SUMMARY
Exercise has been shown to be effective for treating obesity and type 2 diabetes. However, the molecular mechanisms for adaptation to exercise training are not fully understood. Endoplasmic reticulum (ER) stress has been linked to metabolic dysfunction. Here we show that the unfolded protein response (UPR), an adaptive response pathway that maintains ER homeostasis upon luminal stress, is activated in skeletal muscle during exercise and adapts skeletal muscle to exercise training. The transcriptional coactivator PGC-1α, which regulates several exercise-associated aspects of skeletal muscle function, mediates the UPR in myotubes and skeletal muscle through coactivation of ATF6α. Efficient recovery from acute exercise is compromised in ATF6α−/− mice. Blocking ER-stress related cell death via deletion of CHOP partially rescues the exercise intolerance phenotype in muscle-specific PGC-1α KO mice. These findings suggest that modulation of the UPR through PGC1α represents an alternative avenue to improve skeletal muscle function and achieve metabolic benefits.
INTRODUCTION
The molecular mechanisms mediating both the metabolic benefits of exercise and the pathological changes elicited by a sedentary lifestyle are incompletely understood. Skeletal muscle, due to its mass and great capacity to influence metabolism, has a major impact on whole-body metabolic homeostasis. In recent years, many new studies have examined the therapeutic effects of exercise and several novel pathways have been implicated in the regulation of skeletal muscle function during exercise (Bassel-Duby and Olson, 2006; Deshmukh et al., 2008).
The endoplasmic reticulum (ER) is a multifunctional organelle that controls the synthesis, folding, assembly and transport of proteins and also provides a dynamic intracellular Ca2+ storage compartment. Newly synthesized proteins are co-translationally translocated into the ER lumen, where they fold into proper conformations with the aid of molecular chaperones. Under physiological conditions, the majority of ER chaperones, including BiP, calreticulin, and calnexin, store Ca2+ as high capacity Ca2+ binding proteins. Under conditions that perturb protein folding in the ER, such as ER Ca2+ depletion or energy/nutrients deprivations, the expression of ER chaperones is induced through a set of signal transduction cascades that are collectively termed the Unfolded Protein Response (UPR) (Ron and Walter, 2007). The acute UPR activation is thus an important adaptive response to ER stress. Increased expression of chaperones augments the protein folding and processing capacity and restores equilibrium to the ER lumen. However, chronic ER stress leads to sustained UPR activation and failure to suppress the pro-apoptotic aspect of the UPR often leads to ER stress-related cell death (Rutkowski et al., 2006).
To date, there are three well-characterized proximal sensors of the UPR: PERK, IRE1α and ATF6α (Wu and Kaufman, 2006). PERK is a protein kinase that phosphorylates eukaryotic translation initiation factor 2 at Ser51 on the alpha subunit (eIF2α) to attenuate mRNA translation. IRE1α is a bifunctional protein kinase and endoribonuclease that initiates unconventional splicing of Xbp-1 mRNA to produce an active transcription factor. ATF6α transits to the Golgi where it is processed by the site-1 and site-2 proteases to produce a cytosolic fragment that migrates to the nucleus to activate expression of a spectrum of UPR target genes that play important roles in the adaptation of cells to chronic ER stress (Wu and Kaufman, 2006).
The UPR can be provoked by a variety of pathophysiological conditions and ER stress contributes to a number of neurodegenerative, inflammatory and metabolic pathologies (Lin et al., 2008). Recent research has suggested the possibility that skeletal muscle is also sensitive to the physiological stressors that trigger the UPR, including hypoxia, glucose deprivation, anabolic stimulation, and imbalances in calcium homeostasis (Acosta-Alvear et al., 2007; Iwawaki et al., 2004). Therefore, it is possible that the UPR may be involved in modulating skeletal muscle function during physiologically challenging conditions, such as exercise.
The transcriptional coactivator PGC-1α (peroxisome proliferators-activator receptor gamma coactivator-1 alpha) is a key regulator of mitochondrial function, oxidative metabolism and energy homeostasis in a variety of tissues (Lin et al., 2005). Experiments utilizing PGC-1α muscle-specific transgenic mice (MCK-PGC-1α) and muscle-specific knockout mice (MKO-PGC-1α) have revealed a clear role for this transcriptional coactivator in increasing mitochondrial number and function, as well as inducing a switch to oxidative, fatigue-resistant muscle fibers (Handschin et al., 2007a; Lin et al., 2002). The expression of PGC-1α is induced in muscle following both an acute exercise challenge and long-term exercise training (Baar et al., 2002; Russell et al., 2003). Moreover, forced expression of PGC-1α in skeletal muscle of mice is sufficient to improve performance capacity during voluntary and forced exercise (Calvo et al., 2008). In addition (and importantly from a medical perspective), muscle PGC1αprotects this tissue from sarcopenia and metabolic diseases associated with aging (Wenz et al., 2009).
In this study we show that exercise activates the UPR in the skeletal muscle of mice. Gain and loss of function studies in primary myotubes and skeletal muscle in vivo demonstrate that PGC-1α plays an important role in the regulation of the UPR through coactivation of ATF6α, a factor required for recovery from the stress of acute exercise. These findings indicate that PGC-1α acts as an important transcriptional coregulator of the UPR in skeletal muscle during exercise and helps adapt skeletal muscle to exercise training.
RESULTS
Activation of the Unfolded Protein Response (UPR) in skeletal muscle during exercise
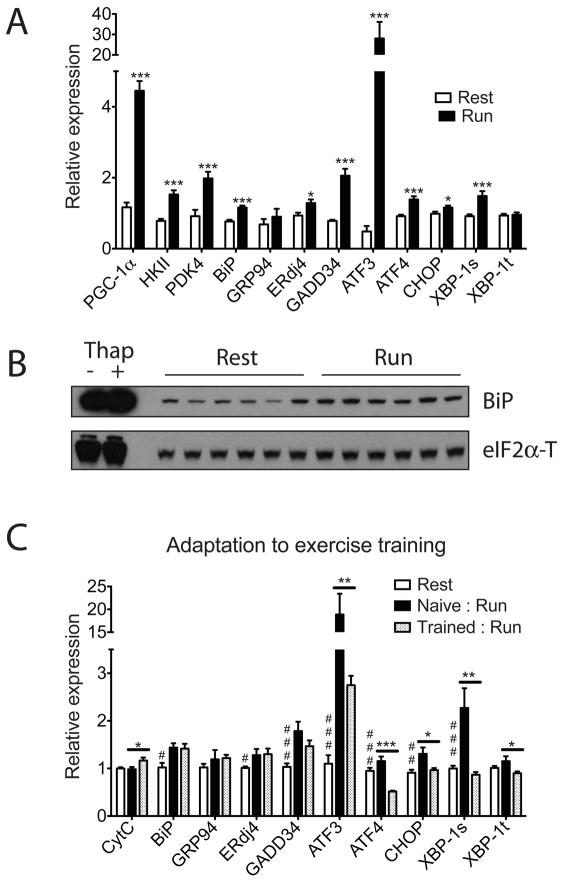
To examine whether the UPR is activated in skeletal muscle during exercise, we investigated the expression of several molecular indicators of ER stress in quadriceps muscle isolated from C57/Bl6 mice following one bout of exhaustive treadmill running. A broad range of known ER stress markers were induced at the mRNA level after a single bout of running, compared to sedentary controls (Figure 1A, male and Figure S1A, female). Molecular components of both the pro-adaptation limb (increase of chaperone/cochaperone: BiP, ERdj4) and anti-adaptation limb of the UPR (induction of cellular stress markers: GADD34, ATF3, ATF4, CHOP and the spliced form of Xbp-1), were increased after exercise. This is consistent with the notion that an acute exercise challenge causes luminal stress in the ER and activates all aspects of the UPR. Expression of genes known to be induced and directly regulate metabolic changes in skeletal muscle upon exercise, including the transcriptional coactivator PGC-1α, Hexokinase II (HKII, the major hexokinase isoform in skeletal muscle) and pyruvate dehydrogenase kinase isozyme 4 (PDK4) are also increased to similar extent as the UPR marker genes; this suggests that the activation of the UPR in this context may have physiological consequences. We also detected increased eIF2α phosphorylation in the quadriceps of mice post-exercise, compared to sedentary controls, and also observed increased accumulation of BiP protein, the most abundant ER chaperone, via immunoblotting and immunostaining (Figure 1B, Figure S1B, Figure S1C and Figure S1D).
Figure 1. The unfolded protein response is activated in skeletal muscle after exercise.
Total RNA and protein lysates from quadriceps were isolated from C57/Bl6 wild-type male mice 5 hrs post one bout treadmill running (n=6) or sedentary control (n=6) as described in experimental procedures. A. Total RNA was analyzed by realtime RT-PCR; Data represented means±SEM. *** indicates p <0.005 vs. control, *, p <0.05. Similar results were observed in more than three independent experiments. B. Protein lysates were probed by immunoblot as indicated, with total eIF2α as a loading control. The first two lanes are positive controls using lysates from primary myotubes treated with 100nM thapsigargin or vehicle for 8hrs. C. Exercise training leads to adaptation in skeletal muscle. Untrained mice (naïve) or trained mice (trained: five sessions of 1hr treadmill run/wk for 4weeks), together with age-matched sedentary control mice were sacrificed 5 hrs after one bout equal distance treadmill running. N=6 animals per group. Total RNA from quadriceps was isolated and analyzed by realtime RT-PCR with CytC as a positive control; Data represented means±SEM. *** indicates p <0.005 trained vs. naïve, **, p <0.01, *, p <0.05. ### indicates p <0.005, rest vs. trained : run, ##, p <0.01, #, p <0.05. Statistical comparison between rest vs. naïve : run is similar to Figure 1A, and not shown in this panel for visual clarity.
Since exercise is a physiological process involving multiple organs and drastically impacts systemic metabolism, it was important to investigate whether the UPR activation post-exercise was specific to skeletal muscle. The heart also experiences significant metabolic changes in response to exercise; however we did not detect activation of the UPR gene program in the hearts of exercised mice under our experimental conditions (Figure S1E), suggesting that skeletal muscle is selectively sensitive to the physiological ER stress caused by this form of exercise. Further analysis shown while the UPR is similarly activated in another muscle type in the limb (gastrocnemius), it is largely unaffected in non-weight bearing back muscle (erector spinae) (Figure S1F and Figure S1G). This implies that mechanical stress exerted by muscle contraction and/or local metabolic changes in the muscle that are directly involved in exercise play a major role in the activation of the UPR pathway in this context.
UPR activation leads to adaptation in skeletal muscle after exercise training
One teleological question regarding UPR activation during exercise is why a stress response pathway, known to be involved in the pathogenesis of a variety of diseases, is activated during a physical process with proven metabolic benefits. Recent studies have suggested that the UPR is as much an adaptive response pathway to maintain day-to-day physiological function in various tissues, as a damage control mechanism to defend cells and tissues against foreign insults and pathological events (Rutkowski and Hegde, 2010). To examine this in the context of chronic exercise training, we compared gene expression in the quadriceps between two groups of age-matched wild-type male mice (naïve versus trained) that were subsequently challenged with equal distance treadmill running. Similar to the results shown in Figure 1A, the naïve, untrained mice show significant activation of the UPR after running. However, in mice that had been trained previously (one hour per day, five days per week for four weeks), a different pattern of UPR activation was observed (Figure 1C). ER chaperones, BiP, GRP94, and the co-chaperone, ERdj4 were increased to a similar extent in both groups. Notably, a stress marker, ATF3, was only induced by 3-fold in the trained group compared to almost 20-fold in mice only run once. Furthermore, CHOP, a transcription factor that regulates ER-stress related cell death, and the spliced form of Xbp-1 mRNA, a sensitive cellular sensor of acute stress, both remain uninduced in trained mice while being significantly elevated in the naïve mice. Interestingly, the expression of ATF4, another stress marker, was significantly lower in trained mice compared to sedentary control mice that had never been exercised. These results strongly suggest that moderate exercise and the accompanying physiological ER stress in skeletal muscle may lead to adaptation and protect skeletal muscle against further stress.
PGC-1α is induced upon ER stress in primary myotubes
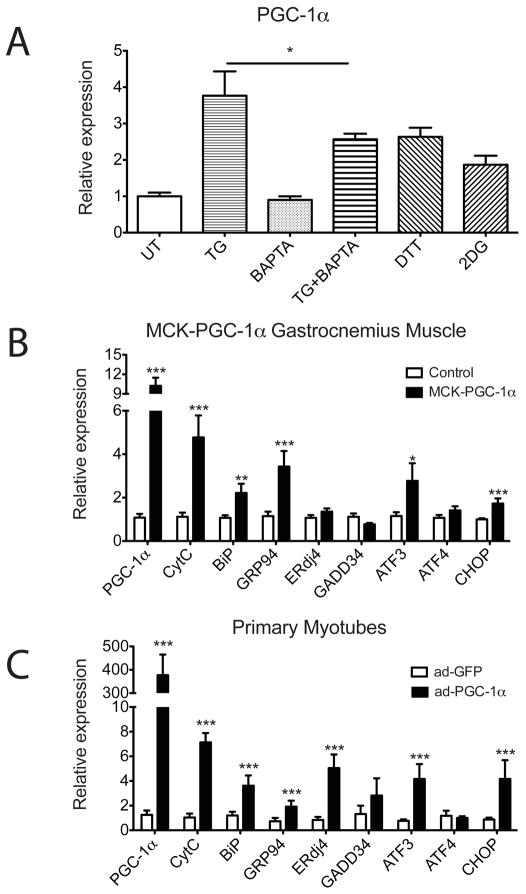
To investigate the molecular mechanisms by which ER stress might affect muscle function, we treated primary myotubes with an array of ER stressors: dithiothreitol (DTT, a reducing reagent that disturbs the oxidizing environment within the ER lumen), 2-deoxy-d-glucose (2DG, a glycolysis-resistant form of glucose that causes an energy crisis during protein folding in the ER), or thapsigargin (TG, Sarco/endoplasmic reticulum Ca2+ ATPase-SERCA inhibitor that depletes ER calcium stores). PGC-1α mRNA levels in myotubes are significantly increased in response to all three ER stressors compared to the untreated control (Figure 2A). TG treatment releases ER luminal Ca2+ into the cytoplasm and intracellular calcium plays a critical role in regulation of PGC-1α expression (Rohas et al., 2007). It is worth noting that the induction of PGC-1α by TG is reduced but not completely blocked by pretreatment with the calcium-specific chelator BAPTA, suggesting that PGC-1α expression may be regulated by other signals emanating from the ER in addition to Ca2+ (Figure 2A).
Figure 2. PGC-1α regulates the UPR in primary myotubes and in skeletal muscle.
(A) PGC-1α is induced upon ER stress in primary myotubes. Wild-type primary myotubes were pretreated with 10μM BAPTA or vehicle for 1hr and followed by vehicle or 100nM Thapsigargin (TG) treatment for 6hrs. Wildtype primary myotubes were treated with 5mM DTT or 10mM 2DG for 6hrs. Total RNA was isolated and the expression of PGC-1α was quantitated by realtime RT-PCR, normalizing against tbp expression. Data represent means±SD from biological triplicates. * indicates p <0.05 TG+BAPTA vs. TG. (B, C) UPR markers are induced by PGC-1α in skeletal muscles and in primary myotubes. B. Total RNA from gastrocnemius muscle isolated from MCK-PGC-1α and littermate control mice was analyzed by real-time RT-PCR for UPR markers expression with CytC as a positive control. N=4–5 animals per group. Data represent means±SEM. *** indicates p <0.005 vs. control, **, p <0.01, *, p <0.05. C. Total RNA from primary myotubes infected with adenovirus expressing PGC-1α or GFP as a control was assayed by realtime RT-PCR for UPR markers expression with CytC as a positive control. Data represent means±SD from biological triplicates. *** indicates p <0.005 vs. control.
PGC-1α induction was not observed in primary myotubes treated with a classic pharmacological ER stressor, tunicamycin (TM, an inhibitor of N-linked glycosylation). Conversely, TM treatment upregulates PGC-1α in differentiated white fat cells (3T3-L1 cells) (Figure S2A and data not shown). Clearly, there is cell-type specific regulation of PGC-1α by specific ER stressors. PGC-1α, as a transcriptional coactivator, has been shown to play different physiological roles in different tissues under various biological contexts. The exact physiological ER stress signal(s) that activates PGC-1α expression in different cell types awaits further study. However, it is conceivable that the optimal environment for posttranslational modification (altered by TM treatment) may not be as crucial in primary myotubes as in fat cells, where adipokines are constantly being folded and secreted through the ER.
Expression of UPR-related genes is induced by PGC-1α in skeletal muscle and primary myotubes
Given the data above, we investigated whether PGC-1αmay be involved in the activation of the UPR in skeletal muscle. First, UPR gene expression levels were investigated in the gastrocnemius muscle from the muscle-specific transgenic mice (MCK-PGC-1α) (Lin et al., 2002). This strain of mice expresses PGC1α in all types of muscle fibers at the levels seen in highly oxidative muscles like the soleus. The chaperones, BiP and GRP94, and the stress markers, ATF3 and CHOP are all significantly induced with transgenic expression of PGC-1α compared to littermate controls, indicating that increased PGC-1α promotes expression of these genes (Figure 2B).
To test whether induction of UPR marker genes by PGC-1α is a cell-autonomous effect, mRNA expression of myotubes infected with adenoviral PGC-1α or a GFP control was examined using Affymetrix arrays. As expected, Ingenuity Pathway Analysis identified “Energy Production” as a major pathway regulated by PGC-1α. It is noteworthy that “protein folding” was the fifth most regulated pathway by PGC-1α in primary myotubes (Figure S2B). Analysis using dChip software showed that 182 “protein folding related” and 46 “response to unfolded proteins” genes are induced by adenoviral expression of PGC-1α (Figure S2C). Importantly, realtime RT-PCR analysis for specific genes, such as BiP, GRP94, CHOP, etc., showed that these genes were induced by 2 to 6 fold by PGC1α, in agreement with the array analyses (Figure 2C). Taken together, these data suggest that PGC-1α regulates a broad range of UPR gene expression in both primary myotubes and in skeletal muscle in vivo. No induction of these UPR marker genes was evident in mouse embryonic fibroblasts (MEFs) or in rat embryonic cardiomyocytes that were similarly infected with adenovirus expressing PGC-1α (Figure S2D and Figure S2E). Thus, this response has some clear cell/tissue selectivity.
PGC-1α-deficient myotubes respond to ER stress, but are defective in the subsequent UPR gene regulation
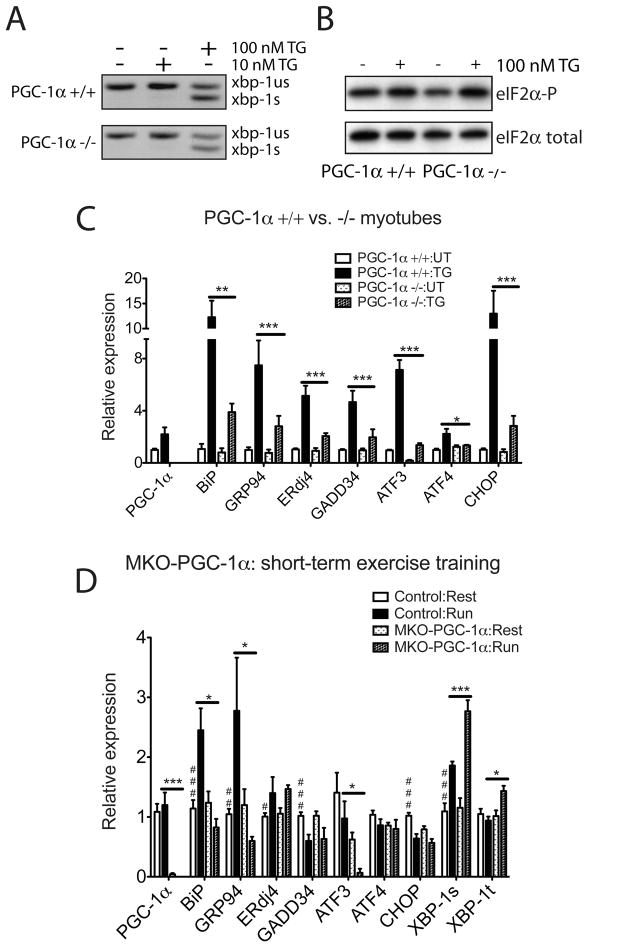
The requirement for PGC-1α in UPR gene regulation was investigated using primary cultures of PGC-1α−/− myotubes and wild-type controls. IRE1α activation (assessed by Xbp1 mRNA splicing; Figure 3A), and PERK activation (measured by eIF2α phosphorylation; Figure 3B), both occurred to similar extents in PGC-1α +/+ and −/− primary myotubes upon ER stress via thapsigargin treatment. We next monitored whether the stress-dependent induction of UPR sentinel genes required PGC-1α (Figure 3C, Figure S3A and Figure S3B). PGC-1α−/− myotubes display no significant alteration in the basal expression of a sampling of stress related genes, with the exception of ATF3 (Figure 3C). Despite ER stress sensing and activation of UPR proximal sensors being largely uncompromised (Figure 3A and 3B), induction of a key set of UPR marker genes such as BiP, GRP94 and CHOP, was defective in PGC-1α−/− myotubes compared to wild-type cells (Figure 3C). Taken together, these data indicate that PGC-1α regulates downstream effector genes of the UPR in primary myotubes.
Figure 3. PGC-1α is necessary to upregulate UPR markers expression in primary myotubes upon ER stress and in skeletal muscle after exercise.
(A, B). PGC-1α-deficient myotubes respond to ER stress. A. Total RNA was isolated from wildtype and PGC-1α−/− primary myotubes treated with 10 or 100nM TG for 8hrs. RT-PCR was used to simultaneously detect both spliced (s) and unspliced (us) Xbp1 mRNA. The image is presented in black and white inverted form for greater visual clarity. B. Wild-type and PGC-1α−/− primary myotubes were treated with 100nM TG for 1hr, followed by cell lysis and immunoblot using antibody that detects phosphorylated form of eIF2α and total eIF2α as a loading control. C. PGC-1α−/− primary myotubes are defective in UPR markers upregulation upon ER stress. Total RNA from wildtype and PGC-1α−/− primary myotubes treated with 100nM TG for 16hrs was assayed by realtime RT-PCR for UPR markers expression, normalizing against tbp expression. Data represent means±SD from biological triplicates. *** indicates p <0.005 +/+:TG vs. −/−:TG, **, p <0.01, *, p <0.05. D. MKO-PGC-1α mice are defective in upregulating ER chaperones and experience exacerbated ER stress after repetitive exercise challenges. Total RNA from quadriceps of MKO-PGC-1α and wildtype control mice either sedentary or after four bouts equal distance treadmill running (once a day for 4 days, recover for one day after the last run) was isolated and analyzed by realtime RT-PCR; N=7–8 animals per group. *** indicates p <0.005 MKO-PGC-1α : Run vs. Control : Run; *, p <0.05. ### indicates p <0.005, control : rest vs. control : run, ##, p <0.01, #, p <0.05.
MKO-PGC-1α mice are defective in upregulating ER chaperones and experience exacerbated ER stress after repetitive exercise challenges
To test whether PGC-1α regulates the UPR activation during exercise in skeletal muscle, we measured UPR target gene expression in skeletal muscle of MKO-PGC-1α and MCK-PGC-1α mice. It has been previously reported that PGC-1α expression level in skeletal muscle correlates with exercise capacity in mice (Handschin et al., 2007a; Wenz et al., 2009). The complex phenotype in skeletal muscle of these mice including differential mitochondrial contents and muscle fiber composition and gross differences in exercise capacity, would confound analysis of data from one bout exhaustive treadmill running. Therefore, we challenged MKO-PGC-1α mice and MCK-PGC-1α mice (together with their wildtype controls) with one bout of equal distance pair-running. Due to the exercise intolerance in MKO-PGC-1α mice (hence the short total-distance run) the activation of the UPR in wildtype mice in this set of experiment is quite moderate (Figure S3C). As in cultured cells, though to lesser extent, we observed defective upregulation of the UPR target genes, BiP and GADD34 in MKO-PGC-1α mice compared to controls. Significantly more Xbp-1 splicing was detected in skeletal muscle of MKO-PGC-1α compared to the wild-type (Figure S3C). In contrast, MCK-PGC-1α mice not only run without signs of fatigue when wild-type control mice were tired, they also displayed less overall stress in skeletal muscle (less Xbp-1 splicing) compared to the control (Figure S3D). It is worth noting that some of the UPR target genes, like BiP, are expressed at a higher basal level in MCK-PGC-1α muscle (as also shown in Figure 2B), whereas others, for example GADD34, is mainly induced after running. Since the mRNA level of PGC-1α is not further induced under this experimental condition, it is possible that additional activation mechanisms exist in regulating the UPR activation during exercise, possibly through regulation of other factors in the transcriptional protein complex or increased PGC-1α activity via translational regulation or posttranslational modifications.
Results in wildtype mice shown above lead to the conclusion that repetitive exercise training induces the adaptive limb of the UPR (ER chaperones/cochaperones) and protects skeletal muscle against further stress caused by future exercise (Figure 1C). The inability of MKO-PGC-1α mice to induce the UPR after one bout running suggests that they might be less able to adapt to repeated exercise training. To test this, we subject MKO-PGC-1α and wildtype control mice to daily pair-running training (once a day for 4 days). The basal level of the UPR target gene expression (after 1 day recovery post the last run) was analyzed in the quadriceps. Exercise training (even short-term for 4 days) significantly elevated expression levels of the ER chaperones BiP, GRP94 and ER cochaperone ERdj4 in the wildtype mice, whereas some of the cellular stress markers including GADD34 and CHOP were decreased. Xbp-1 splicing is higher in the trained wildtype group compared to sedentary controls, suggesting different recovery kinetics of the ER stress anti-adaptive subpathways (Figure 3D). Importantly, upregulation of the ER chaperones after exercise training is defective in the skeletal muscle of MKO-PGC-1α mice. Significant more Xbp-1 splicing is detected in PGC-1α-null muscle, indicating augmented stress in the ER/SR (Figure 3D). These data from MKO-PGC-1α animals subjected to acute exercise challenge and repetitive exercise training provide strong genetic evidence that PGC-1α regulates the UPR in skeletal muscle in response to exercise.
Coactivation of ATF6α by PGC-1α
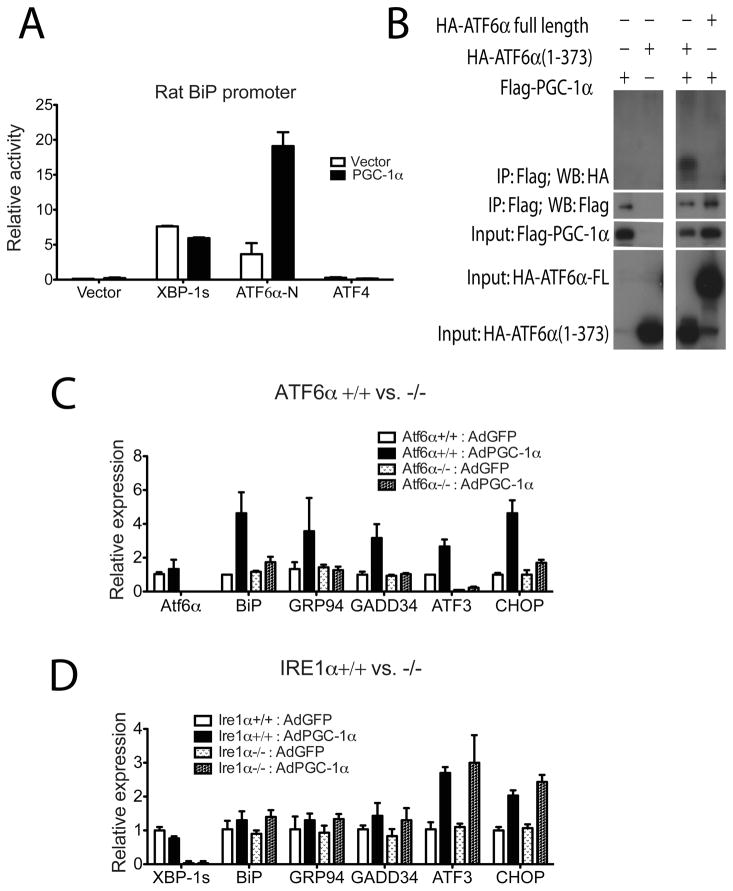
PGC-1α regulates various physiological functions of skeletal muscle and other tissues through the coactivation of nuclear receptors and several other transcription factors (Lin et al., 2005). There are three known major transcription factors that regulate the UPR: XBP-1, ATF6α and ATF4. Since PGC-1α induces BiP expression in myotubes, a reporter construct driven by the BiP promoter was used to test the coactivation of these three factors by PGC-1α. As expected, both XBP-1s (coded by spliced form of Xbp-1 mRNA) and ATF6α-N (N-terminal of cleaved ATF6α) activate the rat BiP promoter in differentiated C2C12 myotubes. However, while the transcriptional activity of XBP-1s was minimally affected by PGC-1α (Figure 4A), this protein greatly augmented the transcriptional activity of ATF6α on this promoter. ATF4 is known not to regulate this region of BiP promoter (−169bp), and does not show any increased regulatory capacity with PGC1α. These results indicate that ATF6α can cooperate with PGC-1α to regulate this region of the BiP promoter.
Figure 4. Coactivation of ATF6α by PGC-1α.
A. C2C12 myoblasts were transfected with expression plasmids for XBP-1 coded by spliced form of Xbp-1 mRNA (XBP-1s), cleaved form of ATF6α (ATF6α-N), ATF4 or expression vector control, respectively, together with expression plasmid for PGC-1α, or vector and rat BiP promoter-luciferase construct. The cells were subsequently differentiated for 2 days and harvested and luciferase activity was measured, normalizing against renilla. Data represent means±SD from biological triplicates. B. Coimmunoprecipitation of ATF6α and PGC-1α. Cultured COS cells were transfected with plasmids as indicated. Total lysates from transfected cells were subjected to immunoprecipitation using beads specific for Flag tag. Both lysates and precipitates were analyzed by immunoblotting with antibodies specific for the Flag and HA epitope tag. (C and D) Total RNA from ATF6α+/+ and −/− primary myotubes (C) and IRE1α+/+ and −/− primary myotubes (D) infected with adenovirus expressing PGC-1α or GFP as a control was assayed by realtime RT-PCR for UPR markers expression. Data represent means±SD from biological triplicates.
Furthermore, these two proteins physically interact in cells, as shown by coimmunoprecipitation assays. PGC-1α is able to interact with and precipitate the activated form of ATF6α (ATF6α 1–373), but not full-length, membrane-embedded ATF6α, when these proteins are co-expressed in cells (Figure 4B). These data suggest that PGC-1α coactivates ATF6α to regulate certain aspects of the UPR. It also implies that prior cleavage of ATF6α is necessary to form this functional complex.
To examine whether ATF6α is required for the PGC-1α-regulated induction of UPR genes, primary myoblasts were isolated from ATF6α −/− and wild-type mice. These cells were then differentiated into primary myotubes in culture and infected with adenovirus expressing PGC-1α or GFP. The induction of UPR genes by PGC-1α was almost completely abolished in the absence of ATF6α (Figure 4C). We next isolated myoblasts from homozygous floxed IRE1α mice and deleted IRE1α in culture by adenoviral CRE expression. In contrast to ATF6α−/− muscle cells, the induction of UPR genes by ectopic PGC-1α expression was similar between IRE1α-deleted primary myotubes and control cells (Figure 4D). Supporting and confirming the role of PGC-1α in the regulation of ATF3 through ATF6α, the ATF3 promoter could be immunopurified by antisera against ectopic expressed PGC-1α protein in myotubes from wildtype mice, but not in ATF6α−/− myotubes. This enrichment can be recovered with ectopic expression of ATF6α (Figure S4). These data indicate that PGC-1α stimulates UPR gene expression, at least in part, through coactivation of ATF6α.
ATF6α−/− mice do not efficiently recover from muscle damage after exercise
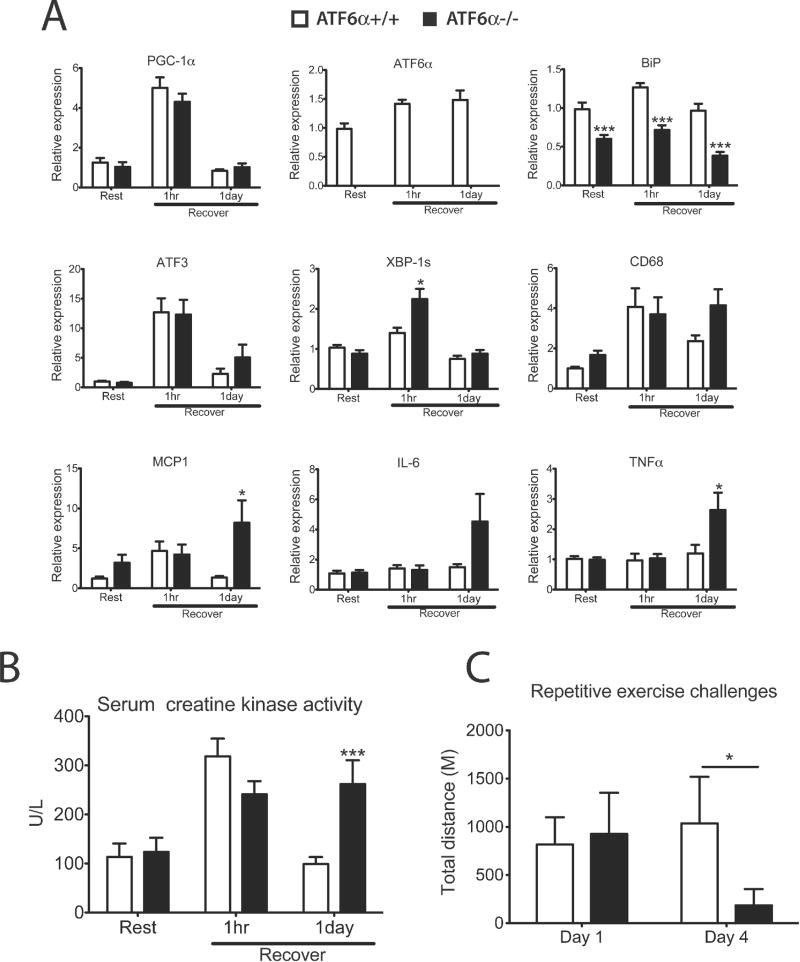
Previous studies have shown that ATF6α modulates ER function to protect cells against chronic ER stress in cultured fibroblasts and in the liver (Wu et al., 2007). Since ATF6 appears to be an important partner for PGC1α in skeletal muscle, we subjected ATF6α−/− mice to treadmill running and examined both exercise performance and tolerance. These mice have similar glycogen content in skeletal muscle and serum lactate level before and after exercise, compared to wildtype control mice (Figure S5A and Figure S5B). There was no statistically significant difference in the total distance run between wild-type and ATF6α−/− mice during one bout exhaustive treadmill running. However, gene expression analysis of RNA isolated from quadriceps showed that the induction of the spliced form of Xbp-1 was more pronounced in the ATF6α−/− mice 1hr after running, indicating a more severe stress experienced in these animals (Figure 5A). Following one day of recovery, muscle from ATF6α−/− mice displayed a greatly increased expression of several proinflammatory genes (especially MCP1 and TNFα). Consistent with ATF6α being a major regulator of the UPR, the abundant ER chaperone BiP, was expressed at a dramatically lower level in ATF6α−/− muscle after exercise (Figure 5A, Figure S5C and Figure S5D).
Figure 5. ATF6α−/− mice do not efficiently recover from muscle damage after exercise.
A. Total RNA from quadriceps of ATF6α +/+ and −/− mice, 1hr or 1 day after equal distance treadmill running and littermate sedentary control mice was isolated and analyzed by realtime RT-PCR; Data represented means±SEM, *** indicates p <0.005 +/+ vs. −/−, *, p <0.05. B. Whole blood was collected by cardiac puncture from mice in (A) and serum creatine kinase activity was determined. Data represented means±SEM, *** indicates p <0.005 +/+ vs. −/−. C. ATF6α−/− mice shown exercise intolerance compared to wild-type control mice after repetitive treadmill runnings. Adult male ATF6α−/− mice and wild-type control (3–4 months old, n=4–12 per group) were subjected to repetitive exhaustive treadmill running once a day for 4 days as described in Experimental Procedures. Distances run on day 1 and day 4 of both genotypes were calculated from the individual performances. Data represented means±SEM. * indicates p <0.05 +/+ vs. −/−.
Intense exercise causes mild muscle damage that leads to higher serum creatine kinase levels in blood leaking from muscle fibers. We therefore measured serum creatine kinase activity in wild-type and ATF6α−/− mice. A similar mild increases of serum creatine kinase activity was detected in both genotypes of mice 1 hr after exercise (Figure 5B). However after one day of recovery, serum creatine kinase activity dropped back to basal levels in wild-type mice whereas the level of serum creatine kinase activity remained elevated in Atf6α-null mice. The impaired ability of ATF6α−/− mice to restore homeostasis after acute exercise predicts that the effects of the absence of ATF6α would be exacerbated by repeated challenges. This hypothesis was confirmed at the organismal level. ATF6α−/− mice show significant exercise intolerance after repetitive exercise challenges (day 4 of daily exhaustive treadmill run for 4 days), while wildtype control mice shown unchanged or even improved exercise capacity, compared to day 1 (Figure 5C). These observations indicate that the UPR activation regulated by ATF6α is important in preserving skeletal muscle integrity following acute exercise and in adaptation to repetitive exercise training.
CHOP deletion partially rescues the exercise intolerance of MKO-PGC-1α mice
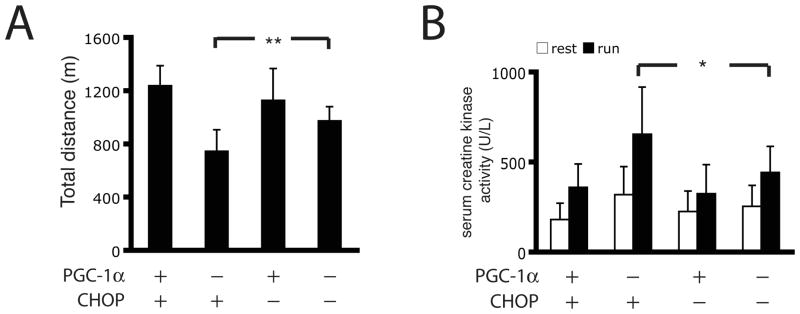
Finally, we examined the functional consequences of this crosstalk between the UPR and PGC1α in vivo. Mice with a muscle-selective KO for PGC1α show a distinct exercise intolerance (Handschin et al., 2007a), and this is due, at least in part, to diminished control of the UPR in skeletal muscle (Figure 3D). Recent studies have shown that the genetic deletion of CHOP preserves tissue function upon pathological ER stress in multiple tissues (Rutkowski et al., 2008; Song et al., 2008; Thorp et al., 2009). We therefore introduced the Chop-null allele into MKO-PGC-1α mice and tested their maximal muscle capacity via exhaustive treadmill running. Importantly, the CHOP−/− mice showed a similar running performance compared to wild-type controls. The MKO-PGC1α mice showed compromised exercise capacity as previously reported (Handschin et al., 2007a). The deletion of CHOP expression in the MKO-PGC-1α background partially rescued the exercise intolerance and improved the total running distance by approximately 50% compared to the MKO-PGC1α mice (Figure 6A). We also detected reduced serum creatine kinase activity in the double knockout mice post-exercise, indicating that blocking ER stress-related cell death through genetic deletion of Chop protects against muscle damage after exercise in MKO-PGC-1α mice (Figure 6B).
Figure 6. Chop deletion partially rescues exercise intolerance observed in MKO-PGC-1α mice.
A. MKO-PGC-1α and control mice either with CHOP-null alleles or not were subject to treadmill running till exhaustion as described in Experimental Procedures. Distance was calculated from individual performances. N=15–22 animals per group. ** indicates p <0.01, PGC-1α MKO:CHOP−/− vs. PGC-1α MKO:CHOP+/+. B. Whole blood was collected by cardiac puncture from mice in (A) and serum creatine kinase activity was determined. Data represented means±SEM, * indicates p <0.05 PGC-1α MKO:CHOP−/− vs. PGC-1α MKO:CHOP+/+.
DISCUSSION
Originally defined as a pathway responding to the accumulation of misfolded proteins in the ER, the UPR has now been shown to bidirectionally communicate with other pathways in energy and nutrient sensing (Ron and Walter, 2007). Skeletal muscle is interesting with respect to the UPR because it has a limited role in protein secretion but it contains an extremely extensive network of specialized ER called the sacroplasmic reticulum (SR). Maintaining the optimal Ca2+ concentration in the SR lumen in the presence of variable ATP concentrations is critical for the regulated SR release of Ca2+ in skeletal muscle contraction during physical activity. We demonstrate here that the UPR is activated in skeletal muscle during excise, and modulates critical adaptations to exercise training. This study further uncovers a molecular cooperation between PGC-1α and ATF6α, one of the main transcription factors regulating the UPR (Figure S6).
The notion that PGC-1α interacts with ATF6α in regulating the UPR is supported by our demonstration that PGC-1α physically associates with ATF6α and increased expression of the same UPR marker genes controlled by PGC-1α is defective in ATF6α−/− myotubes. Although the precise mechanism is not yet clear, PGC-1α appears to regulate the UPR is a tissue-specific manner. This is based on our observation that ectopic expression of PGC-1α does not increase the expression of UPR markers in MEFs and rat embryonic cardiomyocytes (Figure S2). It is thus likely that some skeletal muscle-specific factors may be present in or influence the function of the PGC-1α/ATF6α protein complex. It is tempting to speculate that differences between the PGC-1α/ATF6α crosstalk in skeletal muscle and that in cardiac muscle have evolved to provide an additional layer of regulation in skeletal muscle, where the intensity of cellular stress fluctuates more significantly and frequently, compared to the heart.
The coactivation of ATF6α may not be the only molecular link between PGC-1α and the UPR. It is noteworthy that Montminy and colleagues previously showed that another transcriptional coactivator, CRTC2 mediates crosstalk between the UPR and the gluconeogenic program in the liver via coactivation of ATF6α or CREB (Wang et al., 2009). Interestingly, it has been shown that CRTC2, also referred to as TORC2, together with other TORCs (TORC1 and TORC3) strongly activated PGC-1α transcription through CREB (Wu et al., 2006). Our current study does not exclude a multilevel integration between PGC-1α and the UPR or indirect communication through other signaling pathways.
Mice deficient in muscle PGC-1α have been shown previously to be exercise intolerant (Handschin et al., 2007a). Since PGC-1α controls mitochondrial biogenesis and expression of the neuromuscular injunction (NMJ) gene program, it was suggested that these were likely to be major contributors to the exercise intolerance (Handschin et al., 2007a; Handschin et al., 2007c; Lin et al., 2005). This study shows that at least a substantial part of the function of PGC-1α in exercise tolerance is through control of the response to physiological ER stress. Since PGC1α can block muscular atrophy and muscular dystrophy (Handschin et al., 2007c; Sandri et al., 2006), future studies should illustrate whether blocking detrimental ER stress through deletion of CHOP or decreasing of CHOP expression in skeletal muscle could produce similar therapeutic effects. Chemical inhibitors for ER-stress under development will hopefully provide alternative avenues for treating these muscle pathologies.
EXPERIMENTAL PROCEDURES
Animal Experimentation
Generation of MCK-PGC-1α mice, MKO-PGC-1α mice, Atf6α-null mice and floxed Ire1α mice have been described elsewhere (Handschin et al., 2007b; Lin et al., 2002; Sakaki et al., 2008; Wu et al., 2007). C57/Bl6 wildtype mice and Chop-null mice were obtained from the Jackson laboratory. Mice were maintained under standard conditions. All experiments and protocols were performed in accordance with the respective Animal Facility Institutional Animal Care and Use Committee regulations.
Treadmill Exercise
Animals ran on a treadmill (Columbus Instruments) either on a 0% grade or tilted 10% uphill starting at a warm up speed of 5 m/min for 5 min. Every subsequent 5 min, the speed was increased by 5 m/min until mice were exhausted or a maximal speed of 20 m/min or 25 m/min was reached. Similar results have been obtained via either protocol. Exhaustion was defined as inability of the animal to return to running within 10 seconds after direct contact on an electric stimulus grid. Running time was measured and running distance calculated. Distance is the product of time and speed of the treadmill.
Cell Culture
Primary satellite cells (myoblasts) were isolated from homozygous floxed Ire1α mice, PGC-1α WT and −/−, ATF6α WT and −/− mice as described previously (Megeney et al., 1996). Myoblasts were cultured in F-10 medium supplemented with 20% FBS and basic FGF. For differentiation into myotubes, cells were shifted to DMEM supplemented with 5% horse serum. After 2 days of differentiation, they were transduced with an adenovirus expressing either GFP (green fluorescent protein) or PGC-1α as previously described (St-Pierre et al., 2003). Briefly, the differentiated myotubes were infected with adenoviruses at an MOI of 100 for overnight and the media was refreshed the next morning. The infection efficiencies were confirmed by GFP expression. Cells were harvested 48hrs post infection. Reporter gene assays were performed in a C2C12 muscle cell line. Myoblasts were transfected using Lipofectamine 2000 (Invitrogen) at 80% confluency and subsequently differentiated for 2 days. The cells were then harvested and luciferase activity was measured and normalized to renilla (Dual luciferase reporter assay system; Promega).
Gene Expression Analysis
Total RNA was isolated from cells or tissues using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. 1 μg of total RNA was reverse transcribed and analyzed using Applied Biosystems Real-time PCR System using the ΔΔCt method. Relative gene expression was normalized to TATA box-binding protein (tbp) mRNA levels. The Titan One-Tube RT-PCR kit (Roche) was used for RT-PCR to detect Xbp-1 splicing as previously described (Rutkowski et al., 2006). Affymetrix experiments were performed by the Dana-Farber microarray core facility and subsequent data analysis was conducted with dChip and Ingenuity Pathway Analysis software suites.
Serum Creatine Kinase Assay
Mouse blood was collected and serum isolated using heparin-coated collection tubes (BD Biosciences). Serum creatine kinase activity was then determined with the Stanbio CK Liqui-UV Assay kit (Stanbio Laboratory) according to the manufacturer's protocol.
Statistical Analysis
All results are expressed as means ± SD for cell experiments and ± SEM for animal experiments. Two-tailed Student's t test was used to determine p values. Statistical significance was defined as p < 0.05.
Supplementary Material
Acknowledgments
We thank Dina Laznik and Kevin Brannan for excellent technical assistance. We thank Dr. Umut Ozcan for the adenovirus expressing ATF6αN. We thank members of the Spiegelman laboratory for helpful comments, especially Drs. Patrick Seale and Melin Khandekar. This work was supported by National Institutes of Health Grants (to BMS, DK61562 and DK54477). Portions of this work were supported by the NIH R37 DK042394, R01 HL052173 and P01 HL057346 to RJK and R01 DK084058 from NIDDK to DTR. This work was supported in part by the Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center grant 1 U54 NS053672 to KPC. K.P. Campbell is an investigator of the Howard Hughes Medical Institute. JW was supported by a postdoctoral fellowship from the American Heart Association (Founders Affiliate #09POST2010078). JW and BMS conceived and designed the experiments. JW, JLR, JLE, KAR, JHC, LY, PB, HMT and RWC performed the experiments. JW and BMS analyzed the data and wrote the paper. Work with ATF6α−/− and IRE1α deleted myotubes was done with the laboratory of RJK. Work with ATF6α−/− mice was done with the laboratories of KPC and DTR. The authors have declared that no competing interests exist.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Calvo JA, Daniels TG, Wang X, Paul A, Lin J, Spiegelman BM, Stevenson SC, Rangwala SM. Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J Appl Physiol. 2008;104:1304–1312. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- Deshmukh AS, Hawley JA, Zierath JR. Exercise-induced phospho-proteins in skeletal muscle. Int J Obes (Lond) 2008;32(Suppl 4):S18–23. doi: 10.1038/ijo.2008.118. [DOI] [PubMed] [Google Scholar]
- Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem. 2007a;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- Handschin C, Choi CS, Chin S, Kim S, Kawamori D, Kurpad AJ, Neubauer N, Hu J, Mootha VK, Kim YB, Kulkarni RN, Shulman GI, Spiegelman BM. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest. 2007b;117:3463–3474. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Kobayashi YM, Chin S, Seale P, Campbell KP, Spiegelman BM. PGC-1alpha regulates the neuromuscular junction program and ameliorates Duchenne muscular dystrophy. Genes Dev. 2007c;21:770–783. doi: 10.1101/gad.1525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Rohas LM, St-Pierre J, Uldry M, Jager S, Handschin C, Spiegelman BM. A fundamental system of cellular energy homeostasis regulated by PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:7933–7938. doi: 10.1073/pnas.0702683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, Meier CA, Bell DR, Kralli A, Giacobino JP, Deriaz O. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes. 2003;52:2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki K, Wu J, Kaufman RJ. Protein kinase Ctheta is required for autophagy in response to stress in the endoplasmic reticulum. J Biol Chem. 2008;283:15370–15380. doi: 10.1074/jbc.M710209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci U S A. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Scheuner D, Ron D, Pennathur S, Kaufman RJ. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J Clin Invest. 2008;118:3378–3389. doi: 10.1172/JCI34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- Thorp E, Li G, Seimon TA, Kuriakose G, Ron D, Tabas I. Reduced apoptosis and plaque necrosis in advanced atherosclerotic lesions of Apoe−/− and Ldlr−/− mice lacking CHOP. Cell Metab. 2009;9:474–481. doi: 10.1016/j.cmet.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Vera L, Fischer WH, Montminy M. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature. 2009;460:534–537. doi: 10.1038/nature08111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci U S A. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Wu Z, Huang X, Feng Y, Handschin C, Gullicksen PS, Bare O, Labow M, Spiegelman B, Stevenson SC. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci U S A. 2006;103:14379–14384. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.