) of GlcA residues in N-acetyl heparosan does not involve significant changes from the chain conformation shown in C. Structures A–C are characterized by different dihedral angles between vicinal C–H bonds and distances between nonbonded atoms of the uronic acid residues. Structure D illustrates that glycol-splitting generates extra-degrees of rotational freedom (arrows), allowing abrupt kinks in the oligo/polysaccharide chains where one GlcA residue was periodate-oxidized and borohydride reduced. Similar conformations can be assumed by glycol-split, nonsulfated IdoA residues. (Panels A–C are from Casu, 2005; D from Vlodavsky et al., 2007.)
Fig. 5.
(A) Details of the crystal structure of a 1:1 FGF2-heparin hexasaccharide complex, showing that the two 2-OSO3 residues select different conformations in binding to the growth factor (from Casu and Lindahl, 2001). (B) Partial solution structure of the complex of FGF2 and a synthetic tetrasaccharide, with both the sulfated iduronate residues in 1C4 conformation (Guglieri et al., 2008). Designation of residues as for Fig. 4; ΔU = 4,5-unsaturated uronic acid.
It is increasingly perceived that the flexibility of N-acetylated domains of HS is important in modulating protein binding properties. On the basis of charge repulsions of closely spaced sulfate groups, it is expected that polysulfated chains such as those of S domains are more rigid (“stiff”) than the nonsulfated, or poorly sulfated NA domains. 15N NMR and molecular dynamics simulation studies on N-acetyl heparosan (K5) oligosaccharides demonstrated that nonsulfated chains do indeed behave as relatively disordered random coils, supporting their higher flexibility as compared with reported data on S domains (Mobli et al., 2008).
On the other hand, evidence is being accumulated on the role of the conformational plurality of IdoA rings to facilitate kinking of GAG chains to provide their best fit to protein active sites. A recent study on porcine mucosal HS hexasaccharides highlighted the role of GlcN 6-O-sulfation in determining the balance between the 1C4 and 2S0 conformations of the flanking IdoA2SO3 residues (Murphy et al., 2008). As illustrated for the complex of a tetrasaccharide with FGF2 (Fig. 5B), the conformational flexibility (“plasticity”) of iduronate rings may compensate for a lack of 6-O-sulfation in neighbouring glucosamine residues (Guglieri et al., 2008). It is attractive to think that the bending of NA domains as required to favor dimerization of IL-8 as described in Section 3.2 involves conformational transitions at the level of iduronate residues on both terminals of the S stretches rather than being caused by slight adjustments of the interglycosidic angles along the whole NA stretch. This hypothesis is awaiting experimental proof.
Additional local flexibility can be induced along the chains of heparin/HS by glycol-splitting of C2–C3 bonds of nonsulfated GlcA and IdoA residues, usually performed by periodate oxidation with reductive stabilization of the resulting dialdehydes. By generating additional degrees of freedom at the level of the uronic acid, glycol-split residues act as flexible joints along the heparin/HS chains (Naggi, 2005). This is illustrated in Fig. 4D by the evident kink generated by glycol-splitting of a GlcA residue in an N-acetyl heparin chain (Vlodavsky et al., 2007). Notably, glycol-splitting of all nonsulfated uronic acid residues of heparin/HS generate “nonanticoagulant” species, since it modifies also the GlcA residue in the pentasaccharide sequence of the active site for AT (Conrad, 1998; Casu et al., 2004).
5. Potential applications of non-anticoagulant heparin species in inflammation and cancer
Exogenously administered heparin and low-molecular-weight heparins have been shown to exert beneficial effects in inflammation (reviewed in Lever and Page, 2002; Young, 2008; Rek et al., 2009) and cancer (reviewed in Yip et al., 2006; Lazo-Langner et al., 2007; Casu et al., 2008). Anticancer activities are mostly expressed in animal models through antimetastatic and antiangiogenic properties, these latter being observed mainly for low-molecular weight species (summarized in Casu et al., 2008). The antiinflammatory and antitumor properties of heparin and related species are thought to be largely associated with the influence on pathological functions of HS chains of HSPGs. In fact, heparin-based drugs can conceivably be designed to interfere with HS–protein interactions to compete with (or reinforce) functions of HS chains (Lindahl, 2007; Lindahl and Li, 2009). As exemplified by the intriguing finding that some HS sequences are cryptic promoters while others are inhibitors of cancer progression (Liu et al., 2002), some of the HSPG functions and their interplay with those of other biological effectors are still not completely understood.
The potential antitumor property of heparin species, including oligosaccharides as small as disaccharides, has been most extensively investigated in the context of their effects on angiogenesis. Early studies recognized that full-size heparin displaces growth factors (GFs) stored by the HS chains of HSPGs on cell surfaces and in the EC matrix and is potentially pro-angiogenic because it promotes formation of ternary complexes of heparin-bound GFs (especially FGF2 and VEGF) with GF receptors (Fig. 3)(Bernfield et al., 1999; Mohammadi et al., 2005). As discussed in Section 3.3, small heparin oligosaccharides are unable to promote formation of such ternary complexes and to trigger mitogenic signaling. Octasaccharides are on the borderline of size requirements and may be potentially anti-angiogenic by blocking GFs in a way to prevent their dimerization and/or their interaction with GFRs. However, they might function like longer heparin-like oligosaccharides and induce mitogenic signaling. In fact, heparin octasaccharides act as inhibitors in four in vivo models of angiogenesis that are progressively less dependent on FGF2, indicating that they can inhibit angiogenesis through other mechanisms (Hasan et al., 2005). These results have significant implications for clinical trials of low molecular weight heparins (LMWH) in cancer. In fact, the overall effect of a LMWH on angiogenesis as observed in vivo is probably a balance between the influence of 8-mer or smaller components and those of larger size (Hasan et al., 2005).
Full-length heparins that have been depleted of substituents, that are essential for interaction with antithrombin as well as with GF receptors, such 6-O-desulfated heparin, are antiangiogenic without being significantly anticoagulant and are potential antiangiogenic drugs (Lundin et al., 2000; Hasan et al., 2005). A large number of undersulfated, glycol-split heparins, including their derivatives with different degrees of N-acetylation, can be obtained chemically by generating additional (nonsulfated IdoA) residues susceptible to glycol-splitting. Some of these non-anticoagulant heparins, including one in which about half of the total uronic acid residues were glycol-split (Casu et al., 2002), are good complexing agents for FGF2 (Casu et al., 2004) and VEGF (Pisano et al., 2005) and are endowed with antiangiogenic properties in models in which heparin is either inactive or pro-angiogenic (Casu et al., 2004). At least in part, these properties can be explained in terms of conformational transitions at the level of glycol-split residues, rendering dimerization of the growth factors unfavorable (Casu et al., 2004).
Inhibition of heparanase is a most promising target for the inhibition of both angiogenesis and metastasis (Vlodavsky et al., 2008), as well as of some inflammatory pathologies (Parish, 2006; Vlodavsky et al., 2008; Li and Vlodavsky, 2009). Since the identification of the active site and two heparin/HS binding sequences of the enzyme (reviewed in Fux et al., 2009), heparanase has been targeted with several inhibitors that block either the active site of the enzyme or the heparin/HS binding sites, or both (Ferro et al., 2004; Vlodavsky et al., 2007). Being close HS mimics, heparin-related inhibitors are promising leads for heparanase inhibition (Naggi et al., 2005; Vlodavsky et al., 2007). A N-acetylated, glycol-split heparin was shown to be a potent inhibitor of heparanase, while exerting little or no release of growth factors from the ECM (Naggi et al., 2005; Vlodavsky et al., 2007). This heparin derivative has strong anti-myeloma activity in vivo (Yang et al., 2007b). It is thought to effectively interact with the heparin/HS binding regions of heparanase through its sulfated sequences, as well as with the active site of the enzyme through its glycol-split GlcA residues (Naggi, 2005; Vlodavsky et al., 2007). Another class of heparanase inhibitors consists of heparin oligosaccharides terminating at their reducing end with an aza sugar and this is thought to interact with the active site of the enzyme (Petitou and Driguez, 2006).
Most of the non-anticoagulant heparins are less sulfated than unmodified heparin and consequently expected to have more favorable pharmacokinetic properties than unmodified heparin. Based on early findings that association with corticosteroids induces antiangiogenic properties in heparin (Folkman et al., 1983), several steroid-substituted heparin derivatives have been described, among which a low-molecular weight heparin-deoxycholic acid conjugate which is antiangiogenic in different types of cancer cells even when administered by the oral route (Lee et al., 2009).
In conclusion, heparin oligosaccharides and some non-anticoagulant heparin derivatives are promising candidates as safe and effective antiangiogenic, antimetastatic, and antiinflammatory drugs. However, a deeper knowledge of their mechanisms of action is needed to optimize the drug candidates and to ensure that they hit the selected targets. Although the therapeutic efficacy is likely to be increased by multi-target approaches, the apparent redundancy in sequence-to-activity relationships (Lindahl and Li, 2009) makes it difficult to apply the typical drug discovery tools based on computational surveys of binding proteins to heparin-derived molecules (Mulloy and Forster, 2008). The possibility of the — apparently paradoxical — reversal of biological effects (i.e., from antiangiogenic to pro-angiogenic) depending on interactions with HS in different organs or different physiopathological conditions, requires extensive in vivo experimentation. In fact, HS mimics targeted at a given HSPG could deal with a complex network of mutually interacting species and may also promote interdomain interactions (Forsten-Williams et al., 2008) and exogenous heparin may act at different levels at cell surfaces and in the ECM. However, heparin species designed for specific targets are expected to act preferentially at the level of those proteins (e.g., cytokines, growth factors, or heparanase) that are overexpressed in inflammation and cancer.
In order to design precisely tailored antagonists or agonists of HS (or heparanase) functions, more subtle details of the 3D structure of their relevant complexes still need to be defined. Much of the information available to date is based on biochemical, crystallographic and structural studies on a still limited number of complexes of heparin/HS oligosaccharides with heparin-binding proteins and complexes with their corresponding receptors. With the exception of some examples (a few of them covered in this mini review), GAG sulfate groups involved in relevant biological interactions have been defined only in statistical terms, principally using heparin and heparin derivatives and their fragments instead of the corresponding species derived from HS. A more detailed understanding requires the use of HS species obtained from the HSPGs specifically involved in the interaction (Powell et al., 2004; Lindahl and Li, 2009). However, continuous refinement of tools for structural analysis of HSPGs and their interactions (Sasisekharan et al., 2006; Guglieri et al., 2008; Schenauer et al., 2009; Rek et al., 2009; Korir and Larive, 2009; Guimond et al., 2009) and the expected availability of extended libraries of synthetic heparin/HS oligosaccharides (Seeberger and Werz, 2008; Arungundram et al., 2009), semi-biotechnological (Lindahl et al., 2005) and fully biotechnological heparins/HSs (Martin et al., 2009; Peterson et al., 2009) is expected to provide additional structure–activity information required for further development of heparin-based GAGs as potential drugs in inflammation and cancer, as well as in other therapeutic fields.
6. Uncited reference
Acknowledgment
The authors are grateful to Drs. Giuseppe Cassinelli and Edward A. Yates for critical reading of the manuscript.
Abbreviations
- AT
antithrombin III
- ECM
extracellular matrix
- FGF1
fibroblast growth factor-1
- FGF2
fibroblast growth factor-2
- FGFR
fibroblast growth factor receptor
- GAG
glycosaminoglycan
- GlcA
glucuronic acid
- GlcNAc
N-acetyl glucosamine
- GlcNSO3
glucosamine N-sulfate
- GF
growth factor
- HS
heparan sulfate
- HSPG
heparan sulfate proteoglycan
- IL-8
interleukin-8
- VEGF
vascular endothelial growth factor
References
- Ai X, Do A-T, Kusche-Gullberg M, Lindahl U, Lu K, Emerson CP. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases. QSulf1 and QSulf2. J. Biol. Chem. 2006;281:4969–4976. doi: 10.1074/jbc.M511902200. [DOI] [PubMed] [Google Scholar]
- Angulo J, Ojeda R, de Paz J-L, Lucas R, Nieto PM, Lozano RM, Recondo-Horcajo M, Giménez-Gallego G, Martín-Lomas M. The activation of fibroblast growth factors (FGFs) by glycosaminoglycans: influence of the sulfation pattern on the biological activity of FGF1. ChemBioChem. 2004;5:55–61. doi: 10.1002/cbic.200300696. [DOI] [PubMed] [Google Scholar]
- Arungundram S, Al-Mafraji K, Asong J, Leach FEIII, Amster J, Venot A, Thurnbull JE, Boons G-J. Modular synthesis of heparan sulfate oligosaccharides for structure–activity relationship studies. J. Am. Chem. Soc. 2009;131:17394–17405. doi: 10.1021/ja907358k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada M, Shinomiya M, Suzuki M, Honda E, Sugimoto R, Ikekita M, Imamura T. Glycosaminoglycan affinity of the complete fibroblast growth factor family. Biochim. Biophys. Acta. 2009;1790:40–48. doi: 10.1016/j.bbagen.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Ashikari-Hada S, Habuchi H, Karyia Y, Ytoh H, Reddi AH, Kimata K. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J. Biol. Chem. 2004;279:12346–12354. doi: 10.1074/jbc.M313523200. [DOI] [PubMed] [Google Scholar]
- Bernfield M, Götte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Bisio A, Mantegazza A, Urso E, Naggi A, Torri G, Viskov C, Casu B. High-performance liquid chromatographic/mass spectrometric studies on the susceptibility of heparin species to cleavage by heparanase. Semin. Thromb. Hemost. 2007;33(5):488–495. doi: 10.1055/s-2007-982079. [DOI] [PubMed] [Google Scholar]
- Borsig L. Selectins facilitate carcinoma metastasis and heparin can prevent them. News Physiol. Sci. 2004;19:16–21. doi: 10.1152/nips.01450.2003. [DOI] [PubMed] [Google Scholar]
- Borsig L, Wong R, Hynes RO, Varki NM, Varki A. Synergistic effects of L- and P-selectin in facilitating tumor metastasis can involve non-mucin ligands and implicate leukocytes as enhancers of metastasis. Proc. Natl. Acad. Sci. 2002;99:2193–2198. doi: 10.1073/pnas.261704098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capila I, Linhardt RJ. Heparin–protein interactions. Angew. Chem. Int. Ed. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Casu B. Structure and active domains of heparin. In: Garg HG, Linhardt RJ, Hales CA, editors. Chemistry and Biology of Heparin and Heparan Sulfate. Elsevier; Amsterdam: 2005. pp. 1–28. [Google Scholar]
- Casu B, Lindahl U. Structure and biological interactions of heparin and heparan sulfate. Adv. Carbohydr. Chem. Biochem. 2001;57:159–206. doi: 10.1016/s0065-2318(01)57017-1. [DOI] [PubMed] [Google Scholar]
- Casu B, Petitou M, Provasoli M, Sinaÿ P. Conformational flexibility: a new concept for explaining binding and biological properties of iduronic acid-containing glycosaminoglycans. Trends Biochem. Sci. 1998;13:221–225. doi: 10.1016/0968-0004(88)90088-6. [DOI] [PubMed] [Google Scholar]
- Casu B, Guerrini M, Naggi A, Perez M, Torri G, Ribatti D, Carminati P, Giannini G, Penco S, Pisano C, Belleri M, Rusnati M, Presta M. Short heparin sequences spaced by glycol-split uronate residues are antagonists of fibroblast growth factor 2 and angiogenesis inhibitors. Biochemistry. 2002;41:10519–10528. doi: 10.1021/bi020118n. [DOI] [PubMed] [Google Scholar]
- Casu B, Guerrini M, Guglieri S, Naggi A, Perez M, Torri G, Cassinelli G, Ribatti D, Carminati P, Giannini G, Penco S, Pisano C, Belleri M, Rusnati M, Presta M. Undersulfated and glycol-split heparins endowed with antiangiogenic activity. J. Med. Chem. 2004;47:838–848. doi: 10.1021/jm030893g. [DOI] [PubMed] [Google Scholar]
- Casu B, Vlodavsky I, Sanderson RD. Non-anticoagulant heparins and inhibition of cancer. Pathophysiol. Haemost. Thromb. 2008;36:195–203. doi: 10.1159/000175157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad HE. Heparin Binding Proteins. Academic Press; San Diego: 1998. [Google Scholar]
- Coombe DR. Biological implications of glycosaminoglycan interactions with haemopoietic cytokines. Immunol. Cell Biol. 2008;86:598–607. doi: 10.1038/icb.2008.49. [DOI] [PubMed] [Google Scholar]
- Coombe DR, Kett WC. Heparan sulfate–protein interactions: therapeutic potential through structure-function insights. Cell. Mol. Life Sci. 2005;62:410–424. doi: 10.1007/s00018-004-4293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paz JL, Moseman EA, Noti C, Polito L, van Andrian UH, Seeberger PH. Profiling heparin–chemokine interactions using synthetic tools. ACS Chem. Biol. 2007;2:735–744. doi: 10.1021/cb700159m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JA, Blaum BS, Gallagher JT, Lyon M. The binding properties of minimal oligosaccharides reveal a common heparan sulfate-binding site in hepatocyte growth factor/scatter factor that can accommodate a variety of sulfation patterns. J. Biol. Chem. 2009;284:6311–6321. doi: 10.1074/jbc.M807671200. [DOI] [PubMed] [Google Scholar]
- Delehedde M, Lyon M, Gallagher JT, Rudland PS, Fernig DG. Fibroblast growth factor-2 binds to small heparin-derived oligosaccharides and stimulates a sustained phosphorylation of p42/44 mitogen-activated protein kinase and proliferation of rat mammary fibroblasts. Biochem. J. 2002;366:235–244. doi: 10.1042/BJ20011718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss JL, Regatieri C, Jarrouge TR, Cavalheiro RP, Sampajo LO, Nader HB. Heparan sulfate proteoglycans: structure, protein interactions and cell signaling. Ann. Braz. Acad. Sci. 2009;81:409–4299. doi: 10.1590/s0001-37652009000300007. [DOI] [PubMed] [Google Scholar]
- Fears CY, Woods A. The role of syndecans in diseases and wound healing. Matrix Biol. 2006;25:443–456. doi: 10.1016/j.matbio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Ferro V, Hammond E, Fairweather JK. The development of inhibitors of heparanase, a key enzyme in tumor metastasis, angiogenesis and inflammation. Mini Rev. Med. Chem. 2004;4:672–693. doi: 10.2174/1389557043403729. [DOI] [PubMed] [Google Scholar]
- Folkman J, Langer R, Linhardt RJ, Haudenschild C, Taylor S. Angiogenesis inhibition and tumor regression caused by heparin or a heparin fragment in the presence of cortisone. Science. 1983;221:719–725. doi: 10.1126/science.6192498. [DOI] [PubMed] [Google Scholar]
- Forsten-Williams K, Chu CL, Fannon M, Buczek-Thomas JA, Nugent MA. Control of growth factor networks by heparan sulfate proteoglycans. Ann. Biochem. Eng. 2008;36:2134–2148. doi: 10.1007/s10439-008-9575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fux L, Ilan N, Sanderson RD, Vlodavsky I. Heparanase: busy at the cell surface. Trends Biochem. Sci. 2009;34:511–519. doi: 10.1016/j.tibs.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JT. Multiprotein signaling complexes: regional assembly on heparan sulphate. Biochem. Soc. Trans. 2006;34:341–348. doi: 10.1042/BST0340438. [DOI] [PubMed] [Google Scholar]
- Gandhi NS, Mancera RL. The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- Goger B, Halden Y, Rek R, Mösl D, Pye J, Gallagher J, Kungl AJ. Different affinities of glycosaminoglycan oligosaccharides for monomeric and dimeric interleukin-8: a model for chemokine regulation at inflammatory sites. Biochemistry. 2002;41:1640–1646. doi: 10.1021/bi011944j. [DOI] [PubMed] [Google Scholar]
- Goodger SJ, Robinson CJ, Murphy KJ, Gasiunas N, Harmer NJ, Blundell TL, Pye DA, Gallagher JT. Evidence that heparin saccharides promote FGF2 mitogenesis through two distinct mechanisms. J. Biol. Chem. 2008;283:13001–13008. doi: 10.1074/jbc.M704531200. [DOI] [PubMed] [Google Scholar]
- Götte M. Syndecans in inflammation. FASEB J. 2003;17:575–591. doi: 10.1096/fj.02-0739rev. [DOI] [PubMed] [Google Scholar]
- Guerrini M, Guglieri S, Casu B, Torri G, Mourier P, Boudier C, Viskov C. Antithrombin-binding octasaccharides and role of extensions of the active pentasaccharide sequence in the specificity and strength of interaction. J. Biol. Chem. 2008;283:26662–26675. doi: 10.1074/jbc.M801102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglieri S, Hricovíni, Raman R, Polito L, Torri G, Casu B, Sasisekharan R, Guerrini M. Minimum FGF2 binding structural requirements of heparin and heparan sulfate oligosaccharides as determined by NMR spectroscopy. Biochemistry. 2008;47:13862–13869. doi: 10.1021/bi801007p. [DOI] [PubMed] [Google Scholar]
- Guimond SE, Puvirajesinghe TM, Skidmore MA, Kalus I, Dierks T, Yates EA, Turnbull JE. Rapid purification and high sensitivity analysis of heparan sulfate from cells and tissues: toward glycomics profiling. J. Biol. Chem. 2009;284:25714–25722. doi: 10.1074/jbc.M109.032755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer NJ, Robinson CJ, Adam LE, Ilag LL, Robinson CV, Gallagher JT, Blundell TL. Multimers of the fibroblast growth factor (FGF)–FGF receptor–saccharide complex are formed on long oligomers of heparin. Biochem. J. 2006;393:741–748. doi: 10.1042/BJ20050985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan J, Shnyder SD, Clamp AR, McGown AT, Bicknell R, Presta M, Bibby M, Double J, Craig S, Leeming D, Stevenson K, Gallagher JT, Yayson GC. Heparin octasaccharides inhibit angiogenesis in vivo. Clin. Cancer Res. 2005;11:8172–8179. doi: 10.1158/1078-0432.CCR-05-0452. [DOI] [PubMed] [Google Scholar]
- Hostetter N, Naggi A, Torri G, Ishai-Michaeli R, Casu B, Vlodavsky I, Borsig L. P-selectin and heparanase-dependent antimetastatic activity of non-anticoagulant heparins. FASEB J. 2007;21:3562–3572. doi: 10.1096/fj.07-8450com. [DOI] [PubMed] [Google Scholar]
- Imberty A, Lortat-Jacob H, Pérez S. Structural view of glycosaminoglycan–protein interactions. Carbohydr. Res. 2007;342:430–439. doi: 10.1016/j.carres.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Iozzo RV. Basement membrane proteoglycans: from cellar to ceiling. Nat. Rev., Mol. Cell. Biol. 2005;6:646–656. doi: 10.1038/nrm1702. [DOI] [PubMed] [Google Scholar]
- Iozzo RV, San Antonio JD. Heparan sulfate proteoglycans: heavy hitters in the angiogenesis arena. J. Clin. Invest. 2001;108:335–349. doi: 10.1172/JCI13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo RV, Zoeller JJ, Nyström A. Basement membrane proteoglycans: modulators par excellence of cancer growth and angiogenesis. Mol. Cells. 2009;27:503–513. doi: 10.1007/s10059-009-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemth P, Kreuger J, Kusche-Gullberg M, Sturiale L, Giménez-Gallego G, Lindahl U. Biosynthetic oligosaccharide libraries for identification of protein-binding heparan sulfate motifs. Exploring the structural diversity by screening for fibroblast growth factor FGF1 and FGF2 binding. J. Biol. Chem. 2002;277:30567–30573. doi: 10.1074/jbc.M203404200. [DOI] [PubMed] [Google Scholar]
- Johnson Z, Proudfoot AE, Handel TM. Interaction of chemokines and glycosaminoglycans: a new twist in the regulation of chemokines function with opportunities for therapeutic intervention. Cytokine Growth Factor Rev. 2005;16:625–636. doi: 10.1016/j.cytogfr.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Koenig A, Norgard-Sumnicht K, Linhardt R, Varki A. Differential interactions of heparin and heparan sulfate glycosaminoglycans with the selectins. Implications for the use of unfractionated and low molecular weight heparins as therapeutic agents. J. Clin. Invest. 1998;101:877–889. doi: 10.1172/JCI1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korir AK, Larive K. Advances in the separation, sensitive detection, and characterization of heparin and heparan sulfate. Anal. Bioanal. Chem. 2009;393:155–169. doi: 10.1007/s00216-008-2412-2. [DOI] [PubMed] [Google Scholar]
- Kreuger J, Salmivirta M, Sturiale L, Giménez-Gallego G, Lindahl U. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J. Biol. Chem. 2001;276:30744–30752. doi: 10.1074/jbc.M102628200. [DOI] [PubMed] [Google Scholar]
- Kreuger J, Spillmann D, Li J-P, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J. Biol. Chem. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Geretti E, Brandner B, Goger B, Wells TN, Kungl AJ. A structural and dynamic model for the interaction of interleukin-8 and glycosaminoglycans: support from isothermal fluorescence titrations. Proteins. 2004;54:768–775. doi: 10.1002/prot.10590. [DOI] [PubMed] [Google Scholar]
- Laremore TN, Zhang F, Dordick JS, Linhardt RJ. Recent progresses and applications in glycosaminoglycan and heparin research. Curr. Opin. Chem. Biol. 2009;13:633–640. doi: 10.1016/j.cbpa.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo-Langner A, Goss GD, Spaans JN, Rodger MA. The effect of low-molecular- weight heparin on cancer survival. A systematic review and meta-analysis of randomized trials. J. Thromb. Haemost. 2007;5:729–737. doi: 10.1111/j.1538-7836.2007.02427.x. [DOI] [PubMed] [Google Scholar]
- Lee YD, Lee SW, Kim SK, Lee M, Chang HW, Moon HT, Byun Y, Kim SY. Antiangiogenic activity of orally absorbable heparin derivative in different types of cancer cells. Pharm. Res. 2009 doi: 10.1007/s11095-009-9989-9. 10.1007/s1105-009-9989-9. [DOI] [PubMed] [Google Scholar]
- Lever R, Page CP. Novel drug development opportunities for heparin. Nat. Rev. Drug Discov. 2002;1:140–148. doi: 10.1038/nrd724. [DOI] [PubMed] [Google Scholar]
- Li J-P, Vlodavsky I. Heparin, heparan sulfate and heparanase in inflammatory reactions. Thromb. Haemost. 2009;102(3):823–828. doi: 10.1160/TH09-02-0091. [DOI] [PubMed] [Google Scholar]
- Lindahl U. Heparan sulfate–protein interactions—a concept for drug design? Thromb. Haemost. 2007;98:109–115. [PubMed] [Google Scholar]
- Lindahl U, Li J-P. Interaction between heparan sulfate and proteins — design and functional implications. Int. Rev. Cell Biol. 2009;276:105–159. doi: 10.1016/S1937-6448(09)76003-4. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Kusche-Gullberg M, Kjellén L. Regulated diversity of heparan sulfate. J. Biol. Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Li J, Kusche-Gullberg M, Salmivirta M, Alaranta S, Veromaa T-, Emeis J, Roberts I, Taylor C, Oreste P, Zoppetti G, Naggi A, Torri G, Casu B. Generation of “neoheparin” from E. coli capsular polysaccharide. J. Med. Chem. 2005;48:349–352. doi: 10.1021/jm049812m. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ. Heparin: structure and activity. J. Med. Chem. 2003;46:2551–2564. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- Liu D, Sasisekharan R. Role of heparan sulfate in cancer. In: Garg AG, Linhardt RJ, Hales CA, editors. Chemistry and Biology of Heparin and Heparan Sulfate. Elsevier; Amsterdam: 2005. pp. 699–725. [Google Scholar]
- Liu D, Shriver Z, Venkataraman G, El Shabrawi Y, Sasisekharan R. Tumor cell surface heparan sulfate as cryptic promoters or inhibitors of tumor growth and metastasis. Proc. Natl. Acad. Sci. U. S. A. 2002;99:568–573. doi: 10.1073/pnas.012578299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lortat-Jacob H. The molecular basis and functional implications of chemokine interactions with heparan sulphate. Curr. Opin. Struct. Biol. 2009;19:543–548. doi: 10.1016/j.sbi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Lundin L, Larsson H, Kreuger J, Kanda S, Lindahl U, Salvimirta M, Claesson-Welsh L. Selectively desulfated heparin inhibits fibroblast growth factor-induced mitogeneity and angiogenesis. J. Biol. Chem. 2000;275:24653–24660. doi: 10.1074/jbc.M908930199. [DOI] [PubMed] [Google Scholar]
- Lyon M, Gallagher JT. Biospecific sequences and domains in heparan sulphate and the regulation of cell growth and adhesion. Matrix Biol. 1998;17:485–493. doi: 10.1016/s0945-053x(98)90096-8. [DOI] [PubMed] [Google Scholar]
- Martin JG, Gupta M, Xu Y, Akella S, Liu J, Dordick JS, Linhardt RJ. Toward an artificial Golgi: redisigning the biological activities of heparan sulfate on a digital microfluid chip. J. Am. Chem. Soc. 2009;131:11041–11048. doi: 10.1021/ja903038d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry CL, Lyon M, Deakin JA, Hopwood JJ, Gallagher JT. Highly sensitive sequencing of the sulfated domains of heparan sulfate. J. Biol. Chem. 1999;274:18455–18462. doi: 10.1074/jbc.274.26.18455. [DOI] [PubMed] [Google Scholar]
- Mobli M, Nilson M, Almond A. The structural plasticity of heparan sulfate NA-domains and hence their role in mediating multivalent interactions is confirmed by high-accuracy 15N-NMR relaxation studies. Glycoconj. J. 2008;25:401–414. doi: 10.1007/s10719-007-9081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Mulloy B, Forster MJ. Application of drug discovery software to the identification of heparin-binding sites on protein surfaces: a computational survey of the 4-helix cytokines. Mol. Simul. 2008;34:381–389. [Google Scholar]
- Mulloy B, Forster MJ, Jones C, Davies DB. NMR and molecular modeling studies in the solution conformation of heparin. Biochem. J. 1993;293:849–858. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KJ, Merry CLR, Lyon M, Thompson JE, Roberts IS, Gallagher JT. A new model for the domain structure of heparan sulfate based on the novel specificity of K5 lyase. J. Biol. Chem. 2004;279:27239–27245. doi: 10.1074/jbc.M401774200. [DOI] [PubMed] [Google Scholar]
- Murphy KJ, McLay N, Pye A. Structural studies of heparan sulfate hexasaccharides: new insights into iduronate conformational behaviour. J. Am. Chem. Soc. 2008;130:12435–12444. doi: 10.1021/ja802863p. [DOI] [PubMed] [Google Scholar]
- Naggi A. Glycol-splitting as a device for modulating inhibition of growth factors and heparanase by heparin and heparin derivatives. In: Garg HG, Linhardt RJ, Hales CA, editors. Chemistry and Biology of Heparin and Heparan Sulfate. Elsevier; Amsterdam: 2005. pp. 461–481. [Google Scholar]
- Naggi A, Casu B, Perez M, Torri G, Cassinelli G, Penco S, Pisano C, Giannini G, Ishai-Michaeli R, Vlodavsky I. Modulation of heparanase-inhibiting activity of heparin through selective desulfation, graded N-acetylation, and glycol-splitting. J. Biol. Chem. 2005;280:12103–12113. doi: 10.1074/jbc.M414217200. [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamada S, Toyoshima M, Dong J, Nakajima M, Sugahara K. Structural recognition by recombinant human heparanase that plays critical roles in tumor metastasis. J. Biol. Chem. 2002;277:42488–42495. doi: 10.1074/jbc.M206510200. [DOI] [PubMed] [Google Scholar]
- Parish CR. The role of heparan sulphate in inflammation. Nat. Rev. Immunol. 2006;6:633–643. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- Peterson S, Frick A, Liu J. Design of biologically active heparan sulfate and heparin using an enzyme-based approach. Nat. Prod. Rep. 2009;26:610–627. doi: 10.1039/b803795g. [DOI] [PubMed] [Google Scholar]
- Petitou M, Driguez PA. Azasugar derivatives, heparanase inhibitors. 2006 Eur. Patent CO7H5/06;CO7H3/06. US Appl. US 20070625994 20070123.
- Pisano C, Aulicino C, Vesci L, Casu B, Naggi A, Torri G, Ribatti D, Belleri M, Rusnati M, Presta M. Undersufated, low-molecular weight glycol-split heparin as an antiangiogenic VEGF antagonist. Glycobiology. 2005;15:1C–6C. doi: 10.1093/glycob/cwi007. [DOI] [PubMed] [Google Scholar]
- Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- Presta M, Dell'Era P, Mitola S, Moroni E, Ronca R, Rusnati M. Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev. 2005;16:159–178. doi: 10.1016/j.cytogfr.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Purushothaman A, Uyama T, Kobayashi F, Yamada S, Sugahara K, Rapraeger AC, Sanderson RD. Heparanase enhanced shedding of syndecan-1 by myeloma cells promotes endothelial invasion and angiogenesis. Blood. 2010 doi: 10.1182/blood-2009-07-234757. 10.1182/blood-2009-07-234757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye DA, Vivès RR, Hyde P, Gallagher JT. Regulation of FGF-1 mitogenic activity by heparan sulfate oligosaccharides is dependent on specific structural features: differential requirements for the modulation of FGF-1 and FGF-2. Glycobiology. 2000;10:1183–1192. doi: 10.1093/glycob/10.11.1183. [DOI] [PubMed] [Google Scholar]
- Raman R, Venkataraman G, Ernst S, Sasisekharan V, Sasisekharan R. Structural specificity of heparin binding in the fibroblast growth factor family of proteins. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2357–2362. doi: 10.1073/pnas.0437842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland J, Sanderson RD, Waguespack M, Barker SA, Long R, Carson DD, Marchetti D. Heparanase degrades syndecan-1 and perlecan heparan sulfate: functional implications for tumor cell invasion. J. Biol. Chem. 2004;279:8047–8055. doi: 10.1074/jbc.M304872200. [DOI] [PubMed] [Google Scholar]
- Rek A, Krenn E, Kungl AJ. Therapeutically targeting protein–glycan interactions. Brit. J. Pharmacol. 2009;157:686–694. doi: 10.1111/j.1476-5381.2009.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CJ, Harmer NJ, Goodger SJ, Blundell TL, Gallagher J. Cooperative dimerization of fibroblast growth factor 1 (FGF1) upon a single heparin saccharide may drive the formation of 2:2:1 FGF1–FGFR2c–heparin ternary complexes. J. Biol. Chem. 2005;280:42274–42282. doi: 10.1074/jbc.M505720200. [DOI] [PubMed] [Google Scholar]
- Robinson CJ, Mulloy B, Gallagher GT, Stringer SE. VEGF 165-binding sites within heparan sulfate encompass two highly sulfated domains and can be liberated by K5 lyase. J. Biol. Chem. 2006;281:1731–1740. doi: 10.1074/jbc.M510760200. [DOI] [PubMed] [Google Scholar]
- Rudd TR, Yates EA, Hricovíni M. Spectroscopic and theoretical approaches for the determination of heparin saccharide structure and the study of protein–glycosaminoglycan complexes in solution. Curr. Med. Chem. 2009;16:4750–4766. doi: 10.2174/092986709789878193. [DOI] [PubMed] [Google Scholar]
- Sandbäck-Pikas D, Li J-P, Vlodavsky I, Lindahl U. Substrate specificity of heparanases from human hepatoma and platelets. J. Biol. Chem. 1998;273:18770–18777. doi: 10.1074/jbc.273.30.18770. [DOI] [PubMed] [Google Scholar]
- Sanderson RD, Yang Y. Syndecan-1: a dynamic regulator of the myeloma microenvironment. Clin. Exp. Metastasis. 2008;25:149–159. doi: 10.1007/s10585-007-9125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure–function relationships of glycosaminoglycans. Ann. Rev. Biomed. Eng. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- Schenauer MR, Yu Y, Sweeney MD, Leary JA. CCR2 chemokines bind selectively to acetylated heparan sulfate octasaccharides. J. Biol. Chem. 2007;282:25182–25188. doi: 10.1074/jbc.M703387200. [DOI] [PubMed] [Google Scholar]
- Schenauer MR, Meissen JK, Seo Y, Ames JB, Leary JA. Heparan sulfate separation, sequencing, and isomeric differentiation: ion mobility spectrometry reveals specific iduronic acid and glucuronic acid-containing hexasaccharides. Anal. Chem. 2009;81:10179–10185. doi: 10.1021/ac902186h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeberger PH, Werz DB. Synthesis and medical applications of oligosaccharides. Nature. 2008;446:1046. doi: 10.1038/nature05819. [DOI] [PubMed] [Google Scholar]
- Skidmore MA, Guimond SE, Rudd TR, Ferning DG, Turnbull JE, Yates EA. The activities of heparan sulfate and its analogue heparin are dictated by biosynthesis, sequence, and conformation. Connective Tissue Res. 2008;49:140–144. doi: 10.1080/03008200802148595. [DOI] [PubMed] [Google Scholar]
- Spillmann D, Witt D, Lindahl U. Defining the interleukin-8-binding domain of heparan sulfate. J. Biol. Chem. 1998;273:15487–15493. doi: 10.1074/jbc.273.25.15487. [DOI] [PubMed] [Google Scholar]
- Stevenson JL, Choi SH, Varki A. Differential metastasis inhibition by clinically relevant levels of heparins — correlation with selectin inhibition, not antithrombotic activity. Clin. Cancer Res. 2005;11:7003–7011. doi: 10.1158/1078-0432.CCR-05-1131. [DOI] [PubMed] [Google Scholar]
- Taylor KL, Gallo RL. Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J. 2006;20:9–22. doi: 10.1096/fj.05-4682rev. [DOI] [PubMed] [Google Scholar]
- Van Boekel CAA, Petitou M. The unique antithrombin III binding domain of heparin: a lead of new synthetic antithrombotics. Angew. Chem. Int. Ed. 1993;32:1671–1690. [Google Scholar]
- Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr. Pharm. Des. 2007;13:2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Elkin M, Abboud-Jarrous G-, Levi-Adams F, Fuks L, Shafat I, Ilan N. Heparanase: one molecule with multiple functions in cancer progression. Connective Tissue Res. 2008;49:207–210. doi: 10.1080/03008200802143281. [DOI] [PubMed] [Google Scholar]
- Waern I, Jia J, Pejler G, Zcharia E, Vlodavsky I, Li J-P, Wernersson S. Accumulation of Ym1 and formation of intracellular crystalline bodies in alveolar macrophages lacking heparanase. Mol. Immunol. 2010 doi: 10.1016/j.molimm.2010.02.004. 10.1016/j.molimm.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Wang L, Fuster M, Sriramarao P, Esko JD. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat. Immunol. 2005;6:902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- Whitelock JM, Iozzo RV. Heparan sulfate: a complex polymer charged with biological activity. Chem. Rev. 2005;105:2745–2764. doi: 10.1021/cr010213m. [DOI] [PubMed] [Google Scholar]
- Whitelock JM, Melrose J, Iozzo RV. Diverse cell signaling events modulated by perlecan. Biochemistry. 2008;47:11174–11183. doi: 10.1021/bi8013938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Mcleod V, Miao HQ, Theus A, Zhan F, Shaughnessy JD, Jr., Sawyer J, Li J-P, Zcharia E, Vlodavsky I, Sanderson RD. Heparanase enhances syndecan-1 shedding: a novel mechanism for stimulation of tumor growth and metastasis. J. Biol. Chem. 2007a;282:13326–13333. doi: 10.1074/jbc.M611259200. [DOI] [PubMed] [Google Scholar]
- Yang Y, McLeod V, Day Y, Khotskaya-Sample Y, Shriver Z, Venkataraman G, Sasisekharan R, Naggi A, Torri G, Casu B, Vlodavsky I, Suva LJ, Epstein J, Yacobby S, Shaughnessy JD, Jr., Bartologie B, Sanderson RD. The syndecan-1 heparan sulfate proteoglycan is a viable target for myeloma therapy. Blood. 2007b;110:2041–2048. doi: 10.1182/blood-2007-04-082495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip GW, Smollich M, Götte M. Therapeutic value of glycosaminoglycans in cancer. Mol. Cancer Ther. 2006;5:2139–2148. doi: 10.1158/1535-7163.MCT-06-0082. [DOI] [PubMed] [Google Scholar]
- Young E. The anti-inflammatory effects of heparin and related compounds. Thromb. Res. 2008;122:743–752. doi: 10.1016/j.thromres.2006.10.026. [DOI] [PubMed] [Google Scholar]
- Zhang F, Zhang Z, Lin X, Mohammadi M, Linhardt RJ. Compositional analysis of heparin/heparan sulfate interaction with fibroblast growth factor·fibroblast growth factor receptor complexes. Biochemistry. 2009;48:8368–8379. doi: 10.1021/bi9006379. [DOI] [PMC free article] [PubMed] [Google Scholar]