Abstract
Nuclear localization influences the expression of certain genes. Chromosomal rearrangements can reposition genes in the nucleus and thus could impact the expression of genes far from chromosomal breakpoints. However, the extent to which chromosomal rearrangements influence nuclear organization and gene expression is poorly understood. We examined mouse progenitor B cell lymphomas with a common translocation, der(12)t(12;15), which fuses a gene-rich region of mouse chromosome12 (Mmu12) with a gene-poor region of mouse chromosome15 (Mmu15). We found that sequences 2.3 and 2.7 Mb on either side of the der(12)t(12;15) breakpoint had different nuclear positions measured relative to the nuclear radius. However, their positions were similar to the same loci on unrearranged chromosomes in the same tumor cells, suggesting that changes to nuclear position imposed by the der(12)t(12;15) translocation are constrained within ~2.5 Mb of the breakpoint. In addition, higher-order chromatin folding marked by three-dimensional gene clustering was not significantly altered for the 7 Mb of Mmu15 sequence distal to this translocation breakpoint. Translocation also did not correspond to significant changes in gene expression in this region. These data contrast with those of certain other chromosomal rearrangements and suggest that significant changes to Mmu15 sequence are structurally and functionally tolerated in the tumor cells examined.
INTRODUCTION
The three-dimensional (3D) organization of chromosomes in interphase mammalian nuclei plays a key role in gene expression (Cremer et al. 2001; Dekker 2008; Fraser et al. 2007; Parada et al. 2002a). An initial indication that chromosome 3D structure might be critically linked to gene function was the observation that chromosomes have non-random positions in the nucleus (Croft et al. 1999; Fraser et al. 2007; Khalil et al. 2007; Kosak et al. 2004; Parada et al. 2004; Parada et al. 2002). For example, gene-poor chromosomes are specifically enriched at the nuclear periphery of mouse and human lymphoblasts (Bolzer et al. 2005; Croft et al. 1999; Ferreira et al. 1997; Kupper et al. 2007). Within 3D chromosome “territories” (CTs), genes also are non-randomly organized, preferentially associating with other sequences in cis and in trans (Dietzel et al. 1999; Dostie et al. 2006; Mahy et al. 2002; Osborne et al. 2004; Shopland et al. 2006; Simonis et al. 2006; Tolhuis et al. 2002). Notably, gene expression can be influenced by these 3D interactions, as in the case of intra-chromosomal looping of long distance enhancers to gene promoters and of regulatory interactions among chromosomes (de Laat et al. 2007; Spilianakis et al. 2005; Wright et al. 2010). In addition, experimentally “tethering” chromatin arrays to the nuclear periphery of mammalian cells represses some genes (Finlan et al. 2008; Reddy et al. 2008). Thus, changes to chromosome sequence have the potential to impact chromosome activity by altering gene-gene interactions in cis and in trans or by relocating genes to new sub-nuclear compartments.
Chromosomal translocation results in the nuclear repositioning of some, but not all, chromosome regions (Barki-Celli et al. 2005; Cremer et al. 2003; Croft et al. 1999; Grasser et al. 2008; Murmann et al. 2005; Parada et al. 2002; Taslerova et al. 2006; Taslerova et al. 2003). Furthermore, correlative changes in nuclear position and gene expression recently have been reported for one chromosomal translocation (Harewood et al. 2010). However, the extent to which sequence rearrangements impact nuclear organization, and the resulting long-range effects on the expression of rearranged genes, are still poorly understood. To further establish the “rules” of the relationships among chromosome sequence rearrangement, gene positioning and gene expression in the nucleus, we evaluated a previously undescribed translocation involving gene-rich and gene-poor chromosomal regions on mouse chromosomes 12 and 15 (Mmu12 and Mmu15), respectively.
Mice deficient for both Trp53 and one of two non-homologous end joining pathway proteins, Ligase IV (Lig4) or Artemis (Dclre1c), invariably develop progenitor B (pro-B) cell lymophoma (Donehower et al. 1992; Frank et al. 2000; Mills et al. 2003; Rooney et al. 2004). These Lig4;Trp53 (LP) and Artemis;Trp53 (AP) tumor cells frequently contain rearrangements between the immunoglobulin heavy chain locus (Igh) in a gene-rich region of Mmu12 and the Myc oncogene in a gene-poor region of Mmu15, resulting in two derivative chromosomes: der(12)t(12;15) (hereafter called t(12;15)) and a complex chromosomal rearrangement with amplification of Myc and Igh, referred to as a complicon (Fig. 1a) (Mills et al. 2003; Rooney et al. 2004). The Myc complicon is likely the driver of tumorigenesis in this model, in fact a subset of tumors lack t(12;15), indicating that this chromosome is not required for lymphomagenesis (Mills et al. 2003; Rooney et al. 2004; Woo et al. 2007; Zhu et al. 2002) Thus, t(12;15) provides a convenient model system in which to study the effects of translocation on higher-order chromosome structure and function without the imposition of selective pressures during tumorigenesis. For this study of t(12;15), we focused on the 7 Mb Mmu15 region distal to the translocation breakpoint. This gene-poor Mmu15 region (5 genes/Mb) resembles another chromosomal region on Mmu14 previously shown to form nuclear “gene hubs” with 3D clusters of genes spanning several megabases (Shopland et al. 2006). Our findings with t(12;15) suggest that, unlike other chromosomal regions, the distal portion of Mmu15 can tolerate significant changes to underlying chromosomal sequence with minimal impact on its nuclear organization and gene expression.
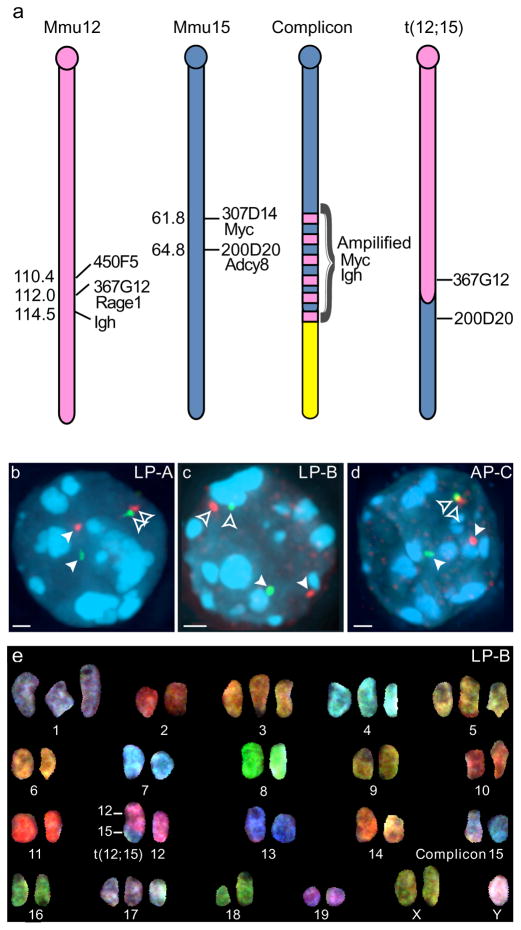
Fig. 1. Three pro-B lymphomas contain one normal Mmu15 and one t(12;15).
(a) Schematic diagrams of Mmu12 and Mmu15 and derivative chromosomes present in LP-A, LP-B, and AP-C tumors. Each cell contains one normal chromosome plus a derivative Mmu15 with complex amplification (complicon) of Myc and Igh loci, and a derivative Mmu12 [der(12)t(12;15) or t(12;15)] with a translocation junction < 2 Mb proximal to the Igh locus on Mmu12 and < 500 kb distal to Myc on Mmu15. Names and positions of BACs used throughout the study are indicated to the right of diagrams and Mb positions (mouse genome build 37 (July 2007)) are to the left. (b–d) Interphase FISH with either Mmu12 BAC 367G12 (green, b & c) or BAC 450F5 (green, d) and Mmu15 BAC 200D20 (red) reveal single, separate foci for each of the probes on intact Mmu12 and Mmu15 (filled arrowheads), and a pair of single, closely positioned foci on t(12;15) (open arrowheads) in (b) LP-A, (c) LP-B, and (d) AP-C tumor nuclei (DAPI, blue). Maximum projections of deconvolved images are shown. Bars, 1.0 μm. (e) Spectral karyotyping confirmed one intact Mmu15 and one t(12;15) in LP-B.
RESULTS
Nuclear positions of Mmu12 and Mmu15 sequences unaltered by translocation
In t(12;15), gene-poor Mmu15 sequences are placed adjacent to a very gene-rich Mmu12 region (5 vs 27 genes/Mb, 7 Mb window) (Fig. 1a). Therefore, we determined whether t(12;15) causes the nuclear repositioning of genes in these flanking regions. We isolated three tumors for analysis, two from LP mice (here called LP-A and LP-B) and one from AP mice (called AP-C). To confirm the presence of t(12;15) in these samples, interphase tumor cells were probed by two-color fluorescence in situ hybridization (FISH) with bacterial artificial chromosomes (BACs), one from Mmu12 and one from Mmu15 (Fig. 1a–d). In all tumor samples, nuclei contained spatially separate signals for each probe, representing unrearranged Mmu15 and Mmu12, as well as a single set of spatially paired signals, corresponding to t(12;15) (Fig. 1b–d). Spectral karyotyping (SKY) and/or whole chromosome painting in interphase preparations verified that the chromosomes detected in interphase nuclei correspond to Mmu12, Mmu15 and t(12;15) (Fig. 1e and Online Resource 1).
To determine whether t(12;15) chromatin repositions in the nuclei of these tumor samples, we localized Mmu12 and Mmu15 sequences 2.3 and 2.7 Mb on each side of the translocation breakpoint (Fig. 1a–e). These probes include the genes Rage1 and Adcy8, respectively. Paired Mmu12 and Mmu15 FISH signals were classified as parts of t(12;15) if the signals were within 1.5 μm of each other and the nucleus also contained an additional unpaired signal of each probe, indicating the unrearranged Mmu12 and Mmu15 (Fig. 1b–d). Signals from both t(12;15) and unrearranged chromosomes were mapped relative to the nuclear radius in these spherical tumor nuclei. The nuclear edge was delineated by lamin B1 and nuclear pore immunostaining as well as DAPI counterstaining. Distances between the centers of BAC signals and the nuclear center were measured in 3D image stacks and were normalized to the entire nuclear radius (Fig. 2a).
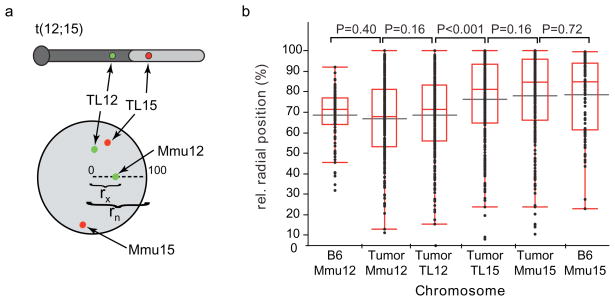
Fig. 2. Nuclear positions of t(12;15) loci are similar to those on unrearranged chromosomes.
(a) Relative radial positions of single BACs from Mmu12 and Mmu15 were determined by measuring the distance from the nuclear center to the locus (rx) and normalizing to the length of the total nuclear radius (rn), as depicted. Thus 0 represents the center of the nucleus and 100 the outer nuclear edge. Relative radial positions were measured for Mmu12 sequences in BAC 367G12 and Mmu15 sequences in BAC 200D20 on both unrearranged chromosomes (Mmu12 and Mmu15) and t(12;15) (TL12 and TL15). BACs were co-hybridized to the same cells by two color FISH (Fig. 1b–d). t(12;15) was discriminated from un-rearranged chromosomes by the close positioning of the two BACs (< 1.5 μm). (b) Cumulative distributions of relative radial positions of indicated loci in all three tumors (LP-A (N = 85), LP-B (N = 87), AP-C (N = 96)) and normal B6 Pro-B cells (N = 96). Horizontal grey bars indicate means, boxes represent median and quartile values, and whiskers indicate the remaining points in the distribution. Indicated P-values were derived from Mann-Whitney U tests.
Radial position measurements of the unrearranged chromosomes in LP-A, LP-B, and AP-C nuclei indicated that the Mmu12 sequence was more centrally positioned than the Mmu15 sequence (P < 0.001, Mann Whitney U-tests; Fig. 2b, Online Resource 2). This difference also was detected in normal pro-B cells from C57BL/6J (B6) mice, suggesting that these loci are infrequently near each other prior to translocation (P < 0.001, Mann Whitney U-test, Fig. 2b). Measured positions were similar in all three tumor samples and were unaffected by methods of cell preparation (Online Resource 2; Materials and Methods). For the t(12;15) in tumor cells, radial position measurements indicated that the Mmu12 and Mmu15 sequences (TL12 and TL15, respectively) positioned similarly to the same loci on unrearranged chromosomes (P > 0.16, Fig. 2b, Online Resource 2), but were different from one another (P < 0.001, Fig. 2 and Online Resource 2), with one exception (TL12 in APC, P ≥ 0.22, Online Resource 2). These data suggest that translocation did not specifically change the nuclear radial position of the probed Mmu12 and Mmu15 loci. Since the Mmu12 and Mmu15 probes are connected by the t(12;15) translocation junction, these findings also suggest that any nuclear repositioning due to this translocation is restricted to sequences within ~2.5 Mb of the breakpoint.
Higher order chromosome folding is insulated from translocation
We next tested whether or not translocation alters higher-order folding marked by the 3D association of genes on t(12;15). The derivative t(12;15) has lost Myc, a powerful oncogene that physically associates with other sequences in the nucleus (Osborne et al. 2007; Wright et al. 2010). In addition, the 7 Mb of Mmu15 sequence immediately distal to the translocation breakpoint (and Myc on intact Mmu15) has genes that are arranged into clusters separated by gene “deserts” (Fig. 3a), similar to Mmu14 sequences previously found to form higher-order nuclear gene hubs (Shopland et al. 2006). The region thus provides a convenient system to determine the effects of translocation and Myc loss on chromosome higher-order folding and the 3D association of multiple, linked genes.
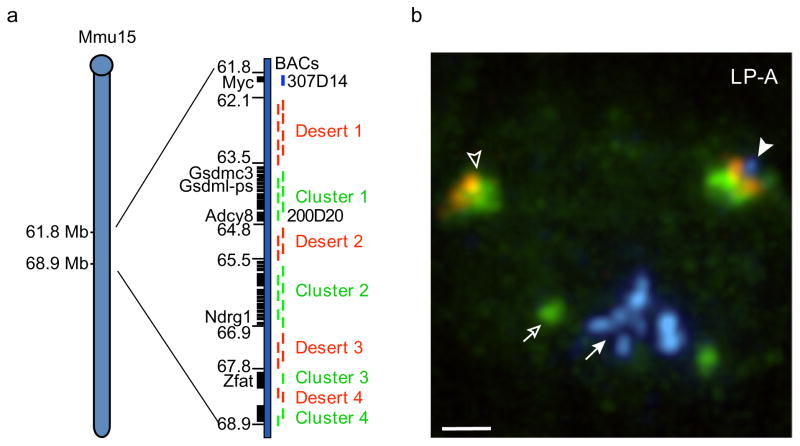
Fig. 3. Scheme for probing higher-order folding distal to the t(12;15) breakpoint.
(a) The Mmu15 region immediately distal to Myc (61.6–68.9 Mb) contains four clusters of genes (black rectangles) separated by large (> 500 kb) gene “deserts” (right). Select genes and Mb positions are indicated to the left. BACs from gene clusters (green bars, right) were pooled and nick translated with digoxigenin-dUTP, whereas pooled desert BACs (red bars, right) were labeled with Cy3-dUTP. To discriminate intact Mmu15 from t(12;15), Myc BAC 307D14 was labeled with Cy5-dUTP (blue). (b) LP-A nucleus probed by DNA-FISH using Mmu15 region gene cluster (green), desert (red), and Myc (blue) probes indicated the presence of one Mmu15 region (with Myc signal, filled arrowhead), one t(12;15) (without Myc signal, open arrowhead), and amplified copies of Myc on a separate, derivative complicon (filled arrow). Additional BAC probe for Agrn 139J18 (green, open arrow), an early replicating gene, was used to identify G1 cells based on the appearance of a singlet rather than doublet FISH signal. A maximum projection of a deconvolved image stack is shown with counter stain omitted. Bar, 1.0 μm.
To differentially label the Mmu15 gene clusters and gene deserts, we generated two pools from 34 BAC probes spanning this 7 Mb region, one corresponding to gene cluster sequences and the other to gene deserts (Fig. 3a, Online Resource 3). Each pool was labeled with a different fluorochrome. An additional BAC corresponding to Myc was labeled with a third fluorochrome (Fig. 3a). Myc is present on unrearranged Mmu15 but missing from t(12;15) and thus allows one to discriminate between the two chromosomes. This FISH labeling scheme detected a Myc-containing Mmu15 (Fig. 3b, filled arrowhead), a Myc-lacking t(12;15) chromosome (Fig. 3b, open arrowhead), and amplified copies of Myc on the complicon chromosome (Fig. 3b, filled arrows) in intact interphase cells from each tumor. A probe corresponding to the early replicating gene, Agrn, on Mmu4 also was included to identify and exclude nuclei that had entered S-phase (Fig. 3b, open arrow) (Shopland et al. 2006).
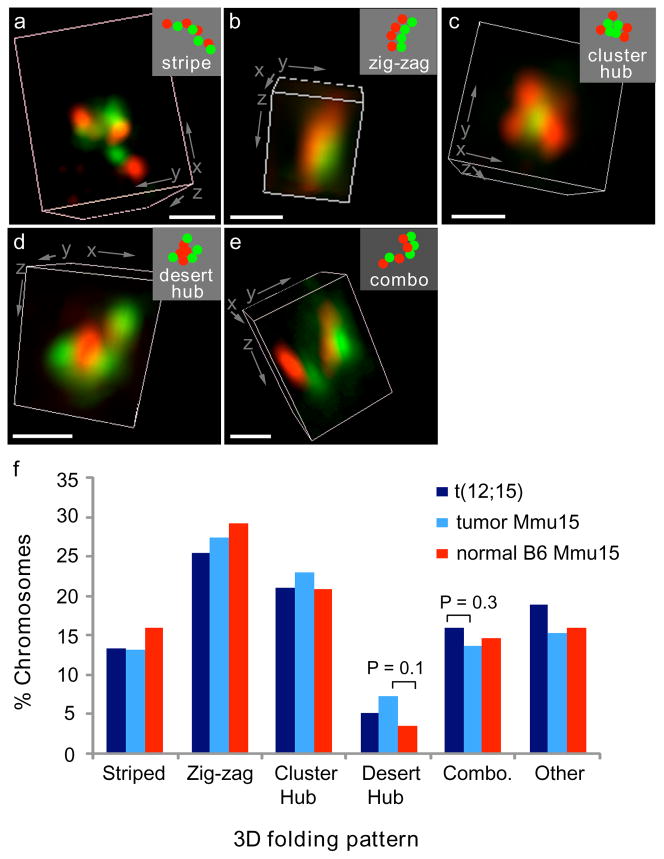
Patterns of 3D higher order folding for translocated and unrearranged Mmu15 were scored according to the strict criteria used to classify 3D folding patterns of the Mmu14 region analyzed previously (Materials and Methods, (Shopland et al. 2006)). Chromosomes with individual gene cluster and desert signals alternated in a linear arrangement were categorized as “striped fibers” (Fig. 4a). Chromosomes with gene deserts aligned parallel to gene clusters were scored as “zig-zags” to reflect the jogging back and forth of the underlying DNA sequence (Fig. 4b). Tight groupings of gene clusters in a central location surrounded by gene deserts were categorized as “gene cluster hubs” (Fig. 4c). Similarly, tight groupings of central gene deserts were scored as “gene desert hubs” (Fig. 4d). Finally, some chromosomes were classified as “combinations” because they displayed two patterns, for example half of the clusters and deserts were in the striped fiber pattern and the other half in a zig-zag (Fig. 4e).
Fig. 4. Patterns of higher-order chromatin folding are not significantly altered by translocation or tumorigenesis.
(a–e) The 7 Mb of Mmu15 distal to Myc and/or the t(12;15) breakpoint were labeled by FISH as in Fig. 3. The relative positions of its gene clusters (green) and deserts (red) were classified as one of five morphological patterns: (a) striped fiber, (b) zigzags, (c) gene cluster hub, (d) gene desert hub, or (e) a combination (combo) of two of the other four patterns. 3D renderings of deconvolved images are shown. Insets (top right) are schematic diagrams of probe patterns. Bars, 500 nm. (f) Scoring the frequency of 3D folding patterns of normal Mmu15 (N = 234) and t(12;15) (N = 232) from the three tumors analyzed indicated no significant difference between the translocated and normal chromosomes (P ≥ 0.3, χ2 tests). Normal pro-B cells from B6 mice (N = 144) also had a similar distribution of folding patterns (P ≥ 0.1, χ2 tests).
In cells from all three tumors, the unrearranged Mmu15 sequences formed into all four of the 3D folding patterns previously detected with Mmu14 (Fig. 4f, Online Resource 4). Furthermore, gene cluster hubs were more frequently noted than gene desert hubs, demonstrating the non-random organization of these structures, also similar to Mmu14 (Shopland et al. 2006). When the translocated and non-translocated chromosomes were compared, the distributions of 3D patterns were not statistically different (P > 0.3, χ2 test). Thus, translocation resulting in t(12;15) did not significantly alter the 3D arrangements of gene clusters and deserts, despite the loss of Myc and its replacement by gene-rich Mmu12 sequence.
We also examined whether the 3D folding patterns for Mmu15 were generally disrupted in tumor cells compared to normal pro-B cells. No significant differences in chromatin folding were found in tumor cells versus normal cells (P > 0.1, χ2 test; Fig. 4f). In addition, for all cell types examined, specific folding patterns were not consistently enriched in either the nuclear interior or at the nuclear periphery (P > 0.3, χ2 tests; Online Resource 5). Together the data strongly suggest that neither translocation nor other changes of tumorigenesis altered the higher order folding patterns of the Mmu15 region under study.
Translocation-independent gene expression of Mmu15 sequences
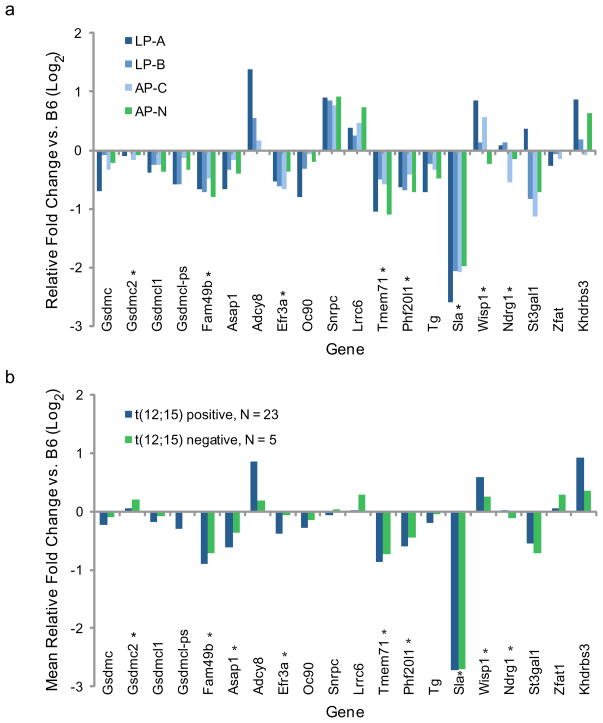
While nuclear compartmentalization (radial positioning) and the 3D clustering of genes are known to impact gene expression, there are several other features of chromatin higher-order structure that we did not assay (e.g. heterochromatin spreading) that could be altered by translocation (de Laat et al. 2007; Finlan et al. 2008; Reddy et al. 2008; Spilianakis et al. 2005). To determine whether the t(12;15) translocation had any long-range impacts on gene expression from these alternative mechanisms, we used microarray analysis to measure mRNA levels of the Mmu15 genes in the 7 Mb region immediately distal to the breakpoint. For this analysis we also analyzed an additional tumor, AP-N, that did not contain a t(12;15) (Online Resource 1) (Woo et al. 2007). AP-N thus provided a baseline measurement of gene expression of the Mmu15 genes in a similar tumor background, but without the t(12;15). Similar expression profiles were found in the t(12;15)–containing and –lacking tumors (Fig. 5a). Indeed, only one gene (Sla) had a consistent 2-fold change in expression relative to normal pro-B cells from B6 mice, and this change also was detected in the t(12;15)-lacking AP-N tumor. Similar expression profiles also were found for additional sets of t(12;15)-containing and -lacking tumors that were not examined cytologically (Fig. 5b, N= 23 and 5, respectively). These data strongly suggest that the t(12;15) translocation does not have a widespread affect on the expression of Mmu15 genes distal to the breakpoint.
Fig. 5. Similar gene expression profiles for 7 Mb Mmu15 region in tumors with and without t(12;15).
(a) Microarray analyses of gene expression in three t(12;15)-containing tumors (LP-A, LP-B, and AP-C) relative to normal pro-B cells from B6 mice show transcript profiles similar to a t(12;15)-lacking tumor (AP-N) for genes in the 7Mb Mmu15 region distal to the t(12;15) breakpoint. Genes are ordered according to Mmu15 sequence, from 63.5 Mb (left) to 68.9 Mb (right). (b) Microarray analysis of gene expression for 23 additional pro-B lymphomas with t(12;15) chromosomes and 5 lymphomas without. Mean fold change relative to normal pro-B cells from 5 age and sex matched B6 controls is shown. Asterisks indicate probe sets with Fs P values < 0.01.
DISCUSSION
Here we show that rearrangement of Mmu15 sequence by translocation does not necessarily have long-range effects on chromosome nuclear structure or gene expression. Potential long-range effects on gene expression would most likely be mediated by changes to higher-order chromatin structure in the nucleus. However, in three pro-B lymphomas we found little change in the radial positioning of sequences 2.3 and 2.7 Mb from the t(12;15) breakpoint, suggesting that any changes to radial positioning are constrained to ~2.5 Mb from the translocation junction. Patterns of higher-order 3D folding also were unaffected in the 7 Mb region of Mmu15 distal to the breakpoint. These findings suggest that the frequency of gene-gene interactions within this region was not substantially impacted by loss of the master regulator, Myc, or juxtaposition of gene-rich Mmu12 chromatin. Finally, expression of the genes in this 7 Mb region was unaltered by other potential changes to chromatin structure. Together, our data suggest that significant changes to chromosome sequence can be structurally and functionally tolerated by distal Mmu15.
Limited changes to radial positioning on t(12;15)
The lack of t(12;15) nuclear repositioning that we detected is consistent with reports of some translocations (Cremer et al. 2003; Croft et al. 1999; Grasser et al. 2008; Taslerova et al. 2006). However, other rearrangements do result in nuclear repositioning, suggesting that certain chromosomes are “dominant” over others in terms of nuclear positioning (Barki-Celli et al. 2005; Cremer et al. 2003; Murmann et al. 2005; Taslerova et al. 2006; Taslerova et al. 2003). These dominating influences could be related to the amount of sequence contributed by each translocation partner or by differences in their gene densities (Croft et al. 1999; Federico et al. 2008; Kupper et al. 2007; Murmann et al. 2005). For t(12;15), approximately 100 Mb of Mmu12 was fused to 40 Mb of Mmu15. Our findings suggest that neither size nor gene density had a substantial impact on the position of the translocation partner, but, rather, they had a limited “bystander effect” of < 2.5 Mb (Morey et al. 2009). As an alternative to size and gene density, specific sequences lost upon translocation might be key for maintaining nuclear positions. For t(12;15), these lost sequences include the Mmu12 locus, Igh, and the Mmu15 locus, Myc. Myc has been found to associate with other seuqences in the nucleus and with “transcription factories” and thus might influence the nuclear positioning of adjacent Mmu15 genes (Osborne et al. 2007; Wright et al. 2010). In addition, specific sequences required for normal Igh positioning have been identified, and these are lost in t(12;15) (Kosak et al. 2002; Yang et al. 2005). Our studies of t(12;15) argue that neither of these key loci are required to maintain the relative positioning of adjacent sequences on their respective chromosomes.
Translocation independent higher-order folding of Mmu15 chromatin
Gene-gene associations in the nucleus can influence gene expression, and the genes that contact each other the most are genes closely linked on the same chromosome (Lieberman-Aiden et al. 2009; Simonis et al. 2006). Thus, translocation has the potential to disrupt functional gene-gene interactions. We previously found that a gene-poor region on Mmu14 folds into predominant 3D structures defined by the degree of aggregation of genes across the region (Shopland et al. 2006). Here we show similar structures for the 7 Mb gene-poor region of Mmu15 adjacent to the t(12;15) breakpoint. These findings also indicate that the nuclear aggregation of these genes did not depend on the master regulatory gene, Myc, or its adjacent gene desert sequences, even though Myc was spatially associated with the gene clusters and deserts in this region on at least half of unrearranged chromosomes (data not shown). Moreover, we also find that the gene-rich Mmu12 sequences in t(12;15) do not interfere with the formation of these 3D structures, further suggesting that the Mmu15 region distal to Myc is structurally insulated from adjacent chromatin. Together with our previous discovery that similar 3D structures formed by completely different genes on Mmu14, our findings of Mmu15 suggest that the marked 3D aggregation of multiple genes is not due to the specific identity of the genes, but it is consistent with the presence of gene deserts in gene-poor genomic regions.
Chromosome specific long-range effects of translocation on gene expression
Changes in the nuclear position of a gene have the potential to change gene expression, either by altering functional associations with a specific nuclear compartment or by altering the set of other sequences with which a gene can interact (de Laat et al. 2007; Dekker 2008; Fraser et al. 2007). Correlative changes in both nuclear position and gene expression have been reported recently for one human translocation (Harewood et al. 2010). In contrast, we did not find significant positional changes or gene expression changes for t(12;15) in mouse lymphomas. Thus, only certain genes may be impacted by chromosomal rearrangement. The data presented here indicate that the distal portion of Mmu15 is relatively robust in maintaining its nuclear structure and function. Thus, our findings suggest that not all translocations are detrimental or confer a selective advantage during tumorigenesis. However, other translocations have exhibited changes in gene expression and thus have the potential to impact cell state. Now the challenge is to identify which nuclear architecture changes impact tumorigenesis.
MATERIALS AND METHODS
Mouse models
Mice heterozygous for a null allele of Ligase IV (Lig4N/+; (Frank et al. 1998)) were crossed with p53 heterozygous mice (Trp53N/+; (Donehower et al. 1992)) to generate doubly deficient mice. These mice were then intercrossed to produce Lig4N/N; Trp53N/N (LP) mice. A similar breeding scheme was applied to generate Dclre1cN/N (Rooney et al. 2004); Trp53 N/N (AP) mice. Each mutation had been previously backcrossed to C57BL/6J (B6) (JAX Mice and Services) four to six generations, generating ~95% B6 genetic content. Mice were regularly monitored and euthanized at the time of overt tumor onset in accordance with Institutional Animal Care and Use Committee guidelines.
Cell isolation and preparation
Lymphoma cells were isolated from two LP mice, LP3026 (LP-A) and LP3438 (LP-B), and two AP mice, AP3450 (AP-C) and AP1899 (AP-N)(Woo et al. 2007), at the time of overt tumor onset. Cervical lymph node tumors were removed from the surrounding tissue. For RNA extraction, a portion of each tumor was immediately immersed in RNALater (Ambion), stored at room temperature for 1 day and then frozen at −80 °C. For DNA extraction, a portion of the tumor along with mouse tail samples were immediately frozen on dry ice and stored at −80 °C. The remaining tumor tissue for each mouse was then placed in 5 ml warm RPMI (Hyclone RPMI-1640) containing 15% fetal bovine serum (FBS, Lonza BioWhittaker), 200 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, Sigma), 2 mM L-glutamine (Invitrogen), 1 mM sodium pyruvate (Stem Cell Technologies), 0.1 mM minimal essential media (MEM) non-essential amino acid solution (Stem Cell Technologies), 1x penicillin/streptomycin (Sigma) and 0.007 mM 2-mercaptoethanol (Sigma) (supplemented RPMI) and physically dispersed into single cell suspension using a polished glass rod and passing the cells through a 70 μm cell strainer (BD Falcon). After washing, tumor cells were resuspended in supplemented RPMI at approximately 1×106 cells per ml. For tumors LP-A, LP-B and AP-N, a portion of the tumor cells were incubated with 0.05 μg/ml KaryoMAX® Colcemid™ solution (Invitrogen) for 3–5 hours at 37°C, swollen in 0.075 M KCl for 15–20 minutes and subsequently fixed in a 3:1 methanol: glacial acetic acid solution on ice. These cells were then dropped onto glass slides to make metaphase chromosome spreads. For tumors LP-A, LP-B, and AP-N, aliquots of viable tumor cells were resuspended in supplemented RPMI, 10% dimethyl sulfoxide (Sigma) and frozen in liquid nitrogen for later use.
For interphase analyses, cells from tumors LP-A and LP-B were processed as follows: Frozen cells were thawed quickly at 37 °C, washed, and resuspended at a concentration of approximately 1×106 cells/ml. Tumor cells were then allowed to recover and grow for 1 to 1.75 hours at 37°C, 5% CO2 and attach to poly-L-lysine (0.01%, Sigma) coated coverslips. This allowed sufficient time for the tumor cells to reconstitute a spherical morphology and actively attach to coverslips with kinetics that were similar to freshly isolated tumors. Cells were then permeabilized for 5 minutes in freshly made cytoskeletal (CSK) buffer containing 10mM ribonucleoside vanadyl complex (New England Biolabs) and 1.2 mM phenylmethylsulfonyl fluoride (PMSF, Sigma) on ice and then fixed in 4% formaldehyde (Ted Pella)/PBS at room temperature as previously described (Shopland et al. 2006; Tam et al. 2002). Cells from tumor AP-C were used without intermediary freezing steps and allowed to settle onto poly-L-lysine coated coverslips as above. Tumor cells from AP-C were then fixed in 4% formaldehyde, permeabilized with 0.5% polyethylene glycol p-tert-octylphenyl ether (Triton X-100, Fisher Scientific), and then repeatedly frozen in liquid nitrogen as described (Solovei et al. 2002). These two cell preparation methods produced no differences in radial positioning or 3D folding of unrearranged chromosomes indicating they are comparable for the purposes of this study (Fig. 4, Online Resources 2 & 4). Normal pro-B cells (IgM−, B220+) were isolated from B6 mice between 8 and 14 weeks of age using MACS® (Miltenyi Biotec) magnetic bead cell separation according to the manufacturer’s protocol (Miltenyi Biotech). Briefly, cells were labeled with biotin conjugated antibodies against specific cell surface antigens, incubated with streptavidin conjugated magnetic MicroBeads (Miltenyi Biotec) and passed through a separation column under high magnetic fields to enrich or deplete for the target cell population. Total bone marrow was first depleted of IgM positive cells (R6-60.2BD, IgM-biotin, Pharmingen™), then enriched for CD45R/B220 positive cells (RA3-6B2, CD45R/B220-biotin, BD Pharmingen™) and resuspended in supplemented RPMI. Pro-B cells were allowed to attach to coverslips and then were fixed in the same manner as tumors LP-A and LP-B (Tam et al. 2002).
Spectral karyotyping
Spectral karyotyping (SKY) was performed as previously described (Woo et al. 2007; Wright et al. 2009). After hybridization with SKY paints (Applied Spectral Imaging), DNA was counterstained with 1 μg/ml of DAPI, and imaged using an ASI SKY Workstation equipped with HiSKY software (Applied Spectral Imaging).
Interphase fluorescence in situ hybridization and immunofluorescence
All BACs used for FISH probes were obtained from the RPCI-23 library (CHORI). Probes were generated by nick-translation with digoxigenin-16 dUTP (Roche), Cy3-dUTP or Cy5-dUTP (GE). For DNA FISH, fixed cells were base-hydrolyzed, heat denatured, and pre-hybridized with 40 μg salmon testes DNA (Sigma-Aldrich) and tRNA (Sigma) for 1 hour at 37°C, washed, and then incubated with either 50 ng of each BAC probe for single BACs or 200ng for pools as well as 10 μg each of mouse Cot-1 DNA (Invitrogen), salmon testes DNA, and tRNA as described (Luo et al. 2009; Shopland et al. 2006; Tam et al. 2002). Digoxigenin-labeled probes were detected with 1:250 FITC anti-digoxigenin antibodies (Roche) in 4xSSC/1% bovine serum albumin (BSA, Roche) at 37°C for 1 hour. For hybridization with Mmu15 paint (Applied Spectral Imaging), the amount of Cot-1 DNA, salmon testes DNA and tRNA were doubled in the probe mix. Where indicated, hybridized cells were post-fixed in 4% formaldehyde/PBS and immunostained with a 1:250 dilution of goat anti-lamin B (M20, Santa Cruz Biotechnology) and 1:100 dilution of mouse anti-Mab414 (ab24609, Abcam) antibodies in 1xPBS/1% BSA for 2 hours at 37°C. Primary antibodies were then detected with 1:250 AlexaFluor 488-donkey anti-goat antibody (Invitrogen) and 1:250 FITC-donkey anti-mouse antibody (715-096-150, Jackson ImmunoResearch) in 1xPBS/1% BSA. Cells were then counter stained with 1 μg/ml DAPI (Sigma-Aldrich) and mounted in vectashield (Vector Laboratories) or 5 mM 1,4-phenylaminediamine hydrochloride (Sigma-Aldrich), 90% glycerol, 1xPBS.
Microscopy and image analysis
Images were obtained using either using a Zeiss Axiovert 200M fluorescence microscope equipped with a 100x, 1.4 NA oil objective lens and an Orca ER cooled CCD camera (Hamamatsu) or a Zeiss Axioplan2 fluorescence microscope similarly equipped with a 100x, 1.4 NA oil objective lens and CoolSnapHQ CCD camera (Photmetrics), both using Axiovision 4.6 software (Carl Zeiss). Image stacks were taken at 0.2 μm intervals, deconvolved and rendered in 3D using Autodeblur software (Media Cybernetics, Inc.) or Imaris software (Bitplane, version 7.0.0).
Chromosomes were classified for 3D folding patterns as described (Shopland et al. 2006). Briefly, for striped fibers, at least 6 clearly alternating cluster-desert signals were detected along a linear fiber; for zig-zags, at least 3 gene clusters aligned parallel to 3 gene deserts; for gene cluster hubs, at least 3 gene signals grouped together in a core with at least 3 gene desert signals surrounding the core at >180°; for gene desert hubs, gene deserts formed the core with surrounding gene clusters as above; for combo, 50% of one pattern and 50% of another pattern (e.g. stripe plus zig-zag) were detected; and for other, patterns that did not fit into any of the above categories. An additional probe for Agrn, BAC 139J18, an early replicating gene, was used to identify cells that had entered S phase based on the appearance of a doublet FISH signal (Shopland et al. 2006). Cells with Agrn doublets were excluded from 3D folding analysis. Co-localization with the nuclear periphery was determined by the close association (≤5% of the relative nuclear radius or ≤ 200 nm) of any probe within the 7 Mb region with the edge of the DAPI counterstain. Distributions of folding patterns were evaluated using χ2 tests (Miller et al. 1965).
Nuclear radial positions of single BAC probe signals were determined in deconvolved, 3D reconstructed images rendered in Imaris (Bitplane, version 7.0.0). Relative radial positions were determined by measuring the distance from the geometric center of the spherical nuclei (based on DAPI counterstain, lamin B1, and nuclear pore immunostaining) to the intensity weighted center of the FISH probe signal and the nuclear radius through the signal using the spot and measurement functions in Imaris.
Gene expression microarray analysis
Primary tumor tissue in RNAlater (Ambion) was lysed in TRIzol™ (Invitrogen) and total RNA isolated according to manufacturer’s directions. B6 pro-B cells were sorted using antibodies directed against IgM (low) and B220 (high) cell surface antigens with a fluorescence activated cell sorter (FACSVantageSE/DiVa, BD Biosciences) equipped with FACSDiVa software into RLT buffer (Qiagen). For this analysis, five independent preparations of pro-B cells from total bone marrow of at least two age and sex matched B6 mice were used. Total RNA was extracted from these cells using RNeasy® Micro kit (Qiagen) according to the manufacturer’s protocol. RNA quality was assessed using an Agilent 2100 Bioanalyzer instrument and RNA 6000 Nano LabChip assay (Agilent Technologies). For both tumor and normal cells, RNAs were reverse transcribed with random T7 primers (Affymetrix). Double stranded cDNA was synthesized with dUTP incorporation using the GeneChip® WT cDNA Synthesis and Amplification Kit (Affymetrix). cDNAs were fragmented and labeled with biotin using terminal deoxynucleotidyl transferase (Affymetrix Labeling Reagent) according to manufacturer’s directions. Ten μg of biotin-labeled cDNA was then hybridized to either GeneChip® Mouse Gene 1.0 ST Arrays (Affymetrix) or to MOE430v2.0 GeneChip® arrays (Affymetrix) for 16 hours at 45 °C. Post hybridization staining and washing were performed according to manufacturer’s protocols using the Fluidics Station 450 instrument (Affymetrix). Arrays were scanned with a GeneChip® Scanner 3000 laser confocal slide scanner and quantified using the GeneChip® Operating Software (GCOS) v1.2.
Microarray data were filtered using the “Mouse 430 to Mouse Exon 1.0 ST Best Match” data set (Affymetrix). Hybridization values were normalized and corrected using the quantiles normalization package, limma, in R (http://www.bioconductor.org/packages) and analyzed by ANOVA using R/maanova statistical tests (http://www.r-project.org, (Churchill 2004)). Relative fold change was calculated relative to the average of 5 independent biological replicates of B6 pro-B cells from two age- and sex-matched mice.
Supplementary Material
(a–c) Interphase nuclei were evaluated by FISH with a Mmu15 whole chromosome paint (green) plus three BACs across Mmu15 (BACs 285N22, 43.5 Mb, red; 187M22, 62.1 Mb, blue; and 438G15, 73.6 Mb, red), as diagrammed in (a). 3D images reveal the presence of an intact Mmu15 (filled arrowhead), a t(12;15) containing only distal Mmu15 sequences (open arrowhead) and the complicon, which contains the proximal portion of Mmu15 (open arrow), in the (b) LP-A and (c) AP-C cells shown. 3D renderings of deconvolved 3D image stacks are shown. Bars, 1.5μm. (d) SKY of LP-A cells confirms the presence of one normal Mmu15, one t(12;15) and one complicon, all containing Mmu15 sequences as seen in (b). (e) SKY of AP-N cells confirmed the absence of t(12;15), the presence of normal copy numbers of Mmu15, and a different translocation involving Mmu12.
Relative radial positions of single BACs from Mmu12 and Mmu15 were determined by measuring the distance from the nuclear center to the locus (rx) and normalizing to the length of the total nuclear radius (rn), as depicted, where 0 represents the center of the nucleus and 100 the outer nuclear edge. Mmu12 BAC 367G12 and Mmu15 BAC 200D20 were differentially labeled and co-hybridized to the same cells by two color FISH. These loci on t(12;15) (TL12 and TL15) were discriminated from un-rearranged chromosomes (Mmu12 and Mmu15) by the close positioning of the two BACs (< 1.5 μm). (b–d) Similar relative radial positions were observed for the unrearranged chromosomes in all 3 tumor samples ((b) LP-A (N=85), (c) LP-B (N=87), and (d) AP-C (N=96), P-values, Mann-Whitney U tests), irrespective of cell preparation method (P ≥ 0.09 between tumors for all chromosomes, Mann-Whitney U test) (Materials & Methods). TL12 and TL15 also mapped to different radial positions in LP-A and LP-B. A statistically significant difference between these two loci was not detected in AP-C, but subtle shifts in their positions relative to unrearranged chromosomes and to each other are apparent in the plot (e). Horizontal grey bars indicate means, boxes represent median and quartile values, and whiskers indicate the remaining points in the distribution.
Thrity-four BACs (right column) from the RPCI-23 Library were used to detect Mmu15 sequence distal to Myc and are listed in genomic order. Cluster and desert assignments also are indicated.
3D folding patterns of the 7 Mb region distal to Myc and the translocation breakpoint are not significantly different for unrearranged Mmu15 and t(12:15) in LP-A (blue) (N = 58), LP-B (green) (N = 83), or AP-C cells (red) (N = 91), when the patterns are considered as a group (P ≥ 0.09, χ2 tests), or among individual patterns (*, P < 0.1, χ2 tests). Comparison of folding between tumor cells that were prepared differently (AP-C vs. LP-A and LP-B) were not significantly different (P > 0.6, χ2 test).
FISH-labeled Mmu15 gene clusters and deserts were scored. No 3D folding pattern was specifically enriched at the nuclear periphery (P ≥ 0.54 for comparisons of all patterns, P ≥ 0.27 for comparisons of any individual patterns, χ2 tests). Error bars represent the standard deviations between tumors LP-A, LP-B and AP-C. Number of chromosomes evaluated for each folding pattern are indicated.
Acknowledgments
We thank Drs. Mary Ann Handel, Anne Peaston, Casey Fox, and Laura Reinholdt for critical reading of this manuscript. We also thank the contributions of The Jackson Laboratory Imaging Sciences, Flow Cytometry, Gene Expression, and Computational Biology Resource services, and Dr. David Serreze for protocols and reagents to isolate primary progenitor B cells. This work was supported by grants from the National Science Foundation (MCB 0817787 (LSS), EPScOR 0132384, and IGERT 0221625), the National Institutes of Health National Center for Research Resources (INBRE 5P20RR016463-08, P20 RR018789) and National Cancer Institute (R01 CA115666 (KDM), CA034196 ), and the Maine Cancer Foundation (LSS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NSF or NIH. Experiments conducted in this report comply with current laws of the United States of America.
Footnotes
The authors declare that there are no financial conflicts of interest associated with this work.
LITERATURE CITED
- Barki-Celli L, Lefebvre C, Le Baccon P, Nadeau G, Bonnefoix T, Usson Y, Vourc’h C, Khochbin S, Leroux D, Callanan M. Differences in nuclear positioning of 1q12 pericentric heterochromatin in normal and tumor B lymphocytes with 1q rearrangements. Genes Chromosomes Cancer. 2005;43:339–349. doi: 10.1002/gcc.20179. [DOI] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA. Using ANOVA to analyze microarray data. Biotechniques. 2004;37:173–175. 177. doi: 10.2144/04372TE01. [DOI] [PubMed] [Google Scholar]
- Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol. 2003;162:809–820. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Grosveld F. Inter-chromosomal gene regulation in the mammalian cell nucleus. Curr Opin Genet Dev. 2007;17:456–464. doi: 10.1016/j.gde.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel S, Schiebel K, Little G, Edelmann P, Rappold GA, Eils R, Cremer C, Cremer T. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity. Exp Cell Res. 1999;252:363–375. doi: 10.1006/excr.1999.4635. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico C, Cantarella CD, Di Mare P, Tosi S, Saccone S. The radial arrangement of the human chromosome 7 in the lymphocyte cell nucleus is associated with chromosomal band gene density. Chromosoma. 2008;117:399–410. doi: 10.1007/s00412-008-0160-x. [DOI] [PubMed] [Google Scholar]
- Ferreira J, Paolella G, Ramos C, Lamond AI. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J Cell Biol. 1997;139:1597–1610. doi: 10.1083/jcb.139.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Grasser F, Neusser M, Fiegler H, Thormeyer T, Cremer M, Carter NP, Cremer T, Muller S. Replication-timing-correlated spatial chromatin arrangements in cancer and in primate interphase nuclei. J Cell Sci. 2008;121:1876–1886. doi: 10.1242/jcs.026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harewood L, Schutz F, Boyle S, Perry P, Delorenzi M, Bickmore WA, Reymond A. The effect of translocation-induced nuclear reorganization on gene expression. Genome Res. 2010;20:554–564. doi: 10.1101/gr.103622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A, Grant JL, Caddle LB, Atzema E, Mills KD, Arneodo A. Chromosome territories have a highly nonspherical morphology and nonrandom positioning. Chromosome Res. 2007;15:899–916. doi: 10.1007/s10577-007-1172-8. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Groudine M. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kupper K, Kolbl A, Biener D, Dittrich S, von Hase J, Thormeyer T, Fiegler H, Carter NP, Speicher MR, Cremer T, Cremer M. Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma. 2007;116:285–306. doi: 10.1007/s00412-007-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Gassman KL, Petell LM, Wilson CL, Bewersdorf J, Shopland LS. The nuclear periphery of embryonic stem cells is a transcriptionally permissive and repressive compartment. J Cell Sci. 2009;122:3729–3737. doi: 10.1242/jcs.052555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J Cell Biol. 2002;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller I, Freund J. Probability and Statistics for Engineers. Prentice-Hall, Inc; Englewood Cliffs, NJ: 1965. [Google Scholar]
- Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- Morey C, Kress C, Bickmore WA. Lack of bystander activation shows that localization exterior to chromosome territories is not sufficient to up-regulate gene expression. Genome Res. 2009;19:1184–1194. doi: 10.1101/gr.089045.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmann AE, Gao J, Encinosa M, Gautier M, Peter ME, Eils R, Lichter P, Rowley JD. Local gene density predicts the spatial position of genetic loci in the interphase nucleus. Exp Cell Res. 2005;311:14–26. doi: 10.1016/j.yexcr.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada L, Misteli T. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 2002;12:425–432. doi: 10.1016/s0962-8924(02)02351-6. [DOI] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Munson PJ, Misteli T. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol. 2002;12:1692–1697. doi: 10.1016/s0960-9822(02)01166-1. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Whitlow S, Eckersdorff M, Manis JP, Lee C, Ferguson DO, Alt FW. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc Natl Acad Sci U S A. 2004;101:2410–2415. doi: 10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J, Stein S, Vincent S, Molloy KR, Kreth G, Cremer C, Bult CJ, O’Brien TP. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol. 2006;174:27–38. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Solovei I, Cavallo A, Schermelleh L, Jaunin F, Scasselati C, Cmarko D, Cremer C, Fakan S, Cremer T. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH) Exp Cell Res. 2002;276:10–23. doi: 10.1006/excr.2002.5513. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Tam R, Shopland LS, Johnson CV, McNeil JA, Lawrence JB. Applications of RNA FISH for visualizing gene expression and nuclear architecture. In: Beatty B, Mai S, Squire J, editors. FISH: A practical approach. Oxford University Press; Oxford: 2002. pp. 93–118. [Google Scholar]
- Taslerova R, Kozubek S, Bartova E, Gajduskova P, Kodet R, Kozubek M. Localization of genetic elements of intact and derivative chromosome 11 and 22 territories in nuclei of Ewing sarcoma cells. J Struct Biol. 2006;155:493–504. doi: 10.1016/j.jsb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Taslerova R, Kozubek S, Lukasova E, Jirsova P, Bartova E, Kozubek M. Arrangement of chromosome 11 and 22 territories, EWSR1 and FLI1 genes, and other genetic elements of these chromosomes in human lymphocytes and Ewing sarcoma cells. Hum Genet. 2003;112:143–155. doi: 10.1007/s00439-002-0847-7. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Woo Y, Wright SM, Maas SA, Alley TL, Caddle LB, Kamdar S, Affourtit J, Foreman O, Akeson EC, Shaffer D, Bronson RT, Morse HC, 3rd, Roopenian D, Mills KD. The nonhomologous end joining factor Artemis suppresses multi-tissue tumor formation and prevents loss of heterozygosity. Oncogene. 2007;26:6010–6020. doi: 10.1038/sj.onc.1210430. [DOI] [PubMed] [Google Scholar]
- Wright JB, Brown SJ, Cole MD. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol. 2010;30:1411–1420. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SM, Woo YH, Alley TL, Shirley BJ, Akeson EC, Snow KJ, Maas SA, Elwell RL, Foreman O, Mills KD. Complex oncogenic translocations with gene amplification are initiated by specific DNA breaks in lymphocytes. Cancer Res. 2009;69:4454–4460. doi: 10.1158/0008-5472.CAN-08-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Riblet R, Schildkraut CL. Sites that direct nuclear compartmentalization are near the 5′ end of the mouse immunoglobulin heavy-chain locus. Mol Cell Biol. 2005;25:6021–6030. doi: 10.1128/MCB.25.14.6021-6030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a–c) Interphase nuclei were evaluated by FISH with a Mmu15 whole chromosome paint (green) plus three BACs across Mmu15 (BACs 285N22, 43.5 Mb, red; 187M22, 62.1 Mb, blue; and 438G15, 73.6 Mb, red), as diagrammed in (a). 3D images reveal the presence of an intact Mmu15 (filled arrowhead), a t(12;15) containing only distal Mmu15 sequences (open arrowhead) and the complicon, which contains the proximal portion of Mmu15 (open arrow), in the (b) LP-A and (c) AP-C cells shown. 3D renderings of deconvolved 3D image stacks are shown. Bars, 1.5μm. (d) SKY of LP-A cells confirms the presence of one normal Mmu15, one t(12;15) and one complicon, all containing Mmu15 sequences as seen in (b). (e) SKY of AP-N cells confirmed the absence of t(12;15), the presence of normal copy numbers of Mmu15, and a different translocation involving Mmu12.
Relative radial positions of single BACs from Mmu12 and Mmu15 were determined by measuring the distance from the nuclear center to the locus (rx) and normalizing to the length of the total nuclear radius (rn), as depicted, where 0 represents the center of the nucleus and 100 the outer nuclear edge. Mmu12 BAC 367G12 and Mmu15 BAC 200D20 were differentially labeled and co-hybridized to the same cells by two color FISH. These loci on t(12;15) (TL12 and TL15) were discriminated from un-rearranged chromosomes (Mmu12 and Mmu15) by the close positioning of the two BACs (< 1.5 μm). (b–d) Similar relative radial positions were observed for the unrearranged chromosomes in all 3 tumor samples ((b) LP-A (N=85), (c) LP-B (N=87), and (d) AP-C (N=96), P-values, Mann-Whitney U tests), irrespective of cell preparation method (P ≥ 0.09 between tumors for all chromosomes, Mann-Whitney U test) (Materials & Methods). TL12 and TL15 also mapped to different radial positions in LP-A and LP-B. A statistically significant difference between these two loci was not detected in AP-C, but subtle shifts in their positions relative to unrearranged chromosomes and to each other are apparent in the plot (e). Horizontal grey bars indicate means, boxes represent median and quartile values, and whiskers indicate the remaining points in the distribution.
Thrity-four BACs (right column) from the RPCI-23 Library were used to detect Mmu15 sequence distal to Myc and are listed in genomic order. Cluster and desert assignments also are indicated.
3D folding patterns of the 7 Mb region distal to Myc and the translocation breakpoint are not significantly different for unrearranged Mmu15 and t(12:15) in LP-A (blue) (N = 58), LP-B (green) (N = 83), or AP-C cells (red) (N = 91), when the patterns are considered as a group (P ≥ 0.09, χ2 tests), or among individual patterns (*, P < 0.1, χ2 tests). Comparison of folding between tumor cells that were prepared differently (AP-C vs. LP-A and LP-B) were not significantly different (P > 0.6, χ2 test).
FISH-labeled Mmu15 gene clusters and deserts were scored. No 3D folding pattern was specifically enriched at the nuclear periphery (P ≥ 0.54 for comparisons of all patterns, P ≥ 0.27 for comparisons of any individual patterns, χ2 tests). Error bars represent the standard deviations between tumors LP-A, LP-B and AP-C. Number of chromosomes evaluated for each folding pattern are indicated.