Abstract
Bacteroides spp. are the predominant organisms in the intestinal tract, and they also are important opportunistic pathogens. Antibiotic therapy of Bacteroides infections often is complicated by the prevalence of drug-resistant organisms which acquire resistance genes from a variety of mobile genetic elements including conjugative transposons (CTns) and mobilizable transposons (MTns). Tn4555 is an MTn that encodes β-lactam resistance, and it is efficiently mobilized by the Bacteroides CTns via a tetracycline (TET)-inducible mechanism. In this study a model system with CTn341 and a Tn4555 minielement was used to examine Tn4555 excision from the chromosome. Using PCR and mobilization assays it was established that excision was stimulated by TET in the presence of CTn341. In order to determine which Tn4555 genes were required for excision, int, tnpA, tnpC, xis, and mobA mutants were examined. The results indicated that int plus two additional genes, tnpC and xis, were required for optimal excision. In addition, there was no requirement for the mobA gene, as had been shown for another MTn, NBU1. The Xis protein sequence is related to a family of plasmid excisionases, but the TnpC gene product did not match anything in the sequence databases. Evidence also was obtained that suggested that Xis is involved in the control of TET-induced excision and in control of mobilization by CTn341. Overall, these results indicate that excision of MTns is a complex process that requires multiple gene products.
Bacteroides spp. are gram-negative, obligate anaerobic bacteria that inhabit the gastrointestinal tracts of humans and animals, where they constitute up to one-third of the total indigenous microflora (12, 18). As part of this normal flora, Bacteroides spp. play a number of significant roles that contribute to the complex intestinal physiology. In addition, they are important in the defense of their host against colonization by pathogens and they have critical input into the normal development of the gastrointestinal tract (7). The close proximity of Bacteroides spp. to their host provides numerous chances for infection, and thus they also are significant opportunistic pathogens that cause a variety of infections ranging from intra-abdominal abscesses to life-threatening septicemia (11, 18). Successful antimicrobial treatment of these infections has become increasingly difficult due to the emergence of resistant strains that have acquired resistance genes located on mobile genetic elements (20).
Bacteroides spp. possess a variety of transmissible genetic elements, including plasmids and transposons. The most common Bacteroides transposons are the large conjugative transposons (CTns), 50 to 100 kb, that encode tetracycline (TET) resistance and smaller (5 to 12 kb) mobilizable transposons (MTns) (13, 20). CTns are self-transmissible, and they are responsible for the high rate (>75%) of Bacteroides TET resistance (14). CTns appear to be the “drivers” of Bacteroides antibiotic resistance transfer since they can also mediate the transfer of coresident plasmids and MTns such as Tn4555 and NBU1 (13).
MTns are a diverse group of elements that can transfer to and integrate into phylogenetically divergent species. All MTns share the common property that they are mobilized by a conjugation-like mechanism when in the presence of a coresident CTn, and this transfer is stimulated more than 1,000-fold by treatment of the donor strains with low concentrations of TET. This requires that the MTns coordinate their excision with the expression of the CTn conjugation apparatus. To understand how excision and transfer are coordinated, we have examined the properties of Tn4555, a 12-kb MTn containing the β-lactamase gene, cfxA (Fig. 1). In the presence of a CTn, TET treatment induces Tn4555 excision and a covalently closed circular intermediate is formed and transferred by conjugation. The transposon then integrates into the chromosome by a site-specific mechanism in which a phage-like tyrosine recombinase (Int) mediates recombination between the joined ends of the transposon (attTn) and a primary target site in the chromosome (attB) (21).
FIG. 1.
Genetic map of the Tn4555 circular form. Genes are shown by the block arrows, and those genes used in this study are hatched; oriT and the transposon termini (attTn) are indicated by black symbols. DNA fragments used for the construction of the pFD660 family of plasmids and their mobA-oriT derivatives are shown by gray blocks inside the circular map. The location of the 485-bp PCR amplicon used to detect the circular form is shown by the hatched box, and the region of Tn4555-NBU1 homology is indicated by the gray box outside of the circular map.
Previously, the minimal requirements for Tn4555 integration were established and showed that tnpA, int, and tnpC were needed for normal integration at wild-type frequencies (23). In the current study a mini-Tn4555 element was used to determine if the same genes were required for TET-induced excision. Results showed int was necessary but not sufficient for excision and that wild-type levels of excision required two additional genes, xis and tnpC. Xis has some homology to several recombination directionality factors (RDFs) which typically play a role in excision reactions, but TnpC seems to be a new RDF which had no matches in the public databases. The results suggest that Xis and TnpC together with Int form a novel, complex excision system.
MATERIALS AND METHODS
Organisms and growth conditions.
Strains used in this study are described in Table 1. Bacteroides strains were grown at 37°C in an anaerobic chamber in brain heart infusion medium supplemented with hemin (16). Escherichia coli EC100 (Epicentre, Madison, Wis.) was used for routine cloning and conjugation experiments. E. coli was grown in Luria-Bertani broth at 37°C in an environmental rotary shaker.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Bacteroides strains | ||
| B. thetaiotaomicron BT5482 | Rfr | 15 |
| B. thetaiotaomicron BT5482::341 | Rfr Tcr; BT5482 containing CTn341 from mating with CLA341 | This work |
| B. fragilis ADB77 | Rfr Tpr ΔthyA | 1 |
| B. fragilis ADB77::Tn4555 | Rfr Tpr Fxr ΔthyA | This work |
| B. vulgatus CLA341 | Tcr Fxr; clinical strain containing CTn341 and Tn4555 | 10 |
| E. coli strains | ||
| EC100 | Str F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ−rpsL nupG | Epicentre, Madison Wis. |
| HB101::RK231 | Kmr Tcr Str; HB101 containing RK231 | 5 |
| Plasmids | ||
| pFD516 | Spr Emr; 7.7-kb suicide vector | 19 |
| pYT102 | Cmr Tcr; 8.2 kb, containing the thyA gene | 1 |
| pFD851 | Apr Fxr; 10.7-kb shuttle vector | This work |
| pFD852 | Apr Fxr; 12.1-kb derivative of pFD851 containing the xis gene | This work |
| pFD859 | Apr Fxr; 12.1-kb derivative of pFD851 containing the tnpC gene | This work |
| pFD861 | Apr Fxr; 13-kb derivative of pFD851 containing the tnpC xis genes | This work |
Antibiotic resistance phenotypes at the following concentrations: Rf, rifampin (20 μg/ml); Tc, tetracycline (5 μg/ml); Tp, trimethoprim (100 μg/ml); Fx, cefoxitin (20 μg/ml); St, streptomycin (25 μg/ml); Sp, spectinomycin (40 μg/ml); Cm, chloramphenicol (25 μg/ml); Ap, ampicillin (50 μg/ml); Em, erythromycin (15 μg/ml).
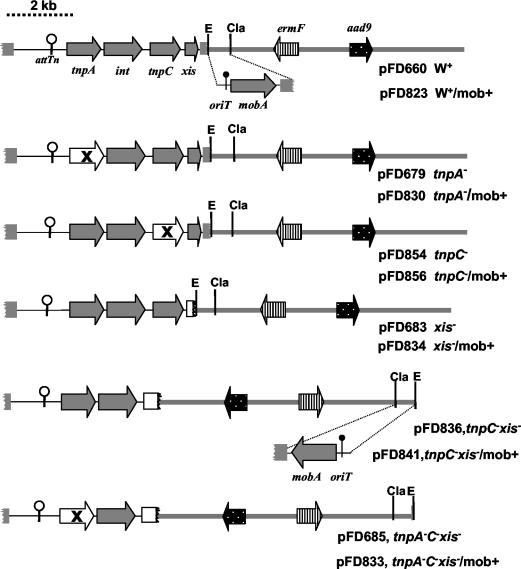
Mutant and plasmid construction.
Mutations were verified by DNA sequence analysis, and Tn4555 base pair coordinates described below refer to GenBank accession number U75371. Mutant constructs are shown in Fig. 2, and all encoded spectinomycin resistance for E. coli selection and erythromycin resistance for Bacteroides selection. Strains containing constructs integrated in the chromosome are designated with an Ω followed by the construct number. The integrating construct pFD660 and the tnpA, xis, and tnpA xis tnpC mutants were described previously (23). The internal deletion mutation in tnpC was made by cloning the 4.7-kb NdeI/SstI fragment of pFD660 into pUC19. This plasmid was used in a PCR mutagenesis protocol with the QuikChange system (Stratagene, La Jolla, Calif.) adapted for the creation of deletions according to Wang and Malcom (24). The resulting mutation was an in-frame deletion mutation of TnpC L28 to N228 (Δ2834 to 3485 of Tn4555). Once the mutation was verified, the mutated NdeI/SstI fragment was exchanged for the parental pFD660 NdeI/SstI fragment by standard cloning methods.
FIG. 2.
Genetic maps of the Tn4555 minitransposons used in this study. All constructs are suicide plasmids that do not replicate in Bacteroides. Genes are indicated by the block arrows, and specific mutations referred to in the text are indicated by the white block arrows containing an X. attTn is indicted by the symbol with the white circle. For each plasmid construct the site of mobA-oriT gene insertion is indicated. The E. coli plasmid sequences are shown by the thick black line, and selection of the elements was for spectinomycin resistance (aad9) in E. coli. The Bacteroides Tn4555 sequences are shown by the thin black line, and erythromycin resistance (ermF) was used for selection in Bacteroides.
The tnpC xis mutant construct (pFD836) was made by first cloning the 4.8-kb Sau3AI fragment from Tn4555 (bp 10,629 to 3,203; Fig. 1) into the BamHI site of pBR322. A DNA fragment containing only the tnpA and int genes was isolated from this plasmid by digestion with SphI and EcoRV and ligated to the pFD516 SphI and SmaI sites. In order to make versions of the integrating constructs that were mobilizable by a CTn, the Tn4555 mobA and oriT genes were amplified by PCR (Tn4555 bp 6224 to 8604; Fig. 1) using primers with EcoRI and ClaI sites engineered into their sequences. The amplified product was cloned into constructs as shown in Fig. 2.
A Tn4555 int deletion mutant was constructed using the positive-selection, two-step double-crossover technique described previously (1). Briefly, an int gene fragment containing an in-frame 744-bp deletion of the int gene (between bp 1538 and 2283) was cloned into BamHI- plus HindIII-digested pYT102 and conjugated into strain ADB77ΩTn4555 where it inserted by homologous recombination into the int gene. This cointegrate was resolved by selection of the second recombination event by plating overnight cultures on minimal medium with glucose, thymine, and trimethoprim to select for thymine auxotrophy. The resulting colonies were screened by PCR to discriminate between isolates containing the deletion and the wild-type int.
The complementation vector pFD851 was a pSC101-based vector derived from pWSK29 (25). For pFD851, the RK2 oriT region was PCR amplified from pFD288 (bp 704 to 1259) (19), using primers with Sau3AI engineered into their sequences. The product was digested with Sau3AI and cloned into the BglII site of pWSK29. A Bacteroides replication origin from pBI143 (19) was inserted into the EcoRV and ClaI sites. The cfxA β-lactamase gene was excised as an EcoRI/BamHI fragment from pFD351 (10) and cloned into the pWSK29 EcoRI and BamHI sites, resulting in pFD851. The xis, tnpC, and xis tnpC genes were PCR amplified (Tn4555 bp 3192 to 4565, 2285 to 3200, and 2285 to 4565, respectively) using primers with SstI and BamHI engineered into their sequences and cloned into pFD851 (Table 1). Shuttle plasmids based on pBI143 have a low- to medium copy number (ca. 20 copies per cell), and each of the clones appeared by inspection of agarose gels containing total DNA to have a similar copy number (19).
Bacterial matings.
Standard filter mating procedures were used for plasmid transfer between E. coli and Bacteroides strains (17). Mating conditions were selected to favor the donors, that is, aerobic for E. coli to Bacteroides and anaerobic for Bacteroides to E. coli. Matings with E. coli donors included RK231 as the conjugation helper plasmid (5). Matings with Bacteroides donors were either induced with 1 μg of TET/ml or not induced (17). For the mating-out assays Bacteroides donors were combined with E. coli EC100 recipients, placed on filters, and incubated for 18 h in an anaerobic chamber. Then cells were washed from the filters and plated on Luria-Bertani agar containing 40 μg of spectinomycin/ml to select for E. coli transconjugants.
PCR excision assays.
Total genomic DNA isolated from TET-induced and noninduced cultures was quantified and then used as templates in PCRs. Diluted templates were amplified with Taq DNA Polymerase (InvitroGen, Carlsbad, Calif.) in 50-μl reaction mixtures according to the manufacturer's instructions. The program used for amplification was 30 cycles of 55°C for 60 s, 72°C for 60 s, and 95°C for 30 s. This program was designed to be an endpoint assay and was not intended to be strictly quantitative. Samples of 10 μl were electrophoresed on 1.0% agarose gels, and results shown are typical of those obtained in at least three independent assays. The Tn4555 primer pair (attL, GGAATATCGGAAACGAATAGC; and attR, GGATGTGAACGGAAGTCAACC) were used for the excision assays. These primers flanked the “joined ends” (attTn) of the transposon and amplified a 485-bp fragment (Fig. 1). Primers derived from the Bacteroides thetaiotaomicron susC gene were used in PCRs to verify template concentrations. The susC primers (sus1, ATCGTTATCCGTTTCCGTCTG; and sus2, GTTCATCCATTGTCCGTAGTG) amplified a 1,044-bp fragment from the B. thetaiotaomicron chromosome. Results from these PCR assays were verified with a second set of primers for both attTn and susC. These primer pairs amplified 127- and 124-bp fragments, respectively. The primers were as follows: for attTn, TGTGCAATTAAAAATAACAACCA, and rev attTn, TCGGATATTTCTGATTAGTTTTGG; for susC, GTTTGGCGTTGATGGAAAAT, and rev susC, CGTAAAACGGATGACCTGCT. Reactions were analyzed under the following PCR conditions: 95°C for 3 min 40 cycles of 95°C for 10 s, 58.5° for 15 s, and 72°C for 15 s. Melting point curves and agarose gels verified that only a single product was formed in these reactions.
RESULTS
Experimental Tn4555 excision system and the role of Int.
In a previous study a mini-Tn4555 element was used to show that three genes, tnpA, int, and tnpC, were required to reproduce a normal transposition phenotype (23). To determine if excision required the same genes, a similar mini-Tn4555 system was used (Fig. 2) together with two excision assays. The first assay was based on the observation that the ends of the excised transposon were joined by a 6-bp coupling sequence to form attTn. Hence, PCR primers were designed that flanked attTn and gave a 485-bp product only when the element was excised. The second assay was a mating-out assay based on the observation that in the presence of a CTn, the integrating construct pFD428, which contains the intact Tn4555 genome, was able to excise from Bacteroides and transfer to E. coli, where it replicated as a plasmid (21).
The excision system was tested with pFD823, which contained the mobA-oriT region cloned into pFD660. Results from mating-out assays with ΩFD823 showed that excision and transfer of this element were similar to those of Tn4555 in that it was highly induced (105-fold) by the addition of TET (Table 2). This finding was supported by the PCR assay, where there was an obvious increase in the amount of amplified product in the TET-induced sample compared to the noninduced samples (Fig. 3A).
TABLE 2.
Determination of Tn4555 excision using mating-out assays
| Strain (relevant genotype)a | Transfer frequencyb
|
|
|---|---|---|
| −Tetracycline | +Tetracycline | |
| ΩFD823 (tnpA+tnpC+xis+) | 3.6 × 10−8 (± 3.5) | 1.2 × 10−3 (± 0.6) |
| ΩFD830 (tnpA tnpC+xis+) | 1.7 × 10−8 (± 3.0) | 2.3 × 10−4 (± 2.1) |
| ΩFD833 (tnpA tnpC xis) | <10−9 | <10−9 |
| ΩFD834 (tnpA+tnpC+xis) | 1.1 × 10−8 (± 1.0) | 1.3 × 10−8 (± 2.0) |
| ΩFD841 (tnpA+tnpC xis) | <10−9 | <10−9 |
| ΩFD856 (tnpA+tnpC xis) | <10−9 | <10−9 |
| ΩFD834/pFD852 (xis+) | <10−9 | <10−9 |
| ΩFD841/pFD861 (tnpC+xis+) | <10−9 | <10−9 |
| ΩFD856/pFD859 (tnpC+) | 4.2 × 10−8 (± 0.8) | 1.4 × 10−4 (± 0.9) |
The strains listed are the B. thetaiotaomicron donor strains. All constructs contained the intact int gene, and their structures are shown in Fig. 2. The relevant mutations are shown in bold and underlined.
Transfer frequencies were determined from the number of E. coli transconjugants obtained per input B. thetaiotaomicron donor, and the results are means from three independent trials. Cultures were induced with 1 μg/ of TET/ml as described in the text.
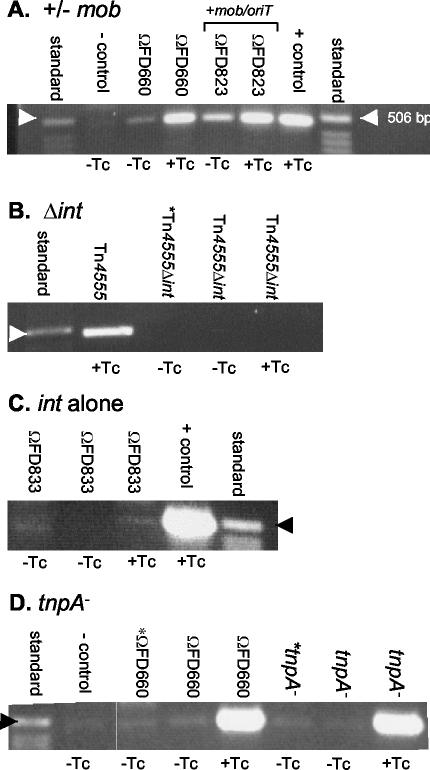
FIG. 3.
Int but not TnpA is required for Tn4555 excision. Shown are PCR excision assays showing agarose gels of PCRs using the attTn primers as described in Materials and Methods. Standard, 1-kb ladder (InvitroGen, Carlsbad, Calif.); +control, BT5482::341 containing Tn4555 induced with TET; −control, BT5482::341 containing Tn4555 not induced with TET. PCRs were from cultures either induced with 1 μg of tetracycline (+Tc) or not induced (−Tc). Lanes marked with an asterisk are from strains that did not contain CTn341. Arrowheads on the sides of the panels indicate the location of the 506-bp marker which aligns close to the expected PCR product.
In a previous study it was shown that the mob-oriT region was required for excision of the Bacteroides MTn NBU1 (15). NBU1 has a mobilization region that is 78% identical (∼1,600-bp region; Fig. 1) to Tn4555, and in fact the two mobilization regions can complement one another (17). Since excision and transfer are linked, it seemed possible that the mob-oriT region would mediate this linkage in both NBU1 and Tn4555. PCR assays of samples from TET-induced and noninduced cultures of ΩFD660 (no mobA-oriT region) revealed excised product, with more product present following TET induction (Fig. 3A). These data suggest that the mobilization region is not required for Tn4555 excision.
The Tn4555 integrase is most closely related to the XerD family of tyrosine recombinases which are required for both insertion and excision of their respective genetic elements. To test the role of Int in excision, we constructed an in-frame int deletion in an integrated copy of Tn4555 present in the Bacteroides chromosome. PCR assays of TET-induced and noninduced samples of Tn4555Δint were negative for excision, whereas product was obtained from the intact transposon (Fig. 3B). Also, there was no transfer of the element to other Bacteroides recipients (data not shown).
In order to determine if Int was sufficient for integration and excision, we used the construct pFD833, which contains int and mobA-oriT but no other intact Tn4555 genes (Fig. 2). Mating-out assays with ΩFD833 were negative within the limits of detection, and PCR assays did not reveal the presence of appreciable product even when excess sample was loaded onto agarose gels (Table 2 and Fig. 3C). Taken together these results show that Int is necessary for excision but that at least one of the other proteins, TnpA, TnpC, or Xis also is required.
A requirement for TnpA in excision was ruled out by testing the two tnpA mutants ΩFD679 and ΩFD830. In mating-out assays ΩFD830 transferred in a TET-inducible manner, although at a slightly lower frequency than the wild-type construct (104-fold induction; Table 2). Likewise the PCR assays revealed the appearance of a strong TET-inducible amplification product in samples from both tnpA mutants (Fig. 3D and data not shown).
Xis plays a role in excision and mobilization.
Xis was predicted to play a role in excision based on sequence homology and is distantly related to several excisionases (9, 22). Assays with the xis mutants provided evidence that it strongly stimulated excision. The mating-out assay revealed a low level of transfer from ΩFD834, but it was not obviously TET inducible. Data from this mating-out assay were consistent with PCR assays, where a low level of amplified product was observed in both induced and noninduced samples (Fig. 4A). A PCR using a second set of attTn primers verified this low level of excised transposon (data not shown).
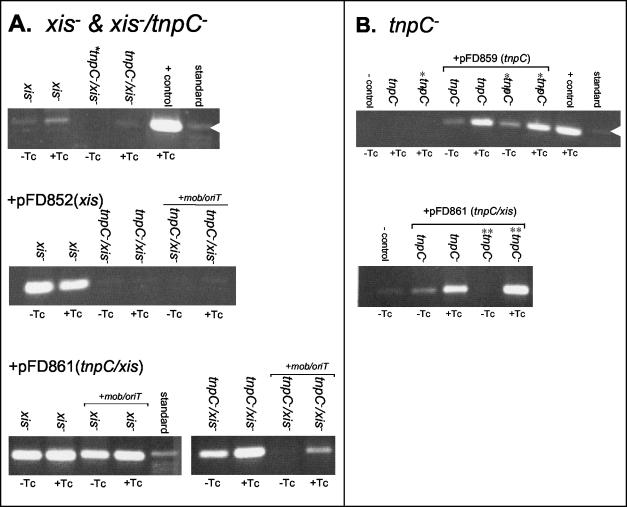
FIG. 4.
The role of Xis and TnpC in excision as shown by PCR assays. (A) Analysis of xis mutant (ΩFD683) and tnpC xis double mutants (ΩFD836 and ΩFD841). Strains complemented with a plasmid containing only the xis gene (pFD852) are shown on the panel labeled +pFD852, and strains complemented with a plasmid containing both xis and tnpC are labeled +pFD861. (B) Analysis of tnpC mutants and derivatives complemented with a plasmid containing only tnpC (pFD859) or a plasmid containing both tnpC and xis (pFD861). Lanes marked with a double asterisk are samples from the mutant ΩFD856 containing mobA-oriT. See the legend to Fig. 3 for all other label designations.
To show that excision depended on xis, the mutant was complemented with the cloned gene. The xis gene has its own transcription promoter (22), so the gene was cloned into a low-copy-number shuttle vector (ca. 20 copies per cell) and transferred into the xis mutant construct ΩFD834. Results from the PCR assay indicated that the mutant excised in the presence of the complementing plasmid, pFD852, and this result was verified with a second PCR assay using a different primer set (Fig. 4A and data not shown). Based on this PCR assay result it was expected that mating-out assays with ΩFD834 would show a high level of transfer; however, there was no transfer of ΩFD834 from the complemented strains (Table 2). This result suggests that there may be some interaction between mobilization and excision.
TnpC is the third gene product required for Tn4555 excision.
A role for TnpC was first inferred from PCR assays with a double tnpC xis mutant in which there was very little to no excised transposon present in either induced or noninduced samples (Fig. 4A). Consistent with these data, the mating-out assays with ΩFD841 also were negative and did not show the low level of constitutive transfer seen with the xis mutant ΩFD834 (Table 2). Excision of the tnpC xis mutants was not complemented with pFD852, which contains only xis, but excision was restored with a plasmid containing both the tnpC and xis genes (pFD861; Fig. 4A). To verify the PCR results, a mating-out assay was performed with the pFD861-complemented mutant ΩFD841. As shown in Table 2 no transfer was detected in induced or noninduced samples. This is the same result observed when xis alone was present on a plasmid, suggesting that it is overexpression of xis that leads to the nonmobilizable phenotype.
TnpC does not match any protein of known function in the public databases; therefore, it was important to document its role in excision independently of the xis mutation. This was accomplished with a tnpC internal deletion mutant. The results shown in Fig. 4B and Table 2 show that tnpC mutants were negative in PCR and mating-out assays. Plasmids containing tnpC alone or tnpC xis were able to complement the excision-less phenotype, and this complementation appeared to be TET inducible (Fig. 4B). The question remained whether pFD859 (tnpC only) could complement the negative phenotype observed in the mating-out assay. The data in Table 2 indicate that not only was transfer restored in the complemented mutant but it was TET inducible (>103-fold) at nearly the wild-type levels. These results show that in contrast to xis, tnpC overexpression did not affect transfer.
DISCUSSION
Conjugal transfer of an MTn from Bacteroides requires excision of the integrated transposon and then circularization prior to processing by the conjugation apparatus (13, 20). This excision process is tightly regulated by TET induction so that it can be coordinated with expression of the CTn transfer genes. In order to understand how the CTn and MTn coregulate their activities, the requirements for Tn4555 excision were determined. Previous work showed that integration of the element can be mediated entirely by Int, but results presented here show that the reverse reaction required Int plus two proteins. The complexity of Tn4555 excision is unusual for genetic elements using a tyrosine recombinase pathway but may reflect the need for coregulation with the CTn. The two accessory proteins required for excision, TnpC and Xis, appear to function as RDFs in that they are small proteins which modulate Int-mediated recombination, pushing the equilibrium toward excision of the element.
Xis is small, 124 amino acids, with a basic pI of 8.02, properties which are common to most bacterial excisionases. Except for Xis-like open reading frames found in the genomes of other Bacteroides species, Tn4555 Xis was most similar (34% identity) to a putative excisionase found in the SGI1 genomic island, which harbors multiple antibiotic resistance determinants in Salmonella enterica (2). It has been observed that Xis proteins often appear to coevolve with their cognate integrases (9). In this regard, it is of interest that the SGI1 integrase is more similar (30% identity) to Int-Tn4555 than any other integrase in the databases; thus, Xis-Tn4555/Int-Tn4555 and Xis-SG1/Int-SGI1 are a matched pair that may have a common ancestor.
In a survey of Xis and Cox proteins, both Xis-Tn4555 and Xis-SGI1 have been assigned to a miscellaneous group of tyrosine RDFs based on their primary amino acid sequences (9). This group is part of the SLP1 clade, which are mostly phage-associated proteins characterized by the presence of a helix-turn-helix (HTH) motif. Xis-Tn4555 does have an HTH motif at M47 to S68 (score of 3.49 predicted using the Dodd and Egan algorithm at http://npsa-pbil.ibcp.fr/cgi-bin/primanal_hth.pl; [3]), and this aligns with the Xis-SGI1 HTH motif. Database searches revealed that this HTH is part of an AlpA domain of a family of transcriptional regulators (8). Other excisionases have been shown to be regulators, such as the Cox protein, which can repress transcription of the bacteriophage HP1 PL promoter (4). Therefore, Xis may be a regulatory gene in the Tn4555 system. Consistent with this idea, it was found that xis mutants had a low level of constitutive excision and mobilization, suggesting that this gene somehow affects TET induction. On the other hand, overproduction of Xis resulted in constitutive up-regulation of excision in the PCR assays (Fig. 4A). Both of these results could be explained if Xis was acting as an activator of excision. Also, results in Table 2 suggest that Xis may have a regulatory role in the control of mobilization since there was a suppression of transfer when xis was present on a multicopy plasmid.
Results from mating-out assays and PCR assays showed that tnpC mutants did not excise, indicating that TnpC is functionally an RDF. In contrast to Xis, the sequence of TnpC was not similar to anything in the public databases and the only conserved domain found in the TnpC sequence was an 80% match to the basic leucine zipper motif. Leucine zippers typically play a role as dimerization domains in many eukaryotic transcriptional regulators, and they have been found to mediate oligomerization of transposase components for some bacterial insertion sequence elements (6). Thus, TnpC may act to facilitate formation of the nucleoprotein excision complex by mediating protein-protein interactions that bring attR and attL into proximity so they can be acted on by Int. In contrast to results with Xis, overproduction of TnpC had no effect on TET-induced excision, suggesting that it has no role in regulation.
Only two MTns have been studied in detail, and it is instructive to point out that in both cases TET-inducible excision has been shown to be a complex phenomenon. Yet even with this need to interact specifically with the CTn, the two MTns do not share any common excision proteins. Tn4555 requires Int plus two accessory proteins. An Int requirement for NBU1 excision has not been shown, but it requires at least three other gene products plus a cis-acting region containing oriT (15). Two required NBU1 excision genes encode a DNA primase-like protein and a helicase-like protein, neither of which are like any domain or motif associated with Xis or TnpC. One additional NBU1 gene product in the excision region, Orf2X, was not previously studied, but it has some similarity to Tn4555-Xis in that it is a small, basic protein (105 amino acids, pI 9.3). Even more intriguing is that Orf2X has an AlpA domain and HTH motif similar to those of Xis.
Unlike Tn4555, NBU1 excision has a strict requirement for oriT and part of the mobA-NBU1 gene in cis. NBU1 may use this cis-acting region as a loading site for the excision complex, and it is thought that the excision protein PrmN1 binds here to prevent premature nicking by Mob (15). Although Tn4555 does not have this specific requirement, we did observe that overproduction of Xis lead to excision of the transposon but inhibited its transfer. One possible explanation for this phenotype is that Xis binds to oriT and prevents it from participating in transfer. Thus, in both MTns mobilization and excision are linked but by entirely different mechanisms. It is likely that the need to coordinate excision with expression of transfer leads to the complexity of MTn excision, but there must be multiple avenues to achieve this coordination.
Acknowledgments
This work was supported by Public Health Service grant AI28884 to C.J.S.
We thank M. Bacic and C. Sund for critical reading and discussion of themanuscript.
REFERENCES
- 1.Baughn, A. D., and M. H. Malamy. 2002. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis:implications for the evolution of the mitochondrial Krebs cycle. Proc. Natl. Acad. Sci. USA 99:4662-4667. [DOI] [PMC free article] [PubMed]
- 2.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esposito, D., J. C. Wilson, and J. J. Scocca. 1997. Reciprocal regulation of the early promoter region of bacteriophage HP1 by the Cox and Cl proteins. Virology 234:267-276. [DOI] [PubMed] [Google Scholar]
- 5.Guiney, D. G., P. Hasegawa, and C. E. Davis. 1984. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc. Natl. Acad. Sci. USA 81:7203-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haren, L., P. Polard, B. Ton-Hoang, and M. Chandler. 1998. Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. J. Mol. Biol. 283:29-41. [DOI] [PubMed] [Google Scholar]
- 7.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 8.Kirby, J. E., J. E. Trempy, and S. Gottesman. 1994. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J. Bacteriol. 176:2068-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis, J. A., and G. F. Hatfull. 2001. Control of directionality in integrase-mediated recombination:examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 29:2205-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker, A. C., and C. J. Smith. 1993. Genetic and biochemical analysis of a novel Ambler class A β-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 37:1028-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redondo, M. C., M. D. Arbo, J. Grindlinger, and D. R. Snydman. 1995. Attributable mortality of bacteraemia associated with the Bacteroides fragilis group. Clin. Infect. Dis. 20:1492-1496. [DOI] [PubMed] [Google Scholar]
- 12.Salyers, A. A. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38:293-313. [DOI] [PubMed] [Google Scholar]
- 13.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 2000. Multiple gene products and sequences required for excision of the mobilizable integrated Bacteroides element NBU1. J. Bacteriol. 182:928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith, C. J. 1985. Characterization of Bacteroides ovatus plasmid pBI136 and structure of its clindamycin resistance region. J. Bacteriol. 161:1069-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith, C. J., and A. C. Parker. 1996. A gene product related to Tral is required for the mobilization of Bacteroides mobilizable transposons and plasmids. Mol. Microbiol. 20:741-750. [DOI] [PubMed] [Google Scholar]
- 18.Smith, C. J., E. R. Rocha, and B. J. Paster. 2003. The medically important Bacteroides spp. in health and disease. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, N. Y.
- 19.Smith, C. J., L. A. Rollins, and A. C. Parker. 1995. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143. Plasmid 34:211-222. [DOI] [PubMed] [Google Scholar]
- 20.Smith, C. J., G. D. Tribble, and D. P. Bayley. 1998. Genetic elements of Bacteroides species: a moving story. Plasmid 40:12-29. [DOI] [PubMed] [Google Scholar]
- 21.Tribble, G. D., A. C. Parker, and C. J. Smith. 1997. The Bacteroides mobilizable transposon Tn4555 integrates by a site-specific recombination mechanism similar to that of the gram-positive bacterial element Tn916. J. Bacteriol. 179:2731-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tribble, G. D., A. C. Parker, and C. J. Smith. 1999. Genetic structure and transcriptional analysis of a mobilizable, antibiotic resistance transposon from Bacteroides. Plasmid 42:1-12. [DOI] [PubMed] [Google Scholar]
- 23.Tribble, G. D., A. C. Parker, and C. J. Smith. 1999. Transposition genes of the Bacteroides mobilizable transposon Tn4555: role of a novel targeting gene. Mol. Microbiol. 34:385-394. [DOI] [PubMed] [Google Scholar]
- 24.Wang, W., and B. A. Malcolm. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions, and insertions using QuikChange site-directed mutatgenesis. BioTechniques 26:680-682. [DOI] [PubMed] [Google Scholar]
- 25.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]