Abstract
The emergence of class D β-lactamases with carbapenemase activity presents an enormous challenge to health practitioners, particularly with regard to the treatment of infections caused by Gram negative pathogens such as Acinetobacter baumanii. Unfortunately, class D β-lactamases with carbapenemase activity are resistant to β-lactamase inhibitors. To better understand the details of the how these enzymes bind and hydrolyze carbapenems, we have determined the structures of two deacylation-deficient variants (K84D and V130D) of the class D carbapenemase OXA-24 with doripenem bound as a covalent acyl-enzyme intermediate. Doripenem adopts essentially the same configuration in both OXA-24 variant structures, but varies significantly when compared to the non-carbapenemase class D member OXA-1/doripenem complex. The alcohol of the 6α hydroxyethyl moiety is directed away from the general base carboxy-K84, with implications for activation of the deacylating water. The tunnel formed by the Y112/M223 bridge in the apo form of OXA-24 is largely unchanged by the binding of doripenem. The presence of this bridge, however, causes the distal pyrrolidine/sulfonamide group to bind in a drastically different conformation compared to doripenem bound to OXA-1. The resulting difference in the position of the side-chain bridge sulfur of doripenem is consistent with the hypothesis that the tautomeric state of the pyrroline ring contributes to the different carbapenem hydrolysis rates of OXA-1 and OXA-24. These findings represent a snapshot of a key step in the catalytic mechanism of an important class D enzyme, and may be useful for the design of novel inhibitors.
Keywords: class D β-lactamase, acyl-enzyme, carbapenem, deacylation-deficient, tautomerization
Introduction
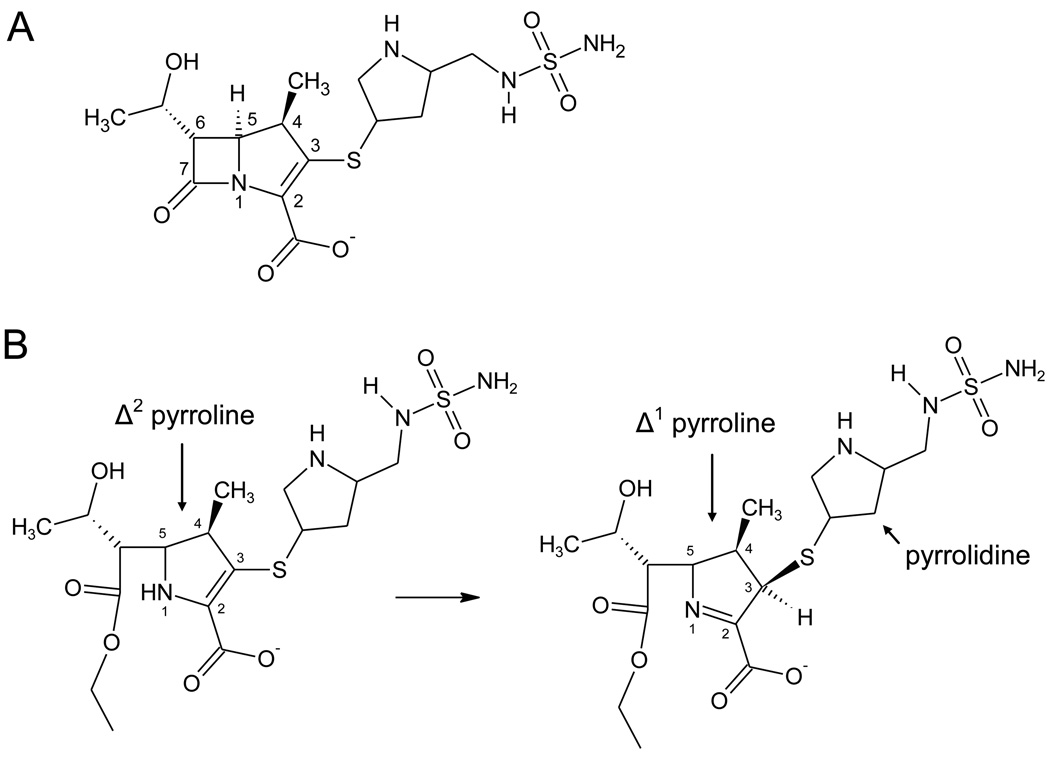
β-lactamase enzymes hydrolyze the β-lactam rings of penicillin, cephalosporin, carbapenem and cephamycin antibiotics, and in doing so provide bacteria with resistance to these drugs.1 There are four classes of β-lactamases (A–D), with class A, C and D using a serine-based covalent catalysis mechanism.2 Class D enzymes possess a very high degree of sequence diversity and are particularly troublesome in Gram negative pathogens such as Acinetobacter baumannii and Pseudomonas aeruginosa.3–5 All β-lactamases in this class are named “OXA” enzymes because many early-discovered members hydrolyzed the isoxazolylpenicillin oxacillin more efficiently than other penicillins. Some class D enzymes, most notably those that show sequence similarity to the OXA-2, −10 and −13 subfamilies, have the ability to hydrolyze extended spectrum cephalosporins (e.g. cefotaxime, ceftazidime, cefepime).5 Other class D members confer resistance to carbapenems, such as imipenem, meropenem and doripenem (Figure 1a), and are referred to as carbapenem hydrolyzing class D β-lactamases (CHDL).6 To date, no class D β-lactamase has been shown to possess both carbapenemase activity and the ability to hydrolyze extended spectrum cephalosporins.7
Figure 1.
(A) The structure of doripenem. (B) Pyrroline tautomerization after doripenem acylation of some β-lactamase enzymes.
The discovery that a conserved active site lysine (K73 in OXA-10) is carboxylated on the ε-amino group greatly advanced our understanding of the mechanism of class D β-lactamases. This unusual modification has now been observed in high-resolution crystal structures of OXA-10,8–10 OXA-111 and OXA-48.12 The carbamate group forms a number of hydrogen bonds with other active site residues, and is therefore bound with micromolar affinity.10,13 Golemi et al. proposed that the carboxy-lysine residue serves as a general base to first activate the serine nucleophile during the acylation event, and then later to activate a water molecule for attack on the ester carbonyl to release product.8 This hypothesis has been supported by mutagenesis studies showing that lysine carboxylation and enzyme activity are highly correlated.8,13–15
β-lactamases in all classes vary in their ability to hydrolyze carbapenems. Narrow spectrum enzymes from class A (e.g. TEM-1, SHV-1, BlaC),16–18 class C (AmpC, P99)19,20 and class D (OXA-1, OXA-13)21,22 can proceed through the acylation step but are unable to effectively hydrolyze the acyl-intermediate. Other enzymes in class A (GES, NPC, KPC-2),23 class C (CMY-10, BER)24,25 and class D (OXA-24*, OXA-23, OXA-48, OXA-51)26 can complete the full catalytic cycle. In most cases, kcat values for carbapenem hydrolysis by these enzymes are quite modest (< 2 s−1),23 compared to > 200 s−1 for typical penicillin substrates. Nonetheless, the acquisition of these carbapenemases can result in significant resistance to carbapenem therapy.26
There have been many theories proposed to explain why some β-lactamases are not able to hydrolyze carbapenems. Soon after the discovery of these drugs, Easton and Knowles suggested that hydrolysis of the lactam ring could lead to tautomerization of the pyrroline ring (Figure 1b) and that the resulting tautomer might be resistant to hydrolysis.27 Indeed, time-dependent tautomerization was recently observed structurally for ertapenem bound to BlaC from Mycobacterium tuberculosis17 Tautomerization is not universal among β-lactamases however, as the crystal structures of TEM-1 and AmpC bound to imipenem show no evidence of this process.16,20 Instead, these structures reveal that upon acylation, the acyl-carbonyl flips ∼180° away from the two main-chain amide nitrogens that make up the oxyanion hole. In these enzymes, this conformation prevents the stabilization of the deacylation transition state, leading to irreversible inhibition. The acyl-carbonyl of meropenem was found in a mixture of both orientations (in and out of the oxyanion hole) when that carbapenem was bound to SHV-1.18 Interestingly, Kalp et al. used Raman crystallography to show that meropenem also undergoes tautomerization when bound to SHV-1, and suggested that, at least for that enzyme, the orientation of the acyl carbonyl and the isomerization event might be linked.28
The crystal structures of two different class D β-lactamase enzymes with a carbapenem antibiotic bound in the acyl-enzyme form were recently determined. First, the OXA-13/meropenem structure21 revealed that the acyl-carbonyl was bound in the oxyanion hole and also showed the mode by which the carboxylate of the drug is stabilized by the active site. A carboxylated-lysine was not observed in the active site, though its absence is likely the result of the low pH of the crystallization conditions. More recently, the 1.4 Å structure of OXA-1 with doripenem bound as an acyl-enzyme intermediate was reported.22 In this structure, K70 was carboxylated and the resulting carbamate formed a hydrogen bond with the hydroxyethyl side-chain of doripenem. There were no water molecules in the active site near the acyl carbonyl, suggesting that inability to recruit water is yet another mechanism by which carbapenems might prevent deacylation.
Two class D carbapenemase structures (OXA-24 and OXA-48) in the absence of any bound ligand have been determined.12,29 In the case of OXA-24, Santillana et al. note the presence of a “bridge” across the top of the active site formed by residues Y112 and M223 (OXA-24 numbering for all residues, except as noted).29 The authors suggested that this hydrophobic barrier likely contributes to the very strong binding affinity of OXA-24 for the carbapenem meropenem. Interestingly, OXA-24 has a very low efficacy toward hydrolysis of the penicillin oxacillin, a fact which has been attributed to steric clash between the bridge residues and the bulky isoxazolyl side-chain of the β-lactam.29 More recently, the structures of OXA-24 with several novel penicillin sulfone inhibitors have also been determined.30 These structures reveal key interactions between the acyl-inhibitor moieties, the oxyanion hole, and the bridge residues (particularly Y112).30 In the case of OXA-48, Docquier et al. note the absence of a similar bridge, and suggest that an active site-induced rotation of the carbapenem hydroxyethyl moiety allows a more efficient approach of the deacylating water.12 The structure of a CHDL in complex with a carbapenem would greatly help evaluate the merit of these hypotheses. In this article, we report two independent structures of the carbapenemase OXA-24 (K84D and V130D) with the carbapenem doripenem bound, revealing key structural details that help illuminate the mechanistic differences among class D β-lactamases and may lead to better inhibitor design.
Results
Because of the hydrolytic activity that OXA-24 possesses for carbapenems (kcat ≥ 0.3 s−1),29 we reasoned that capturing doripenem as an acyl intermediate with this enzyme required an active site substitution that would greatly slow down deacylation rates. Based on our previous work with OXA-1 in which substitution for the carboxylated lysine led to deacylation-deficiency, we altered the homologous lysine in OXA-24 (K84) to aspartate.14 We also prepared OXA-24 V130D, speculating that the proximity of the side-chain of residue 130 to the ε-amino group of K84 would render this variant deacylation-deficient as well. Incubation of both variants with the fluorescent penicillin bocillin followed by SDS-PAGE demonstrated accumulation of the acyl-intermediate, substantiating this prediction (data not shown).14
The proteins were expressed, purified by carboxy-methyl cellulose chromatography and crystallized in 100 mM TRIS-HCl, pH 8.5, 2.0 M ammonium sulfate. Subsequently, the structures of each of these mutants were determined in acyl-enzyme complexes with the carbapenem, doripenem (Table 1). The resolution for the K84D/doripenem structure was 2.1 Å and for V130D/doripenem, 2.25 Å.
Table 1.
Crystallographic summary of the OXA-24 mutants K84D and V130D in complexes with doripenem.
| K84D/doripenem |
V130D/doripenem |
|
|---|---|---|
| Cell constants (Å; °) | a=b=102.33 c=85.45; α=β=γ=90 |
a=b=102.27 c=85.45; α=β=γ=90 |
| Spacegroup | P 41 | P 41 |
| Resolution (Å) | 2.10 (2.18-2.10)a | 2.25 (2.33-2.25)a |
| Unique reflections | 50,780 | 41,616 |
| Total observations | 212,181 | 128,525 |
| Rmerge (%) | 7.4 (35.2) | 7.5 (33.9) |
| Completeness (%)b | 98.8 (94.2) | 99.0 (95.5) |
| <I>/<σ I> | 18.7 (3.9) | 17.1 (3.1) |
| Resolution range for refinement (Å) | 50-2.1 | 50-2.25 |
| Number of reflections used in refinement |
48197 | 39512 |
| Number of protein residues | 488 | 488 |
| Number of water molecules | 314 | 197 |
| rmsd bond lengths (Å) | 0.015 | 0.017 |
| rmsd bond angles (°) | 1.5 | 1.6 |
| Rcryst (%) | 18.9 | 18.9 |
| Rfree (%)c | 23.1 | 23.3 |
| Ramachandran plot statistics | ||
| Favored (%) | 95.6 | 96.7 |
| Allowed (%) | 4.2 | 3.3 |
| Disallowed (%) | 0.2 | 0 |
| Average B-factor, protein atoms (Å2, molecule A) |
37.0 | 36.4 |
| Average B-factor, protein atoms (Å2, molecule B) |
36.8 | 36.3 |
| Average B-factor, doripenem atoms (Å2, molecule A) |
40.2 | 47.6 |
| Average B-factor, doripenem atoms (Å2, molecule B) |
41.3 | 47.5 |
| Average B-factor, water molecules (Å2) |
44.9 | 39.8 |
Values in parentheses are for the highest resolution shell.
Fraction of theoretically possible reflections observed.
Rfree was calculated with 5% of reflections set aside randomly.
Comparison of these two structures to the apo enzyme (PDB entry 2JC7) showed that the K84D and V130D substitutions do not introduce any major changes to the overall fold of OXA-24.29 Structural alignments between the α-carbons of each mutant and the apo-structure result in rmsd values of 0.21 Å (comparing 230 C-α atoms from the A monomer of K84D to structure 2JC7) and 0.16 Å (232 C-α atoms from the A monomer of V130D). For seven key active site residues (S81, Y112, S128, W167, L168, K218 and M223), the rmsd values were 0.214 Å (52 atoms aligned for K84D and wild-type apo) and 0.445 Å (54 atoms aligned for V130D and wild-type apo).
Analysis of the final models of these structures was performed with Procheck.31 In the K84D/doripenem structure, 99.8% of the non-proline, non-glycine residues are in the preferred or allowed region of the Ramachandran plot (100% for the V130D/doripenem complex).
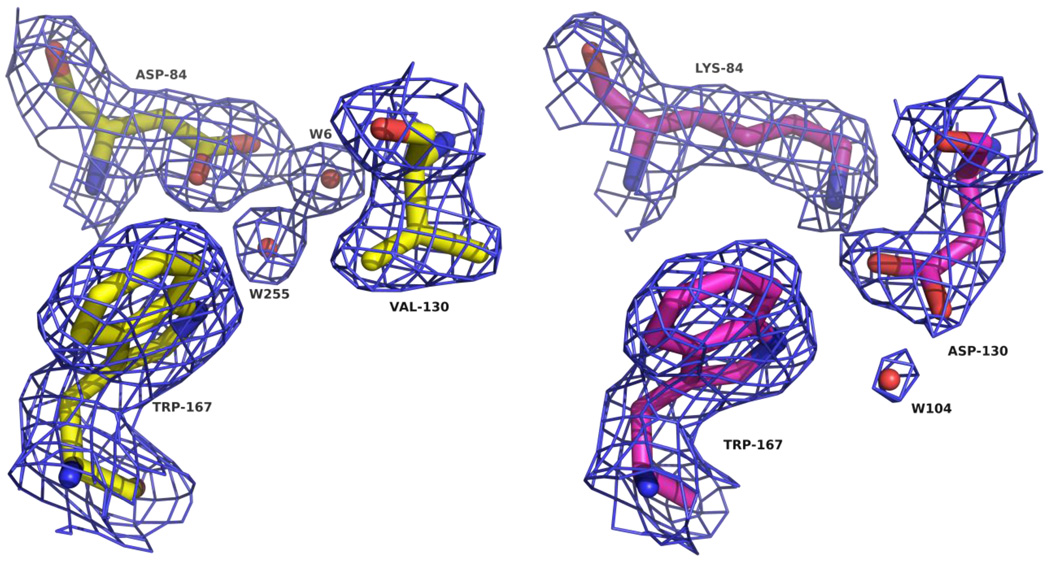
In both variants, the most obvious changes observed were in the interactions between the side-chains of positions 84 and 130 themselves, reflecting the close relationship of these residues in the active site. For the K84D variant, each oxygen atom of the aspartate carboxylate attracts a water molecule to fill the area normally occupied by the rest of the carboxy-lysine side-chain (Figure 2). Interestingly, one of these waters acts as a hydrogen bond acceptor with the side-chain Nε1 of W167, which is normally bonded to the carbamate moiety in all carboxylated OXA enzymes observed to date.8,10–12,22 The V130D substitution results in the formation of a novel salt-bridge (or possibly a hydrogen-bond) between the aspartate side-chain and the ε-amino group of K84 (Figure 2). We observed only very weak electron density beyond the K84 ε-amino nitrogen, suggesting that the V130D mutation stabilizes the decarboxylated form of the enzyme.
Figure 2.
Arrangement of the active site residues of OXA-24 mutants (A) K84D (yellow; D84, V130 and W167) and (B) V130D (magenta; K84, D130 and W167). Blue cages represent 2Fo-Fc electron density maps contoured at 1.0 σ. This and all subsequent figures were prepared with PyMOL.41
A small but significant difference between the two variant OXA-24 structures is in the area of the substituted residue of V130D. The extra bulk provided by the aspartate causes a slight rotation of the W167 side chain, and causes the L168 side chain to be tilted back ∼1.5 Å. These changes have very little effect on the rest of the active site and the position of the bound ligand. Due to these subtle changes in the V130D mutant, the structure of the K84D mutant more closely matches the published apo structure (2JC7)29 at these residues.
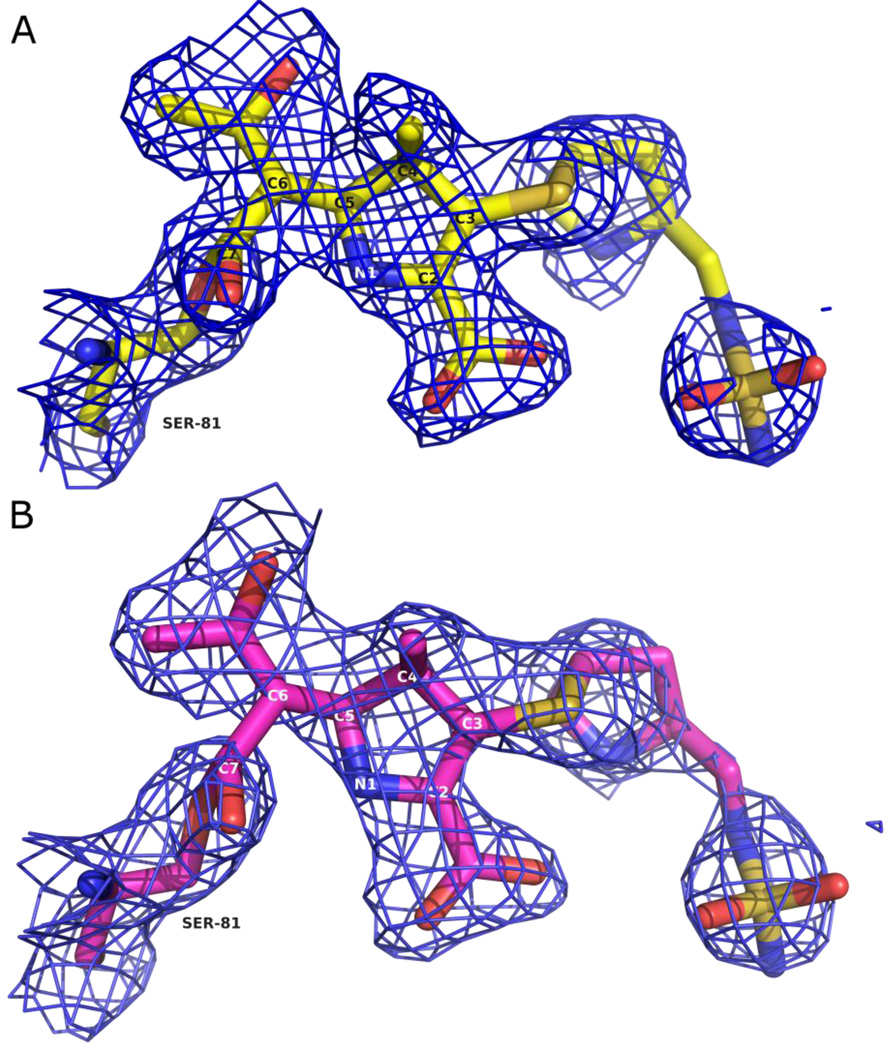
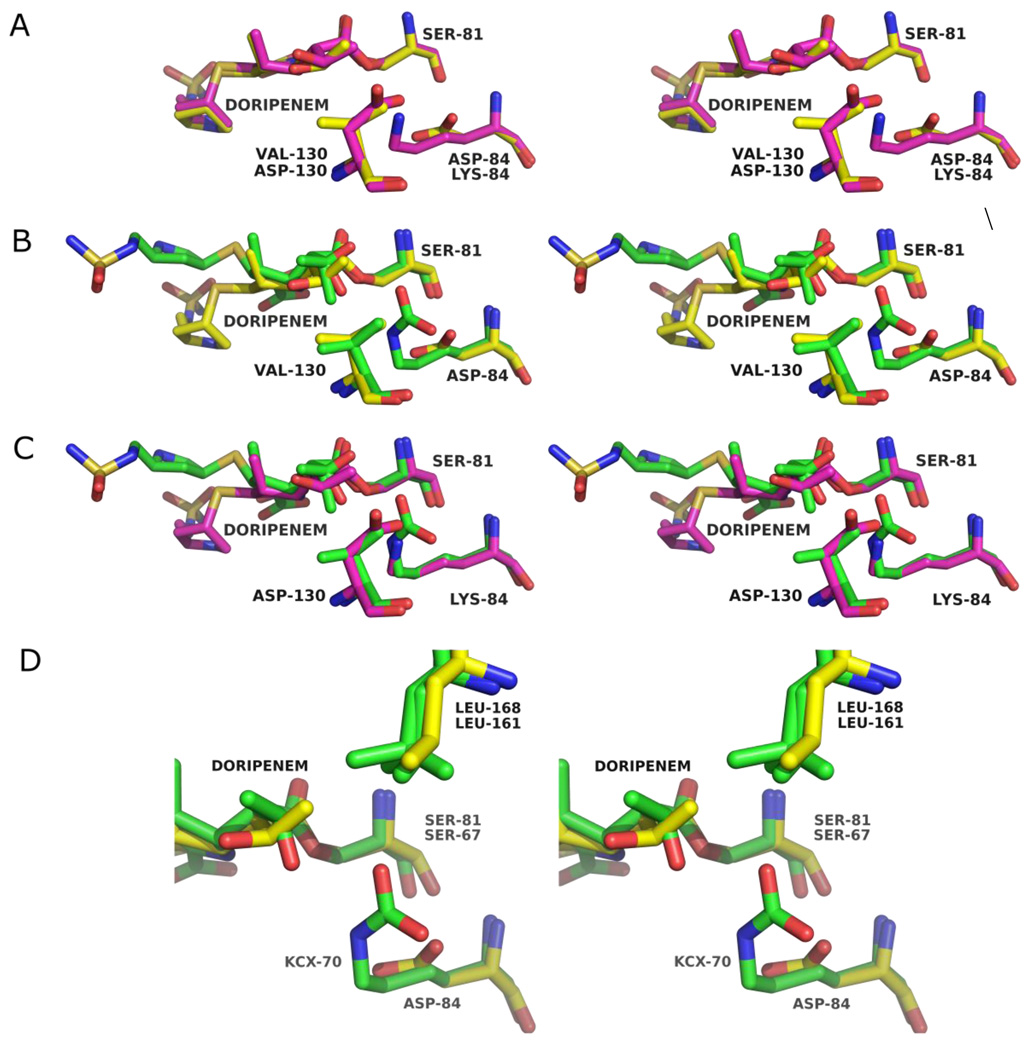
In both structures, unambiguous electron density was observed in the active sites in the initial Fo-Fc difference maps contoured at 3σ. Omit maps were calculated from the final refined models and confirm the conformation and orientation of the modeled ligand in these structures (Figure 3a and 3b).32 Electron density was continuous with the Oγ atom of the catalytic serine residue, S81, indicating a covalent bond between this residue and atom C7 of the acylated doripenem. As with the OXA-1/doripenem structure, both variant OXA-24/doripenem structures show strong electron density for all atoms of the doripenem molecule indicating that fragmentation of the bound ligand does not occur for the duration of the soak. The structures of doripenem bound to both OXA-24 mutants are very similar to each other, with an rmsd of 0.15 Å for 23 non-hydrogen ligand atoms in the A monomers of each mutant (Figure 4a). Given the very similar structures observed, each individual amino acid substitution appears to have minimal impact on the structure of the acyl-intermediate itself.
Figure 3.
Structures of doripenem bound to OXA-24 mutants (A) K84D (yellow) and (B) V130D (magenta). Blue cages represent omit electron density maps contoured at 1.0 σ for K84D and V130D.
Figure 4.
Stereoviews showing structural alignments between the doripenem-bound forms of (A) OXA-24 mutants K84D (yellow) and V130D (magenta), (B) OXA-1 (PDB entry 3ISG; green) and OXA-24 K84D (yellow) and (C) OXA-1 (green) and OXA-24 V130D (magenta). (D) Expanded view of the hydroxyethyl moieties of doripenem bound to OXA-24 K84D (yellow) and OXA-1 (green) as well as their interaction with an active site leucine (L168 in OXA-24; L161 in OXA-1). L161 of OXA-1 adopts two partial occupancy rotamers.
Comparisons of the doripenem structure in the two variant OXA-24 complexes with the same carbapenem bound to OXA-1 show some expected areas of high similarity (Figure 4b and 4c). For instance, the position of the serine side-chain and the ester linkage match closely between the two enzymes. Most notably, the acyl-carbonyl of the ester linkage is positioned in the oxyanion hole in the two OXA-24 variants (not shown) in the same way that it is in the OXA-1 complex.22 Similarly, the acyl-carbonyls of several novel penicillin sulfone inhibitors were firmly bound in the oxyanion hole of OXA-24.30
Beyond the proximity of the covalent acyl-enzyme attachment to the catalytic serine, the conformation of doripenem bound to OXA-24 K84D and V130D differs drastically from that observed in the OXA-1/doripenem complex. Unlike the OXA-1/doripenem structure in which two different positions could be modeled for the hydroxyethyl moiety of the bound drug,22 we did not find evidence of multiple conformations for any part of doripenem in either variant OXA-24 structure. There are three common rotamers available to the hydroxyethyl group of doripenem, and the conformation for this group when bound to OXA-24 K84D and V130D does not match either of the two distinct positions observed in the OXA-1/doripenem complex.22 This hydroxyethyl position is stabilized in part by a hydrophobic interaction between its methyl group and the side-chain of L168 (3.2 Å and 3.5 Å for K84D and V130D, respectively), as suggested by Docquier et al. for OXA-48 (Figure 4d). Assuming this group adopts a similar orientation in the wild-type OXA-24 complex, this hydroxyethyl conformer would preclude a potentially inhibitory hydrogen bond with the carbamate of carboxy-K84.
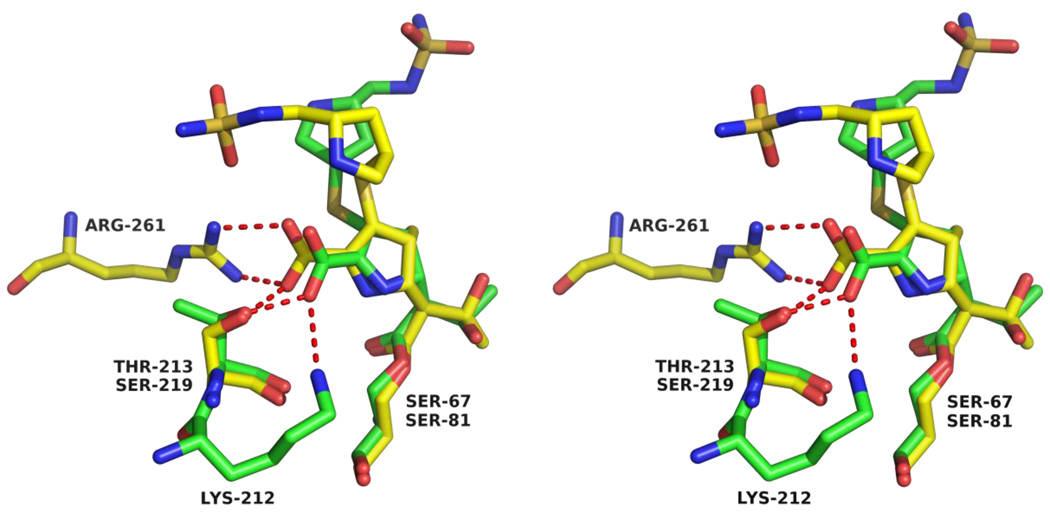
The pyrroline ring assumes a slightly different pitch between OXA-1 and the OXA-24 variants and is also shifted laterally approximately 0.7 Å (Figure 5). These observations suggest that structural variations arise from a difference in the mode by which the carboxylate moiety attached to the pyrroline ring is stabilized in OXA-1 and the OXA-24 variants. In the former, the side-chains of K212 and T213 form a salt-bridge and hydrogen bond respectively with the carboxylate (Figure 5). In the latter, S219 and R261 provide the stabilizing side-chain interactions as predicted by Santillana et al.29 Residues T213 (OXA-1) and S219 (OXA-24) are sequentially and positionally homologous, but K212 (OXA-1) and R261 (OXA-24) occupy quite different places in the active site and thereby pull the carboxylate in different directions.
Figure 5.
Stereoview of the alignment of OXA-1/doripenem (green) and OXA-24 K84D/doripenem (yellow) active sites showing the different mode of stabilization of the carboxylate. Residues R261, S81 and S219 are from OXA-24 K84D, while K212, S67 and T213 are from OXA-1. In the OXA-1/doripenem structure, only one of the two modeled hydroxyethyl rotamers is shown for clarity.
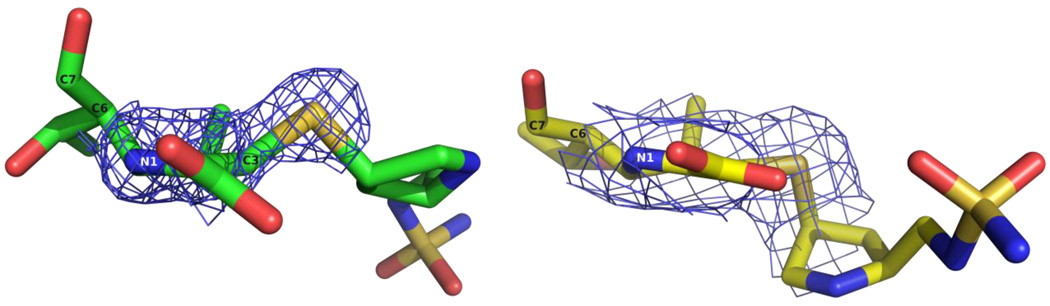
Beyond the pyrroline ring, the structure of doripenem diverges even more between the OXA-1 and variant OXA-24 bound structures. As seen in Figure 6 (left panel), the sulfur atom that bridges the pyrroline and pyrrolidine rings is angled above the former approximately 30° in the OXA-1 structure.22 The electron density of this sulfur in the OXA-24 variant structure adopts a position much closer to the plane of the pyrroline ring (right panel). The planar orientation of this sulfur suggests that in OXA-24, doripenem remains in the Δ2 tautomeric form, rather than isomerizing to the Δ1 form as observed in OXA-1 (Figure 1b). Additionally, with the sulfur in the planar position, the more distal pyrrolidine ring is forced approximately 4.5 Å closer to M114. Simultaneously, this ring rotates along the S-C1A bond to lie more deeply in the active site compared to its orientation toward the top of the active site in OXA-1. In this position, the pyrrolidine is cradled in a hydrophobic pocket composed of residues M114, Y112, M223 and W221. Notably, the loss of the hydrophobic interaction between Y112 and the pyrrolidine ring helps explain the lower MIC and higher KM observed for meropenem with the OXA-24 mutant Y112A.29
Figure 6.
Position of the bridge sulfur of doripenem bound to OXA-1 (PDB entry 3ISG; green) and OXA-24 K84D (yellow). Blue cages represent 2Fo-Fc electron density maps contoured at 1.0 σ for OXA-1/doripenem and OXA-24 K84D/doripenem.
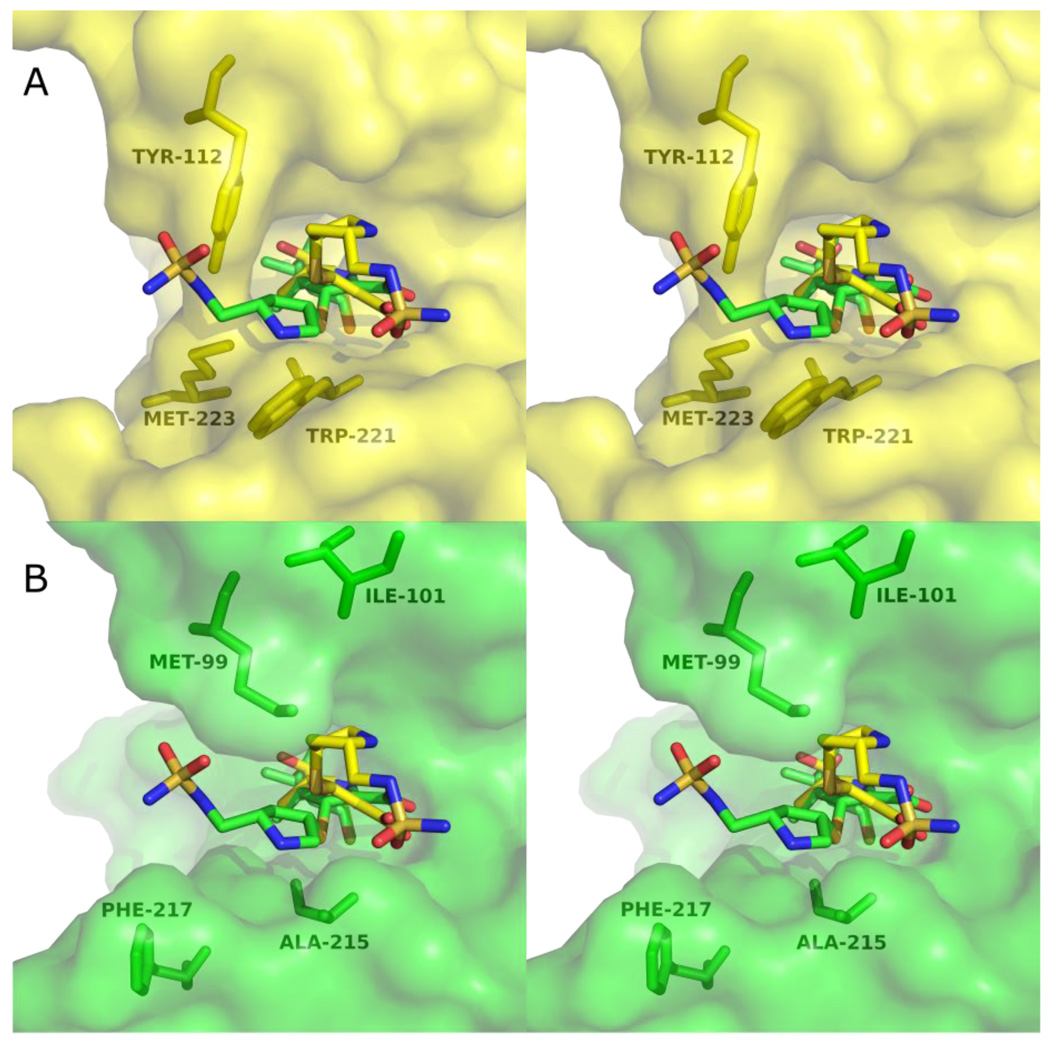
The architectural differences between the active sites of OXA-1 and OXA-24 help explain the different doripenem side-chain orientations we observe. Most notably, the bridge formed by Y112 and M223 of OXA-24 would clash with the pyrrolidine ring and sulfonamide moiety of the doripenem conformation observed in OXA-1 (Figure 7a). In OXA-1, the positions homologous to Y112 and M223 in OXA-24 are occupied by a methionine (M99) and phenylalanine (F217), respectively. Although these residues take up approximately the same volume as Y112 and M223, their side-chains are oriented quite differently (Figure 7b). This effectively peels back the two halves of the bridge and opens up a large space for the pyrrolidine/sulfonamide portion of doripenem in OXA-1. Moreover, the position of the pyrrolidine ring in the OXA-1 complex is facilitated by the small side-chain volume of A215. This orientation would not be possible in OXA-24, which contains a tryptophan at the same position (W221). This same tryptophan side-chain causes a major deviation in OXA-24 compared to OXA-1, pushing out the loop containing residues 252–258 and causing a rotation in helix α9 (helix α 10 in OXA-1) of approximately 13°. The movement of this loop away from the active site in the OXA-24 variants precludes the hydrogen bond between the main-chain amide of L255 and the pyrrolidine nitrogen that we observed in OXA-1/doripenem.22
Figure 7.
Stereoviews of the position of doripenem in the active sites of OXA-24 K84D and OXA-1. (A) Doripenem and four active site residues of OXA-24 K84D are shown in yellow. The structure of doripenem as it is bound to OXA-1 (green sticks) is shown after aligning the C-α carbons of OXA-1 and OXA-24 K84D. (B) Doripenem bound to the active site of OXA-1 is shown in green, with the aligned doripenem from OXA-24 K84D overlaid (yellow sticks). For clarity, only one of the two modeled side-chain conformers of M99 (OXA-1) is shown.
Conversely, the orientation of M99 in OXA-1 (sequentially homologous to Y112 in OXA-24) makes it sterically incompatible with the pyrrolidine ring of doripenem in the OXA-24 variant conformation (Figure 7b). To avoid this clash, the entire pyrrolidine/sulfonamide side-chain is pushed away from M99 in OXA-1. The presence of M99 therefore likely contributes to the movement of the sulfur out of the plane of the pyrroline ring and thereby encourages formation of the Δ1 tautomer in OXA-1. In its OXA-24 variant position, the pyrrolidine ring makes a favorable non-polar contact with M114 of OXA-24 (3.8 Å), which would not be possible with the equivalent residue (I101) of OXA-1.
Discussion
The complex of doripenem and the carbapenemase OXA-24 (K84D and V130D) differs from its structure bound to the narrow spectrum OXA-1 in critical ways. These differences likely contribute to the substrate specificity divergence between the two enzymes. Most strikingly, key active site changes force the pyrrolidine/sulfonamide side-chain of doripenem to adopt quite different trajectories between OXA-1 and the OXA-24 variants. These different trajectories correlate to, and may be responsible for, the alternate positions assumed by the bridge sulfur with respect to the plane of the pyrroline ring. A bridge sulfur atom close to the plane of the ring, as it is in the OXA-24 variants, suggests that the pyrroline double bond remains between carbons 2 and 3 (Δ2 isomer) as it is in the intact substrate molecule (Figure 1b). This contrasts with what is observed for OXA-1/doripenem, in which the movement of the sulfur out of the plane of the ring (to form a novel R stereocenter at carbon 3) indicates tautomerization of the pyrroline to the Δ1 form.22 This kind of post-acylation tautomerization has been proposed as an explanation of why carbapenems inhibit many β-lactamases.27,33,34 Is it therefore possible that the lack of doripenem tautomerization on OXA-24 is responsible for this enzyme’s carbapenemase activity?
One intriguing possibility is that carbapenem tautomerization leads to other conformational changes that preclude recruitment or activation of the deacylating water. A recent report by Tremblay et al. describing the structure of ertapenem bound to the endogenous β-lactamase of Mycobacterium tuberculosis (BlaC) shows that the drug is in the Δ2 tautomer immediately post-acylation, but rearranges to the Δ1 tautomer over time.17 In this structure the tautomerization is accompanied by rotation of the drug’s hydroxyethyl moiety. This movement changes the hydrogen bonding pattern such that the final Δ1 tautomer closely interacts with the general base Oε2 atom of E166, pulling this atom away from the deacylating water.17 In our structure of OXA-1/doripenem, we argued that a similar hydrogen bond between the hydroxyl group and carboxy-K70 (OXA-1 numbering) resulted in a stable doripenem acyl-enzyme intermediate by preventing the recruitment of the deacylating water.22 We find it intriguing that in all four monomers of OXA-24/doripenem (i.e. both monomers of the V130D and K84D mutants), we observe the hydroxyl group rotated directly away from the area normally occupied by the carbamate. With the hydroxyl group pointing away, the carbamate is perhaps more capable of activating water and proceeding through deacylation. While this interesting correlation between hydroxyethyl conformational changes and tautomerization occurs in β-lactamases from two different classes, confirmation that it is not an artifact of the K84D and V130D substitutions will have to wait for a structure of doripenem bound to a fully-carboxylated OXA-24 enzyme. That said, this particular rotamer of the hydroxyethyl group seems to be stabilized by a close nonpolar interaction between the doripenem methyl group and the side-chain of L168 in OXA-24 K84D and V130D. In the OXA-24 K84D structure, the distance between the methyl group of doripenem’s hydroxyethyl and the closest carbon atom of L168 is fully compatible with a stabilizing van der Waals interaction (3.2 Å for monomer A and 3.1 Å for monomer B; see Figure 4d). In the OXA-1 structure, however, the homologous leucine (L161) is shifted ∼ 0.7 Å closer to the doripenem, and thereby precludes the adoption of the same hydroxyethyl rotamer. Interestingly, Docquier et al. use molecular dynamics simulations to show that a similar methyl group/leucine interaction likely occurs in OXA-48.12 The authors suggest that this interaction and the hydroxyethyl rotation that it causes would not occur in the narrow spectrum OXA-10 and may be responsible for the carbapenemase activity of OXA-48.12 Because this leucine is highly conserved across both narrow-spectrum and carbapenemase members of the class D subfamily, only further carbapenem-bound crystal structures will illuminate whether the distance between it and the hydroxyethyl moiety is correlated to carbapenemase activity.
Another important possibility involves the protonation of substrate upon cleavage of the lactam bond. Sun et al. proposed that protonation of the lactam nitrogen leaving group is achieved via S115 (OXA-1 numbering), which shuttles a proton from the carbamate of carboxy-K70.11 By removing this proton from carboxy-K70, the carbamate is regenerated as a general base that can recruit and deprotonate the deacylating water. For carbapenems, however, tautomerization moves the site of protonation from N1 to C3 of the pyrroline ring. This carbon, 4.8 Å from S115 in OXA-1, would not likely use that residue as a general acid, even with the rotation of the serine’s hydroxyl that has been proposed in class D β-lactamases.11,21 Loss of the proton shuttle could potentially leave the carboxy-K70 carbamate protonated and unable to recruit or activate the deacylating water, resulting in the strong inhibition of OXA-1 by carbapenems. Bou et al. have proposed a similar mechanism for the inhibition of OXA-24 by penicillin sulfone inhibitors.30 Conversely, the Δ2 tautomer of doripenem when bound to OXA-24 requires protonation of N1, and thus could maintain the regenerative proton shuttle. Although S128 in the OXA-24 variant structures reported here is not within hydrogen bonding distance to the N1 nitrogen, minor rotation would bring its alcohol to within 2.8 Å of it.
There has been some speculation that the arginine residue that binds the β-lactam carboxylate in some enzymes plays a key role in carbapenem turnover. Zafarrala and Mobashery suggest that this arginine encourages Δ2→Δ1 tautomerization of imipenem in TEM-1, and thus discourages deacylation.34 Residue R261 of the OXA-24 variants binds the doripenem carboxylate, in a manner similar to that seen in class A enzymes such as TEM-1 and SHV-1, as well as class D members OXA-10 and OXA-13.15,16,18,21 The class A enzyme BlaC, class C enzyme AmpC and class D enzyme OXA-1 on the other hand, do not have an arginine in this area.17,20,22 All seven of these enzymes are strongly inhibited by carbapenems, suggesting that the presence or absence of the arginine is not correlated with the turnover of these drugs. Tremblay et al. suggest that the presence of the arginine may encourage the ∼180° flip of the acyl carbonyl (as seen in TEM-1 and AmpC, but not BlaC or OXA-1).17 The OXA-1 and OXA-24 variant doripenem complexes both maintain the acyl-carbonyl in the oxyanion hole despite the arginine difference, and argue against this.
The differences in the active sites between OXA-1 and OXA-24 are extensive enough that we hypothesize that substitutions at one or two residues will not be enough to convert OXA-1 into an OXA-24 type carbapenemase. Although the OXA-24 bridge residues Y112 and M223 in our deacylation-deficient variants strongly affect the orientation of the pyrrolidine/sulfonamide side chain (and thus the pyrroline tautomer form), many other residues unique to each structure likely cause major effects as well (e.g. M99 in OXA-1 and W221 in OXA-24). The bulk of W221 is disruptive enough to cause a rotation of helix α9 (compared to the homologous helix α10 in OXA-1), and this helix makes key contacts with doripenem in both enzymes. Docquier et al. suggested that the mechanism of carbapenem hydrolysis within class D is likely to vary, as OXA-48 and OXA-24 have very different architectures.12 Thus it is possible that OXA-1 could undergo minor changes and still become a carbapenemase via some other alteration of the active site architecture.
The different modes of doripenem binding to OXA-1 and the OXA-24 variants only add to the remarkable diversity of carbapenem conformations in acyl-enzyme complexes. What is even more striking is that the diversity does not correlate along simple phylogenetic lines or active site features. Although some class A and class C lactamases (e.g. TEM-1, SHV-1 and AmpC) flip the acyl carbonyl away from the oxyanion hole, others do not (BlaC). Additionally, some lactamases that are inhibited by carbapenems tautomerize the drug upon acylation (OXA-1 and BlaC), while others do not (OXA-13, TEM-1, AmpC). Even the chirality of tautomerization varies between OXA-1 (R isomer) and BlaC (S isomer). This diversity suggests that β-lactamase/carbapenem interactions, whether leading to productive turnover events or not, are remarkably plastic and respond to subtle changes in the active sites of otherwise very similar enzymes. Lastly, our structural analysis reveals insights into the future design of carbapenem-like β-lactamase inhibitors of this important class of resistance determinants. Specifically, the atomic-level structural information we have obtained can be used to direct the iterative modification of a basic carbapenem scaffold. The resulting derivatives can be screened for those that maintain the tight binding of doripenem and possess much slower deacylation rates.
Materials and Methods
Mutagenesis and protein purification
The blaOXA-24 gene was the kind gift of Dr. John Quinn.4 Mutant OXA-24 genes were prepared by the PCR overlap extension method35 and cloned into the NdeI and BamHI sites of pET24a. After transformation of the plasmids into BL21(DE3) E. coli cells, cultures were grown in lysogeny broth (LB) containing 25 µg/mL kanamycin and induced with 100 µM isopropyl β-D-1-thiogalactopyranoside (IPTG) (Calbiochem) for 90 min. Cells were collected by centrifugation (9500g for 20 min at 25°C, Sorvall SLA-3000 rotor) and frozen at −20°C overnight. The cell pellet was thawed in 20 mL of 50 mM NaH2PO4 and 1 mM EDTA (pH 7.0) supplemented with 100 µL of HALT Protease Inhibitor Cocktail (Thermo Fischer Scientific, Rockford, IL). Lysozyme (Sigma, St. Louis, MO), at a final concentration of 0.5 mg/mL, was added to break the cell walls and DNA was eliminated using a final concentration of 2.5 µg/mL DNAse I and 5 mM MgCl2. After centrifugation of the lysate (26900g for 30 min, 4°C, Sorvall SS-34 rotor), the supernatant was dialyzed against 4 L of 5 mM NaH2PO4, pH 5.8 at 4°C. The lysate was applied to a CM-32 carboxymethyl-cellulose (Whatman, Kent, UK) column (1.5 cm × 5 cm) equilibrated with 5 mM NaH2PO4, pH 5.8. Protein was eluted with a linear gradient of 5 mM NaH2PO4, pH 5.8 to 50 mM NaH2PO4, pH 7.0. Fractions of the OXA-24 variants (> 95% purity by SDS-PAGE) were combined and concentrated to ∼ 8 mg/mL using Amicon Ultra 10 kDa MWCO centrifugal filtration devices (Millipore, Billerica, MA).
Crystal growth and structure determination
Crystals of the OXA-24 β-lactamase mutants K84D and V130D were grown using hanging drop vapor diffusion in a 12 µL drop containing 4.5 mg/mL of the mutant β-lactamase mixed in a 1:1 ratio with well buffer (100 mM TRIS-HCl, pH 8.5, 2.0 M ammonium sulfate). Microseeding techniques were employed to obtain diffraction quality crystals, which appeared within 1–2 days at 25°C. Diffraction data were collected on the LS-CAT beamline (21-ID-D) at the Advanced Photon Source (Argonne, IL) at 100 K using a MarCCD detector. Before data collection the crystals were soaked in a solution containing > 20 mM doripenem for 10 min and then transferred into well buffer containing 5–7% sucrose for 30 sec before flash-cooling in liquid nitrogen.
Reflections were indexed, integrated, and scaled using HKL-2000.36 The space group for both deacylation-deficient variants was P41 with two OXA-24 monomers in the asymmetric unit. The structures of the OXA-24 K84D/doripenem and OXA-24 V130D/doripenem complexes were determined with Phaser37 using the A monomer of wild-type apo OXA-24 (PDB entry 2JC729) as the initial phasing model (all water and ligand molecules removed). Refinement and electron density map calculations were performed with REFMAC538 in the CCP4 Program Suite.39 Manual rebuilding of the model was done with Coot.40 The final model of the K84D/doripenem complex contained 244 residues in each monomer, 314 water molecules, and three sulfate molecules, and the final model for V130D/doripenem contained 244 residues in each monomer, 197 water molecules, and six sulfate molecules. Doripenem was modeled into each of the active sites based on initial Fo-Fc difference electron density maps and further refined with REFMAC5.
Accession Numbers
The coordinates and structure factors for the complexes have been deposited with the Protein Data Bank as 3PAE and 3PAG for the K84D/doripenem and V130D/doripenem structures, respectively.
Acknowledgments
This research was supported by National Institutes of Health grant 1R15AI082416-01 (D.A.L.), the Veterans Affairs Merit Review Program, Geriatric Research Education and Clinical Care VISN 10 (R.A.B.), and the National Institutes of Health grant RO1 AI072219 (R.A.B). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
Abbreviations
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- EDTA
ethylenediaminetetraacetic acid
- PDB
protein data bank
- rmsd
root mean square deviation
- KCX
carboxy-lysine residue
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
OXA-24 and OXA-40 are identical enzymes
Contributor Information
Kyle D. Schneider, Email: k-schneider@northwestern.edu.
Caleb J. Ortega, Email: cjortega87@gmail.com.
Robert A. Bonomo, Email: Robert.Bonomo@va.gov.
Rachel A. Powers, Email: powersra@gvsu.edu.
David A. Leonard, Email: leonardd@gvsu.edu.
References
- 1.Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chemical Reviews. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 2.Bush K, Jacoby GA. Updated Functional Classification of β-lactamases. Antimicrob. Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Livermore DM. Has the era of untreatable infections arrived? J. Antimicrob. Chemother. 2009;64:i29–i36. doi: 10.1093/jac/dkp255. [DOI] [PubMed] [Google Scholar]
- 4.Lolans K, Rice TW, Munoz-Price LS, Quinn JP. Multicity Outbreak of Carbapenem-Resistant Acinetobacter baumannii Isolates Producing the Carbapenemase OXA-40. Antimicrob. Agents Chemother. 2006;50:2941–2945. doi: 10.1128/AAC.00116-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Livermore DM, Woodford N. The β-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends in Microbiology. 2006;14:413–420. doi: 10.1016/j.tim.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H. OXA-143, a Novel Carbapenem-Hydrolyzing Class D β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009;53:5035–5038. doi: 10.1128/AAC.00856-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Naas T, Nordmann P. Diversity, Epidemiology, and Genetics of Class D β-lactamases. Antimicrob. Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S. Critical involvement of a carbamylated lysine in catalytic function of class D β-lactamases. Proc Natl Acad Sci U S A. 2001;98:14280–14285. doi: 10.1073/pnas.241442898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maveyraud L, Golemi D, Kotra LP, Tranier S, Vakulenko S, Mobashery S, Samama JP. Insights into class D β-lactamases are revealed by the crystal structure of the OXA10 enzyme from Pseudomonas aeruginosa. Structure. 2000;8:1289–1298. doi: 10.1016/s0969-2126(00)00534-7. [DOI] [PubMed] [Google Scholar]
- 10.Maveyraud L, Golemi-Kotra D, Ishiwata A, Meroueh O, Mobashery S, Samama JP. High-resolution X-ray structure of an acyl-enzyme species for the class D OXA-10 β-lactamase. J Am Chem Soc. 2002;124:2461–2465. doi: 10.1021/ja016736t. [DOI] [PubMed] [Google Scholar]
- 11.Sun T, Nukaga M, Mayama K, Braswell EH, Knox JR. Comparison of β-lactamases of classes A and D: 1.5-Å crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci. 2003;12:82–91. doi: 10.1110/ps.0224303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Docquier JD, Calderone V, De Luca F, Benvenuti M, Giuliani F, Bellucci L, Tafi A, Nordmann P, Botta M, Rossolini GM, Mangani S. Crystal structure of the OXA-48 β-lactamase reveals mechanistic diversity among class D carbapenemases. Chem Biol. 2009;16:540–547. doi: 10.1016/j.chembiol.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Leonard DA, Hujer AM, Smith BA, Schneider KD, Bethel CR, Hujer KM, Bonomo RA. The role of OXA-1 β-lactamase Asp(66) in the stabilization of the active-site carbamate group and in substrate turnover. Biochem J. 2008;410:455–462. doi: 10.1042/BJ20070573. [DOI] [PubMed] [Google Scholar]
- 14.Schneider KD, Bethel CR, Distler AM, Hujer AM, Bonomo RA, Leonard DA. Mutation of the active site carboxy-lysine (K70) of OXA-1 β-lactamase results in a deacylation-deficient enzyme. Biochemistry. 2009;48:6136–6145. doi: 10.1021/bi900448u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baurin S, Vercheval L, Bouillenne F, Falzone C, Brans A, Jacquamet L, Ferrer JL, Sauvage E, Dehareng D, Frere JM, Charlier P, Galleni M, Kerff F. Critical role of tryptophan 154 for the activity and stability of class D β-lactamases. Biochemistry. 2009;48:11252–11263. doi: 10.1021/bi901548c. [DOI] [PubMed] [Google Scholar]
- 16.Maveyraud L, Mourey L, Kotra LP, Pedelacq J-D, Guillet V, Mobashery S, Samama J-P. Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A β-lactamases from the antibiotic-resistant bacteria. Journal of the American Chemical Society. 1998;120:9748–9752. [Google Scholar]
- 17.Tremblay LW, Fan F, Blanchard JS. Biochemical and Structural Characterization of Mycobacterium tuberculosis β-lactamase with the Carbapenems Ertapenem and Doripenem. Biochemistry. 2010;49:3766–3773. doi: 10.1021/bi100232q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nukaga M, Bethel CR, Thomson JM, Hujer AM, Distler A, Anderson VE, Knox JR, Bonomo RA. Inhibition of class A β-lactamases by carbapenems: crystallographic observation of two conformations of meropenem in SHV-1. J Am Chem Soc. 2008;130:12656–12662. doi: 10.1021/ja7111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galleni M, Amicosante G, Frere JM. A survey of the kinetic parameters of class C β-lactamases. Cephalosporins and other β-lactam compounds. Biochem J. 1988;255:123–129. doi: 10.1042/bj2550123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beadle BM, Shoichet BK. Structural basis for imipenem inhibition of class C β-lactamases. Antimicrob Agents Chemother. 2002;46:3978–3980. doi: 10.1128/AAC.46.12.3978-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pernot L, Frenois F, Rybkine T, L'Hermite G, Petrella S, Delettre J, Jarlier V, Collatz E, Sougakoff W. Crystal structures of the class D β-lactamase OXA-13 in the native form and in complex with meropenem. J Mol Biol. 2001;310:859–874. doi: 10.1006/jmbi.2001.4805. [DOI] [PubMed] [Google Scholar]
- 22.Schneider KD, Karpen ME, Bonomo RA, Leonard DA, Powers RA. The 1.4 Å crystal structure of the class D β-lactamase OXA-1 complexed with doripenem. Biochemistry. 2009;48:11840–11847. doi: 10.1021/bi901690r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mammeri H, Poirel L, Nordmann P. Extension of the hydrolysis spectrum of AmpC β-lactamase of Escherichia coli due to amino acid insertion in the H-10 helix. Journal of Antimicrobial Chemotherapy. 2007;60:490–494. doi: 10.1093/jac/dkm227. [DOI] [PubMed] [Google Scholar]
- 25.Kim JY, Jung HI, An YJ, Lee JH, Kim SJ, Jeong SH, Lee KJ, Suh PG, Lee HS, Lee SH, Cha SS. Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C β-lactamase. Molecular Microbiology. 2006;60:907–916. doi: 10.1111/j.1365-2958.2006.05146.x. [DOI] [PubMed] [Google Scholar]
- 26.Walther-Rasmussen J, Hoiby N. OXA-type carbapenemases. J. Antimicrob. Chemother. 2006;57:373–383. doi: 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- 27.Easton CJ, Knowles JR. Inhibition of the RTEM β-lactamase from Escherichia coli. Interaction of the enzyme with derivatives of olivanic acid. Biochemistry. 1982;21:2857–2862. doi: 10.1021/bi00541a008. [DOI] [PubMed] [Google Scholar]
- 28.Kalp M, Carey PR. Carbapenems and SHV-1 β-lactamase form different acyl-enzyme populations in crystals and solution. Biochemistry. 2008;47:11830–11837. doi: 10.1021/bi800833u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santillana E, Beceiro A, Bou G, Romero A. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc Natl Acad Sci U S A. 2007;104:5354–5359. doi: 10.1073/pnas.0607557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bou G, Santillana E, Sheri A, Beceiro A, Sampson JM, Kalp M, Bethel CR, Distler AM, Drawz SM, Pagadala SR, van den Akker F, Bonomo RA, Romero A, Buynak JD. Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase. J Am Chem Soc. 2010;132:13320–13331. doi: 10.1021/ja104092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 32.Bhat T. Calculation of an OMIT map. Journal of Applied Crystallography. 1988;21:279–281. [Google Scholar]
- 33.Charnas RL, Knowles JR. Inhibition of the RTEM β-lactamase from Escherichia coli. Interaction of enzyme with derivatives of olivanic acid. Biochemistry. 1981;20:2732–2737. doi: 10.1021/bi00513a005. [DOI] [PubMed] [Google Scholar]
- 34.Zafaralla G, Manavathu EK, Lerner SA, Mobashery S. Elucidation of the role of arginine-244 in the turnover processes of class A β-lactamases. Biochemistry. 1992;31:3847–3852. doi: 10.1021/bi00130a016. [DOI] [PubMed] [Google Scholar]
- 35.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallographica Section D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC. Schrödinger, LLC; The PyMOL Molecular Graphics System Version 1.3. [Google Scholar]