Abstract
Bacteriophage D3112 represents one of two distinct groups of transposable phage found in the clinically relevant, opportunistic pathogen Pseudomonas aeruginosa. To further our understanding of transposable phage in P. aeruginosa, we have sequenced the complete genome of D3112. The genome is 37,611 bp, with an overall G+C content of 65%. We have identified 53 potential open reading frames, including three genes (the c repressor gene and early genes A and B) that have been previously characterized and sequenced. The organization of the putative coding regions corresponds to published genetic and transcriptional maps and is very similar to that of enterobacteriophage Mu. In contrast, the International Committee on Taxonomy of Viruses has classified D3112 as a λ-like phage on the basis of its morphology. Similarity-based analyses identified 27 open reading frames with significant matches to proteins in the NCBI databases. Forty-eight percent of these were similar to Mu-like phage and prophage sequences, including proteins responsible for transposition, transcriptional regulation, virion morphogenesis, and capsid formation. The tail proteins were highly similar to prophage sequences in Escherichia coli and phage Phi12 from Staphylococcus aureus, while proteins at the right end were highly similar to proteins in Xylella fastidiosa. We performed phylogenetic analyses to understand the evolutionary relationships of D3112 with respect to Mu-like versus λ-like bacteriophages. Different results were obtained from similarity-based versus phylogenetic analyses in some instances. Overall, our findings reveal a highly mosaic structure and suggest that extensive horizontal exchange of genetic material played an important role in the evolution of D3112.
Bacteriophages play central roles both in the shaping of natural populations of bacteria and in the development of genetics research. Historically, they were profoundly important for seminal studies illuminating the nature of the gene and in early studies of gene regulation. Bacteriophages have, of course, also been essential tools for molecular genetic research. Recently, these phages have again come to prominence due to the discovery of the remarkable dynamics underlying their evolution.
One of the best-studied bacteriophages is the temperate, transposable phage Mu (51). This predator of Escherichia coli has the ability to integrate randomly into the host genome, often leading to mutations in the host, and to transduce variable amounts of heterogeneous host DNA (53). These traits, characteristic of the transposable phages, make it an extraordinary resource for genetic research. Unfortunately, very few other transposable phages have been isolated from the gram-negative Enterobacteriaceae.
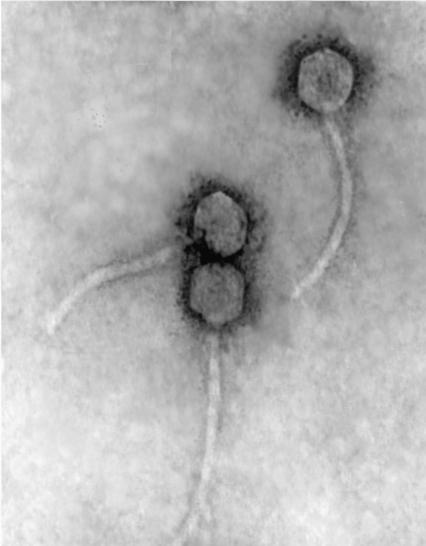
In contrast, over 60 distinct temperate, transposable bacteriophages have been isolated from Pseudomonas aeruginosa, a gram-negative bacterium belonging to the family Pseudomonadaceae (2). D3112-like phages (30, 31, 45) and B3-like phages (1) represent two major groups of P. aeruginosa transposable phages which have been shown to recombine with low frequency (35). D3112 was found to have a genomic structure and life cycle that closely approximate those of Mu (5, 10, 36, 40, 45). Like Mu, D3112 is also a temperate phage. Upon infection, the phage genome is integrated into the host genome via transposition, and the c protein represses the lytic cycle and maintains stable integration of the genome (45). Surprisingly, D3112 and Mu share no similarity at the DNA level (40), and the virion structure of bacteriophage D3112 is morphologically more similar to the λ-like phages (Fig. 1). This has resulted in D3112 being classified as a member of the λ-like Siphoviridae family (phages with long, noncontractile tails) by the International Committee on Taxonomy of Viruses. Bacteriophage Mu, on the other hand, is classified in the Mu-like family of Myoviridae (phages with long, contractile tails).
FIG. 1.
Electron micrograph of D3112 bacteriophages, negatively stained with 2% uranyl acetate.
D3112's bacterial host, P. aeruginosa, is an opportunistic human pathogen that causes a variety of severe diseases in immunocompromised hosts. P. aeruginosa is the most-common cause of pneumonia in intensive care units, the second most common cause of nosocomial pneumonia, and the leading cause of death among cystic fibrosis (CF) patients. This highly adaptable bacterium is resistant to most antibiotics and causes persistent biofilm-mediated infections that are of serious medical concern (15). The persistence of P. aeruginosa and the constant emergence of new bacterial pathogens have led to a renewed interest in transposable phages, for both research and therapeutic purposes. D3112 serves as a promising model to aid in the study of this type of phage. The transposable nature of D3112 has already been exploited to create mini-D3112 transposable elements capable of introducing new genetic material stably into the chromosome of P. aeruginosa (16, 28, 56).
The need to further our understanding of temperate, transposable phages has prompted the genomic sequencing of D3112. Here we report the shotgun cloning, genome sequencing, and annotation of the D3112 genome. Comparative studies with other Mu-like phages and prophages were performed. Phylogenetic analyses of D3112 were also carried out to elucidate the evolutionary history of the phage. Our findings will contribute to the establishment of a phylogenetically based taxonomic framework for bacteriophages.
MATERIALS AND METHODS
Bacterial strains, phage, and culture conditions.
P. aeruginosa strains PAS429, which carries the D3112cts prophage with a temperature-sensitive c repressor (cts), and wild-type strain PAO1 (kindly provided by M. S. DuBow, Department of Microbiology and Immunology, McGill University) were grown in Luria-Bertani broth (LB) supplemented with kanamycin (50 μg/ml) in a 30°C shaking incubator (46). Wild-type P. aeruginosa strain PAK, also known as PAO6 (kindly provided by L. Burrows, Department of Surgery, The Hospital for Sick Children, Toronto, Ontario, Canada), was grown in the same manner. A highly virulent P. aeruginosa strain, PA14 (kindly provided by F. M. Ausubel, Department of Molecular Biology, Massachusetts General Hospital), was grown in LB supplemented with rifampin (50 μg/ml). E. coli strain DH5α was grown in LB at 37°C, supplemented with ampicillin (50 μg/ml) and kanamycin (50 μg/ml).
Preparation of D3112cts lysates.
PAS429 was grown overnight without antibiotics. The culture was diluted 1:50 in LB and grown for an additional 3 h or until the optical density at 600 nm reached 0.6. The culture was shifted to 42°C for roughly 2 h or until the cells lysed. MgSO4 was added to a final concentration of 2 mM, CaCl2 was added to a final concentration of 0.2 mM, and chloroform was added to a final concentration of 1% of the lysate volume. Cell debris was removed by centrifugation, and the cleared lysates were stored at 4°C.
Extraction of D3112cts DNA.
Phage DNA was prepared as described previously (48). DNase and RNase were added at a final concentration of 100 μg/ml each to 1.5 ml of a high-titer cleared liquid lysate, and this mixture was incubated at 37°C for 30 min. Twenty microliters of 2 M ZnCl2 was added per milliliter of lysate, and the mixture was incubated at 37°C for 5 min and then centrifuged for 1 min. The supernatant was discarded, and the pellet containing the phage was resuspended in 500 μl of TES (0.1 M Tris-HCl, pH 8; 0.1 M EDTA; 0.3% sodium dodecyl sulfate). This was followed by lysis at 65°C for 15 min. The suspension was mixed thoroughly after the addition of 60 μl of 3 M potassium acetate, pH 4.8, and then incubated on ice for 20 min. Proteins were removed by centrifugation, and the supernatant was transferred to a new tube. An equal volume of isopropanol was added, and the mixture was incubated on ice for 5 min. The DNA was recovered by centrifugation, washed with 70% ethanol, air dried, and resuspended in a small volume of TE (10 mM Tris-HCl, pH 8.0; 1 mM EDTA, pH 8.0).
D3112 infection.
D3112 infection of bacterial cells was performed using a modification of the method described by Roncero et al. (44). Host bacteria were mixed with phage at a high multiplicity of infection. After addition of soft agar, the cultures were transferred to LB plates supplemented with 1 mM MgSO4. Bacterial phenotypes resistant to infection were visualized as confluent lawns, while sensitive phenotypes were distinguishable as completely lysed plates.
Shotgun cloning of D3112cts.
Partial digestion of bacteriophage DNA was performed using the DNase Shotgun cleavage kit (Novagen, Madison, Wis.). A minimum of 1 μg of purified phage DNA was cleaved with 0.01 U of DNase I. DNase I concentrations were optimized according to the manufacturer's protocol. DNA fragments between 500 and 2,000 bp were size selected from a 1% (wt/vol) TAE agarose gel (21). DNA fragments were purified and concentrated using the QIAquick PCR Purification kit (Qiagen Inc., Valencia, Calif.). Staggered ends of the DNA fragments were repaired and dephosphorylated using T4 DNA polymerase and alkaline phosphatase, respectively (New England Biolabs, Beverly, Mass.). The resulting fragments were ligated into the pCR4Blunt-TOPO vector (Invitrogen Corp., Carlsbad, Calif.) and transformed into E. coli DH5α. Transformants were transferred to plates with selective media containing ampicillin and kanamycin.
PCR amplification and sequencing.
Direct colony-PCR amplification of the inserts was performed using modified vector primers M13F (5′-GTAAAACGACGGCCAGT-3′) and M13R (5′-CAGGAAACAGCTATGACCATG-3′). The PCR conditions used were denaturation at 94°C for 4 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 2 min, with a final extension at 72°C for 5 min. Amplified products were sized by gel electrophoresis, and fragments >500 bp were selected for sequencing. The selected PCR mixtures were enzymatically cleaned for sequencing with the addition of a 1/100 reaction volume of alkaline phosphatase (10 U/μl) and exonuclease I (20 U/μl) (New England Biolabs). The reaction mixtures were incubated at 37°C for 30 min, followed by 85°C for 15 min to destroy the enzymes. Sequencing was performed using modified T3 (5′-GCCAAGCTCAGAATTAACCCTCACTAAAGG-3′) and T7 (5′-CGACGGCCAGTGAATTGTAATACGACTC-3′) primers. Ten-microliter reaction mixtures were prepared with the CEQ Dye Terminator Cycle Sequencing (DTCS) Quick Start kit (Beckman Coulter Canada, Inc., Mississauga, Ontario, Canada) and run on the Beckman CEQ2000XL sequencer (6). DNA sequences were edited using the BioEdit Sequence Alignment Editor software (22). Sequence alignments and the assembly of continuous overlapping sequences into contigs were performed with Sequencher 4.0 (Gene Codes Corp., Ann Arbor, Mich.). Gaps between contigs were bridged by designing primers off the end of each contig and randomly pairing them in PCR amplifications with D3112cts DNA as the template. Amplified products were sequenced using the original amplification primers.
Sequence annotation and analysis.
Potential open reading frames (ORFs) were identified using GeneMark.HMM version 2.0 for prokaryotes (http://opal.biology.gatech.edu/GeneMark/heuristic_hmm2.cgi) (9). Potential ORFs were compared against the NCBI protein databases using BLASTP and PSI-BLAST (3, 4). Sequences were also scanned using PROSITE (part of the ExPASy suite) (19) and Pfam (8). Molecular masses were determined using ProtParam (part of the ExPASy suite). The DNA sequence was scanned for putative tRNA genes by tRNAscan-SE (www.genetics.wustl.edu/eddy/tRNAscan-SE/) (33) and FAStRNA (http://bioweb.pasteur.fr/seqanal/interfaces/fastrna.html) (18). Restriction enzyme analysis was determined by Vector NTI (version 6.0; InforMax, Inc., Bethesda, Md.). The moving average G+C content was determined by using the EMBOSS program GEECEE (41) by averaging the G+C content in a 1,000-bp window moved across the genome in 100-bp steps. The sliding window was implemented with the aid of the EMBOSS program SPLITTER (41).
Phylogenetic analysis.
ORFs were picked for analysis if their translated sequences had more than two significant matches to proteins in the NCBI databases. The most-similar sequences were downloaded on the basis of two criteria: expect thresholds below 5e−15 and sequence lengths similar to that of the query ORF. Choosing sequences of similar length helped eliminate those that had significant local alignments due to conserved domains but that could not be globally aligned. Alignments were made with ClustalX (version 1.81) using default parameters (52).
The phylogenies were produced using two techniques: Bayesian and neighbor joining. For the Bayesian analysis, a MrBayes Nexis block was generated for each ORF using the MrBayes Block Form (http://darwin.zoology.gla.ac.uk/∼rpage/mrbayes), and analyzed with MrBayes 2.01 (25). Each inference consisted of four Markov chains starting from random trees and running for 200,000 generations. One tree was sampled every 100 generations. The initial burn-in trees were discarded. Fifty-percent majority rule consensus trees were generated. The numbers at the interior branches represent the percentage of time a clade appears in the sampled trees. Trees were viewed in PAUP* and Treeview (38, 50). Neighbor-joining trees were generated for each ORF using the Prodist and Neighbor modules of PHYLIP (20). The Jones-Taylor-Thornton model was used to compute the protein distance matrices, including 500 bootstrap replicates. All alignments and trees are available at http://www.botany.utoronto.ca/ResearchLabs/GuttmanLab/index.stm.
Nucleotide sequence accession number. The complete nucleotide sequence of bacteriophage D3112 is available under GenBank accession number AY394005.
RESULTS
Sequence of D3112.
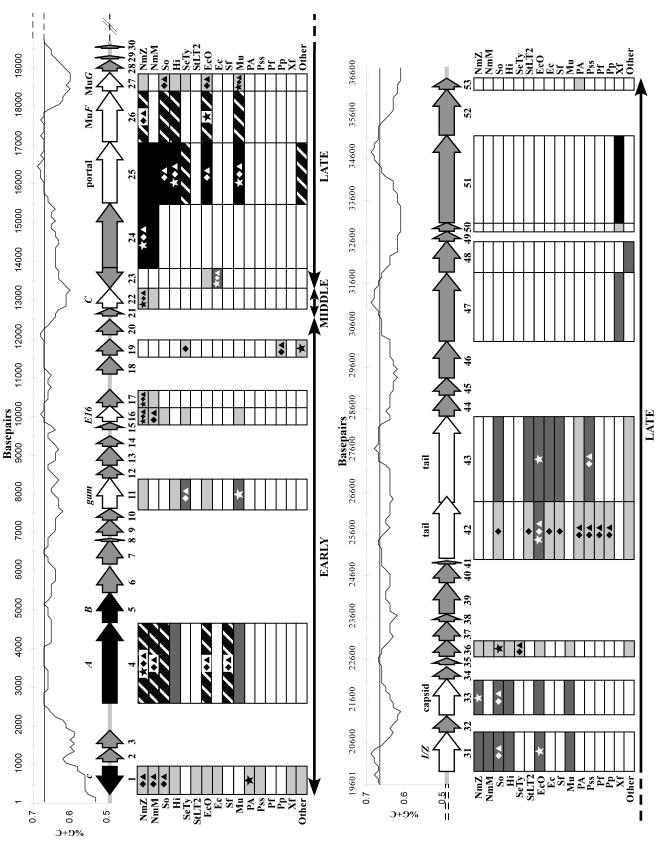
The complete genomic sequence of P. aeruginosa bacteriophage D3112 was obtained using a shotgun sequencing approach. More than 500 DNA fragments were sequenced, providing ninefold coverage of the genome on average. The complete nucleotide sequence is 37,611 bp, which is in agreement with previous estimates (30). We identified 53 potential ORFs, as illustrated in Fig. 2.
FIG. 2.
Genetic map of bacteriophage D3112, along with the G+C content of the genome above the map and a summary of the similarity and phylogenetic analyses below. The map begins in the top panel and continues in the lower panel. The approximate boundaries of the early, middle, and late regions are indicated at the bottom of each panel by solid black lines with double arrowheads. The middle portion of the schematic shows the D3112 predicted ORFs and their orientations, represented by the filled arrows. Numbers below each arrow correspond to the ORF number, while names above each arrow identify known proteins with high similarity. Three previously sequenced genes are indicated by the black arrows. White arrows designate ORFs with similarity to known proteins, and gray arrows correspond to unknown ORFs. Above the genetic map is a graphical representation of the G+C content, drawn to scale with the gene map. The nucleotide number is shown on the x axis, and the G+C content is shown on the y axis. The horizontal line at 67% G+C denotes the average G+C content of P. aeruginosa. Below the genetic map are the results from similarity and phylogenetic analyses. The names bordering the boxes stand for the bacterial host species most commonly found to be highly similar to D3112. The differently shaded boxes illustrate the different degrees of similarity (determined by BLASTP expect values). White boxes represent no significant similarity, light gray boxes represent similar proteins with expect value thresholds from 1e−20 to 1e−60, dark gray boxes represent thresholds from 1e−61 to 1e−100, hatched boxes represent thresholds from 1e−101 to 1e−164, and black boxes represent nearly identical proteins (e = 0.0). The star symbols inside the boxes indicate the species with the highest similarity to the D3112 ORFs on the basis of expect value. The diamonds show the species most closely related to the D3112 ORFs by the Bayesian method, and the triangles show the species most closely related to the D3112 ORFs by the neighbor-joining method. The abbreviations for strain designations are as follows: NmZ, N. meningitidis Z2491; NmM, N. meningitidis MC58; So, S. oneidensis; Hi, Haemophilus influenza; SeTy, S. enterica serovar Typhi; StLT2, S. enterica serovar Typhimurium LT2; EcO, E. coli O157:H7; Ec, E. coli; Sf, S. flexneri; Mu, enterobacteriophage Mu; PA, P. aeruginosa; Pss, P. syringae pv. syringae; Pf, Pseudomonas fluorescens; Pp, Pseudomonas putida; Xf, X. fastidiosa; and Other, other highly similar species not common to most of the ORFs. For ORF19, the most similar species in the “Other” category is Magnetospirillum magnetotacticum.
The G+C content of the D3112 genome averages 64.34%, which is very similar to the 67% G+C content of P. aeruginosa (Fig. 2). Only ORFs 1 and 2 have lower G+C contents of 56 and 53%, respectively. A number of other regions with G+C contents of approximately 60% are also present throughout the genome. This skew is typically seen in the mosaic structure of viruses and is indicative of the acquisition of these genes from organisms with lower G+C contents. Except for these unusual modules, the G+C contents of the majority of temperate phage genomes closely match those of their hosts (29). Another measure of horizontal transfer is a bias in the usage of synonymous codons. The codon usage of D3112 and P. aeruginosa were compared (data not shown) using the Countcodon Program (www.kazusa.or.jp/codon/countcodon.html) (37). Our comparison revealed no significant differences. Several members of the Siphoviridae possess tRNA genes which can increase the overall translation rate by transcribing tRNAs that recognize rare codons (29). No tRNA species were found when the genome of D3112 was scanned with tRNAscan-SE (33) and FAStRNA (18).
Sequencing of the leftmost and rightmost ends of D3112 revealed heterogeneous P. aeruginosa sequences. This supports the earlier finding of Rehmat and Shapiro and indicates the phage is capable of integrating into different locations (16, 40). After trimming the flanking heterogeneous sequences, we determined that the sequence at the left extremity common to all of our phage clones begins with the bases 5′-TGC-3′, followed by the 3′ end of the P. aeruginosa outer membrane protein oprM. Except for the addition of the first three bases, our findings were in agreement with those of Autexier et al. (5). The difference is most likely due to our having a different isolate of the phage. The right extremity has been shown to be significantly more variable in size due to imprecise excision of the phage (16). We have found up to 2 kb of heterogeneous DNA attached to the right end of D3112, which was also trimmed from our final sequence.
Sequence comparisons between the two existing GenBank entries for the left end of D3112 and our sequence revealed 10 discrepancies, 1 of which is the extra bases at the 5′ end (5, 54). Six discrepancies occurred within ORFs. Three of these could effect the resulting proteins and are discussed below. The remaining three differences are located in noncoding regions at positions 1041, 2342, and 2429. Our sequence was confirmed at these positions with at least 10-fold sequencing coverage from both strands. These discrepancies could be due to errors in earlier sequencing efforts or due to different isolates of the phage.
Genome organization of D3112cts.
The genome of bacteriophage D3112 can be divided into three functional regions: the left end or early region, which is responsible for genome integration, modulation of phage gene expression, and modulation of host response; the middle region, which is responsible for control of late gene transcription; and the right end or late region, which is responsible for virion morphogenesis and contains ORFs of largely unknown function. To determine whether any large rearrangements have occurred in our isolate, we generated a restriction map of our genome for comparison to one previously reported by Rehmat and Shapiro (40), using the identical enzymes SalI, KpnI, HpaI, HindIII, and EcoRI (data not shown). Our map was in agreement with the previous map, except for a reversal of the last KpnI and HpaI sites at bases 34025 and 34034, respectively. This region does not appear to be consistent with the invertible tail segment of phage Mu given the lack of similarity to any tail-like sequences and its location in ORF 51 at the right extremity of the genome. The discrepancy could be due to errors in earlier analyses or differences in the right end of our phage isolate. Overall, there do not appear to be any major rearrangements in our isolate.
A transcriptional map of D3112 was recently constructed by Bidnenko et al. (11), suggesting that the genome is comprised of six independent transcriptional units which correspond to a modular organization similar to that of phage Mu. They reported that transcription occurred from left to right in the same order as the genes located on their genetic map, except for the c repressor. As seen in Fig. 2, our findings generally agree with the previous findings, but we identified two ORFs that are transcribed from left to right (ORFs 1 and 23).
Analysis of D3112 gene products.
Three ORFs from the left end of D3112 have been previously identified (those encoding the c repressor and A and B transposases) (45, 54). Five additional genes have been genetically characterized (cip1, kil, C, ts47, and c91) (10-12). We identified a total of 53 ORFs, 7 of which begin with a GUG initiation codon, while the remaining ORFs begin with an AUG codon. Structurally and functionally the genome of D3112 is very similar to that of bacteriophage Mu, although there is no detectable homology at the DNA level (36). A complete list of the D3112 ORFs and their putative functions determined by BLASTP analysis is shown in Table 1. Figure 2 shows a schematic of all the ORFs and their degrees of similarity to known proteins as determined by BLASTP analysis.
TABLE 1.
Bacteriophage D3112cts ORFs and putative genes
| Gene no. | Range (bp) | Strand | Size (aa) | Mass (kDa) | Significant match(es)b | Putative function | BLASTP
|
|
|---|---|---|---|---|---|---|---|---|
| e valuec | Score | |||||||
| 1 | 173-895 | − | 240 | 26.3 | S13498; D3112 repressor cI (P. aeruginosa) | Repressor | 9e−54 | 211 |
| NP_061565; phage D3 cI (P. aeruginosa) | Repressor | 1e−37 | 157 | |||||
| 2 | 1044-1388 | + | 114 | 13.2 | NP_284593; prophage PMN1 Ner-like DNA-binding protein (N. meningitidis Z2491) | Transcriptional regulator | 1e−4 | 46.2 |
| 3 | 1391-1825 | + | 144 | 16.2 | None | |||
| 4 | 2551-4623 | + | 690 | 77.8 | S62728; D3112 gene A (P. aeruginosa) | Transposition | 0 | 659 |
| NP_284035; prophage PNM2 putative transposase (N. meningitidis Z2491) | Transposition | 1e−128 | 461 | |||||
| 5 | 4620-5390 | + | 256 | 27.8 | S62729; phage D3112 gene B (P. aeruginosa) | Transposition | 1e−118 | 425 |
| NP_716277; prophage MuSo1 putative transposon (S. oneidensis MR-1) | Transposition | 6e−85 | 314 | |||||
| 6 | 5387-6055 | + | 222 | 24.6 | None | |||
| 7 | 6127-6762 | + | 211 | 23.6 | None | |||
| 8 | 6747-6863 | + | 38 | 4.3 | None | |||
| 9a | 6863-7276 | + | 137 | 15.1 | NP_716279; hypothetical protein (S. oneidensis MR-1) | Unknown | 0.006 | 40 |
| 10a | 7263-7550 | + | 95 | 10.7 | NP_384293; pit accessory protein (Sinorhizobium meliloti) | Transport of small anions | 0.74 | 33 |
| 11 | 7567-8367 | + | 266 | 29.3 | NP_050614; phage Mu Gam (E. coli) | Host nuclease inhibitor | 3e−63 | 242 |
| NP_456013; phage host nuclease inhibitor (S. enterica subsp. enterica serovar Typhi) | Host nuclease inhibitor | 7e−63 | 241 | |||||
| 12 | 8369-8677 | + | 102 | 11.2 | None | |||
| 13 | 8680-9168 | + | 162 | 17.7 | None | |||
| 14 | 9171-9440 | + | 89 | 9.6 | None | |||
| 15 | 9624-9806 | + | 60 | 6.5 | None | |||
| 16 | 9803-10210 | + | 135 | 15.5 | NP_283952; hypothetical protein (N. meningitidis Z2491) | Unknown | 4e−50 | 196 |
| NP_274021; E16-related protein (N. meningitidis MC58) | Integration host factor | 1e−49 | 195 | |||||
| 17 | 10197-10646 | + | 149 | 17.1 | NP_284576; prophage PNM1 putative integral membrane protein (N. meningitidis Z2491) | Unknown | 2e−56 | 218 |
| NP_274022; hypothetical protein (N. meningitidis MC58) | Unknown | 4e−51 | 200 | |||||
| 18 | 11041-11487 | + | 148 | 16 | None | |||
| 19 | 11510-11926 | + | 138 | 15.4 | ZP_00053720; hypothetical protein (Magnetosptrillum magnetotacticum) | Unknown | 1e−53 | 208 |
| NP_049909; phage PM2 structural protein P5 (Alteromonas) | Structural | 2e−49 | 194 | |||||
| 20 | 12042-12443 | + | 133 | 14 | None | |||
| 21 | 12554-12742 | + | 62 | 6.7 | None | |||
| 22 | 12753-13253 | + | 166 | 18.1 | NP_284564; prophage PNM1 hypothetical DNA-binding protein (N. meningitidis Z2491) | Unknown | 5e−45 | 188 |
| NP_274124; prophage MuMenB hypothetical protein (N. meningitidis MC58) | Unknown | 9e−41 | 174 | |||||
| 23 | 13255-13791 | − | 178 | 19.8 | NP_112049; phage HK620 unknown protein (E. coli) | Unknown | 2e−64 | 245 |
| NP_287260; prophage CP-933N unknown protein (E. coli O157:H7 EDL933) | Unknown | 2e−52 | 206 | |||||
| 24 | 13790-15445 | + | 551 | 61.9 | NP_284562; prophage PMN2 hypothetical protein (N. meningitidis Z2491) | Unknown | 0 | 889 |
| NP_274126; prophage MuMenB hypothetical protein (N. meningitidis MC58) | Unknown | 0 | 844 | |||||
| 25 | 15457-17043 | + | 528 | 57.8 | NP_439651; predicted coding region (Haemophilus influenza Rd) | Unknown | 0 | 713 |
| NP_050633; phage Mu gp29 protein (E. coli) | Portal | 0 | 706 | |||||
| 26 | 17043-18329 | + | 428 | 47.2 | NP_312998; hypothetical protein, similar to Mu gp30 (E. coli O157:H7) | Head assembly cofactor | 1e−147 | 524 |
| NP_050634; phage Mu late F ORF (E. coli) | Virion morphogenesis | 1e−146 | 519 | |||||
| 27 | 18329-18796 | + | 155 | 16.6 | NP_050635; phage Mu protein G (E. coli) | Morphogenesis | 7e−38 | 156 |
| NP_456029; putative protein similar to Mu gpG (S. enterica subsp. enterica serovar Typhi) | Virion morphogenesis | 1e−35 | 149 | |||||
| 28 | 18927-19199 | + | 90 | 9.9 | None | |||
| 29a | 19205-19363 | + | 52 | 6 | NP_539801; DNA gyrase subunit A (Brucella melitensis 16M) | ATP hydrolyzing | 8e−10 | 33.5 |
| 30a | 19470-19574 | + | 34 | 3.9 | AAO59158; hypothetical protein (P. syringae py. tomato DC3000) | Unknown | 1.4 | 32.7 |
| 31 | 19918-20898 | + | 326 | 34.7 | NP_313000; putative protease (E. coli O157:H7) | Protease | 3e−91 | 336 |
| NP_050636; phage Mu gp32 (E. coli) | Protease (I) and scaffold (Z) | 5e−90 | 332 | |||||
| 32 | 20908-21306 | + | 132 | 13.7 | None | |||
| 33 | 21334-22242 | + | 302 | 33 | NP_284557; prophage PNM1 conserved hypothetical protein (N. meningitidis Z2491) | Unknown | 6e−76 | 285 |
| NP_050638; phage Mu major head subunit (E. coli) | Major head subunit | 1e−73 | 277 | |||||
| 34a | 22254-22556 | + | 100 | 10.6 | NP_716309; conserved hypothetical protein (S. oneidensis MR-1) | Unknown | 0.088 | 36.6 |
| 35 | 22619-22795 | + | 58 | 6.3 | None | |||
| 36 | 22792-23208 | + | 138 | 14.7 | NP_716310; conserved hypothetical protein (S. oneidensis MR-1) | Unknown | 3e−45 | 179 |
| NP_439658; conserved hypothetical protein similar to FluMu gp36 (H. influenza Rd) | Unknown | 7e−45 | 178 | |||||
| 37 | 23205-23684 | + | 159 | 17.3 | ZP_00043681; hypothetical protein (Magnetococcus sp. strain MC-1) | Unknown | 1e−48 | 192/PICK> |
| 38 | 23672-23851 | + | 59 | 6.6 | None | |||
| 39 | 23884-24654 | + | 256 | 27 | NP_518984; conserved hypothetical protein (Ralstonia solanacearum GMI1000) | Unknown | 7e−37 | 154 |
| 40a | 24658-25143 | + | 161 | 17.7 | NP_297809; hypothetical protein (X. fastidiosa 9a5c) | Unknown | 1.4 | 33.1 |
| 41 | 25143-25271 | + | 42 | 4.6 | None | |||
| 42 | 25274-26818 | + | 514 | 54.1 | NP_286523; prophage CP-933K putative tail component (E. coli O157:H7 EDL933) | Tail component | 4e−62 | 240 |
| AAL82321; phage phi12 tail fiber (S. aureus 8325) | Tail fiber | 6e−60 | 233 | |||||
| NP_461530; prophage Gifsy-1 phage tail component H (S. enterica serovar Typhimurium LT2) | Tail component | 8e−60 | 232 | |||||
| 43 | 26671-28833 | + | 720 | 76.4 | NP_287594; prophage CP-9330 putative tail component (E. coli O157:H7 EDL933) | Tail component | 6e−99 | 363 |
| NP_459895; prophage Fels-1 putative minor tail protein (S. enterica serovar Typhimurium LT2) | Tail protein | 8e−94 | 346 | |||||
| 44 | 28840-29391 | + | 183 | 19.9 | None | |||
| 45 | 29357-29797 | + | 146 | 16.3 | None | |||
| 46 | 29799-30722 | + | 307 | 33.2 | None | |||
| 47 | 30722-32425 | + | 567 | 62.3 | NP_297812; hypothetical protein (Xylella fastidiosa 9a5c) | Unknown | 1e−173 | 610 |
| 48 | 32460-33236 | + | 258 | 28.3 | NP_540260; hypothetical phage protein (Brucella melitensis 16M) | Unknown | 1e−85 | 317 |
| ZP_00011482; hypothetical protein (Rhodopseudomonas palustris) | Unknown | 3e−78 | 292 | |||||
| 49 | 33245-33484 | + | 79 | 8.8 | None | |||
| 50 | 33481-33699 | + | 72 | 8.2 | NP_299395; hypothetical protein (X. fastidiosa 9a5c) | Unknown | 4e−30 | 130 |
| 51 | 33689-35896 | + | 735 | 79.9 | ZP_00039982; hypothetical protein (X. fastidiosa Dixon) | Unknown | 0 | 892 |
| 52 | 35893-37044 | + | 383 | 40.9 | None | |||
| 53 | 37041-37334 | + | 97 | 10.7 | BAB03224; hypothetical protein (P. aeruginosa PAO1) | Unknown | 7e−13 | 74.4 |
Weak BLAST hits, most likely representing conserved domains.
GenBank accession number is followed by phage or prophage name, protein description, and host in parentheses.
E, expect value.
The leftmost ORF encodes the well-characterized lysogenic c repressor (45). Expression of the leftmost ORF in P. aeruginosa renders the cells immune to superinfection, which supports its role as repressor of the lytic cycle of infection (5). According to the BLASTP analysis, the D3112 c repressor is more similar to the cI repressor of lambdoid phage D3 of P. aeruginosa, with 50% amino acid similarity and 32% amino acid identity, than to the c repressor of phage Mu. Both the D3 and D3112 proteins contain a helix-turn-helix DNA binding motif that is present in a large family of transcriptional regulators including Cro and cI (45).
ORF 2 may encode the cip (for “control of interaction of phages”) protein, which has been shown to be functionally similar to the Ner protein in phage Mu. As such, cip may serve as a negative regulator of c repressor transcription (11). It is expressed only during the prophage stage.
No DNA or protein homologies were found in our analysis for ORF 3, but it contains an insertion of 5′-GGCCGCGTGGC-3′ at position 1709, compared to previously published sequences. This discrepancy causes a frameshift, the functional significance of which is unknown. Our sequence is supported by ninefold sequencing coverage of the region.
The last major sequence discrepancy between our data and the previously submitted sequence is located in ORF 6. Our data report an insertion of 5′-GCC-3′ at position 5442. This insertion does not cause a frameshift and is verified by 10-fold sequence coverage from both strands.
The first half of ORF 11 is similar to the host nuclease inhibitor protein Gam in phage Mu. Conservation of this protein among pathogens suggests that this is an important factor for overcoming host defense and establishing infection. The putative D3112 Gam is 267 amino acids (aa) long, which averages 92 aa longer than other homologues. The latter half of ORF11 (aa 178 to 248) does not align with Gam but displays 44% identity to a hypothetical E. coli O157:H7 protein.
ORF 16 displays 63% similarity (40% identity) to hypothetical protein NMA1186 in prophage PNM1 of Neisseria meningitidis Z2491. NMA1186 has reported similarity to Mu protein E16 (http://www.sanger.ac.uk). ORF 16 also has weak similarity to the E16 homologue in N. meningitidis MC58. This suggests that ORF 16 is similar to phage Mu E16 (gemA) and may be responsible for modulation of host response (36).
ORF 22 is a likely candidate for the C gene or locus ts47 on the basis of its location in the D3112 genome (10, 11). The C gene was mapped between 12 to 16 kb of the D3112 genome and was shown to be a positive regulator of viral late gene transcription. It neighbors the ts47 locus, a second positive regulator of late gene transcription, which was mapped between 13.5 and 21 kb of the D3112 physical map. The presence of both gene products is essential for a normal level of late gene transcription (10). ORF 22 has 74% similarity (56% identity) to a hypothetical DNA-binding protein in prophage PNM1 of N. meningitidis Z2491. This putative DNA-binding activity is suggestive of a nonstructural role, possibly as a transcriptional regulator.
ORFs 25 to 33 are part of the late-region genes responsible for head morphogenesis. The gene order of this segment agrees with those of phage Mu, prophage FluMu, and Mu-like prophage PNM1 (36).
ORF 33 has 63% similarity (42% identity) to a hypothetical protein in prophage PNM1 and 61% similarity (40% identity) to the major head subunit gpT in phage Mu. Interestingly, the beginning and end of ORF 33 align with the PNM1 and Mu proteins, but not a 12-aa stretch in the middle spanning aa 168 to 179. The significance of this short region, which has no matches in the databases, remains to be determined.
ORFs 42 and 43 belong to the late-region genes responsible for tail morphogenesis and show similarity to lambdoid phages as opposed to Mu-like phages. ORF 42 has 42% similarity (23% identity) to a putative tail component of prophage CP-933K in E. coli O157:H7 EDL933. It is also highly similar to a tail fiber protein of phage Phi12 in Staphylococcus aureus. ORF 43 has 35% similarity (21% identity) to a putative tail component of prophage CP-933O of E. coli O157:H7 EDL933.
The ORFs at the right end of the genome have unknown functions, with the majority displaying very high amino acid similarity to hypothetical proteins in Xylella fastidiosa.
Infectivity and host range.
D3112 has been shown to infect many different strains of P. aeruginosa (44). Because the D3112 plaques are small and difficult to see, host bacteria were mixed with phage at a high multiplicity of infection to ensure complete lysis in the plate assay. Using this approach, a resistant phenotype (confluent lawn) is clearly distinguishable from a sensitive phenotype (complete lysis).
Our plate assays showed that D3112 was able to infect P. aeruginosa wild-type strains PA14 and PAK, but not our PAO1 isolate. This is could be due to our isolate already harboring a related prophage belonging to either the D3112 or B3 groups, which would render this host resistant to superinfection by D3112. The isolate could also express different surface receptors not recognized by the phage.
Phylogenetic and similarity analyses.
Separate phylogenetic analyses were performed for each ORF which had at least three significant BLAST hits. The ORFs analyzed were ORFs 1, 4, 11, 16, 17, 19, 22, 23, 24, 25, 26, 27, 31, 33, 36, 42, and 43. Unrooted trees were built using the Bayesian and neighbor-joining methods. With the exceptions of ORFs 19 and 42, the placements of D3112 in the trees generated by both methods were identical, supported by posterior probabilities of at least 94% for Bayesian analyses and bootstrap values of at least 70% for neighbor-joining analyses (summarized in Fig. 2). The results for ORF 19 were inconsistent. The Bayesian analysis placed D3112 ORF 19 in a strongly supported monophyletic group with Salmonella enterica and Pseudomonas putida (posterior probability of 99%). The neighbor-joining analysis placed D3112 in a poorly supported clade with P. putida (bootstrap support of 58%). If the weak node is collapsed, the results agree with the Bayesian tree. A similar situation applies to ORF 42. The neighbor-joining tree placed ORF 42 in a very poorly supported clade with Pseudomonas species (bootstrap of 41%). If the unsupported node is collapsed, the tree becomes identical to the Bayesian tree.
The degree of amino acid similarity between the D3112 ORFs and bacterial proteins with significant similarity to at least one ORF is shown in Fig. 2. The shading of the boxes indicates the degree of similarity based on BLASTP expect values also reported in Table 1. Phylogenetic analysis was used to resolve these relationships further. In the majority of cases, the phylogenetic analysis agreed with the BLASTP similarity search. The majority of D3112 ORFs comprising the early, middle, and late head regions (ORFs 1, 4, 16, 17, 22, 24, 26, and 33) were most closely related to proteins in N. meningitidis. D3112 ORF 4, which encodes A transposase, was also equally similar to proteins in E. coli O157:H7 and Shigella flexneri.
Interspersed throughout the genome are ORFs in clades with host species other than N. meningitidis. For example, ORFs 25, 27, 31, and 33 appear to be part of a genomic segment that is more closely related to S. oneidensis, a bacterium belonging to the Alteromonadaceae. The late tail segment, represented by ORFs 42 and 43, represent a diverse module compared to the first part of the genome. Both proteins display no similarity to phage Mu or N. meningitidis. Of particular interest is the clustering of D3112 ORF 43 with the plant pathogen Pseudomonas syringae. Since phage tails are believed to be involved in host recognition and attachment, one can speculate that this is a host-specific adaptation for a group of transposable phage capable of infecting a wide range of Pseudomonas species.
DISCUSSION
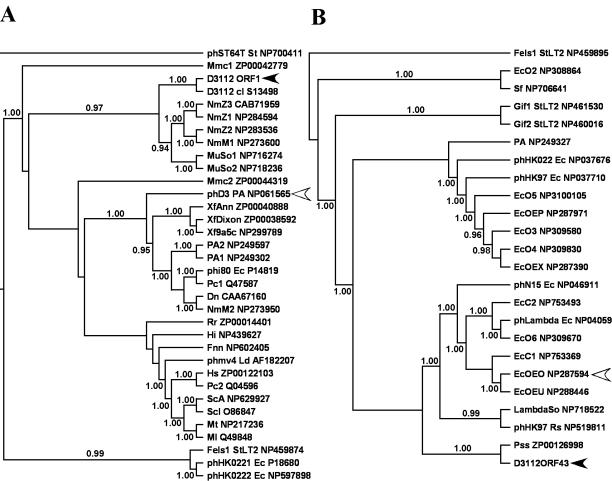
Phage D3112 was characterized by BLASTP local similarity search and phylogenetic analyses. Our results are consistent with previous studies that revealed that the genomic organization of D3112 is similar to Mu, while its morphology is similar to λ (11). The majority of ORFs in the early, middle, and late head regions possess amino acid similarity to, and reflect the organization of, Mu-like prophages from N. meningitidis Z2491. These same conclusions are also reflected in our phylogenetic studies. Since these regions comprise the majority of the D3112 backbone, we hypothesize that they are the most-ancestral part of the genome, suggesting that D3112 may have evolved from a phage which originally infected N. meningitidis. The level of nucleotide sequence divergence was so high that reliable sequence alignments could only be made at the protein level. This divergence may have been due to host adaptation, an idea supported by the similar G+C contents of D3112 and its host P. aeruginosa. An exception is ORF 1, which has an unusually low G+C content of 56%, suggesting a more recent horizontal origin. BLASTP analysis shows that ORF 1 is most similar to a homologous protein in P. aeruginosa phage D3 (29), but our phylogenetic analysis of ORF 1 and its G+C content strongly support a more recent common ancestry with N. meningitidis (G+C content, 51.8%) or S. oneidensis (G+C content, 46.0%), as shown in Fig. 2 and 3A.
FIG. 3.
Bayesian estimates of phylogeny using the Jones-Taylor-Thornton model for bacteriophage D3112 translated ORF 1 (A) and ORF 43 (B). Fifty-percent majority rule consensus trees are shown, with the numbers at the interior branches indicating the Bayesian posterior probabilities for each clade. Only clade probabilities of 94% or more are shown. Black arrowheads indicate the D3112 ORFs, and open arrowheads indicate the taxon with the highest similarity by BLASTP to that ORF. Taxon names beginning with “ph” indicate phages, followed by the phage name and the bacterial host species. The GenBank accession numbers for the taxa are found at the end of the taxon names. Names are as described in Fig. 2 with the following exceptions: Dn, Dichelobacter nodosus; EcOE, E. coli O157:H7 EDL933 followed by the cryptic prophage designation; Fels1, Gif1, and Gif2, prophages in S. enterica serovar Typhimurium LT2; Fnn, Fusobacterium nucleatum subsp. nucleatum; Hs, Haemophilus somnus; λSo, λ-like prophage in S. oneidensis; Ld, Lactobacillus delbrueckii; Ml, Mycobacterium leprae; Mt, Mycobacterium tuberculosis; MuSo, Mu-like prophage in S. oneidensis; Pc, Pectobacterium carotovorum; Rr, Rhodospirillum rubrum; ScA, Streptomyces coelicolor A3; and Scl, Streptomyces clavuligerus.
The tail segment (ORFs 42 and 43) constitutes a diverse set of proteins believed to be responsible for recognition of host cell surface receptors and, thus, host range (47). The tail components of Mu and Mu-like prophages PNM1 and FluMu show strong similarity to each other (34, 36) but differ substantially from D3112. This divergence could be responsible for the differences in host specificity of D3112, with D3112-like phages infecting the Pseudomonadaceae. Interestingly, while BLASTP analyses found the D3112 tail proteins to have the highest similarity to enteric prophage sequences, the more thorough phylogenetic analyses clearly place the D3112 tail proteins in a clade with proteins from Pseudomonas species, as seen in Fig. 2 and 3B. Due to the high similarity between the D3112 tail proteins and prophage sequences in E. coli O157:H7, one can speculate that the genes encoding these proteins originated in E. coli, were transferred into a Pseudomonas phage, and then diversified as the phage adapted to new Pseudomonas hosts. Given the limited knowledge about the existing diversity of transposable phage, horizontal transfer from a third party cannot be ruled out.
The proteins at the right end of the D3112 genome are strikingly similar to hypothetical proteins found in X. fastidiosa, suggesting the recent acquisition of this region from X. fastidiosa. Pieces of host DNA are more easily copackaged with the right end of the phage, accounting for the presence of up to 2 kb of heterogeneous host DNA present in our sequences. Sometimes the proteins encoded in the packaged host DNA can provide adaptive functions for the phage that are not necessary for its life cycle, such as conferring species specificity, enhancing virulence, affecting phage gene expression, or modulating host responses (23, 27, 36, 49, 55). In this case the relevant sequences can become a permanent part of the phage genome, as hypothesized for the X. fastidiosa-like sequences at the right end of the D3112 genome.
Interspersed around the D3112 genome are stretches of sequence that are similar to those from disparate phages, further illuminating the mosaic nature of the phage. The vast majority of these segments appear to have been acquired via horizontal gene transfer between D3112 and other phages with bacterial hosts belonging to the phylum Proteobacteria. These observations suggest that the rate or likelihood of transmission of genetic material via horizontal transfer may be correlated with the relatedness of the host species. In the case of homologous recombination, limits to horizontal gene transfer due to genetic distance have been observed in bacteria (42). Restriction of horizontal gene transfer by their host's species boundaries has also been observed in dairy phage (13).
A more thorough sampling of phage sequences will be needed before we understand the global diversity and evolution of transposable phage. One aspect of phage genomics that has received considerable attention is the development of criteria for taxonomic classification (32, 43). Recent attempts at viral taxonomy have used a range of methods, including the conservation of morphological structures and the comparison of whole viral genome structure, organization, and similarity (7, 17, 43). There are an ever-growing number of examples of incongruence between the morphology-based taxonomic classification systems and sequence-based systems. Sequence similarity-based taxonomy, the basis of phage comparative genomics, has been seen as a powerful and promising alternative (13, 14, 17, 39). Yet, our analyses clearly demonstrate that the results of local similarity-based analyses (e.g., BLASTP searches) and evolutionary (phylogenetic) analyses do not necessarily agree. Comparisons of these two methods for our 17 analyzed D3112 ORFs found that only 65% of the ORFs showed complete agreement between the similarity-based and phylogenetic approaches or at least placed the ORF of interest in a clade with multiple taxa, one of which also displayed the highest similarity. The remaining 35% of the ORFs (ORFs 1, 11, 26, 31, 36, and 43) showed significant inconsistencies between the most similar sequence identified by BLASTP and the closely related sequences identified by phylogenetic analyses. Figure 3 presents two representative examples of this similarity and phylogenetic inconsistency.
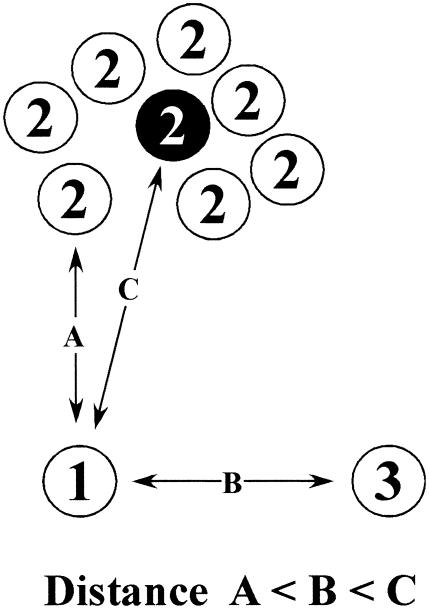
Why do the conclusions from similarity-based analyses differ from those drawn from phylogenetic analyses? We do not believe that the inconsistencies can be attributed to failings of the phylogenetic methods. We took the conservative analytical approach of trimming regions of questionable homology out of the multiple sequence alignments and performing the phylogenetic analyses by two independent methods (neighbor-joining and Bayesian analyses). The differences are most likely due to the inherent limitations of the BLAST algorithm. BLAST is a pairwise local alignment algorithm which excels at rapidly identifying sequences of local similarity from a sea of unrelated sequence. This is clearly an extremely powerful method for identifying matches to a query sequence in a large database, but the obvious drawback of this algorithm from the perspective of phage taxonomy is it preferentially identifies relatively short regions of high similarity, such as that seen between conserved domains, over regions of global similarity. Since BLAST uses pairwise comparisons, it is also unable to identify clusters of related sequences, such as multiple isolates from the same species. This may be a problem if one member of this cluster of related sequences is substantially more related to the query sequence than the average of the cluster (Fig. 4). Lastly, similarity analyses provide no basis for distinguishing homology from homoplasy (identity due to parallel or convergent evolution). Consequently, evolutionary inferences drawn from similarity-based approaches must be evaluated carefully.
FIG. 4.
Example of inconsistency between similarity-based and phylogenetic analyses. Three taxa (1, 2, and 3) are presented in a conceptual genotypic space. Taxon 1 is the query, while taxa 2 and 3 are related taxa. There are seven representatives of taxon 2 (e.g., different isolates from the same species), with the black circle representing the taxon average. The distance (distance = 1 − similarity) between two taxa is proportional to their distance in the genotypic space. Taxon 1 is most similar to an individual of taxon 2 (distance A). The next shortest distance is to taxon 3 (distance B), and then finally, the next shortest distance is to the average of all members of taxon 2 (distance C). Similarity analysis (e.g., BLAST) would find the strongest relationship between taxa 1 and 2. Phylogenetic analysis would cluster the individual members of taxon 2 and reassess their relationship to taxon 1 based on some measure of the average of taxon 2 (this is strictly true only for distance-based methods but is true in practice for other phylogenetic methods). As a result, the phylogenetic tree would reveal the evolutionarily more realistic conclusion that taxa 1 and 3 are more closely related, since the distance to taxon 3 (distance B) is less than the distance to the average of taxon 2 (distance C).
Phylogenetic analyses, on the other hand, are built upon multiple sequence alignments. We used ClustalX, a global sequence alignment algorithm, which assumes that sequences are similar over their entire length and tries to align them from end to end (with end gaps allowed). Multiple sequence alignments are built progressively upon a guide tree, thereby avoiding biases inherent in the pairwise local alignment. The danger with global alignments is seen if certain regions of the alignment are very poorly conserved, or not homologous. These problems can be addressed by judicious trimming of the sequence to remove unalignable regions. Finally, phylogenetic methods are vastly superior in dealing with homoplasy.
The current trend towards using sequence data to resolve viral taxonomical issues has raised the question of identifying the most-appropriate level for analysis. There are four levels of resolution that can be used for the analysis of genomes. The first is the comparison of whole phage genomes, such as in the study of lambdoid and dairy phages (14, 17, 24, 26). The second entails the use of smaller segments of phage genomes, such as the structural segment containing the head and tail genes (17). The third is the level of an individual gene or ORF. The fourth is at the level of conserved motifs within genes. Lawrence et al. (32) discuss the problems associated with mosaicism of viral genomes and how these invalidate approaches based on strictly pairwise local alignments for the first two levels of comparison. One exception to this conclusion is the work with the dairy phages, because they constitute an unusually homogeneous group of phages that display significant similarity at the nucleotide level (13). Similarity-based analyses at the level of a gene or smaller are biased by the tendency to align conserved motifs. Based on our results with D3112, we propose that phylogenetic analyses should be performed at the level of individual genes, since these represent the functional units of these highly mosaic systems. Phylogenetic approaches are clearly superior to approaches based on similarity and local alignments. They are less prone to be biased by conserved motifs and use more of the evolutionary information of a sequence, thereby permitting more power to disentangle homology from homoplasy.
There is ongoing debate as to whether there exists a single gene which can be used to build viral phylogenies (32, 39). We argue that the evolutionary history of a phage should be reflected in the distinct phylogenies of all of its individual genes. Thus, building a multigenic phylogenetic framework for each phage moves us away from the failings of phenetic approaches (morphological, structural, or sequence similarity) towards a cladistic basis for viral taxonomy. Lawrence et al. (32) have addressed related issues and proposed similar guidelines for a viral taxonomy. These gene-by-gene phylogenetic methods more realistically reflect the relationships between phages by taking into account the mosaic nature of these important organisms.
Acknowledgments
We thank Michael DuBow, Lori Burrows, and Frank M. Ausubel for generously supplying the P. aeruginosa bacterial strains. We also thank Michael DuBow for his helpful advice in handling phage D3112 and Jean Marc Moncalvo for his expert advice with difficult sequence alignments. We thank John Stavrinides for his expertise with computers.
We greatly appreciate the generous support of Beckman Coulter, Inc. This work was supported by grants from the National Science and Engineering Research Council of Canada, and the Canadian Foundation for Innovation.
REFERENCES
- 1.Akhverdian, V. Z., E. A. Khrenova, M. A. Reulets, T. V. Gerasimova, and V. N. Krylov. 1985. The properties of transposable phages of Pseudomonas aeruginosa belonging to 2 groups distinguished by DNA-DNA homology. Genetika 21:735-747. [PubMed] [Google Scholar]
- 2.Akhverdyan, V. Z., E. A. Khrenova, V. G. Bogush, T. V. Gerasimova, N. B. Kirsanov, and V. N. Krylov. 1984. Wide distribution of transposable phages in natural populations of Pseudomonas aeruginosa. Genetika 20:1612-1619. [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., and E. V. Koonin. 1998. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 23:444-447. [DOI] [PubMed] [Google Scholar]
- 5.Autexier, C., S. Wragglegare, and M. S. Dubow. 1991. Characterization of the Pseudomonas aeruginosa transposable phage D3112 left-end regulatory region. Biochim. Biophys. Acta 1088:147-150. [DOI] [PubMed] [Google Scholar]
- 6.Azadan, R. J., J. C. Fogleman, and P. B. Danielson. 2002. Capillary electrophoresis sequencing: maximum read length at minimal cost. BioTechniques 32:24-26, 28. [DOI] [PubMed] [Google Scholar]
- 7.Bamford, D. H., R. M. Burnett, and D. I. Stuart. 2002. Evolution of viral structure. Theor. Popul. Biol. 61:461-470. [DOI] [PubMed] [Google Scholar]
- 8.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bidnenko, E. M., V. Z. Akhverdian, and V. N. Krylov. 2000. Phenotypic effect of mutation ts47 in the Pseudomonas aeruginosa phage transposon D3112 on expression of late phage genes. Genetika 36:1721-1724. [PubMed] [Google Scholar]
- 11.Bidnenko, E. M., V. Z. Akhverdyan, and V. N. Krylov. 2000. Transcriptional mapping and studying the control of transcription of the Pseudomonas aeruginosa transposable phage D3112. Russ. J. Genet. 36:1385-1394. [Google Scholar]
- 12.Bourkaltseva, M. V., O. V. Shabourova, E. A. Pleteneva, and V. N. Krylov. 1999. x811, a mutation of Pseudomonas aeruginosa transposable phage D3112 with a pleiotropic effect. Russ. J. Genet. 35:233-237. [PubMed] [Google Scholar]
- 13.Brussow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 14.Brussow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 15.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 16.Darzins, A., and M. J. Casadaban. 1989. Mini-D3112 bacteriophage transposable elements for genetic analysis of Pseudomonas aeruginosa. J. Bacteriol. 171:3909-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desiere, F., S. Lucchini, C. Canchaya, M. Ventura, and H. Brussow. 2002. Comparative genomics of phages and prophages in lactic acid bacteria. Antonie Leeuwenhoek 82:73-91. [PubMed] [Google Scholar]
- 18.el-Mabrouk, N., and F. Lisacek. 1996. Very fast identification of RNA motifs in genomic DNA. Application to tRNA search in the yeast genome. J. Mol. Biol. 264:46-55. [DOI] [PubMed] [Google Scholar]
- 19.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. A. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felsenstein, J. 1989. PHYLIP-Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 21.Frohlich, M. W., and D. S. Parker. 2001. Running gels backwards to select DNA molecules larger than a minimum size. BioTechniques 30:264-266. [DOI] [PubMed] [Google Scholar]
- 22.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 23.Hattman, S., and W. Sun. 1997. Escherichia coli OxyR modulation of bacteriophage Mu mom expression in dam+ cells can be attributed to its ability to bind hemimethylated Pmom promoter DNA. Nucleic Acids Res. 25:4385-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrix, R. W. 2002. Bacteriophages: evolution of the majority. Theor. Popul. Biol. 61:471-480. [DOI] [PubMed] [Google Scholar]
- 25.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 26.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 27.Kamp, D., and R. Kahmann. 1981. The relationship of two invertible segments in bacteriophage Mu and Salmonella typhimurium DNA. Mol. Gen. Genet. 184:564-566. [DOI] [PubMed] [Google Scholar]
- 28.Karkhoffschweizer, R. R., and H. P. Schweizer. 1994. Utilization of a mini-Dlac transposable element to create an alpha-complementation and regulated expression system for cloning in Pseudomonas aeruginosa. Gene 140:7-15. [DOI] [PubMed] [Google Scholar]
- 29.Kropinski, A. M. 2000. Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J. Bacteriol. 182:6066-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krylov, V. N., V. G. Bogush, and J. Shapiro. 1980. Pseudomonas aeruginosa phages, which DNA structure is similar to Mu1 phage DNA. 1. General description, localization of endonuclease-sensitive sites in DNA, and the structure of D3112 phage homoduplexes. Genetika 16:824. [Google Scholar]
- 31.Krylov, V. N., V. G. Bogush, A. S. Yanenko, and N. B. Kirsanov. 1980. Pseudomonas aeruginosa phages which DNA is similar in structure with Mu1 phage DNA. 2. The evidence of relationship of D3112, B3 and B39 phages—analysis of restriction with endonucleases, isolation of recombinant between D3112 and B3 phages. Genetika 16:975-984. [PubMed] [Google Scholar]
- 32.Lawrence, J. G., G. F. Hatfull, and R. W. Hendrix. 2002. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol. 184:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masignani, V., M. M. Giuliani, H. Tettelin, M. Comanducci, R. Rappuoli, and V. Scarlato. 2001. Mu-like prophage in serogroup B Neisseria meningitidis coding for surface-exposed antigens. Infect. Immun. 69:2580-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mit'kina, L. N., and V. N. Krylov. 1999. Natural interspecific hybrids of transposable phages of Pseudomonas aeruginosa. Russ. J. Genet. 35:1015-1022. [PubMed] [Google Scholar]
- 36.Morgan, G. J., G. F. Hatfull, S. Casjens, and R. W. Hendrix. 2002. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317:337-359. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 39.Proux, C., D. van Sinderen, J. Suarez, P. Garcia, V. Ladero, G. F. Fitzgerald, F. Desiere, and H. Brüssow. 2002. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J. Bacteriol. 184:6026-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rehmat, S., and J. A. Shapiro. 1983. Insertion and replication of the Pseudomonas aeruginosa mutator phage D3112. Mol. Gen. Genet. 192:416-423. [DOI] [PubMed] [Google Scholar]
- 41.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, M. S., and F. M. Cohan. 1993. The effect of DNA sequence divergence on sexual isolation in Bacillus. Genetics 134:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roncero, C., A. Darzins, and M. J. Casadaban. 1990. Pseudomonas aeruginosa transposable bacteriophage D3112 and bacteriophage B3 require pili and surface growth for adsorption. J. Bacteriol. 172:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salmon, K. A., O. Freedman, B. W. Ritchings, and M. S. DuBow. 2000. Characterization of the lysogenic repressor (c) gene of the Pseudomonas aeruginosa transposable bacteriophage D3112. Virology 272:85-97. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sandmeier, H. 1994. Acquisition and rearrangement of sequence motifs in the evolution of bacteriophage tail fibres. Mol. Microbiol. 12:343-350. [DOI] [PubMed] [Google Scholar]
- 48.Santos, M. A. 1991. An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. Nucleic Acids Res. 19:5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swinton, D., S. Hattman, P. F. Crain, C. S. Cheng, D. L. Smith, and J. A. McCloskey. 1983. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2′-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc. Natl. Acad. Sci. USA 80:7400-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony and other methods. Sinauer Associates, Sunderland, Mass.
- 51.Symonds, N., A. Toussaint, P. van de Putte, and M. M. Howe. 1987. Phage Mu. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 52.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toussaint, A., M. J. Gama, J. Laachouch, G. Maenhautmichel, and A. Mhammedialaoui. 1994. Regulation of bacteriophage Mu transposition. Genetika 93:27-39. [DOI] [PubMed] [Google Scholar]
- 54.Ulycznyj, P. I., K. A. Salmon, H. Douillard, and M. S. DuBow. 1995. Characterization of the Pseudomonas aeruginosa transposable bacteriophage D3112 A and B genes. Biochim. Biophys. Acta 1264:249-253. [DOI] [PubMed] [Google Scholar]
- 55.van de Putte, P., S. Cramer, and M. Giphart-Gassler. 1980. Invertible DNA determines host specificity of bacteriophage Mu. Nature 286:218-222. [DOI] [PubMed] [Google Scholar]
- 56.Yanenko, A. S., N. B. Kirsanov, T. V. Gerasimova, A. O. Bekkarevitch, and V. N. Krylov. 1988. Mini-genomes of the transposable phage D3112 Pseudomonas aeruginosa and their properties. Genetika 24:956-959. [PubMed] [Google Scholar]