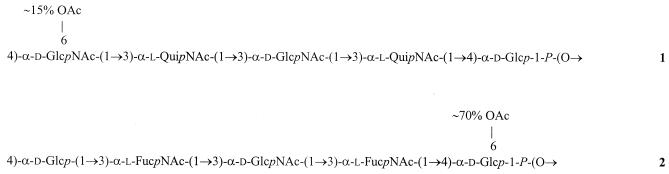Abstract
Shigella is an important human pathogen. It is generally agreed that Shigella and Escherichia coli constitute a single species; the only exception is Shigella boydii type 13, which is more distantly related to E. coli and other Shigella forms and seems to represent another species. This gives S. boydii type 13 an important status in evolution. O antigen is the polysaccharide part of the lipopolysaccharide in the outer membrane of gram-negative bacteria and plays an important role in pathogenicity. The chemical structure and genetic organization of the S. boydii type 13 O antigen were investigated. The O polysaccharide was found to be acid labile owing to the presence of a glycosyl phosphate linkage in the main chain. The structure of the linear pentasaccharide phosphate repeating unit (O unit) was established by nuclear magnetic resonance spectroscopy, including two-dimensional COSY, TOCSY, ROESY, and H-detected 1H,13C and 1H,31P HMQC experiments, along with chemical methods. The O antigen gene cluster of S. boydii type 13 was located and sequenced. Genes for synthesis of UDP-2-acetamido-2,6-dideoxy-l-glucose and genes that encode putative sugar transferases, O unit flippase, and O antigen polymerase were identified. Seven genes were found to be specific to S. boydii type 13. The S. boydii type 13 O antigen gene cluster has higher levels of sequence similarity with Vibrio cholerae gene clusters and may be evolutionarily related to these gene clusters.
Shigella is a well-known human pathogen and causes diseases such as diarrhea and bacilliary dysentery (46). Shigella and Escherichia coli strains are generally thought to be sufficiently similar to be placed in the same species. However, it has been suggested that Shigella boydii type 13 should be classified as neither Shigella nor E. coli (10, 11, 17, 34). Recent analysis of sequence variation in eight housekeeping genes showed that most Shigella serotypes fell into three clusters of E. coli and that there were five outliers, including S. boydii type 13 (38). Four of the outlier strains also fell within the range of variation of E. coli, but S. boydii type 13 is markedly more divergent (38) and seems to be the sole representative of another species. Therefore, S. boydii type 13 may have important status and may play a role in the evolution of Shigella and E. coli.
Since Shigella strains lack flagellar (H) and capsular (K) antigens, subdivision of these organisms into different serotypes is solely based on the O antigens. There are 46 Shigella serotypes; however, there are only 33 distinct O antigen forms, and the other serotypes are variants that have resulted from phage modification (2, 4, 21, 32). Of the 33 distinct O antigen forms, 13 overlap known E. coli O antigens, and 20 are unique to Shigella clones (9, 16, 17). There are 166 O serotypes in the E. coli scheme (33), and it appears that altogether there are 186 distinct O antigen forms in E. coli and Shigella combined.
The O antigen (O polysaccharide) is part of the lipopolysaccharide (LPS) in the outer membrane of gram-negative bacteria. It consists of oligosaccharide repeating units (O units), which usually contain two to eight residues of a broad range of sugars, including both common and rarely occurring sugars and their derivatives. The O antigen is one of the most variable cell constituents due to the variation in the types of sugars present, the arrangement of the sugars within the O unit, and the linkages between O units. The surface O antigen is subject to intense selection by the host immune system and other environmental factors, such as bacteriophages, which may account for maintenance of diverse O antigen forms in species such as E. coli. Among the 186 O antigen forms of Shigella and E. coli, the chemical structure of the O antigen is known for 23 Shigella O serotypes and more than 60 E. coli O serotypes, including 3 of 13 O antigen forms found in serotypes of both taxa (22, 27).
Genes for O antigen synthesis are normally located in a gene cluster, which maps between galF and gnd in E. coli and Salmonella. Our studies of the genetic basis of O antigen variation have shown that differences among the diverse forms of the O antigen are almost entirely due to genetic variation in this gene cluster.
Seven Shigella O antigen gene clusters have been sequenced (http://www.microbio.usyd.edu.au/BPGD/default.htm). Analysis of the sequences showed that the four gene clusters which encode O antigens that are unique to Shigella strains all have features which indicate that they were recently formed (47). It seems that there has been a rapid expansion of O antigen forms in Shigella strains. The O antigen is an important factor in pathogenicity, and it has been proposed that the new O antigen forms are forms that improve fitness in the recently acquired intracellular mode of colonization of Shigella strains (38). Since S. boydii type 13 is only distantly related to E. coli and other Shigella strains, studies of the structure and genetics of its O antigen should help us understand the evolutionary history of this unique pathogenic clone.
In this paper we show that S. boydii type 13 has a nonstoichiometrically O-acetylated linear pentasaccharide phosphate O unit containing two residues each of 2-acetamido-2,6-dideoxy-l-glucose (l-QuiNAc) and 2-acetamido-2-deoxy-d-glucose and one d-glucose 1-phosphate group. We also demonstrated that the O antigen gene cluster of S. boydii type 13 is located between galF and gnd and includes genes for synthesis of UDP-l-QuiNAc, genes that encode putative sugar transferases, and O antigen processing genes. By screening with other E. coli strains (including Shigella strains), we identified several genes specific to S. boydii type 13.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All plasmids used in this study were maintained in E. coli K-12 strain DH5α, which was purchased from Beijing Dingguo Biotechnology Development Center (Beijing, People's Republic of China). The S. boydii type 13 strain used was M1350 (38), which was supplied by Johanne Lefebvre of the Laboratoire de Santé Publique du Québec, Sainte-Anne-de-Bellevue, Quebec, Canada. All other Shigella and E. coli strains used are listed in Table 1.
TABLE 1.
Bacterial strains and PCR pools used for testing S. boydii type 13
| Pool | Strains whose chromosomal DNA were included in the pool | Source |
|---|---|---|
| 1 | E. coli type strains for O serotypes 1, 2, 3, 4, 10, 16, 18, 39 | IMVSa |
| 2 | E. coli type strains for O serotypes 40, 41, 48, 49, 71, 73, 88, 100 | IMVS |
| 3 | E. coli type strains for O serotypes 102, 109, 119, 120, 125, 126, 137 | IMVS |
| 4 | E. coli type strains for O serotypes 138, 139, 149, 7, 5, 6, 11, 12 | IMVS |
| 5 | E. coli type strains for O serotypes 13, 14, 15, 17, 19ab, 20, 21, 22 | IMVS |
| 6 | E. coli type strains for O serotypes 23, 24, 25, 26, 27, 28, 29, 30 | IMVS |
| 7 | E. coli type strains for O serotypes 32, 33, 34, 35, 36, 37, 38, 42 | IMVS |
| 8 | E. coli type strains for O serotypes 43, 44, 45, 46, 50, 51, 52, 53 | IMVS |
| 9 | E. coli type strains for O serotypes 54, 55, 56, 57, 58, 59, 60, 61 | IMVS |
| 10 | E. coli type strains for O serotypes 62, 63, 64, 65, 66, 68, 69, 70 | IMVS |
| 11 | E. coli type strains for O serotypes 74, 75, 76, 77, 78, 79, 80, 81 | IMVS |
| 12 | E. coli type strains for O serotypes 82, 83, 84, 85, 86, 87, 89, 90 | IMVS |
| 13 | E. coli type strains for O serotypes 91, 92, 95, 96, 97, 98, 99, 101 | IMVS |
| 14 | E. coli type strains for O serotypes 112, 162, 113, 114, 115, 116, 117, 118 | IMVS |
| 15 | E. coli type strains for O serotypes 123, 165, 166, 167, 168, 169, 170, 171 | —b |
| 16 | E. coli type strains for O serotypes 172, 173, 127, 128, 129, 130, 131, 132 | —c |
| 17 | E. coli type strains for O serotypes 133, 134, 135, 136, 140, 141, 142, 143 | IMVS |
| 18 | E. coli type strains for O serotypes 144, 145, 146, 147, 148, 150, 151, 152 | IMVS |
| 19 | E. coli type strains for O serotypes 153, 154, 155, 156, 157, 158, 159, 164 | IMVS |
| 20 | E. coli type strains for O serotypes 160, 161, 163, 8, 9, 124, 111 | IMVS |
| 21 | E. coli type strains for O serotypes 103, 104, 105, 106, 107, 108, 110 | IMVS |
| 22 | S. boydii type strains for O serotypes 4, 5, 6, 8, 9, 11, 12, 14 | IEMd |
| 23 | S. boydii type strains for O serotypes 1, 3, 7, 8, 10, 13, 15, 16, 17, 18 | IEM |
| 24 | S. dysenteriae type strains for O serotypes 1, 2, 3, 4, 5, 6, 8 | IEM |
| 25 | S. dysenteriae type strains for O serotypes 7, 9, 10, 11, 12, 13 and S. sonnei type strains | IEM |
| 26 | S. flexneri type strains for O serotypes 6a, 1a, 1b, 2a, 2b, 3, 4a, 4b, 5(v:7), 5(v:4) | IEM |
| 27 | Like pool 23, but no S. boydii serotype 13, used as a positive control | IMVS |
IMVS, Institute of Medical and Veterinary Science, Adelaide, Australia.
O123 was obtained from the Institute of Medical and Veterinary Science; the rest of the strains were obtained from the Statens Serum Institute, Copenhagen, Denmark.
O172 and O173 were obtained from the Statens Serum Institute, Copenhagen, Denmark; the rest of the strains were obtained from the Institute of Medical and Veterinary Science.
IEM, Institute of Epidemiology and Microbiology, Chinese Academy of Preventive Medicine, Beijing, People's Republic of China.
Construction of a random DNase I shotgun bank.
Chromosomal DNA was prepared as previously described (5). Primers 1523 (5′-ATTGTGGCTGCAGGGATCAAAGAAATC-3′) and 1524 (5′-TAGTCGCGCTGNGCCTGGATTAAGTTCGC-3′), based on the galF and gnd genes, respectively, were used to amplify the S. boydii type 13 O antigen gene cluster by using the Expand Long Template PCR system (Roche). Each PCR cycle consisted of denaturation at 94°C for 10 s, annealing at 60°C for 30 s, and extension at 68°C for 15 min. To limit any PCR errors, five individual PCR products were pooled. The PCR products were digested with DNase I, and the resulting DNA fragments were cloned into pGEM-T Easy to produce a bank by using the method described previously (49).
Sequencing and analysis.
The plasmid DNA template used for sequencing was prepared by the method of Sambrook et al. (41). Sequencing was carried out by Shanghai Sangon Biological Engineering Technology and Service Co. Ltd. (Shanghai, People's Republic of China) with an ABI PRISM 377-96 automated DNA sequencer. The Staden package (43) and the Artemis program (40) were used for sequence assembly and gene annotation, respectively. The BLOCKMAKER program (20) was used for searching conserved motifs. BLAST and PSI-BLAST (3) were used for searching databases, including the GenBank database and the Pfam protein motif database (6), for possible gene functions. The algorithm that was described previously (15) was used to identify potential transmembrane segments. Sequence alignment and comparison were done with the ClustalW program (45).
Deletion of the wbwH gene from an S. boydii type 13 strain.
The wbwH gene was replaced by a chloramphenicol acetyltransferase (CAT) gene by using the RED recombination system of phage lambda (12, 51). The CAT gene was PCR amplified from plasmid pKK232-8 (Pharmacia) by using primers binding to the 5′ and 3′ ends of the gene, and each primer carried 36 bp based on the S. boydii DNA which flanks wbwH (upstream primer, 5′-TGATGACTATCTCCCCGGTAGTACTCGAGTTAGTGCATGGAGAAAAAAATCACTGG; downstream primer, 5′-AGTGCTTCATTATATTGTCAATAGCAGAAGTAACAGTTACGCCCCGCCCTGCCAC). The PCR product was transformed into an S. boydii type 13 strain carrying pKD20, and chloramphenicol-resistant transformants were selected after induction of the RED genes by the protocol described by Datsenko and Wanner (12). PCR with primers specific for the CAT gene and S. boydii type 13 DNA flanking the wbwH gene was carried out to confirm the replacement.
Preparation of LPS.
Bacteria were grown to the late log phase in 8 liters of Luria-Bertani medium by using a 10-liter fermentor (BIOSTAT C-10; B. Braun Biotech International, Melsungen, Germany) with constant aeration at 37°C and pH 7.0. Bacterial cells were washed and dried as described by Robbins and Uchida (39). The LPS (860 mg) was isolated from dried cells (15.8 g) by the phenol-water method (50) and was purified by precipitation of nucleic acids and proteins with CCl3CO2H as described previously (52).
Degradation of the LPS.
The LPS (150 mg) was hydrolyzed with aqueous 2% acetic acid at 100°C for 30 min, and a lipid precipitate was removed by centrifugation at 13,000 ×g for 20 min. The water-soluble carbohydrate portion was fractionated by gel permeation chromatography on a column (56 by 2.6 cm) of Sephadex G-50 (S) in 0.05 M pyridinium acetate buffer (pH 4.5) with monitoring with a Knauer differential refractometer to obtain two oligosaccharides, OS-I (30 mg) and OS-II (53 mg).
OS-II (45 mg) in water (1 ml) was reduced with NaBH4 (50 mg) at 20°C for 16 h, and after neutralization with concentrated acetic acid, OS-III (38 mg) was isolated by gel permeation chromatography on a column (80 by 1.6 cm) of TSK HW-40 in aqueous 1% acetic acid.
The LPS (100 mg) was heated with aqueous 12% ammonia (4 ml) at 37°C for 16 h, and after evaporation, a polysaccharide (28.5 mg) was isolated by gel permeation chromatography on a Sephadex G-50 (S) column as described above.
Sugar analysis.
OS-II was hydrolyzed with 2 M CF3CO2H at 120°C for 2 h, and sugars were identified as the alditol acetates (42) by gas-liquid chromatography (GLC) with a Hewlett-Packard model 5890 Series II instrument equipped with a 30-m capillary column containing SPB-5 (Supelco) by using a temperature gradient in which the temperature increased from 150 to 320°C at a rate of 5°C · min−1. The absolute configurations of the monosaccharides were determined by GLC of the acetylated (S)-2-octyl glycosides as described previously (31).
Methylation analysis.
OS-II (3 mg) was dephosphorylated with aqueous 48% HF at 4°C for 16 h, and the reagent was lyophilized with absorption of HF in a trap with solid NaOH. OS-II and dephosphorylated OS-II (3 mg each) were methylated with CH3I in dimethyl sulfoxide in the presence of sodium methylsulfinylmethanide (19). Partially methylated monosaccharides were derived by hydrolysis under the conditions that were used in the sugar analysis, conventionally reduced with NaBD4, acetylated, and analyzed by GLC-mass spectrometry (MS) with a Hewlett-Packard HP 5989A instrument equipped with a 30-m HP-5MS column (Hewlett-Packard) under the chromatographic conditions that were used for GLC (see above).
NMR spectroscopy.
Samples were deuterium exchanged by freeze-drying them twice from D2O and then examined as solutions in 99.96% D2O at 27°C. One-dimensional 1H nuclear magnetic resonance (1H-NMR) and 31P-NMR and two-dimensional NMR spectra were recorded with Bruker DRX-500 or DRX-600 MHz spectrometers, and one-dimensional 13C-NMR spectra were recorded with a Bruker AMX-360 MHz spectrometer by using internal acetone (δH 2.225, δC 31.45) or aqueous 85% H3PO4 (δP 0.0) as the reference. Two-dimensional NMR spectra were obtained by using standard Bruker software, and the Bruker XWINNMR 2.6 program was used to acquire and process the NMR data. Mixing times of 100 and 200 ms were used in total correlation spectroscopy (TOCSY) and rotating-frame nuclear Overhauser effect spectroscopy (ROESY) experiments, respectively.
MS.
Electrospray ionization MS was performed in the negative ion mode by using a Fourier transform ion cyclotron resonance mass analyzer (ApexII; Bruker Daltonics) equipped with a 7-T actively shielded magnet and an Apollo electrospray ion source. Samples were dissolved in a mixture of 2-propanol, water, and triethylamine (30:30:0.01, vol/vol/vol) at a concentration ∼20 ng · μl−1 and sprayed with a flow rate of 2 μl · min−1.
Other methods.
Membrane preparation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and silver staining for visualizing the LPS were carried out as described by Wang and Reeves (48).
PCR specificity assay.
Chromosomal DNA were prepared from 186 E. coli strains, including Shigella strains of different O antigen serotypes. The quality of DNA was examined by PCR amplification of the mdh gene (coding for malate dehydrogenase and present as a housekeeping gene in E. coli) by using primers as described previously (37). A total of 26 pools of E. coli and Shigella O serotypes were made, and each pool contained between 6 and 10 strains (Table 1). The pools were screened by PCR by using primers based on specific genes of S. boydii type 13. Each PCR was carried out in a 25-μl (total volume) mixture, and 15 μl of the mixture was loaded on an agarose gel to check for amplified DNA.
Nucleotide sequence accession number.
The DNA sequence of the S. boydii type 13 O antigen gene cluster has been deposited in the GenBank database under accession number AY369140.
RESULTS AND DISCUSSION
Elucidation of the structure of the S. boydii type 13 O antigen.
The LPS was isolated from dried bacterial cells of S. boydii type 13 by the phenol-water procedure (50). No high-molecular-mass polysaccharide was observed by gel permeation chromatography with Sephadex G-50 (S) after mild acid hydrolysis of the LPS with dilute acetic acid at 100°C. In contrast, O deacylation of the LPS with aqueous 12% ammonia at 37°C resulted in a polysaccharide. However, this compound produced a poorly resolved 1H-NMR spectrum, which precluded detailed structural analysis by two-dimensional NMR spectroscopy. Therefore, the products of mild acid degradation of the LPS were separated by gel permeation chromatography with Sephadex G-50 into two fractions, OS-I and OS-II, which represented a mixture of a core oligosaccharide and a fragment of the O polysaccharide containing two or more O units (OS-I) and a mixture of a core oligosaccharide and a smaller fragment consisting of one O unit (OS-II).
Sugar analysis by GLC of the alditol acetates showed that OS-II contained Glc, GlcN, and 2-amino-2-deoxyglucose (QuiN) at a ratio of 1:1.05:1.1 (detector response), as well as a minor amount of a heptose from LPS core contamination. GLC of the acetylated (S)-2-octyl glycosides confirmed the identities of the monosaccharides and revealed the d configuration of Glc and GlcN and the l configuration of QuiN. These findings are in agreement with data from a previous sugar analysis of the O polysaccharide of S. boydii type 13, including isolation and identification of l-QuiN as the crystalline N-acetyl derivative (13).
Methylation analysis of OS-II revealed 4-substituted Glc, 3-substituted GlcNAc, and 3-substituted QuiNAc. OS-II was dephosphorylated with aqueous 48% HF. Methylation analysis of the dephosphorylated OS-II revealed the same monosaccharides and, in addition, terminal GlcNAc.
OS-II was reduced with borohydride to convert Glc to glucitol (Glc-ol), and the resulting oligosaccharide (OS-III) was studied by electrospray ionization MS and NMR spectroscopy. The mass spectrum of OS-III had a mass peak for a compound with a molecular mass of 1,040.37 Da, which corresponds to a (GlcN)2(QuiN)2(Glc-ol)Ac4P oligosaccharide phosphate (whose calculated monoisotopic molecular mass is 1,040.36 Da).
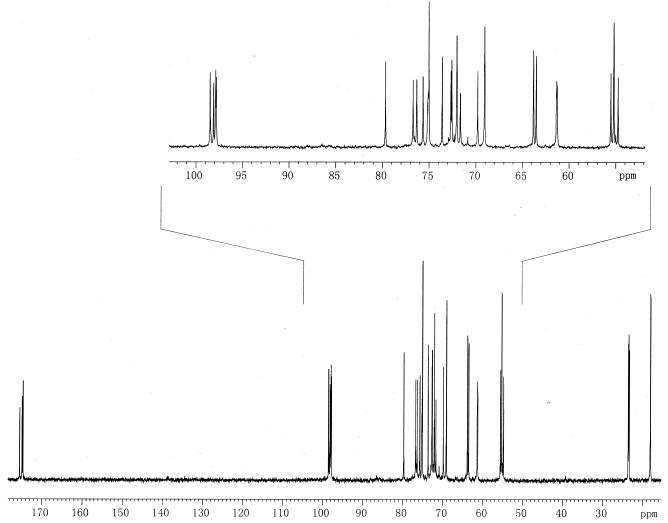
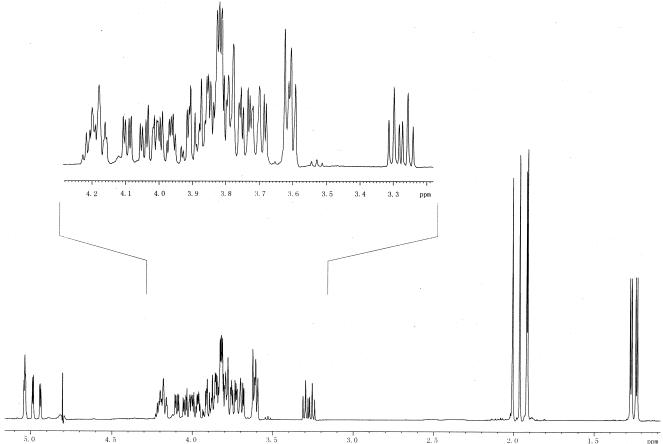
The 13C-NMR spectrum of OS-III (Fig. 1 and Table 2) had signals for four anomeric carbons at δ 97.8 to 98.5, four HOCH2-C groups (C-6 of GlcN and C-1 and C-6 of Glc-ol) at δ 61.2 to 63.8, two CH3-C groups (C-6 of QuiN) at δ 17.9 and 18.0, four nitrogen-bearing carbons (C-2 of GlcN and QuiN) at δ 54.7 to 55.5, and 16 other sugar carbons at δ 69.1 to 79.7. The signal at δ 75.1 was split due to coupling to phosphorus. The 31P-NMR spectrum of OS-III had one signal for a phosphate group at δ 0.8. The 1H-NMR spectrum of OS-III (Fig. 2 and Table 3) had signals for four anomeric protons at δ 4.94 to 5.04, two CH3-C groups (H-6 of QuiN) at δ 1.24 and 1.27, and other protons at δ 3.26 to 4.20.
FIG. 1.
90-MHz 13C-NMR spectrum of OS-III.
TABLE 2.
13C-NMR data for OS-IIIa
| Sugar residue | δ (ppm)
|
|||||
|---|---|---|---|---|---|---|
| C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | |
| α-d-GlcpNAcII-(1→ | 98.1 | 54.7 | 71.7 | 75.1b | 72.0 | 61.2 |
| →3)-α-l-QuipNAcII-(1→ | 97.9 | 55.2 | 76.7 | 75.0 | 69.1 | 17.9 |
| →3)-α-d-GlcpNAc1-(1→ | 97.8 | 55.2 | 76.3 | 69.1 | 72.7 | 61.3 |
| →3)-α-k-QuppNAC1-(1→ | 98.5 | 55.5 | 75.6 | 75.0 | 69.8 | 18.0 |
| →4)-d-Glc-ol | 63.8 | 72.0 | 72.6 | 79.7 | 73.6 | 63.5 |
Signals for N-acetyl groups are at δ 23.3, 23.4, 23.5, and 23.6 (all CH3), as well as δ 174.7 (2C), 174.9, and 175.5 (CO). The signals with a chemical shift difference of <0.1 ppm could be interchanged.
The signal is split due to coupling to phosphorus.
FIG. 2.
600-MHz 1H-NMR spectrum of OS-III.
TABLE 3.
1H-NMR data for OS-IIIa
| Sugar residue | δ (ppm)
|
||||||
|---|---|---|---|---|---|---|---|
| H-1 | H-2 | H-3 | H-4 | H-5 | H-6(6a) | H-6b | |
| α-d-GlcpNAc4PII-(1→ | 5.03 | 3.92 | 3.89 | 4.01 | 4.19 | 3.71 | 3.87 |
| →3)-α-l-QuipNAcII-(1→ | 4.94 | 4.04 | 3.78 | 3.26 | 4.20 | 1.24 | |
| →3)-α-d-GlcpNac1-(1→ | 5.04 | 4.00 | 3.83 | 3.61 | 4.17 | 3.77 | 3.84 |
| →3)-α-l-QuipNAc1-(1→ | 4.99 | 4.10 | 3.82 | 3.30 | 3.86 | 1.27 | |
| →4)-d-Glc-ol | 3.61b | 3.82 | 3.83 | 3.79 | 3.97 | 3.61 | 3.74 |
Signals for N-acetyl groups are at δ 1.91, 1.92, 1.96, and 2.01.
H-1a; H-1b at δ 3.69.
Together, these data indicated that OS-III is a linear monophosphorylated pentasaccharide containing two residues each of d-GlcNAc and l-QuiNAc and one residue of Glc-ol.
The 1H- and 13C-NMR spectra of OS-III were assigned by performing two-dimensional 1H,1H correlation spectroscopy (COSY), TOCSY, and H-detected 1H,13C heteronuclear multiquantum coherence (HMQC) experiments (Tables 2 and 3), and spin systems for the four amino sugars and Glc-ol were identified. Based on J1,2 coupling constant values of 3 to 4 Hz, it was inferred that all monosaccharide residues are α-linked.
A two-dimensional ROESY experiment showed the following correlations between the anomeric protons and protons at the linkage carbons: GlcNAcII H-1 and QuiNAcII H-3, QuiNAcII H-1 and GlcNAcI H-3, GlcNAcI H-1 and QuiNAcI H-3, and QuiNAcI H-1 and Glc-ol H-4 at δ 5.03 and 3.78, 4.94 and 3.83, 5.04 and 3.82, and 4.99 and 3.79, respectively. These data defined the modes of the linkages and the monosaccharide sequence in OS-III. An H-detected 1H,31P HMQC experiment showed that there was a correlation between the signals for phosphorus and H-4 of GlcNAcII at δ 0.8 and 4.01, and hence, GlcNAcII is phosphorylated at position 4. The substitution pattern in OS-III was confirmed by downfield displacements of the signals for C-3 of GlcNAcI, QuiNAcI, and QuiNAcII, as well as C-4 of GlcNAcII and Glc-ol, compared with the positions in the corresponding unsubstituted monosaccharides (8, 28).
These data showed that OS-III has the following structure:
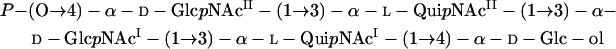 |
In addition to the major signals, the NMR spectra of OS-II, but not the NMR spectra of OS-III, contained minor signals for an O-acetyl group (δC 21.6, δH 2.13 for CH3) and a number of minor signals from an O-acetylated sugar residue. By using COSY, TOCSY, and 1H,13C HMQC spectroscopy, these signals were assigned to GlcNAcII that is O acetylated at position 6. This conclusion was based on characteristic changes in NMR chemical shifts of the minor 6-O-acetylated GlcNAcII residue compared with those of the major non-O-acetylated GlcNAcII residue. In particular, the signal for H-6 shifted downfield from δ 3.79 to 4.31 and the signal for C-6 shifted downfield from δ 61.2 to 64.2 due to a deshielding effect of the O-acetyl group. In contrast, the signal for C-5 shifted upfield from δ 72.0 to 69.5 (β-effect of O acetylation) (23). Comparison of the integral intensities of the minor signal for the O-acetyl group at δ 2.13 and each of the four major signals for the N-acetyl groups at δ 1.92 to 2.01 showed that GlcNAcII is O acetylated in only ∼15% of the O units. The absence of any O-acetyl group from OS-III can be accounted for by elimination of this group under the alkaline borohydride reduction conditions.
In order to elucidate the mode of the linkage between O units, OS-I was studied by two-dimensional COSY, TOCSY, and 1H,13C and 1H,31P HMQC spectroscopy. In addition to the cross-peaks that were present in the spectra of OS-II and OS-III, the 1H,13C HMQC spectrum had an H-1-C-1 cross-peak for α-Glc at δ 5.59 and 96.6. The 1H,31P HMQC spectrum showed a correlation of the signals for phosphorus and H-1 of Glc at δ −1.5 and 5.59. Together, these data demonstrated that an α-glucopyranosyl phosphate group (α-d-Glcp-1-P) was present. Therefore, the O units were connected via the phosphate group, and the lability of the O polysaccharide during acid hydrolysis is accounted for by easy cleavage of the glycosyl phosphate linkage, as reported previously for other bacterial polysaccharides with oligosaccharide phosphate repeating units.
Based on these data, we concluded that the O polysaccharide of S. boydii type 13 has structure 1 shown in Fig. 3. This structure exhibits marked similarity to structure 2 of the O polysaccharide of E. coli O172 (29) (Fig. 3).
FIG. 3.
Structures of the O polysaccharides of S. boydii type 13 (structure 1) (this study) and E. coli O172 (structure 2) (29). The structures show chemical repeating units, which may be any cyclic permutation of the biological repeating units that are assembled and then polymerized in the biosynthesis of the O antigens.
Sequencing of the region between galF and gnd from S. boydii type 13.
The sequence for 14,504 bases that included galF and gnd was obtained. Eleven open reading frames, not including galF and gnd, were identified, and all of them had the same transcriptional direction (from galF to gnd) (Fig. 4). The open reading frames were assigned functions on the basis of their similarity to open reading frames in the databases (Table 4).
FIG. 4.
O antigen gene cluster of S. boydii type 13.
TABLE 4.
Putative functions of proteins encoded by genes in the S. boydii type 13 O antigen gene cluster
| Protein | G+C content of gene (%) | No. of residues | Putative function | Related protein(s)
|
||
|---|---|---|---|---|---|---|
| Protein (accession no.) | No. of residues | % Identity (% similarity) | ||||
| Wzx | 31.9 | 427 | O unit flippase (Wzx) | Wzx of Yersinia pestis KIM (AAM84654) | 436 | 28 (47) |
| Wzx of Yersinia pestis CO92 (CAC92346) | 436 | 28 (47) | ||||
| Wzx of Yersinia pseudotuberculosis type O:1b (CAB63295) | 436 | 27 (47) | ||||
| Wzy | 29.1 | 393 | O antigen polymerase (Wzy) | Wzy of Escherichia coli O7 (AAD44158) | 370 | 22 (47) |
| Wzy of Escherichia coli O157:H7 (AAC32340) | 367 | 24 (47) | ||||
| WbwE | 32.8 | 262 | Glycosyltransferase | WbeiB of Edwardsiella ictaluri isolate 93-146 (AAL25629) | 256 | 23 (45) |
| Lic2B of Haemophilus influenzae strain G622 (AAM12037) | 266 | 28 (50) | ||||
| WbwF | 28.4 | 357 | Glycosylphosphotransferase (d-Glc-1-P transferase) | TarF of Bacillus subtilis strain W23 (CAC86113) | 234 | 29 (50) |
| TarL of Bacillus subtilis strain W23 (CAC86106) | 206 | 24 (46) | ||||
| WbwG | 32.7 | 345 | Glycosyltransferase | WbbK of Escherichia coli K-12 (AAC31636) | 288 | 29 (50) |
| FnlA | 37.9 | 345 | l-QuiNAc synthesis | WbvB of Vibrio cholerae O37 (AAM22595) | 343 | 76 (88) |
| QnlA | 34.6 | 292 | l-QuiNAc synthesis | WbvR of Vibrio cholerae O37 (AAM22596) | 284 | 51 (71) |
| QnlB | 36.0 | 382 | l-QuiNAc synthesis | WbvD of Vibrio cholerae O37 (AAM22597) | 374 | 68 (81) |
| WbwH | 35.2 | 406 | Glycosyltransferase (l-QuiNAc transferase) | WbuB of Escherichia coli O26 (AAN60464) | 398 | 48 (66) |
| Orf10 of Vibrio cholerae O37 (AAM22598) | 404 | 41 (64) | ||||
| Orf10 | 34.9 | 173 | Unknown | WaaK of Escherichia coli K-12 (AF019375) | 380 | 10 (28) |
| WbwI | 30.1 | 374 | O-Acetyltransferase | Mlr8454 of Mesorhizobium loti strain MAFF303099 (BAB54335) | 368 | 34 (52) |
| IS element | 41.9 | 639 | Remnant of IS1004 | IS1004 of V. cholerae O1 classical (Z67733) | 628 | 54 |
(i) Sugar biosynthetic pathway genes.
orf6, orf7, and orf8 exhibited 76, 51, and 68% sequence identity to wbvB (fnlA), wbvR (qnlA), and wbvD (qnlB), respectively, of the O antigen gene cluster of Vibrio cholerae O37. In V. cholerae O37, these genes are responsible for conversion of UDP-l-GlcNAc to UDP-l-QuiNAc by encoding a multifunctional epimerase/dehydrase, a dTDP-4-dehydrorhamnose reductase, and a UDP-N-acetylgucosamine 2-epimerase, respectively (25, 26). The process begins with dehydration of UDP-l-GlcNAc by FnlA to form UDP-2-acetamido-2,6-dideoxy-l-lyxo-hex-4-ulose, which is then reduced by QnlA to form UDP-2-acetamido-2-deoxy-l-rhamnose. UDP-2-acetamido-2-deoxy-l-rhamnose is epimerized by QnlB to form UDP-2-acetamido-2-deoxy-l-quinovose (UDP-l-QuiNAc) (25, 26). wbvB was identified as the first gene of the UDP-2-acetamido-2,6-dideoxy-l-galactose(l-FucNAc) pathway, and after discussion with other workers we designated it fnlA; the other genes are the additional genes in the UDP-l-QuiNAc pathway, which, again after discussion, we designated qnlA and qnlB. Therefore, orf6, orf7, and orf8 can be confidently identified as genes that encode enzymes for synthesis of UDP-l-QuiNAc and are designated fnlA, qnlA, and qnlB, respectively.
The other two sugar residues in the O antigen of S. boydii type 13, GlcNAc and Glc, are common sugars which are synthesized by enzymes encoded by genes outside the O antigen gene cluster.
(ii) Sugar transferase genes.
Glycosyltransferases are specific for sugar donors and sugar accepters and the linkage between them. To synthesize the O unit of the S. boydii type 13 O antigen, which contains five sugars, five individual glycosyltransferases are required. It is known that WecA transfers GlcNAc-1-phosphate or GalNAc-1-phosphate to the carrier lipid undecaprenol phosphate (UndP) to initiate O unit synthesis in the E. coli strains whose antigens have GlcNAc or GalNAc as the first sugar in the O unit (1). The wecA gene is located outside the O antigen gene cluster in E. coli. In S. boydii type 13, WecA is responsible for transfer of either GlcNAcI-1-phosphate or GlcNAcII-1-phosphate to UndP. Accordingly, one of the following sugar sequences should occur in the properly assembled O unit of S. boydii type 13:
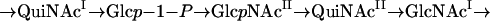 |
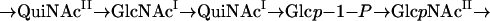 |
Four additional sugar transferase genes are expected to be present in the O antigen gene cluster of S. boydii type 13. The orf9 product exhibited 48% amino acid identity and 66% amino acid similarity to WbuB encoded by a gene in the E. coli O26 antigen gene cluster. It has been proposed that WbuB is a transferase responsible for making the linkage in the α-l-FucpNAc-(1→3)-α-d-GlcpNAc disaccharide (14). Based on the high level of similarity of orf9 and wbuB, the same type of linkage between the sugars, and the structural similarity of l-QuiNAc and l-FucNAc, we propose that orf9 encodes an l-QuiNAc transferase for making the linkage in the α-l-QuipNAc-(1→3)-α-d-GlcpNAc disaccharide. orf9 was designated wbwH.
Orf4 has one predicted transmembrane domain and exhibits similarity to TarF and TarL, which are a putative glycerophosphotransferase and a ribitolphosphotransferase, respectively, in Bacillus subtilis (30). Since both TarF and TarL are sugar phosphate transferases, it seems likely that Orf4 mediates transfer of Glc-1-P, the only sugar phosphate present in the O antigen of S. boydii type 13. orf4 was designated wbwF.
The deduced protein encoded by orf3 exhibits similarity to WbeB, a putative glycosyltransferase from Edwardsiella ictaluri, and Lic2B, a putative galactosyltransferase from Haemophilus influenzae (36). Orf5 exhibits 29% amino acid identity to WbbK, a putative glycosyltransferase from E. coli K-12 (44). orf3 and orf5 were proposed to be the remaining glycosyltransferase genes and were designated wbwE and wbwG, respectively.
(iii) O antigen processing genes.
The orf1 product exhibited 27 to 28% amino acid identity to the O unit flippase (Wzx) of Yersinia pestis KIM and CO92 and Yersinia pseudotuberculosis type O:1b. It has 12 predicted transmembrane segments, which is a feature of Wzx proteins. The Orf1 and Wzx proteins of the three Yersinia strains were analyzed by using the BLOCKMAKER program, and 11 conserved motifs were found. The consensus sequence of the conserved motifs was used in the PSI-BLAST program to search the Genpept database. The Wzx proteins of the three Yersinia strains and other distantly related Wzx proteins were retrieved (E value = 2 × e−64) after three iterations. This indicates that orf1 is an O unit flippase gene (wzx), and it was designated accordingly.
Orf2 exhibited 22 and 24% sequence identity to O antigen polymerases (Wzy) of E. coli VW187 (O7:K1) and E. coli C664-1992 (O157:H7), respectively. A transmembrane region search indicated that it had 10 predicted transmembrane segments with a large periplasmic loop of 81 amino acid residues, which is a characteristic topology for O antigen polymerases (Wzy). The Orf2 and Wzy proteins of the two E coli strains were analyzed by using the BLOCKMAKER program, and six conserved motifs were found. The consensus sequence of the conserved motifs was used in the PSI-BLAST program to search the Genpept database. Except for Wzy proteins of the two E. coli strains, only distantly related Wzy proteins were retrieved (E value = 3 × e−39) after three iterations. This indicates that orf2 is an O antigen polymerase gene (wzy), and it was designated accordingly.
Orf11 exhibited 34% amino acid identity to Mlr8454, an O-acetyltransferase from Mesorhizobium loti, and belonged to an acyltransferase family (pfam01757; E value = 6 × e−10). Therefore, we suggest that Orf11 mediates O acetylation of GlcNAcII at position 6. orf11 was designated wbwI.
S. boydii type 13 DNA between galF and gnd is the O antigen gene cluster.
O antigen gene clusters are located at different positions in different species (http://www.microbio.usyd.edu.au/BPGD/default.htm). As discussed above, based on sequencing data, we found that all genes necessary for synthesis of the S. boydii type 13 O antigen are located between galF and gnd. We then replaced wbwH of S. boydii type 13 with a CAT gene to make strain G1150. Membrane preparations were electrophoresed on SDS-PAGE gels, and LPS was visualized by silver staining. It was found that while the wild-type S. boydii type 13 strain produced normal LPS, G1150 produced only the lipid A-core part of the LPS without any O antigen (data not shown). This confirms that although S. boydii type 13 does not seem to be the same species as E. coli and other Shigella strains (38), its O antigen gene cluster is located between galF and gnd on the chromosome, which is the usual location in strains of E. coli and Salmonella.
The S. boydii type 13 gene cluster was recently assembled.
There are several components of the gene cluster that appear to be the result of gene substitutions. In addition to the genes expected for synthesis of the S. boydii type 13 O antigen mentioned above, there are two other genes, orf10 and wbwI, located in the O antigen gene cluster. orf10 exhibits 46% nucleotide acid identity with the middle part (positions 232 to 796) of the waaK gene of E. coli K-12. WaaK is a GlcNAc transferase (24). However, there has clearly been deletion in orf10, and this gene is no longer functional. It is interesting that orf10 is adjacent to wbwI, which encodes a transferase for an O-acetyl group that is not likely to be essential for polymerization and is present in only 15% of the GlcpNAcII residues. It is possible that the inactivation of orf10 was related to the acquisition of wbwI. It is also interesting in that a defective waaK homologue is present in the E. coli O26 gene cluster (14). The gene orders in S. boydii type 13 and E. coli O26 are similar at the 3′ end, and the fnlA, fnlB, fnlC, and glycosyl transferase genes and the waaK homologue of the latter organism exhibit homology with the fnlA, qnlA, qnlB, and glycosyl transferase genes and the waaK homologue of the former organism. Another comparison that should be made is with E. coli O172, which has a similar structure with UDP-l-FucNAc residues in place of two UDP-l-QuiNAc residues and a Glc residue in place of one GlcNAc residue, but all linkages in and between the pentasaccharide-phosphate O units are the same (Fig. 3). As mentioned above, UDP-l-FucNAc and UDP-l-QuiNAc have parallel biosynthetic pathways, and it will be interesting to compare the two gene clusters.
It is believed that diseases of humans caused by enteric bacterial infections emerged after agricultural settlement, which occurred about 8000 B.C., because the nature of infection and transmission makes these organisms unlikely to have been successful in the previous hunter-gatherer societies (18, 35). S. boydii type 13 must have emerged as a human pathogen in the last 10,000 years, and its O antigen cluster may have assembled as part of the adaptation to a new niche.
A region which exhibited a high level similarity to the insertion sequence IS1004 was found upstream (positions 812 to 1450) of the first O antigen gene. This region exhibited 54% nucleotide acid identity with IS1004, which is 628 bp long and is found mainly in V. cholerae clones (7). Also, orf6, orf7, orf8, and orf9 exhibit 68, 59, 67, and 49% nucleotide identity with fnlA, qnlA, qnlB, and orf10 of the V. cholerae O37 antigen gene cluster, respectively. Therefore, we suggest that at least part of the S. boydii type 13 O antigen gene cluster originated from V. cholerae or a related species.
Identification of S. boydii type 13-specific genes.
We found that in other strains sugar transferase and O antigen processing genes can be specific to O antigens. Twenty-one pairs of oligonucleotide primers were designed based on the sequences of wbwE, wbwF, wbwG, wbwH, wbwI, wzx, and wzy (Table 5) and were used to screen the genes specific for S. boydii type 13 by performing PCR with DNA from 27 DNA pools (Table 5). These pools contained DNA of strains that represented all 186 O serotypes of E. coli, including Shigella. Each of the 21 primer pairs produced a band of the predicted size from the pool containing S. boydii type 13 DNA, whereas no PCR products of the expected size were obtained from other pools. Thus, these genes are highly specific to S. boydii type 13 and have the potential to be used for detection of this strain.
TABLE 5.
PCR specificity test with S. boydii type 13 genes
| Gene | Base position of gene | Forward primer (base positions) | Reverse primer (base positions) | Length of PCR fragment (bp) | No. of pools | Anealing temp of PCR (°C) |
|---|---|---|---|---|---|---|
| wzx | 1695-3032 | 59 (2012-2030) | 60 (2931-2949) | 938 | 0a | 52 |
| 79 (1887-1904) | 80 (2599-2616) | 729 | 0 | 52 | ||
| 83 (2009-2026) | 84 (2493-2510) | 503 | 0 | 60 | ||
| wzy | 3019-4197 | 61 (3252-3269) | 62 (4081-4099) | 848 | 0 | 55 |
| 85 (3206-3224) | 43 (4160-4176) | 955 | 0 | 55 | ||
| 86 (3565-3582) | 87 (4083-4100) | 535 | 0 | 58 | ||
| wbwE | 4184-4969 | 273 (4399-4417) | 274 (4943-4962) | 564 | 0 | 50 |
| 88 (4368-4385) | 89 (4670-4688) | 321 | 0a | 55 | ||
| 90 (4227-4244) | 91 (4821-4840) | 614 | 0a | 55 | ||
| wbwF | 4972-6042 | 261 (5242-5261) | 262 (5677-5694) | 453 | 0b | 52 |
| 263 (5071-5088) | 264 (5874-5791) | 721 | 0 | 60 | ||
| 265 (5600-5617) | 266 (5889-5908) | 309 | 0 | 56 | ||
| wbwG | 6138-7172 | 67 (6089-6106) | 68 (6904-6921) | 833 | 0 | 60 |
| 96 (6306-6324) | 97 (6906-6923) | 617 | 0 | 58 | ||
| 98 (6050-6067) | 99 (6996-7013) | 964 | 0c | 60 | ||
| wbwH | 10193-11410 | 92 (10379-10397) | 93 (10953-10970) | 592 | 0 | 52 |
| 94 (10448-10465) | 95 (10813-10830) | 356 | 0 | 58 | ||
| 275 (10199-10217) | 276 (10098-11115) | 917 | 0 | 52 | ||
| wbwI | 11935-13056 | 69 (11971-11990) | 70 (12872-12891) | 921 | 0 | 62 |
| 96 (11973-11990) | 97 (12998-13015) | 1,043 | 0 | 60 | ||
| 98 (12682-12700) | 99 (12997-13014) | 333 | 0c | 58 |
One pool gave a band that was the wrong size.
One pool gave three bands that were wrong sizes.
Two pool gave bands that were wrong sizes.
Conclusions.
The structure of the S. boydii type 13 O antigen was determined together with the sequence of the O antigen gene cluster. There are two residues of the relatively rare compound N-acetylquinovosamine, two GlcNAc residues, one partially O-acetylated residue, and one glucose residue. The structure is unusual because there is a glycosyl phosphate linkage in the main chain. The sequence showed that the gene cluster consists of 11 open reading frames, 1 of which appears to have suffered a substantial deletion, leaving 10 putative functional genes which account for all functions expected for synthesis of the structure, although only 5 genes could be fully identified on the basis of homology. The presence of the IS remnant and the presence of the defective gene are both thought to indicate that there were recent evolutionary changes in the gene cluster.
Acknowledgments
We thank J. Lefebvre and all the institutes listed in Table 1 for kindly supplying strains. We are grateful to A. Kondakova and H. Moll for MS and GLC-MS analyses.
This study was supported by the Chinese National Science Fund for Distinguished Young Scholars, by the NSFC General Program, by the 863 Program, by funds from the Science and Technology Committee of Tianjin City to L.W., and by the Russian Foundation for Basic Research (grant 03-04-39020 to S.N.S.).
REFERENCES
- 1.Alexander, D. C., and M. A. Valvano. 1994. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J. Bacteriol. 176:7079-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, G. E., and N. K. Verma. 2000. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 8:17-23. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3398-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagdian, G., O. Luderitz, and A. M. Staub. 1966. Immunochemical studies on Salmonella. XI. Chemical modification correlated with conversion of group B by bacteriophage 27. Ann. N.Y. Acad. Sci. 133:405-424. [DOI] [PubMed] [Google Scholar]
- 5.Bastin, D. A., and P. R. Reeves. 1995. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene 164:17-23. [DOI] [PubMed] [Google Scholar]
- 6.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam Protein Families Database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bik, E. M., R. D. Gouw, and F. R. Mooi. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J. Clin. Microbiol. 34:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bock, K., and C. Pedersen. 1983. Carbon-13 nuclear magnetic resonance spectroscopy of monosaccharides. Adv. Carbohydr. Chem. Biochem. 41:27-65. [Google Scholar]
- 9.Brenner, D. J. 1984. Enterobacteriaceae, p. 408-420. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams and Wilkins, Baltimore, Md.
- 10.Brenner, D. J., G. R. Fanning, G. V. Miklos, and A. G. Steigerwalt. 1973. Polynucleotide sequence relatedness among Shigella species. Int. J. Syst. Bacteriol. 23:1-7. [Google Scholar]
- 11.Brenner, D. J., A. G. Steigerwalt, H. G. Wathen, R. J. Gross, and B. Rowe. 1982. Confirmation of aerogenic strains of Shigella boydii 13 and further study of Shigella serotypes by DNA relatedness. J. Clin. Microbiol. 16:432-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dmitriev, B. A., L. V. Backinowsky, N. K. Kochetkov, and N. A. Khomenko. 1973. Immunochemical studies on Shigella boydii lipopolysaccharides. Eur. J. Biochem. 34:513-518. [DOI] [PubMed] [Google Scholar]
- 14.D'Souza, J. M., L. Wang, and P. R. Reeves. 2002. Sequence of the Escherichia coli O26 antigen gene cluster and identification of O26 specific genes. Gene 297:123-127. [DOI] [PubMed] [Google Scholar]
- 15.Eisenberg, D., E. Schwarz, M. Komaromy, and R. Wall. 1984. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179:125-142. [DOI] [PubMed] [Google Scholar]
- 16.Ewing, W. H. 1986. Edwards and Ewing's identification of the Enterobacteriaceae, 4th ed. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 17.Ewing, W. H., R. W. Reavis, and B. R. Davis. 1958. Provisional Shigella serotypes. Can. J. Microbiol. 4:89-107. [DOI] [PubMed] [Google Scholar]
- 18.Fenner, F. 1970. The effects of changing social organisation on the infectious diseases of man, p. 48-68. In S. W. Boyden (ed.), The impact of civilization on the biology of man. University of Toronto Press, Toronto, Canada.
- 19.Hakomori, S. I. 1964. A rapid permethylation of glycolipids and polysaccharides catalyzed by methylsulfinyl carbanion in dimethylsulfoxide. J. Biochem. 55:205-208. [PubMed] [Google Scholar]
- 20.Henikoff, S., J. G. Henikoff, W. J. Alford, and S. Pietrokovski. 1995. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene 163:GC17-GC26. [DOI] [PubMed]
- 21.Huan, T. P., D. A. Bastin, B. L. Whittle, A. A. Lindberg, and N. Verma. 1997. Molecular characterization of the genes involved in O-antigen modification, attachment, integration and excision in Shigella flexneri bacteriophage SfV. Gene 195:217-227. [DOI] [PubMed] [Google Scholar]
- 22.Jansson, P. E. 1999. The chemistry of O-polysaccharide chains in bacterial lipopolysaccharides, p. 155-178. In H. Brade, S. M. Opal, S. N. Vogel, and D. C. Morrison (ed.), Endotoxin in health and disease. Marcel Dekker, New York, N.Y.
- 23.Jansson, P.-E., L. Kenne, and E. Schweda. 1987. Nuclear magnetic resonance and conformational studies on monoacetylated methyl d-gluco- and d-galacto-pyranosides. J. Chem. Soc. Perkin Trans. 1:377-383. [Google Scholar]
- 24.Klena, J. D., E. Pradel, and C. A. Schnaitman. 1992. Comparison of lipopolysaccharide biosynthesis genes rfaK, rfaL, rfaY, and rfaZ of Escherichia coli K-12 and Salmonella typhimurium. J. Bacteriol. 174:4746-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kneidinger, B., S. Larocque, J. Brisson, N. Cadotte, and J. Lam. 2003. Biosynthesis of 2-acetamido-2,6-dideoxy-l-hexoses in bacteria follows a pattern distinct from those of the pathways of 6-deoxy-l-hexoses. Biochem. J. 371:989-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kneidinger, B., K. O'Riordan, J. Li, J. Brisson, J. Lee, and J. Lam. 2003. Three highly conserved proteins catalyze the conversion of UDP-N-acetyl-d-glucosamine to precursors for the biosynthesis of O antigen in Pseudomonas aeruginosa O11 and capsule in Staphylococcus aureus type 5. Implications for the UDP-N-acetyl-l-fucosamine biosynthetic pathway. J. Biol. Chem. 278:3615-3627. [DOI] [PubMed] [Google Scholar]
- 27.Knirel, I. A., and N. K. Kochetkov. 1994. Structure of lipopolysaccharides from gram-negative bacteria. III. Structure of O-specific polysaccharides. Biokhimiya 59:1784-1851. [PubMed] [Google Scholar]
- 28.Knirel, Y. A., N. A. Kocharova, A. S. Shashkov, N. K. Kochetkov, E. V. Kholodkova, and E. S. Stanislavsky. 1987. Somatic antigens of Pseudomonas aeruginosa. The structure of O-specific polysaccharide chains of the lipopolysaccharides from P. aeruginosa II (Sandvik) and V (IM-1, Verder-Evans). Eur. J. Biochem. 166:189-197. [DOI] [PubMed] [Google Scholar]
- 29.Landersjo, C., A. Weintraub, and G. Widmalm. 2001. Structural analysis of the O-antigen polysaccharide from the Shiga toxin-producing Escherichia coli O172. Eur. J. Biochem. 268:2239-2245. [DOI] [PubMed] [Google Scholar]
- 30.Lazarevic, V., F. X. Abellan, S. B. Moller, D. Karamata, and C. Mauel. 2002. Comparison of ribitol and glycerol teichoic acid genes in Bacillus subtilis W23 and 168: identical function, similar divergent organization, but different regulation. Microbiology 148:815-824. [DOI] [PubMed] [Google Scholar]
- 31.Leontein, K. L., and J. Lönngren. 1993. Determination of the absolute configuration of sugars by gas-liquid chromatography of their acetylated 2-octyl glycosides. Methods Carbohydr. Chem. 9:87-89. [Google Scholar]
- 32.Lindberg, A. A., C. G. Hellerqvist, G. Bagdian-Motta, and P. H. Mäkela. 1978. Lipopolysaccharide modification accompanying antigenic conversion by phage P27. J. Gen. Microbiol. 107:279-287. [Google Scholar]
- 33.Lior, H. 1994. Classification of Escherichia coli, p. 31-72. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Wallingford, United Kingdom.
- 34.Manolov, D. G. 1959. The Shigella genus. J. Hyg. Epidemiol. Microbiol. Immunol. 3:84. [PubMed] [Google Scholar]
- 35.McKeown, T. 1988. The origins of human disease. Basil Blackwell, Oxford, United Kingdom.
- 36.Pettigrew, M. M., B. Foxman, C. F. Marrs, and J. R. Gilsdorf. 2002. Identification of the lipooligosaccharide biosynthesis gene lic2B as a putative virulence factor in strains of nontypeable Haemophilus influenzae that cause otitis media. Infect. Immun. 70:3551-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pupo, G. M., D. K. R. Karaolis, R. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robbins, P. W., and T. Uchida. 1962. Studies on the chemical basis of the phage conversion of O-antigens in the E-group Salmonella. Biochemistry 1:323-335. [DOI] [PubMed] [Google Scholar]
- 40.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualisation and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sawardeker, J. S., J. H. Sloneker, and A. Jeanes. 1965. Quantitative determination of monosaccharides as their alditol acetates by gas liquid chromatography. Anal. Chem. 37:1602-1603. [Google Scholar]
- 43.Staden, R. 1996. The Staden sequence analysis package. Mol. Bio/Technol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson, G., B. Neal, D. Liu, M. Hobbs, N. H. Packer, M. Batley, J. W. Redmond, L. Lindquist, and P. R. Reeves. 1994. Structure of the O-antigen of E. coli K-12 and the sequence of its rfb gene cluster. J. Bacteriol. 176:4144-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specifc gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wachsmuth, K., and G. K. Morris. 1989. Shigella, p. 447-462. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, New York, N.Y.
- 47.Wang, L., W. Qu, and P. R. Reeves. 2001. Sequence analysis of four Shigella boydii O-antigen loci: implication for Escherichia coli and Shigella relationships. Infect. Immun. 69:6923-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, L., and P. R. Reeves. 1994. Involvement of the galactosyl-1-phosphate transferase encoded by the Salmonella enterica rfbP gene in O antigen subunit processing. J. Bacteriol. 176:4348-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5:83-91. [Google Scholar]
- 51.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zych, K., F. V. Toukach, N. P. Arbatsky, K. Kolodziejska, S. N. Senchenkova, A. S. Shashkov, Y. A. Knirel, and Z. Sidorczyk. 2001. Structure of the O-specific polysaccharide of Proteus mirabilis D52 and typing of this strain to Proteus serogroup O33. Eur. J. Biochem. 268:4346-4351. [DOI] [PubMed] [Google Scholar]