Abstract
Background
A common single-nucleotide polymorphism (SNP) in the promoter of the Connexin-40 (Cx40) gene GJA5 was suggested to affect Cx40 promoter activity and the risk of atrial fibrillation (AF), but the role of other common Cx40 polymorphisms is unknown.
Methods and Results
8 SNPs within the Cx40 gene region were tested for association with Cx40 levels measured in atrial tissue from 61 individuals. The previously described Cx40 promoter SNP (rs35594137, −44G→ A) was not associated with Cx40 mRNA levels. However, a common SNP (rs10465885) located in the TATA box of an alternative Cx40 promoter was strongly associated with Cx40 mRNA expression (P<0.0001) and displayed strong and consistent allelic expression imbalance in human atrial tissue. A promoter-luciferase assay in cultured murine cardiomyocytes demonstrated reduced activity of the promoter containing the minor allele of this SNP (P<0.0001). Both rs35594137 and rs10465885 were tested for association with early onset lone AF (≤60 years of age) in 384 cases and 3010 population controls. rs10465885 was associated with the AF phenotype (OR=1.18, P=0.046). This result was confirmed in a meta-analysis including two additional early onset lone AF case-control cohorts (OR=1.16, P=0.022). rs35594137 was not associated with the lone AF phenotype in any of the cohorts studied or in a combined analysis.
Conclusions
A previously described Cx40 promoter SNP was not found to influence Cx40 expression or risk of AF. We describe an alternate promoter polymorphism that directly affects levels of Cx40 mRNA in vivo and is associated with early onset lone AF.
Keywords: atrial fibrillation, ion channels, genetics, allelic expression imbalance
Cardiac connexin-40 (Cx40) is found exclusively in the atria and conduction system. Along with connexin-43, it comprises the gap junctions that electrically couple atrial myocytes.1 As Cx40 is the only connexin found exclusively in the atria, several studies have investigated the effect of genetic alterations in the Cx40 gene gap junction protein, alpha 5 (GJA5) on the development of atrial conduction abnormalities and atrial fibrillation (AF). Complete absence of Cx40 in Cx40−/− mice leads to altered propagation and ectopic rhythms in the atria2 and arrhythmias on surface electrocardiogram.3 In 15 patients with idiopathic AF, Gollob and colleagues discovered three somatic and one germ line mutation in the Cx40 coding region, resulting in dominant-negative Cx40 activity.4
The Cx40 gene GJA5 contains two alternative first exons (exons 1A and 1B), utilizing separate promoters (promoters A and B), resulting in two alternative transcripts (Cx40 transcripts A and B) that share a common second coding exon.5 In 2003, Groenewegen and colleagues identified a common single nucleotide polymorphism (SNP) in the Cx40 promoter A (rs35594137, −44G→ A)6 that was later associated with decreased Cx40 promoter A activity in vitro,6, 7 atrial vulnerability and AF.8 We hypothesized that additional common Cx40 polymorphisms may also affect Cx40 expression in human atrial tissues, and that these polymorphisms may predispose to AF.
We characterized a SNP, rs10465885, in the Cx40 promoter B that alters a putative TATA box element and strongly influences expression of Cx40 transcript B and total Cx40 expression in human atria. We further determined that the promoter B SNP acts in cis, influencing expression of the same-strand transcript. The promoter B SNP was significantly associated with early onset lone AF in a case-control cohort study, and this finding was validated in a meta-analysis including two additional cohorts. In contrast to previous reports, we found that the promoter A SNP was not significantly associated with Cx40 expression in vivo, nor was it associated with the AF phenotype.
Methods
Study Subjects
Atrial appendage tissues were obtained from 61 patients undergoing coronary artery bypass grafting, valve surgery or the Maze surgical AF ablation procedure. Most patients (85.2%) from whom tissues were obtained had a history of AF, and 49.2% of patients had AF at the time of surgery. Supplemental Table 1 (online data supplement) provides a detailed description of this cohort.
Three AF case-control cohorts were used to analyze the association of the Cx40 promoter A and B SNPs with the lone AF phenotype. All case subjects had lone AF, defined as AF in the absence of structural heart disease, and all subjects were of European ancestry. In addition, subjects were deemed to have early onset lone AF if their age at AF diagnosis was ≤60 years.
Subjects from the Cleveland Clinic Lone Atrial Fibrillation GeneBank (CCAF) (n=596, with 384 early onset cases) were used as the discovery cohort along with 3010 population control subjects from the Illumina iControl Database. A replication cohort consisting of AF cases (n=119, with 45 early onset cases) and non-AF controls (n=7395) was obtained from the Atherosclerosis Risk in Communities (ARIC) study9. A second replication case-control cohort consisted of lone AF subjects (n=375, with 335 early onset cases) from the Massachusetts General Hospital Atrial Fibrillation Study (MGH)10 and non-AF controls (n=1101) from the Framingham Heart Study (FHS)11–13. Supplemental Table 2 provides a detailed description of the three cohorts. All subjects gave informed consent, and all studies were approved by their respective Institutional Review Boards.
Other methods are in the online data supplement.
Results
Haplotype analysis of the GJA5 gene
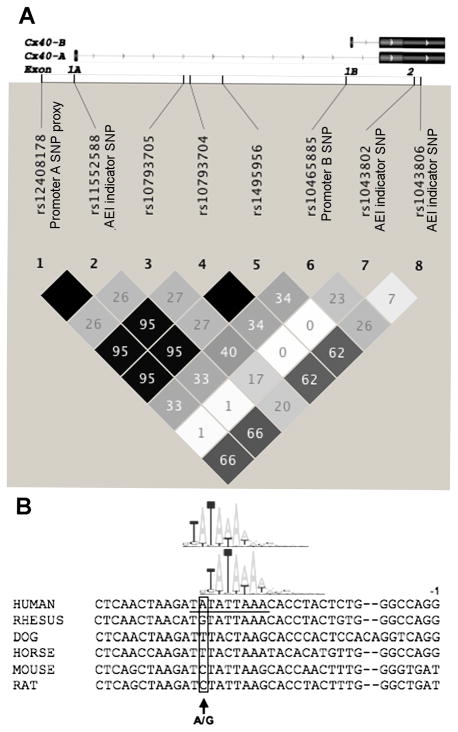
Seven contiguous SNPs from the Illumina Hap550 BeadChip located within the GJA5 gene and proximal promoter region, plus rs11552588, a component of the previously described Cx40 promoter A haplotype6, were studied. Genotypes at these eight SNPs in the 61 atrial tissue samples were loaded into Haploview v.4.1 (Broad Institute) to calculate their LD relationships (Fig. 1a). From these data it was determined that rs12408178 was in perfect LD (r2=1) with rs11552588, the SNP at +71 in exon 1A, which is in perfect LD with the promoter A SNP rs35594137 (−44G→ A).7, 8 This established rs12408178 as a perfect proxy for the promoter A SNP. In contrast, the promoter A SNP proxies were only moderately linked to the promoter B SNP (r2=0.33).
Figure 1.
Cx40 gene region and features of the promoter B SNP. (a) The two alternative transcripts (A and B) of the Cx40 gene GJA5 are illustrated, both sharing a common second coding exon. The positions of the promoter B SNP (rs10465885), the promoter A SNP proxy rs12408178 and each SNP used as an allelic expression imbalance indicator (AEI) are denoted. Linkage disequilibrium relationships (r2) between SNPs are illustrated. (b) Sequence alignment of the promoter region near the promoter B SNP in various mammalian species. The promoter B SNP site (A/G), located at −26 relative to the start site of transcription, is denoted. The overlapping TATA box sequences are underlined, and graphical representations of their respective nucleotide frequency matrices are shown above the sequence.
The promoter B SNP alters a putative TATA box
The sequence of the proximal promoter of human Cx40 transcript B, along with the homologous sequence in other mammalian species, is shown in Fig. 1b. The promoter B SNP resides at −26 bp relative to the start of transcription, where it changes one of two partially-overlapping TATA boxes in the proximal promoter. A summed nucleotide frequency score was calculated for each putative TATA box using the JASPAR14 nucleotide frequency values of each base position in a 7-base window beginning at the first ‘T’ of each sequence. The summed nucleotide frequency score of the core consensus sequence ‘TATAAAA’ was set to 1.0. Relative to the core consensus sequence, the 3’-shifted TATA box sequence had a summed nucleotide frequency score of 0.855. The 5’-shifted TATA box sequence score is dependent upon the genotype of the promoter B SNP. The ‘A’ allele of this SNP results in a summed nucleotide frequency score of 0.776, while the ‘G’ allele results in a poorer score of 0.620.
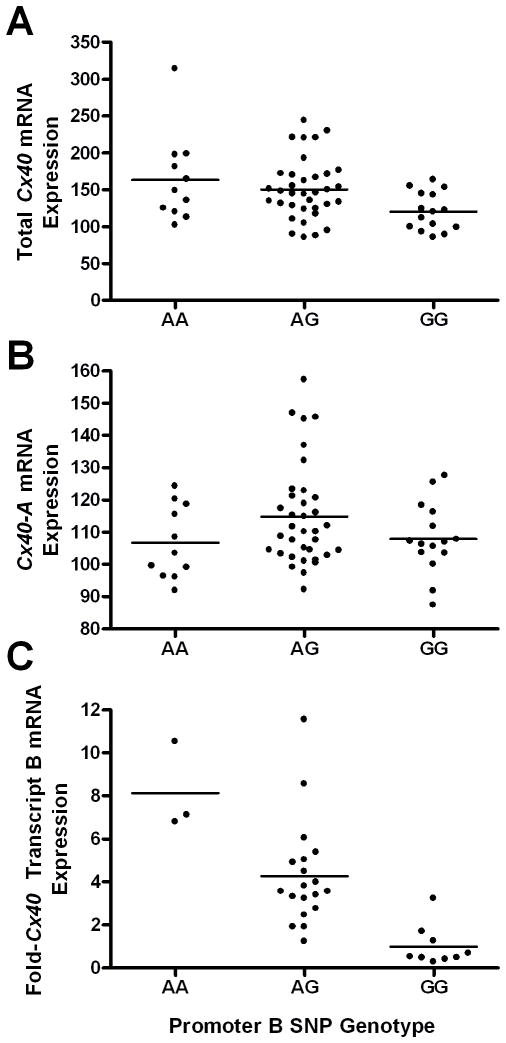
The promoter B SNP genotype is associated with Cx40 transcript levels in vivo
cDNAs from 61 human atrial tissue samples were run on Illumina Human Ref-8 v2 expression bead chips, and the signals for total Cx40 and Cx40 transcript A were analyzed. Genotypes at each of the 8 SNPs shown in Figure 1a were tested for an association with Cx40 expression levels. Only the promoter B SNP (rs10465885) genotype was significantly associated with total Cx40 mRNA levels in the 61 atrial tissue samples (Fig. 2a, P=0.013, additive genetic model), with the presence of one ‘G’ allele at the promoter B SNP genotype associated with a 16% decrease in total Cx40 mRNA expression. As expected, the promoter B SNP was not significantly associated with Cx40 transcript A levels (Fig. 2b, P=0.78, one-way ANOVA). None of the other 7 SNPs within the Cx40 gene region were significantly associated with total Cx40 mRNA levels (one-way ANOVA, data not shown).
Figure 2.
The promoter B SNP genotype is associated with total Cx40 mRNA expression. Levels of total Cx40 and Cx40 transcript A were measured by separate probes on Illumina Human Ref-8 v2 Expression Bead Chips in human atrial tissue from 61 individuals. (a) The promoter B SNP genotype was significantly associated with total Cx40 mRNA levels (P=0.013). (b) The promoter B SNP genotype was not significantly associated with levels of Cx40 transcript A (P=0.78). (c) Relative levels of Cx40 transcript B were quantified by real-time PCR in 31 atrial tissue samples. The promoter B SNP genotype was significantly associated with Cx40 transcript B levels (P<0.0001).
Because none of the probes on the microarray directly measured Cx40 transcript B, qPCR of Cx40 transcript B was performed on cDNA from 31 of the 61 atrial tissue samples. The promoter B SNP genotype was strongly associated with Cx40 transcript B expression in a dose-dependent manner (Fig. 2c, P<0.0001, one-way ANOVA), with the presence of one ‘G’ allele associated with 4.3-fold lower levels of Cx40 transcript B. In a linear regression analysis of the additive genetic model, the promoter B SNP genotype was associated with 49% of the variation in Cx40 transcript B expression in the 31 atrial tissue samples, with a corresponding quantitative LOD score of 4.85.
The promoter A SNP does not affect Cx40 levels in vivo
The genotype at the promoter A SNP proxy was not significantly associated with levels of Cx40 transcript A in the microarray assay of the 61 tissue samples (Fig. 3, P=0.55, additive genetic model). Similarly, qPCR of Cx40 transcript A in the atrial tissue samples failed to show any association of the promoter A SNP proxy with Cx40 transcript A expression (P=0.66, additive genetic model, Supplemental Figure 1). In addition, the promoter A SNP proxy was not significantly associated with levels of total Cx40 mRNA (P=0.82, additive genetic model, data not shown).
Figure 3.
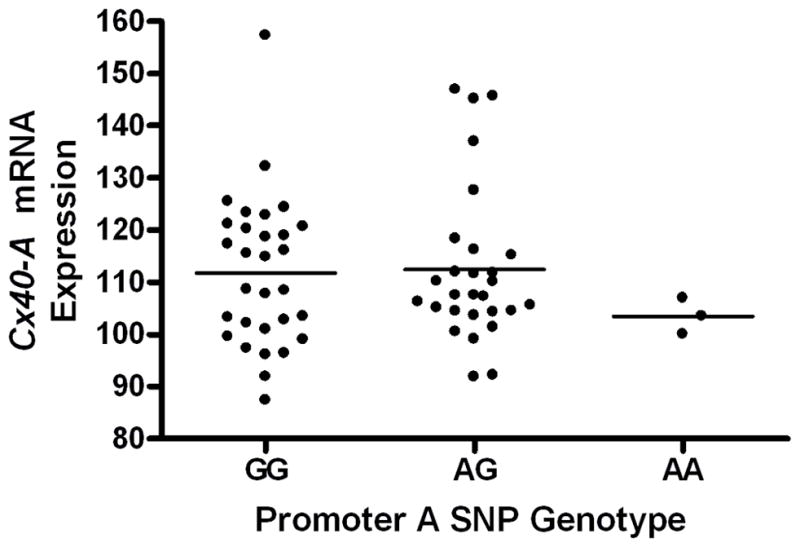
The promoter A SNP is not significantly associated with Cx40 transcript A expression (P=0.55). Levels of Cx40 transcript A were measured on Illumina Human Ref-8 v2 Expression Bead Chips in human atrial tissue from 61 individuals.
The promoter B SNP acts in cis to affect Cx40 transcript B expression in vivo
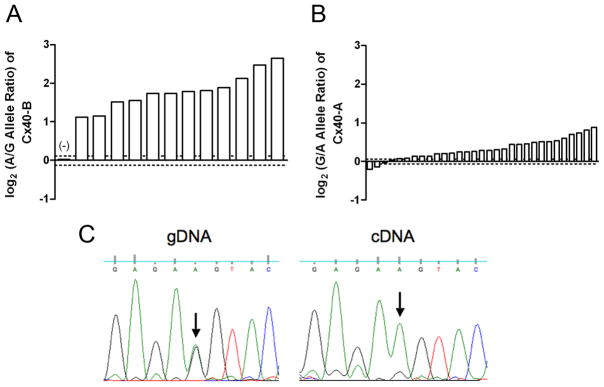
Because the promoter B SNP alters the sequence of one of an overlapping pair of TATA box elements, we hypothesized that the effect of the promoter B SNP on Cx40 transcript B expression occurs in cis through modulation of transcription. Therefore, we tested for allelic expression imbalance, comparing Cx40 transcript B levels produced from each allele in atrial tissue samples from subjects heterozygous for both the promoter B SNP and an indicator SNP within Cx40 transcript B. In all of these atrial tissue samples, the amount of mRNA produced from the minor G allele at the promoter B SNP was strongly and significantly decreased compared to the amount produced from the major A allele, with a mean decrease of 3.3-fold and a range of 2.2 to 6.3-fold (Fig. 4a). Representative sequence tracings from both cDNA and gDNA in the region surrounding the indicator SNP rs1043806 are shown in Figure 4c, illustrating the differing allelic ratios. As a control, one sample homozygous at the promoter B SNP but heterozygous for an indicator SNP was assayed, and was found to have no significant allelic expression imbalance (Fig. 4a, first bar marked as (−)).
Figure 4.
Allelic expression imbalance analysis of Cx40 transcripts A and B. Significant allelic mRNA imbalance occurred outside of a 95% confidence interval created by the gDNA allelic ratios (dotted lines). Each bar represents a different subject. (a) All of the promoter B SNP heterozygotes displayed consistent allelic expression imbalance of Cx40 transcript B favoring the ‘A’ allele. One sample homozygous for the promoter B SNP but heterozygous at an indicator SNP was included as a negative control (-). (b) Most (30) of the promoter A SNP heterozygotes demonstrated weak allelic expression imbalance of Cx40 transcript A favoring the −44A allele. Two samples were not significantly associated with allelic expression imbalance and another two samples exhibited small but significant mRNA expression imbalance in the opposite direction. (c) Representative sequence tracings at the indicator SNP rs1043806 in gDNA and Cx40 transcript B cDNA from a sample heterozygous for the promoter B SNP.
The promoter A SNP is incompletely associated with allelic expression imbalance
We measured allelic expression imbalance of Cx40 transcript A in atrial tissue samples from subjects heterozygous for the promoter A SNP and the indicator SNP rs11552588 in exon 1 of transcript A. Most samples exhibited small but significant allelic expression imbalance favoring the −44A allele, with a mean increase in Cx40 transcript A expression of 1.3-fold (Fig. 4b). However, significant allelic expression imbalance was not observed in 2 samples, and another 2 samples exhibited significant allelic expression imbalance in the opposite direction. This indicates that the promoter A SNP may be in partial LD with a potential causative genetic variant that has a minor effect on the transcription of Cx40 transcript A.
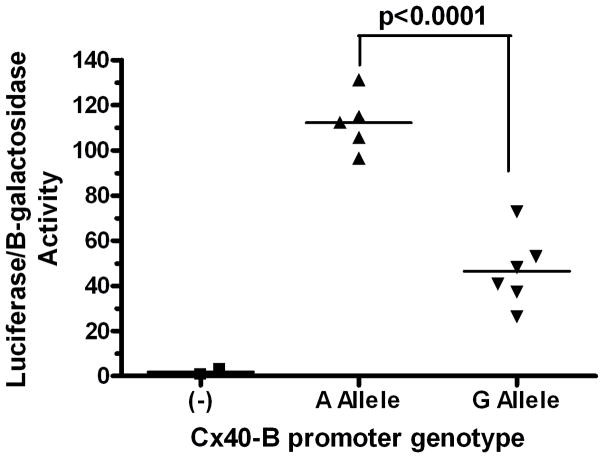
The promoter B SNP affects promoter activity in transfected HL-1 murine cardiomyocytes
To exclude the influence of other potential genetic variations in LD with the promoter B SNP on Cx40 transcript B expression, we utilized a promoter-driven reporter gene assay in cultured cells. HL-1 murine atrial cardiomyocytes were transiently transfected with a promoterless firefly luciferase reporter construct (negative control) or constructs driven by the native Cx40 transcript B promoter containing either the A or G allele at the promoter B SNP site. Each transfection included a plasmid expressing β-galactosidase as a transfection efficiency control. HL-1 cells transfected with the reporter construct containing the G allele at the promoter B SNP site exhibited 2.4-fold lower luciferase activity compared to cells transfected with the construct containing the A allele (Fig. 5, P<0.0001, student’s t-test).
Figure 5.
The promoter B SNP significantly affects Cx40 transcript B promoter activity (P<0.0001, N=6). Firefly luciferase normalized to beta-galactosidase activity is shown after transfection of reporter constructs driven by the native Cx40 B promoter with either A or G genotype at the promoter B SNP, or with a promoter-less construct (-). Data show means of individual transfected wells and the group mean.
The promoter B SNP is significantly associated with atrial fibrillation
The promoter A SNP proxy (rs12408178) and the promoter B SNP (rs10465885) were tested for association with the early onset lone AF phenotype (age of diagnosis <60 years) using 384 CCAF cases and 3010 healthy population controls in a logistic regression analysis under a log-additive genetic model. The promoter B SNP was significantly associated with early onset lone AF status with an OR=1.18 (95%CI 1.00–1.39, P=0.046, Fig. 6), with the under expressed ‘G’ allele over represented in AF cases. In contrast, the promoter A SNP proxy was not significantly associated with early onset lone AF (OR=1.11, P=0.26, Fig. 6).
Figure 6.
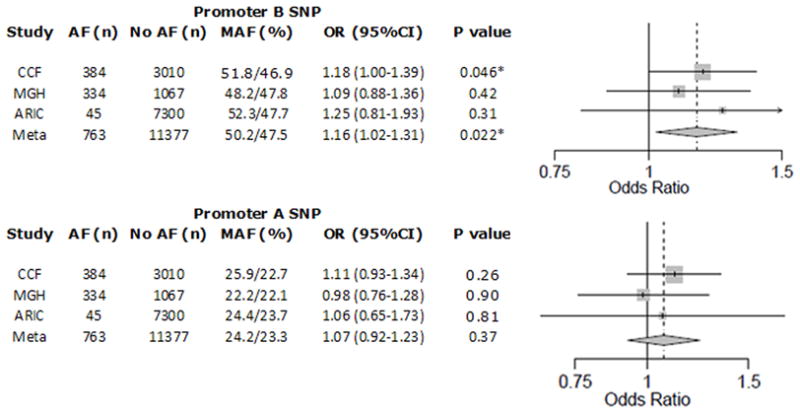
Meta-analysis of promoter B and A SNPs for association with early onset lone AF. Meta-analyses of the results (additive genetic model) from the three cohorts were performed using a fixed effect model. Promoter SNP B (top), but not promoter SNP A (bottom), was significantly associated with early onset lone AF. Grey boxes reflect the weighting (W), based on sample size, of each cohort in the meta-analysis. MAF, minor allele frequency for cases/controls; OR, odds ratio for lone AF with each additional copy of minor allele; CI, confidence interval. *denotes P<0.05.
Two additional case-control cohorts were used to confirm the findings from the discovery CCAF cohort (Fig. 6). The promoter B SNP had an OR of 1.25 for AF in 45 early-onset AF cases and 7300 non-AF controls from the ARIC cohort, but due to the small number of cases this was not statistically significant (P=0.31). The promoter A SNP proxy was not significantly associated with early onset AF in the ARIC cohort (OR=1.06, P=0.81). The promoter B SNP had an OR of 1.09 for lone AF in 334 early-onset AF cases from the MGH cohort and 1067 healthy controls from the FHS cohort, which was in the same direction as the other two studies, but not statistically significant (P=0.42). The promoter A SNP proxy was not significantly associated with early onset AF in this group (OR=0.98, P=0.90).
A meta-analysis including all three cohorts confirmed the finding of the discovery cohort, revealing a significant association between the promoter B SNP and early onset lone AF (OR=1.16, 95%CI 1.02–1.31, P=0.022, Fig. 6). A similar meta-analysis failed to find a significant association between the promoter A SNP proxy and early onset lone AF (OR=1.07, P=0.37, Fig. 6).
The meta-analyses of the early onset lone AF case control studies, including all three cohorts, had 80% power to detect a relative risk of 1.16 for the minor allele of the promoter B SNP and a relative risk of 1.18 for the minor allele of the promoter A SNP in the additive genetic model. None of the meta-analyses displayed any significant heterogeneity.
Neither the promoter A nor the promoter B SNP was significantly associated with the lone AF phenotype in any of the cohorts when subjects of all ages were included in their respective AF case cohorts (Supplemental Table 3), demonstrating the expected stronger genetic effects in younger cases.
Discussion
We used mRNA expression data and analysis of mRNA allelic expression imbalance in human atrial tissue to characterize a novel regulatory polymorphism in the Cx40 gene GJA5. This polymorphism, rs10465885, is located in the promoter region of Cx40 transcript B, where it alters the configuration of one of two partially-overlapping TATA box elements. The strong correlation between the promoter B SNP and Cx40 mRNA levels and its consistent and profound effect on Cx40 transcript B allelic mRNA expression ratios demonstrate that this polymorphism is strongly associated in cis with transcription of Cx40 transcript B in vivo. Its location in a TATA box element and the results of the promoter-luciferase reporter gene assay provide strong evidence that the promoter B SNP directly affects transcription of Cx40 transcript B.
Comparison of the promoter region altered by the promoter B SNP with a nucleotide frequency matrix of the human TATA box sequence revealed two possible overlapping TATA box sequences (Fig. 1b). The similarity of the 5′-shifted TATA box to the consensus TATA sequence is substantially improved by the G→ A transition at the promoter B SNP site. The base affected by the promoter B SNP is not strictly conserved in mammals and, interestingly, the ‘non-A’ allele seems to be present in the sequences of at least 5 other mammalian species. In addition, the chimpanzee genome at this site contains the G allele, suggesting that the A allele could be a gain-of-function mutation unique to humans.
Cx40−/− knockout mice exhibit atrial ectopy and arrhythmias on surface ECG, and rare dominant-negative defects in the Cx40 coding sequence are associated with AF in humans. However, most SNPs associated with complex traits identified by genome-wide association studies to date are not located within gene coding regions or open reading frames and are instead expected to affect gene expression. Therefore, we tested for an association between Cx40 promoter polymorphisms and AF.
We found that the newly characterized promoter B SNP was significantly associated with early onset lone AF in our discovery cohort. Although the association of the promoter B SNP with AF did not reach statistical significance in the two individual replication cohorts, these analyses were based upon smaller sample sizes and were not adequately powered to detect the effect size observed in the discovery cohort. However, the effect of the promoter B SNP observed in each of the replication cohorts was consistent with the effect observed in the discovery cohort with increased risk associated with the G allele. Thus, a meta-analysis including all three cohorts confirmed the significant association between the promoter B SNP and early-onset lone AF. In addition, the G allele resulting in diminished Cx40 expression was over represented in the AF cases, suggesting that lower Cx40 levels may also be associated with AF. This is consistent with previous observations2–4 that have suggested that impaired Cx40 expression or function predisposes to atrial conduction abnormalities and AF. The observed odds ratio of 1.16 per risk allele in the meta-analysis is modest and is expected under natural selection given the high minor allele frequency (0.47) of this SNP. However, the presence of two risk alleles is associated with an odds ratio of 1.35, which underscores the potential importance of this SNP in predicting AF risk.
Since each of the two replication cohorts did not achieve statistical significance on their own, how strong is the evidence supporting the association of rs10465885 with early onset lone AF? Firstly, all three studies had odds ratios in the same direction, showing increased risk with the under expressed G allele. Secondly, the combined meta-analysis with the greatest power was more highly significant than the discovery cohort, demonstrating that as the sample size increased the association grew stronger. Thirdly, other genetic association studies with weak effects (e.g. odds ratio of ~1.2) are often not replicated in the individual validation cohorts, but when combined, the effects are highly significant. For example, the recently indentified SNP associated with abdominal aortic aneurysm was assessed in eight separate follow up cohorts, and six of these failed to find a significant association.15 However, combining these eight cohorts together yielded a significant association.15 A prior genome wide association study meta-analysis for lone AF in subjects 65 years of age or younger detected and validated two genome-wide significant loci, but did not find a genome-wide significant association for the Cx40 gene.16 However, many real associations are present that fail to meet the strict standard of genome-wide significance, which is very stringent due to the correction for hundreds of thousands of SNPs tested. For example, genome-wide significant loci for height, which is a highly heritable trait, account for only 5% of the variance in height in studies with tens of thousands of subjects; but, 45% of the variance can explained by considering all SNPs simultaneously in a smaller cohort.17 Thus, many more SNPs add to the heritability of height that cannot be identified with certainty using current statistical methods. We conclude that the evidence on hand supports the association of the Cx40-B promoter SNP rs10465885 with early onset lone AF, although we cannot definitively rule out that this finding is a false positive.
Interestingly, the promoter B SNP was not significantly associated with lone AF when subjects of all ages were included in the analyses. The prevalence of conditions that predispose to AF such as hypertension, coronary artery disease, valve disease, and heart failure increases with age. Therefore, although these analyses were performed in subjects with lone AF, the inclusion of older subjects (with AF caused by other ‘non-genetic’ factors) in the AF case cohort may dilute the effect of a potential genetic variant. We confirmed the lack of association of the promoter B SNP with lone AF in subjects of all ages in a recent meta-analysis of five case-control cohorts comprising 1,335 lone AF case and 12,844 unaffected subjects, which overlaps the ARIC and MGH/FHS cohorts of the current study.16 In this study rs10465885 was not significantly associated with AF (odds ratio of 1.064, confidence interval 0.93 to 1.22, p=0.64). Age stratified analysis was not performed in this mega-analysis.
A common SNP in the promoter of Cx40 transcript A, first identified co-segregating with an SCN5A mutation in familial atrial standstill,6 has also been associated with atrial vulnerability and AF in a small sample of 30 individuals.8 The association with AF was later replicated in a Taiwanese population of 173 AF patients and 232 controls,7 yielding an odds ratio of 1.54 for the minor allele of promoter A SNP in the additive genetic model. Promoter-luciferase assays in cell culture showed varying (<20%7 to 65%6) reduction in the activity of the promoter containing the minor allele of the promoter A SNP. A further study suggested that the minor allele of this promoter SNP negatively affected binding of GATA4, a major cardiac-specific transcription factor, to its binding site in the Cx40 transcript A promoter.18 We sought to extend these in vitro observations by studying the effect of this SNP on Cx40 mRNA expression levels in vivo in human atrial tissues. However, we were unable to detect a significant effect of the promoter A SNP on Cx40 transcript A or total Cx40 mRNA expression. Similarly, analysis of allelic expression imbalance in individuals heterozygous at the promoter A SNP revealed minimal and inconsistent allelic expression imbalance of Cx40 transcript A. For a candidate polymorphism to be considered a true cis-acting regulatory polymorphism, allelic expression imbalance must be consistent. Thus, our data argues against a true cis-acting regulatory role for the promoter A SNP.
In contrast to previous reports,7, 8 the promoter A SNP was not significantly associated with AF in any of the case-control cohorts, or in the combined meta-analysis. The discordance of our findings with the Taiwanese study could in part be due to race-specific genetic effects or differences in the baseline patient characteristics. However, our meta-analysis had a markedly greater sample size, and was sufficiently powered to detect a relative risk of 1.18 for the minor allele of the promoter A SNP. Our study demonstrated that there was no association between the promoter A SNP and lone AF with an effect size of 1.18 or greater in populations of European ancestry.
In conclusion, we determined that a previously described Cx40 promoter A polymorphism, rs35594137 (−44G→ A), was not associated with Cx40 mRNA levels, allelic expression imbalance in human atrial tissue, or the lone AF phenotype. However, we identified a different SNP, rs10465885, as a common Cx40 promoter B variant with a direct cis-effect on Cx40 gene expression. Importantly, this promoter B SNP was also significantly associated with early onset lone AF. Additional studies are needed to more fully characterize the effects of this SNP in atrial tissue and its link to AF pathophysiology. However, this study represents an important step towards understanding the genetic determinants of Cx40 expression, as well as the potential role of variation in Cx40 expression as a determinant of AF risk.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. William Claycomb (LSU Health Science Center) for his gift of HL-1 cells. The authors thank the staff and participants of the ARIC study for their important contributions.
Funding Sources: This work was supported by NIH grant HL090620 to Drs. Chung, Barnard, Smith, and Van Wagoner. It was also supported in part by NIH/NCRR, CTSA 1UL-RR024989, Cleveland, Ohio. Robert Wirka was a Howard Hughes Medical Institute Medical Research Training Fellow. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. Dr. Alonso was partially supported by NIH grant 1RC1HL099452 and American Heart Association grant 09SDG2280087. Dr. Lubitz was supported by a training grant in the Epidemiology of Cardiovascular Disease from the NIH (T32HL007575). This work was also supported by grants from the NIH: HL092577 to Drs. Ellinor and Benjamin; and RC1-HL101056 to Dr. Benjamin; and DA027021 to Dr. Ellinor.
Footnotes
Conflict of Interest Disclosures: None
References
- 1.van Veen AA, van Rijen HV, Opthof T. Cardiac gap junction channels: modulation of expression and channel properties. Cardiovasc Res. 2001;51:217–29. doi: 10.1016/s0008-6363(01)00324-8. [DOI] [PubMed] [Google Scholar]
- 2.Bagwe S, Berenfeld O, Vaidya D, Morley GE, Jalife J. Altered right atrial excitation and propagation in connexin40 knockout mice. Circulation. 2005;112:2245–53. doi: 10.1161/CIRCULATIONAHA.104.527325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirchhoff S, Nelles E, Hagendorff A, Kruger O, Traub O, Willecke K. Reduced cardiac conduction velocity and predisposition to arrhythmias in connexin40-deficient mice. Curr Biol. 1998;8:299–302. doi: 10.1016/s0960-9822(98)70114-9. [DOI] [PubMed] [Google Scholar]
- 4.Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, Shao Q, Liu X, Veinot JP, Tang AS, Stewart AF, Tesson F, Klein GJ, Yee R, Skanes AC, Guiraudon GM, Ebihara L, Bai D. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med. 2006;354:2677–88. doi: 10.1056/NEJMoa052800. [DOI] [PubMed] [Google Scholar]
- 5.Dupays L, Mazurais D, Rucker-Martin C, Calmels T, Bernot D, Cronier L, Malassine A, Gros D, Theveniau-Ruissy M. Genomic organization and alternative transcripts of the human Connexin40 gene. Gene. 2003;305:79–90. doi: 10.1016/s0378-1119(02)01229-5. [DOI] [PubMed] [Google Scholar]
- 6.Groenewegen WA, Firouzi M, Bezzina CR, Vliex S, van Langen IM, Sandkuijl L, Smits JP, Hulsbeek M, Rook MB, Jongsma HJ, Wilde AA. A cardiac sodium channel mutation cosegregates with a rare connexin40 genotype in familial atrial standstill. Circ Res. 2003;92:14–22. doi: 10.1161/01.res.0000050585.07097.d7. [DOI] [PubMed] [Google Scholar]
- 7.Juang JM, Chern YR, Tsai CT, Chiang FT, Lin JL, Hwang JJ, Hsu KL, Tseng CD, Tseng YZ, Lai LP. The association of human connexin 40 genetic polymorphisms with atrial fibrillation. Int J Cardiol. 2006;27:27. doi: 10.1016/j.ijcard.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 8.Firouzi M, Ramanna H, Kok B, Jongsma HJ, Koeleman BP, Doevendans PA, Groenewegen WA, Hauer RN. Association of human connexin40 gene polymorphisms with atrial vulnerability as a risk factor for idiopathic atrial fibrillation. Circ Res. 2004;95:e29–33. doi: 10.1161/01.RES.0000141134.64811.0a. [DOI] [PubMed] [Google Scholar]
- 9.Alonso A, Agarwal SK, Soliman EZ, Ambrose M, Chamberlain AM, Prineas RJ, Folsom AR. Incidence of atrial fibrillation in whites and African-Americans: the Atherosclerosis Risk in Communities (ARIC) Study. Am Hear J. 2009;158:111–117. doi: 10.1016/j.ahj.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellinor PT, Yoerger DM, Ruskin JN, MacRae CA. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;118:179–84. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 11.Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, D’Agostino RBS, Fox CS, Larson MG, Murabito JM, O’Donnell CJ, Vasan RS, Wolf PA, Levy D. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 12.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 13.Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–4. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gretarsdottir S, Baas AF, Thorleifsson G, Holm H, den Heijer M, de Vries J-P, Kranendonk SR, Zeebregts CJ, van Sterkenburg SM, Greelkerken RH, van Rij AM, Williams MJ, Boll AP, Kostic JP, Jonasdottir A, Jonasdottir A, Walters GB, MAsson G, Sulem P, Saemundsdottir J, Mouy M, Magnusson KP, Tromp G, Elmore JR, Sakalihasan N, Limet R, Defraigne J-O, Ferrell RE, Ronkainen A, Ruigrok YM, Wijmenga C, Grobbee DE, Shah SH, Granger CB, Quyyumi AA, Vaccarino V, Patel RS, Zafari AM, Levey AI, Austin H, Girelli D, Pignatti PF, Olivieri O, Martinelli N, Malerba G, Trabetti E, Becker LC, Becker DM, Reilly MP, Rader DJ, Mueller T, Dieplinger B, Haltmeyer M, Urbonavicius S, Lindblad B, Gottsater A, Gaetani E, Pola R, Wells P, Rodger M, Forgie M, Langlois N, Corral J, Vicente V, Fontcuberta J, Espana F, Grarup N, Jorgensn T, Witte DR, Hansen T, Pedersen O, Aben KK, de Graff J, Holewijn S, Folkersen L, Franco-Cereceda A, Eriksson P, Collier DA, Stefansson H, Steinthorsdottir V, Olafsson K, Magnusson MK, Palmason R, Haraldsdottir S, Andersen K, Onundarson PT, Thorgeirsson G, Kiemeney LA, Powell JT, Carey DJ, Kuivaniemi H, Lindholt JS, Jones GT, Kong A, Blankensteijn JD, Matthiasson SE, Thorsteinsdottir U, Stefansson K. Genome-wide association study identifies a sequence variant within the DAB2IP gene conferring susceptibility to abdominal aortic aneurysm. Nat Genet. 2010;42:692–697. doi: 10.1038/ng.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, Sinner MR, de Bakker PIW, Mueller M, Lubitz SA, Fox E, Darbar D, Smith NL, Smith JD, Schnabel RB, Soliman EZ, Rice KM, Van Wagoner DR, Beckman B-M, van Noord C, Wang K, Ehret GB, Rotter JI, HAzen SL, Steinbeck G, Smith AV, Launer LJ, Harris TB, Makino S, Nelis M, Milan DJ, Perz S, Esko T, Kottgen A, Moebus S, Newton-Cheh C, Li M, Mohlenkamp S, Wang TJ, Kao WHL, Vasan RS, Nothen MM, MacRae CA, Stricker BHC, Hofman A, Uitterlinden AG, Levey D, Boerwinkle E, Metspalu A, Topol EJ, Chakravarti A, Gudnason V, Psaty BM, Roden DM, Meitinger T, Wichmann H-E, Witteman JCM, Barnard J, Arking DE, Benjamin EJ, Heckbert SR, Kaab S. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firouzi M, Bierhuizen MF, Kok B, Teunissen BE, Jansen AT, Jongsma HJ, Groenewegen WA. The human Cx40 promoter polymorphism −44G-->A differentially affects transcriptional regulation by Sp1 and GATA4. Biochim Biophys Acta. 2006;1759:491–6. doi: 10.1016/j.bbaexp.2006.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.