Abstract
A strategy devised to isolate a gene coding for a dihydrofolate reductase from Thermus thermophilus DNA delivered only clones harboring instead a gene (the T. thermophilus dehydrogenase [DHTt] gene) coding for a dihydropteridine reductase which displays considerable dihydrofolate reductase activity (about 20% of the activity detected with 6,7-dimethyl-7,8-dihydropterine in the quinonoid form as a substrate). DHTt appears to account for the synthesis of tetrahydrofolate in this bacterium, since a classical dihydrofolate reductase gene could not be found in the recently determined genome nucleotide sequence (A. Henne, personal communication). The derived amino acid sequence displays most of the highly conserved cofactor and active-site residues present in enzymes of the short-chain dehydrogenase/reductase family. The enzyme has no pteridine-independent oxidoreductase activity, in contrast to Escherichia coli dihydropteridine reductase, and thus appears more similar to mammalian dihydropteridine reductases, which do not contain a flavin prosthetic group. We suggest that bifunctional dihydropteridine reductases may be responsible for the synthesis of tetrahydrofolate in other bacteria, as well as archaea, that have been reported to lack a classical dihydrofolate reductase but for which possible substitutes have not yet been identified.
Dihydrofolate reductase (DHFR; EC 15.1.3) catalyzes the synthesis of tetrahydrofolate (FH4), a key metabolite involved in the synthesis of several amino acids, purines, and deoxythymidylate. The substrate of DHFR, dihydrofolate (FH2), is a product of thymidylate synthase (TS; thyA gene), which uses N5,N10-methylene-FH4 as a substrate. Escherichia coli DHFR-null mutants (with a mutation in dyrA or folA) appear to be nonviable, even on rich medium, unless they are also TS deficient (1, 6); this finding suggests that TS activity is toxic in a dyrA background because it exhausts residual FH4 production due to another enzyme proceeding at a slower pace than DHFR (4). This substitute enzyme could be a dihydropteridine reductase (DHPR; EC 1.6.99.7), since E. coli contains a DHPR able to catalyze the same reaction as DHFR at a comparatively low rate (16, 17). The metabolic role of prokaryotic DHPR is not known; it is possible that nonenzymatic oxidation of FH4 produces a quinonoid FH2 species which is regenerated to FH4 by DHPR but not by DHFR (14). In the few proteobacteria (but not E. coli) in which aromatic amino acid hydroxylases have been reported to occur, a DHPR regenerates the reduced cofactor tetrahydropterin, which is oxidized during the hydroxylase reaction (21). The metabolic role of these hydroxylases, however, is not clear.
Trimethoprim (TMP) is a powerful inhibitor of prokaryotic DHFRs, but when such a DHFR is overexpressed from a plasmid, it may confer TMP resistance on an E. coli host (5, 13, 19, 20). We have used this approach in order to isolate a dyrA gene from Thermus thermophilus. As described below, the gene selected in this way proved, however, to code for a dehydrogenase (T. thermophilus dehydrogenase [DHTt]) of the short-chain dehydrogenase/reductase (SDR) family (12) with both DHPR and DHFR activities. No T. thermophilus dyrA homologue could be isolated by this approach; also, there appears to be no DHFR gene in the Thermus genome (A. Henne, personal communication).
MATERIALS AND METHODS
Culture conditions.
E. coli strains were grown at 37°C in rich liquid medium 853 (19) or in solid medium with added 1.5% agar (Difco). Kanamycin (KAN) at 50 μg ml−1 or KAN plus 10 mM TMP was added to the medium for bacteria harboring recombinant plasmids. T. thermophilus HB27 was grown at 72°C in a medium containing (per liter) 9 g of tryptic soy broth (Difco), 4 g of yeast extract (Difco), and 3 g of NaCl; the pH of this medium was adjusted to 7.5 (11).
Cloning the DHTt gene.
Restriction enzymes and T4 ligase were purchased from Boehringer Mannheim. T. thermophilus HB27 genomic DNA partially digested with the enzyme Sau3A was used to construct a genomic λZAP DNA library in the pBK-CMV vector (Stratagene) according to the manufacturer's instructions. This library was used for the transformation of E. coli strain XL1-Blue MRF (Stratagene) as described in Results.
Enzyme assays.
DHFR activity was assayed as described in reference 19 except that the buffer was 50 mM potassium phosphate (pH 6.5) and the temperature was as indicated in the text. The DHPR assay was run with 50 mM potassium phosphate buffer at pH 7.0 and at 35°C with a 0.1 mM concentration of the cofactor (NADPH or NADH), 0.1 mM pteridine, and 6,7-dimethyl-7,8-dihydropterine in the quinonoid form (qPtH2), which was obtained from the 5,6,7,8-tetrahydro form and an equimolar amount of 2,6-dichloroindophenol. One unit of reductase activity was defined as the amount of activity required to convert 1 nmol of NADPH per min.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by using a Pharmacia PhastSystem with a discontinuous buffer system and a continuous 8 to 25% gradient gel. Gels were stained with Coomassie brilliant blue. Protein standards (0.5 μg each on the gel; Pharmacia) used for the estimation of subunit molecular masses were phosphorylase b (94 kDa), albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa).
Native gel electrophoresis for enzyme activity staining.
The Bio-Rad Prep-Cell system was used at a basic pH. The running buffer was Tris-HCl at pH 10.0. The gel (about 5 ml) was composed of 1 volume of solution A (running buffer supplemented with 0.46 ml of N,N,N′,N′-tetramethylethylenediamine/100 ml), 2 volumes of solution B (10% acrylamide, 2.5% N,N-methylene bisacrylamide), and 4 volumes of solution C (0.14% ammonium persulfate). The loading buffer contained 25 mM Tris, 100 mM glycine, 0.1% bromophenol blue, and 10% saccharose. About 5 μg of protein was loaded on the gel and run for 1 h at 150 V.
RESULTS
Isolation of the DHTt gene and analysis of the nucleotide sequence.
The λZAP T. thermophilus genomic library was plated for titration on rich medium containing KAN and for selection on plates containing KAN and TMP. For ca. 4,000 colonies screened (with a mean insert size of 2.8 kb and thus a fragment sampling covering six times the T. thermophilus genome), there were 20 colonies on the KAN-TMP plates (with insert sizes ranging from 1.5 to 4.0 kb). Crude extracts from overnight cultures of these recombinant strains were cleared by centrifugation, treated for 20 min at 70°C to inactivate resident E. coli DHFR activity, and assayed for DHFR activity at 50°C as described previously (19). All of these extracts displayed similar thermoresistant DHFR activities (on average, 2.5 U/mg of protein), whereas neither the extract from the control E. coli dyrA+ strain nor that from E. coli transformed with a plasmid carrying an unrelated gene (pTAD1GDH [3]) displayed measurable activity.
The smallest DNA insert that could be recovered from the cognate phagemids (pDAT1-20) by EcoRI/PstI digestion and that proved to be able to confer resistance to TMP was 1.7 kb long. It contained only one 702-bp open reading frame (ORF) encoding a putative polypeptide of 234 amino acids, named DHTt. This ORF was found by PCR amplification to be present in all recombinant pDATs. The GC content was high (69%), as expected for a Thermus gene. Surprisingly, the DHTt sequence showed no similarity to that of any DHFR but instead with sequences from representatives of the SDR family (Fig. 1). The sequence contains residues that are highly conserved throughout the SDR family, such as the NAD+/NADP+ cofactor-binding motif (GXXXGXG, positions 8 to 14), the NNAG motif (NNVG in DHTt, positions 85 to 88), and the YXXXK motif (residues 153 to 157), claimed to be crucial for substrate binding and catalysis by SDR family proteins (Fig. 1) (7, 8).
FIG. 1.
Multiple alignment of amino acid sequences of selected reductases belonging to the SDR family, in order of similarity: D. radiodurans 3-oxoacyl-(acyl-carrier-protein) reductase (DrOAR), the homologous protein from E. coli (EcOAR), T. thermophilus DHPR (TtDH), DHPR 1 (nonquinonoid) from L. major (LmPTR1), quinonoid DHPR from L. major (LmqDPR), and rat DHPR. Bold type indicates identical residues in at least four of the six sequences. Sites conserved in the SDR family are underlined.
Overexpression and characterization of the DHTt gene product.
The DHTt-encoding gene was subcloned from pDAT5 into pET24a, an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression vector, yielding plasmid pTTDH. The ORF was amplified by PCR by using oligonucleotides designed to bring the ATG start codon in frame with the vector NdeI cloning site. The highest DHFR specific activity was obtained from cells grown at 37°C up to a density of 4 × 108 cells/ml and then induced by 1 mM IPTG and further incubated at 22°C for 4 h. The specific activity after 15 min of incubation of the extract at 50°C was 14.9 U/mg of protein at 35°C in E. coli pTTDH, compared to 2.9 U/mg of protein in pDAT5. This activity proved insensitive to 600 μM methotrexate and 10 μM TMP, concentrations which are fully inhibitory for E. coli DHFR (5).
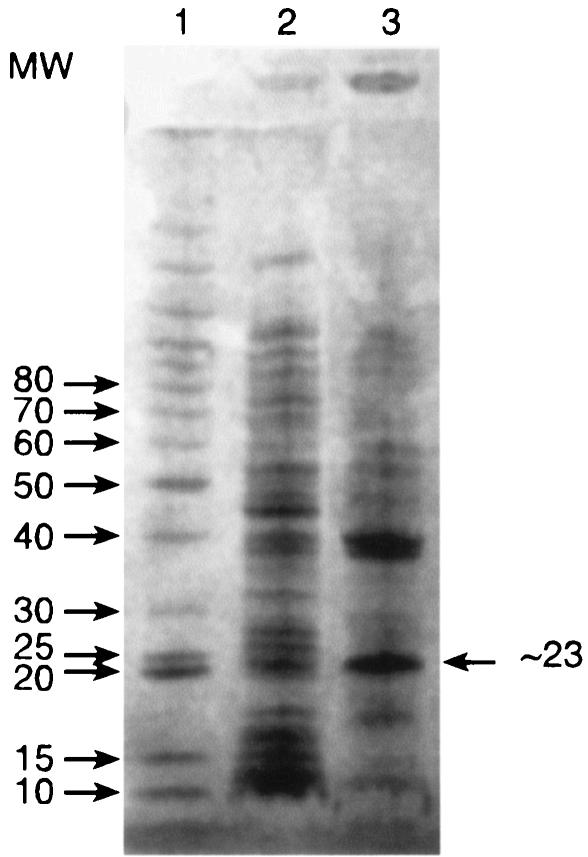
Attempts at purifying the protein were hampered by low solubility, probably due to the high content of hydrophobic residues (50% L+V+I+M+F+A+P+W). E. coli pTTDH extracts kept for a few hours at 4°C formed a white precipitate that contained the activity and could be partly solubilized at pH 10; further dilution of such preparations under assay conditions fully solubilized them. SDS-PAGE of the cold precipitate gave a major band (Fig. 2) with a molecular mass of about 23 ± 3 kDa, consistent with the 25.4-kDa value calculated from the sequence. Moreover, Edman degradation showed this band to consist of only one protein with an N-terminal amino acid sequence exactly like that deduced from the gene: MRTALVTGSAK. Fast protein liquid chromatography (data not shown) gave a broad estimate of 50 to 90 kDa for the native protein; the high pI (>8.9) of this protein prevents quantitative electrophoresis by standard methods. The native enzyme is probably a dimer (like rat DHPR and several other members of the SDR family [15]), but the possibility of the occurrence of tetramers cannot be excluded.
FIG. 2.
Partial purification of DHTt after cold precipitation, as shown by SDS-PAGE. Lane 1, molecular weight (MW) markers (in thousands, indicated on the left); lane 2, E. coli (pTTDH) clarified crude extract; lane 3, cold precipitate resuspended in 25 mM Tris-HCl (pH 10).
Enzymatic specificity of the DHTt protein.
The enzyme (cold precipitate) exhibited no detectable activity with either 6-biopterin (the best substrate for Leishmania PTR1 [10]) or dl-6-methyl-7,8-dihydropterine (a substrate for E. coli DHPR [16]) under all conditions tested (pHs 5.5 to 8.5 with different buffers in the presence of salts) (Table 1). Activity could be detected only with qPtH2. The enzyme was active with NADPH. With NADH, the background oxidation was too elevated for the detection of activity (but see the discussion of the next experiment). In contrast to E. coli DHPR, which is a flavoprotein (16) and exhibits pteridine-independent oxidoreductase activity with potassium ferricyanide, the Thermus enzyme did not exhibit such activity (Table 1); it thus appears similar to mammalian DHPRs, which do not contain a flavin prosthetic group. When assayed under the same conditions, the activities of FH2 (DHFR) and qPtH2 (DHPR) in the presence of NADPH are present at an ∼1:5 ratio (Table 1).
TABLE 1.
Specific activity of DHPR from T. thermophilus with various substratesa
| Substrate | Sp act (U/mg of protein) |
|---|---|
| 6-Biopteridin | <0.05 |
| PtH2 | <0.05 |
| qPtH2 | 26.2 ± 2.7 |
| Potassium ferricyanide | <0.05 |
| FH2 | 5.2 ± 1.1 |
Activity was measured at 35°C in 50 mM potassium phosphate buffer (pH 7.5) in the presence of 0.1 mM NADPH and 0.1 mM pteridine.
Activity staining on native gel electrophoresis gels.
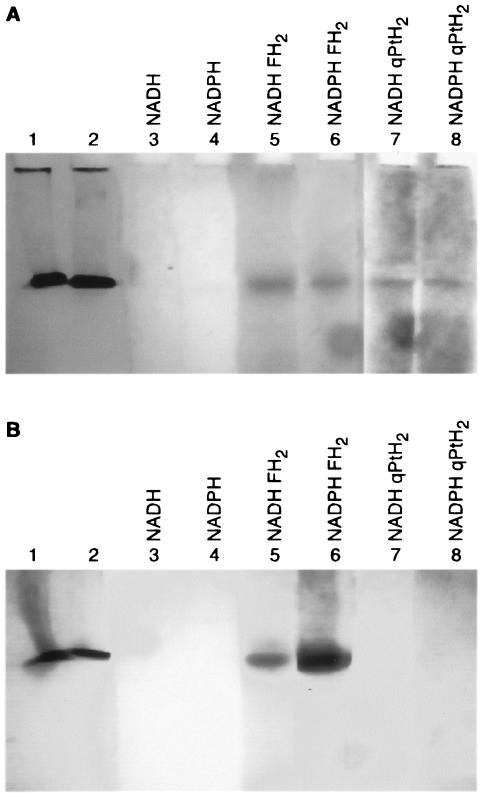
The activity staining method used 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyl tetrazolium bromide (MTT), which is reduced by the product of the enzymatic reaction to a formazan with a maximum λ of 560 nm. To allow the migration of the solubilized protein, basic gels were prepared (see Materials and Methods). Thermotoga maritima DHFR was used as a control, since it also has a high pI, has a rather similar Mr, and possesses DHFR activity but no DHPR activity. One gel was loaded with DHTt in lane 1 and with T. maritima DHFR in lanes 2 to 8; the second gel was loaded inversely. After the electrophoresis of both gels in parallel, gel pieces corresponding to the different lanes were cut (Fig. 3). For each gel, pieces 1 and 2 were stained with Coomassie blue and pieces 3 to 8 were incubated at 37°C in a solution containing 50 mM phosphate buffer (pH 7.0), 0.5 mM NADH or NADPH, and 5 mg of MTT/ml. In addition, incubation solutions for lanes 5 and 6 contained 0.5 mM FH2, but lanes 7 and 8 contained 0.5 mM qPtH2 (formed from 0.5 mM PtH4 and 0.5 mM 2,6-dichloroindophenol). After 30 min of incubation, an intensely stained band appeared for T. maritima DHFR (Fig. 3A) incubated with FH2 and NADPH (lane 6). Two hours later, a faint T. maritima DHFR band appeared on the gel pieces incubated with FH2 and NADH (lane 5). At that time, bands were also detectable on the gel pieces containing DHTt and incubated either with FH2 and NADH or NADPH (Fig. 3B, lanes 5 and 6) or with qPtH2 and NADH or NADPH (Fig. 3B, lanes 7 and 8). DHTt exhibited DHFR and DHPR activities with both NADH and NADPH, but no preference for one of the cofactors was detectable under these conditions.
FIG. 3.
Activity staining of DHTt (A) and T. maritima DHFR (B) on electrophoresis gels. After electrophoresis of the proteins (see the text), gel pieces were either stained with Coomassie blue (lanes 1 and 2) or incubated as described in the text and as indicated above the lanes in the presence of substrates and/or cofactors and 5 mg of MMT/ml.
DISCUSSION
The main points of this study can be summarized as follows. The T. thermophilus genome harbors a gene coding for a reductase (DHTt) which displays both DHPR and DHFR activities with NADH and NADPH as cofactors and possesses specific features of members of the SDR protein family. The DHFR activity of this reductase is considerable, i.e., about 20% of the DHPR activity detected with qPtH2 as the substrate. DHTt has no pteridine-independent oxidoreductase activity and is insensitive to concentrations of methotrexate and TMP that fully inhibit E. coli DHFR. Moreover, none of the clones that we could isolate from an apparently representative Thermus library expressed a true DHFR, and most significantly, no corresponding dyrA gene appears to exist in the T. thermophilus genome; indeed, BLAST probing of the complete genome (A. Henne, personal communication) did not reveal any sequence homologous to the corresponding genes from T. maritima and Deinococcus radiodurans (which is the closest known relative of Thermus [18]). It seems that the essential role of DHFR in cellular metabolism is challenged by our findings, since an unrelated dehydrogenase of the SDR family may fulfill the function classically assumed by a DHFR. It has already been reported that a number of bacteria and some archaea appear to lack a DHFR gene (9), but the substitute function had not yet been identified in any of these organisms. A bifunctional DHPR/DHFR such as DHTt is clearly a good candidate for the agent responsible for this substitute function. Moreover, just as thymidylate may be synthesized by a classical ThyA protein or by a quite unrelated ThyX flavoprotein (9), it appears that the concomitant reduction of FH2 or various pteridines may be carried out by an enzyme without a flavin prosthetic group (like DHTt or the nonquinonoid pteridine reductase PTR1 of Leishmania major [2]) or by an unrelated flavoprotein, as in E. coli. The evolutionary origin of these functional redundancies is an intriguing question; temperature by itself is not likely to be the discriminating evolutionary factor, since T. maritima DHFR and DHTt are both thermophilic enzymes.
The role of DHTt as dihydropteridine reductase in Thermus is not known. In particular, whether Thermus possesses aromatic amino acid hydroxylases using a tetrahydropterin as a cofactor (see the introduction and reference 21) remains to be investigated.
Acknowledgments
This work was supported by concerted actions of the Belgian State and the Free University of Brussels.
We thank J. Van Beeumen (University of Ghent) for DHPR N-terminal sequence determination and A. Henne for a personal communication. We also thank Jean-Pierre ten Have for the layout of the figures.
REFERENCES
- 1.Ahrweiler, P. M., and C. Frieden. 1988. Construction of a fol mutant strain of Escherichia coli for use in dihydrofolate reductase mutagenesis experiments. J. Bacteriol. 170:3301-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bello, A. R., B. Nare, D. Freedman, L. Hardy, and S. M. Beverley. 1994. PTR1: a reductase mediating salvage of oxidized pteridines and methotrexate resistance in the protozoan parasite Leishmania major. Proc. Natl. Acad. Sci. USA 91:11442-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Fraia, R., W. Wilquet, M. A. Ciardello, V. Carratore, A. Antignani, L. Camardella, N. Glansdorff, and G. di Prisco. 2000. NADP+-dependent glutamate dehydrogenase in the Antarctic psychrotolerant bacterium Psychrobacter sp. TAD1. Characterization, protein and DNA sequence, and relationship to other glutamate dehydrogenases. Eur. J. Biochem. 267:121-131. [DOI] [PubMed] [Google Scholar]
- 4.Hamm-Alvarez, S. F., A. Sancar, and K. V. Rajagopalan. 1990. The presence and distribution of reduced folates in Escherichia coli dihydrofolate reductase mutants. J. Biol. Chem. 265:9850-9856. [PubMed] [Google Scholar]
- 5.Hitchings, G. H., Jr. 1989. Nobel lecture in physiology or medicine, 1988: selective inhibitors of dihydrofolate reductase. In Vitro Cell. Dev. Biol. 25:303-310. [DOI] [PubMed] [Google Scholar]
- 6.Howell, E. E., P. G. Foster, and L. M. Foster. 1988. Construction of a dihydrofolate reductase-deficient mutant of Escherichia coli by gene replacement. J. Bacteriol. 170:3040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jörnvall, H., B. Persson, M. Krook, S. Atrian, R. Gonzàlez-Duarte, J. Jeffery, and D. Gosh. 1995. Short-chain dehydrogenases/reductases (SDR). Biochemistry 34:6003-6013. [DOI] [PubMed] [Google Scholar]
- 8.Lye, L.-F., M. L. Cunningham, and S. M. Beverley. 2002. Characterization of quinonoid-dihydropteridine reductase (QDPR) from the lower eukaryote Leishmania major. J. Biol. Chem. 277:38245-38253. [DOI] [PubMed] [Google Scholar]
- 9.Myllykallio, H., G. Lipowski, D. Leduc, J. Filee, P. Forterre, and U. Liebl. 2002. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 297:105-107. [DOI] [PubMed] [Google Scholar]
- 10.Nare, B., L. W. Hardy, and S. M. Beverley. 1997. The roles of pteridine reductase 1 and dihydrofolate reductase-thymidylate synthase in pteridine metabolism in the protozoan parasite Leishmania major. J. Biol. Chem. 272:13883-13891. [DOI] [PubMed] [Google Scholar]
- 11.Oshima, T., and K. Imahori. 1974. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Bacteriol. 24:102-112. [Google Scholar]
- 12.Persson, B., M. Krook, and H. Jörnvall. 1991. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur. J. Biochem. 200:537-543. [DOI] [PubMed] [Google Scholar]
- 13.Rood, J. I., A. J. Laird, and J. M. Williams. 1980. Cloning of the Escherichia coli K-12 dihydrofolate reductase gene following mu-mediated transposition. Gene 8:255-265. [DOI] [PubMed] [Google Scholar]
- 14.Shiman, R. 1984. Phenylalanine hydroxylase and dihydropteridin reductase, p. 179-249. In R. L. Blakley and S. J. Benkovic (ed.), Folates and pterins, vol. 2. John Wiley and Sons, New York, N.Y.
- 15.Varughese, K. I., M. M. Skinner, J. M. Whiteley, D. A. Matthews, and N. H. Xuong. 1992. Crystal structure of rat liver dihydropteridine reductase. Proc. Natl. Acad. Sci. USA 89:6080-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasudevan, S. G., D. C. Shaw, and W. L. F. Armarego. 1988. Dihydropteridine reductase from Escherichia coli. Biochem. J. 255:581-588. [PMC free article] [PubMed] [Google Scholar]
- 17.Vasudevan, S. G., B. Paal, and W. L. F. Armarego. 1992. Dihydropteridine reductase from Escherichia coli exhibits dihydrofolate reductase activity. Biol. Chem. Hoppe-Seyler 373:1067-1073. [DOI] [PubMed] [Google Scholar]
- 18.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilquet, V., J. A. Gaspar, M. Van De Lande, M. Van de Casteele, C. Legrain, E. M. Meiering, and N. Glansdorff. 1998. Purification and characterization of recombinant Thermotoga maritima dihydrofolate reductase. Eur. J. Biochem. 255:628-637. [DOI] [PubMed] [Google Scholar]
- 20.Xu, Y., G. Feller, C. Gerday, and N. Glansdorff. 2003. Moritella cold-active dihydrofolate reductase: are there natural limits to optimization of catalytic efficiency at low temperature? J. Bacteriol. 185:5519-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao, G., X. Tianhui, S. Jian, and R. A. Jensen. 1994. Pseudomonas aeruginosa possesses homologues of mammalian phenylalanine hydroxylase and 4α-carbinolamine dehydratase/DCoH as part of a three-component gene cluster. Proc. Natl. Acad. Sci. USA 91:1366-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]