SUMMARY
DnaK is a molecular chaperone responsible for multiple aspects of proteostasis. The intrinsically slow ATPase activity of DnaK is stimulated by its co-chaperone, DnaJ, and these proteins often work in concert. To identify inhibitors, we screened plant-derived extracts against a re-constituted mixture of DnaK and DnaJ. This approach resulted in the identification of flavonoids, including myricetin, which inhibited activity by up to 75%. Interestingly, myricetin prevented DnaJ-mediated stimulation of ATPase activity, with minimal impact on either DnaK’s intrinsic turnover rate or its stimulation by another co-chaperone, GrpE. Using NMR, we found that myricetin binds DnaK at an unanticipated site between the IB and IIB subdomains and that it allosterically blocked binding of DnaJ. Together, these results highlight a “gray box” screening approach, which approximates a limited amount of the complexity expected in physiological, multi-protein systems.
INTRODUCTION
Many emerging drug targets operate as part of multi-protein complexes in vivo (Gavin et al., 2002). Often, these ensembles have a single enzymatic output, such as ATP consumption, that is catalyzed by one core component. The non-enzyme partners typically regulated this activity, by impacting subcellular localization, substrate selection and/or turnover rates. Because the protein partners are often important for tuning biological function and integrating information from signaling pathways, there is interest in identifying molecules that specifically target the protein-protein interactions in complexes (Arkin and Wells, 2004; MacBeath and Schreiber, 2000). Such inhibitors are often identified by either screening for protein-protein contacts in purified systems or by structure-guided design (Horswill et al., 2004; Kortemme and Baker, 2002; Magliery et al., 2005; Vassilev et al., 2004; Yin and Hamilton, 2005). These strategies are increasingly useful, but all in vitro screens are prone to attrition when the active compounds are taken into cell-based or animal systems. An alternative approach is to use “black box” cell-based, phenotypic screens. Cell-based approaches are powerful because they incorporate multiple parameters, such as an intact plasma membrane, a full complement of regulatory pathways and native multi-protein complexes, which better approximate physiological states. However, post-screen target identification can often be challenging. We have been pursuing an approach that might be considered a compromise between these platforms. In this strategy, multiple components of a known protein complex are purified and re-constituted in vitro (Chang et al., 2008a; Miyata et al., 2010b). Only one component typically has measurable activity, yet the other, ancillary partners are likely to impact the turnover rate, approximating a more physiological enzymatic output. In addition, protein-protein interactions between the core enzyme and its partners might mask some potential drug-binding sites, while inducing conformers that reveal other, latent sites. We term this approach “gray box”, because it is expected to find the middle ground between biochemical and cell-based assays, providing some limited amount of physiological complexity while reducing investment in downstream target identification.
As a model system, we have principally focused on the Escherichia coli DnaK protein, which belongs to the highly conserved heat shock protein 70 (Hsp70) family. These molecular chaperones participate in many aspects of cellular proteostasis (Genevaux et al., 2007; Powers et al., 2009) and are emerging drug targets (Evans et al., 2010). Like all Hsp70 family members, DnaK consists of two domains: a 41-kDa nucleotide binding domain (NBD), which provides energy for the chaperone machine by hydrolyzing ATP, and a 26-kDa substrate binding domain (SBD) which binds to the exposed hydrophobic regions of unfolded or partially folded proteins (Bertelsen et al., 2009; Chang et al., 2008b; Genevaux et al., 2007; Harrison et al., 1997; Zhu et al., 1996). The NBD is further divided into four subdomains, IA, IIA, IB and IIB, which constitute a deep, ATP-binding cleft (Harrison et al., 1997) (Figure 1A). The NBD is connected to the SBD by a hydrophobic linker, which is thought to regulate inter-domain allosteric crosstalk (Han and Christen, 2001; Swain et al., 2007) and linking nucleotide state to substrate binding affinity. Briefly, the ATP-bound form of DnaK has relatively poor affinity for its protein substrates, while the ADP-bound state binds tightly (Buchberger et al., 1995; Szabo et al., 1994).
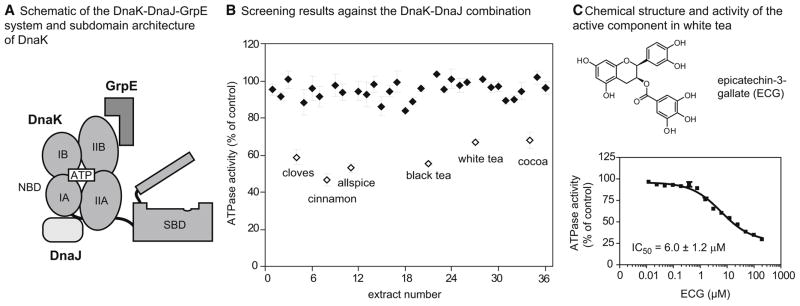
Figure 1. Screening plant extracts against the DnaK-DnaJ system reveals epicatechin-3-gallate as the major inhibitor in white tea.
(A) Schematic of the DnaK chaperone, with the nuceotide-binding domain (NBD), substrate-binding domain (SBD) and the NBD subdomains (IA, IIA, IB, IIB) highlighted. The co-chaperones DnaJ and GrpE are shown near their approximate binding sites on DnaK’s NBD. (B) Results of the screen of natural product extracts (40 μg/mL) against the ATPase activity of the DnaK (0.6 μM) and DnaJ (1 μM). Each extract was screened in duplicate and the range is shown relative to a solvent control. The active compounds (> 25% inhibition) are shown in open symbols (Table S1). (C) Chemical structure of ECG and its dose dependent inhibition of DnaK-DnaJ. The active component of white tea was identified by bio-assay guided fractionation and the structure confirmed by comparing the NMR spectra to an authentic sample (Figure S1). The dose dependence experiment was performed against DnaK-DnaJ in triplicate and the error bars represent standard error of the mean.
Nucleotide turnover by DnaK is tightly controlled by co-chaperones, including DnaJ and GrpE (Liberek et al., 1991). DnaJ belongs to the Hsp40 family of co-chaperones and it is thought to bind the IA and IIA subdomains of DnaK (Gassler et al., 1998; Jiang et al., 2007; Suh et al., 1998) (Figure 1A). In turn, this protein-protein interaction allosterically stimulates the ATPase activity of DnaK. In the presence of DnaJ, ADP release becomes rate-limiting and another co-chaperone, GrpE, assists nucleotide exchange via protein-protein interactions with the IB and IIB subdomains (Harrison et al., 1997; Liberek et al., 1991). Thus, the combination of DnaK, DnaJ and GrpE efficiently turns over ATP and allows control over chaperone activity (Liberek et al., 1991; Russell et al., 1998; Schroder et al., 1993). While only the core chaperone, DnaK, has any enzymatic activity, knockouts of DnaJ or GrpE share some phenotypes, such as temperature-sensitivity, in common with ΔdnaK cells (Ang et al., 1986; Sell et al., 1990). These results suggest that the entire network is required for efficient chaperone activity. Further, mutants that specifically disrupt interactions between DnaK and DnaJ give similar phenotypes (Gassler et al., 1998), further suggesting that protein-protein contacts among the components are required.
Using this well characterized system, we sought to develop screens to specifically target its emergent properties. Namely, because ATP turnover is greatly accelerated by the DnaJ co-chaperone, we developed an ATPase assay that measures turnover by the DnaK-DnaJ system (Chang et al., 2008a; Miyata et al., 2010b). Here, we screened a library of plant-derived, natural product extracts and identified myricetin as an inhibitor. Consistent with the screening design, this compound selectively inhibited the DnaK-DnaJ complex and had little activity against either DnaK alone or the DnaK-GrpE combination. Unexpectedly, we found that myricetin bound to the IB and IIB subdomains, approximately 20–30 Å away from the DnaJ-DnaK interacting site, yet it appeared to influence the DnaK-DnaJ interaction. Finally, we discuss how this approach might provide a template for “gray box” screening against other re-constituted multi-protein complexes in vitro.
RESULTS
Screening for Natural Products that Inhibit the DnaK-DnaJ Complex
To initiate a screen for potential inhibitors of the DnaK-DnaJ combination, we assembled a collection of organic extracts from 36 commercial spices and crude plant materials. For this specific study, we chose to use common plant materials, containing largely known compounds, to facilitate downstream identification of active components. Also, because the Hsp70-Hsp40 family is highly conserved and expressed in many plant pathogens, we were interested in whether natural product extracts might be particularly enriched for inhibitors of this system.
This collection was arrayed in 96-well plates and screened at 40 μg/mL using a previously established, high throughput ATPase assay (Chang et al., 2008a). The key design criteria included the use of high ATP concentrations (1 mM, ≫ Km) to minimize discovery of nucleotide-competitive inhibitors. Secondly, we employed an optimized ratio of DnaK (0.6 μM) to DnaJ (1 μM) that gives robust ATP turnover and good Z′ scores (~ 0.6), to favor identification of inhibitors (Chang et al., 2008a; Miyata et al., 2010b). Using this approach, we screened each crude extract in triplicate and found that only 6 of 36 extracts (allspice, black tea, cocoa, cinnamon, cloves, and white tea) inhibited ATPase activity by more than 30% (Figure 1B and Table S1). Using activity-guided fractionation on reverse phase HPLC columns, we identified the major active component from the white tea extract as epicatechin-3-gallate (ECG) (Figures 1C and S1). ECG is known to be prevalent in this plant (Friedman, 2007). In dose dependence experiments, the IC50 of authentic ECG was 6.0 ± 0.9 μM against the DnaK-DnaJ system (Figure 1D).
Identification of the Structure-Activity Relationships (SAR) of Flavonoid-Based Inhibitors
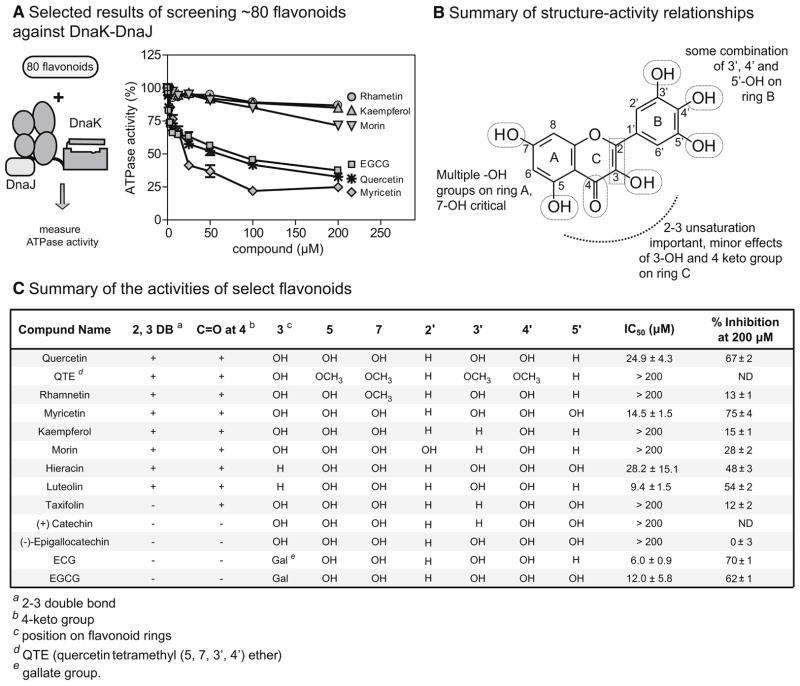
ECG is a member of a large family of plant-derived flavonoids that possess a variety of anti-oxidation and other activities (Friedman, 2007; Khan and Mukhtar, 2008). Although these well-known scaffolds often have modest selectivity, they can be powerful probes for identifying new drug-binding sites (Cuccioloni et al., 2009; Shapiro, 2004). Therefore, we chose to further explore the structure-activity relationships (SAR) of flavonoids in the specific context of the DnaK-DnaJ system. Towards that goal, we collected 80 related flavonoids (structures available in Figure S2) and determined their activity against DnaK-DnaJ (Figure 2A). This approach provided a number of key SAR observations, which are summarized in Figure 2B. For example, we found that the free phenolic groups on ring A were required for activity; quercetin provided up to 67% inhibition (IC50 = 27.6 μM), whereas its tetramethoxy derivative, quercetin tetramethyl (5,7,3′,4′) ether (QTE), had no significant activity (IC50 > 200 μM) (Figure 2C). Most strikingly, rhamnetin, which differs from quercetin only by a 7-methoxy substitution on ring A, was also inactive, suggesting a particularly important role for the C7 phenolic group (Figure 2C). On ring B, hydroxyl groups at the 3′, 4′ and 5′ positions appeared to improve activity. For example, myricetin, with hydroxyls at all three positions, provided the highest percent inhibition (75 % at 200 μM; IC50 = 14.5 μM), while the 4′ mono-hydroxylated kaempferol lacked activity (IC50 > 200 μM) (Figure 2C). On ring C, we found that, although the alkene was clearly required, the hydroxyl group on position 3 made only minor contributions to activity, as revealed by comparing myricetin and quercetin to their counterparts lacking this modification, hieracin (IC50 = 28.2 μM) and luteolin (IC50 = 9.4 μM) (Figure 2C). It is worth noting that epicatechin-3-gallate (ECG) and epigallocatechin 3-gallate (EGCG), which contain an extra gallate group appended to ring C did not fit the general SAR patterns, which might suggest that they have a different binding mode (Figure 2C). Because myricetin provided the best overall inhibition, we chose to further characterize its binding and mechanism.
Figure 2. The structure activity relationship of flavonoid-based inhibitors of DnaK-DnaJ.
(A) Approximately 80 flavonoids were chosen based on their structural similarity to EGC (Figure S2) and screened against the DnaK-DnaJ ATPase activity. The activities of select examples are shown in relation to a DMSO solvent control. The ATPase assays were performed in triplicate and the error bars represent standard error of the mean. (DnaK, 0.6 μM; DnaJ, 1 μM) (B) A summary of the structure-activity relationships, highlighting the key structural features of the flavonoid scaffold important for activity against DnaK-DnaJ. (C) Summary of the functional group decoration and activity of select flavonoids. These examples were selected to highlight key SAR observations.
Myricetin Binds to the IB and IIB Subdomains of DnaK
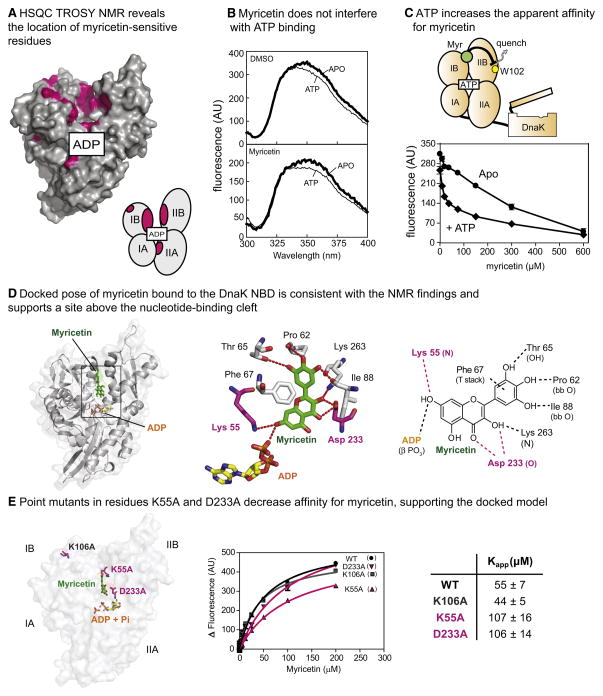
To gain insight into how flavonoids inhibit ATPase activity, we first performed nuclear magnetic resonance (NMR) experiments to examine whether the compound might bind DnaK. In these studies, 1H-15N labeled DnaK NBD (250 μM, in the presence of 1 mM ADP) was titrated with myricetin and a two dimensional 1H-15N HSQC-TROSY experiment was performed. These experiments revealed a set of chemical shift perturbations mainly in the IB and IIB subdomains facing the nucleotide binding cleft, along with scattered residues of the IA and IIA (Figure 3A and S3A). Interestingly, these results suggested that myricetin did not bind to the same sites as ATP or DnaJ; instead, the binding site appeared to be in a distinct location “above” the nucleotide-binding cleft (Figure 3A).
Figure 3. Myricetin binds to the IB and IIB subdomains of DnaK NBD and does not compete with ATP.
(A) Mapping of the chemical shifts above 0.02 ppm (pink; Figure S3A) onto DnaK suggests that myricetin binds to the upper nucleotide cleft, between the IB and IIB subdomains. (PDB: 1DKG). The approximate position of the nucleotide is shown for orientation and the cartoon schematic is used to identify the subdomains. (B) Saturating myricetin (100 μM) did not block tryptophan fluorescence (excitation 295 nm) in response to ATP. DnaK = 5 μM, ATP = 100 μM. Results are representative of experiments performed in triplicate. (C) ATP (2 mM) enhanced the apparent affinity of myricetin for DnaK, as measured by tryptophan fluorescence (Ex 295/Em. 342 nm). DnaK = 5 μM. Results are the average of triplicates and the error bars represent standard error of the mean. Error bars are often smaller than the symbols. (D) A representative conformation of myricetin in DnaK, determined by LD simulations of the myrecitin+ADP+Pi DnaK NBD (see the Methods section for details). Contacts observed are in good agreement with observed NMR shifts and SAR data. (E) Point mutations in K55A and D233A (highlighted in purple) significantly reduce binding of myricetin to DnaK, as measured by tryptophan fluorescence, compared to wild type and a control mutant (K106A). Results are the average of triplicate experiments and the error is standard error of the mean.
To further explore this model, we used intrinsic tryptophan fluorescence to study myricetin-induced, structural changes in DnaK. This chaperone has a single tryptophan (Trp102), which is present at the NBD-SBD interface, which has been extensively used as a probe for nucleotide-induced structural transitions (Buchberger et al., 1995). Briefly, ATP is known to induce a blue shift (349 to 345 nm), with a corresponding 18% decrease in intensity (Buchberger et al., 1995; Theyssen et al., 1996). When we saturated DnaK with myricetin (100 μM), tryptophan fluorescence responded normally to ATP binding, suggesting that the compound did not strongly influence the nucleotide-induced structural changes (Figure 3B). Additionally, it was reported that when flavonoids bind near a tryptophan residue, they quench its fluorescence (Conseil et al., 1998). Based on our NMR results, myricetin appeared to interact near (~10 Å) Trp102; therefore, we expected to see a fluorescence quench as myricetin bound to DnaK. Indeed, myricetin quenched the Trp fluorescence of nucleotide-free (Apo) DnaK (Figure 3C). Further, when we added saturating levels of ATP (2 mM), the apparent affinity (Kapp) of myricetin decreased from ~600 μM to 38 μM, suggesting that myricetin had even better binding affinity when ATP was present (Figure 3C). These results support a model in which myricetin binds to a site on DnaK that is distinct from the ATP-binding site and that ATP might even improve its affinity for DnaK.
The NMR chemical shift results were initially puzzling because the distance between the responsive residues on the IB and IIB subdomains in the available crystal structure (PDB code: 1DKG; (Harrison et al., 1997)) seemed unlikely to be filled by myricetin. However, the 1DKG crystal structure likely represents an “open” form of DnaK’s NBD because it is a co-complex with GrpE and the IB and IIB subdomains have been proposed to be closer to each other during the ATPase cycle (Bhattacharya et al., 2009; Woo et al., 2009). To explore possible conformational changes in DnaK and how they might impact myricetin binding, we turned to computer simulations. First, a more dynamic structure of DnaK’s NBD was examined using unrestrained all-atom Langevin Dynamics (LD) simulations. In these simulations, the presence of ATP or ADP/Pi significantly altered the conformation of DnaK, bringing the IB and IIB subdomains into proximity. Docking calculations based on those “closed” forms of DnaK revealed that myricetin could occupy a position that satisfied the observed NMR shifts (data not shown). While it was satisfying that the observed contacts were consistent with the NMR studies, the conformations were obtained without myricetin present. Therefore, to determine a conformation for the system that could incorporate any induced-fit phenomena we next conducted additional LD simulations of DnaK’s NBD with myricetin, ADP, and Pi bound. The simulations started from the open form, and the protein was allowed to collapse to a closed structure that best incorporated myricetin. These simulations also supported the binding site between the IB and IIB subdomains that was suggested by docking (Figure 3D). There was a wide degree of sampling seen for myricetin, but the conformation shown in the figure represents over half of the observed conformations. Importantly, the observed binding mode was consistent with the SAR. For example, the critical 7-hydroxyl on ring A could form hydrogen bonds to the β-phosphate of ADP and Lys55. Further, the three hydroxyls of the B ring formed hydrogen bonds with the side-chain hydroxyl of Thr65 and the backbone carbonyl oxygens of Pro62 and Ile88. Finally, the unsaturation in ring C is most likely required for aligning ring B to form those hydrogen bonds and van der Waals contacts with Pro90, and a T-shaped aromatic stacking interaction with Phe67. Together, these contacts appeared to assist the closing of the upper nucleotide-binding cleft, with myrectin interacting with both the IB and IIB subdomains.
To test this model, we generated point mutants designed to disrupt the interaction. Specifically, we targeted Lys55 and Asp233, which are proposed to make contacts with the 7-position hydroxyl on ring A and oxygens on ring C. In addition, we selected another residue far from the proposed myricetin-binding site (Lys106) as a negative control (Figure 3E). Substituting alanine in these location generated the mutants K55A, D233A and K106A, which were well behaved by circular dichroism (Chang et al., 2010). Using tryptophan fluorescence, we then measured their apparent affinity for myricetin. Consistent with the previous studies, the K55A and D233A mutants bound myricetin only weakly (Kapp >100 μM), while the control mutant, K106A, was similar to the wild type (Kapp 43 ± 5 and 55 ± 7 μM, respectively) (Figure 3E). These results further supported the proposed flavonoid interaction site.
Thus far, these studies have focused on potential binding of myricetin to DnaK. However, it also seemed possible that this compound might directly interact with DnaJ, so we performed isothermal calorimetry (ITC) studies to test this idea. Briefly, we titrated myricetin into a solution of DnaJ (10 μM) and found that it had no apparent affinity (Figure S3B). However, these titrations were “noisy”, which precluded a definitive conclusion. Therefore, we performed similar titrations into the truncated J domain (residues 2–108), which is better behaved in ITC experiments (unpublished observations and Figure S3B). In these studies, myricetin clearly failed to bind the J domain (Figure S3B). Although these findings do not preclude weak binding of myricetin to full length DnaJ, they support a model in which the major partner for myricetin is DnaK.
Myricetin Specifically Blocks DnaJ’s Ability to Stimulate ATP Turnover
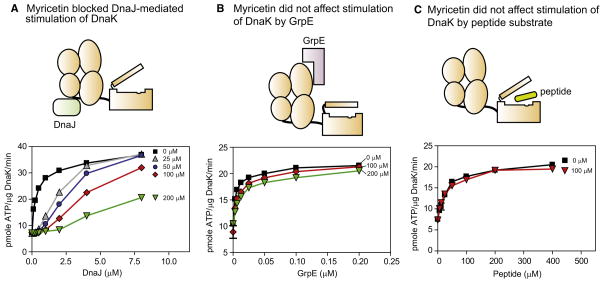
To better understand the implications of these interactions, we next measured whether myricetin could inhibit the ability of DnaJ to stimulate DnaK’s ATPase activity. In the absence of compound, DnaJ stimulated ATPase activity by approximately 4-fold, with a half-maximal value (KDnaJ) of 0.37 ± 0.03 μM (Figure 4A). Addition of myricetin at 200 μM completely suppressed the stimulatory activity of DnaJ (Figure 4A) and, even at 50 μM, this compound dramatically weakened DnaJ’s activity (Figure 4A). Conversely, it had no effect on the intrinsic ATPase activity of DnaK (Figure S4D) and it did not influence the stimulation of DnaK by the other co-chaperone, GrpE (Figures 4B and S4C). Finally, peptide substrates are known to stimulate ATP turnover (Buchberger et al., 1994), so we tested whether myricetin could block this activity. Using the high affinity model substrate, NRLLLTG, we found that myricetin had no effect on peptide-mediated stimulation (Figure 4C and S4B). Together, these results strongly suggest that myricetin selectively blocks the co-chaperone activity of DnaJ, without impacting the function of other DnaK partners.
Figure 4. Myricetin specifically blocked DnaJ cochaperone activities.
(A) In ATPase assays, myricetin blocked DnaJ-mediated stimulation. In all the ATPase experiments, the results are the average of triplicates and the error bars represent standard error of the mean. (B) Even at the highest concentrations, myricetin did not impact GrpE-mediated stimulation of ATP turnover. (C) Similarly, myricetin was unable to block stimulation by a model substrate peptide. Additional tests of this idea can be found in Figure S4.
As mentioned above, we found that myricetin did not bind to the truncated J domain by ITC (see Figure S3B). Therefore, we wanted to specifically test whether this compound could block J domain activity. Consistent with previous reports (Liberek et al., 1991), we found that the J domain stimulates DnaK’s ATP turnover (Figure S4A). Similar to what we observed with full length DnaJ, myricetin was able to block this stimulatory activity (Figure S4A).
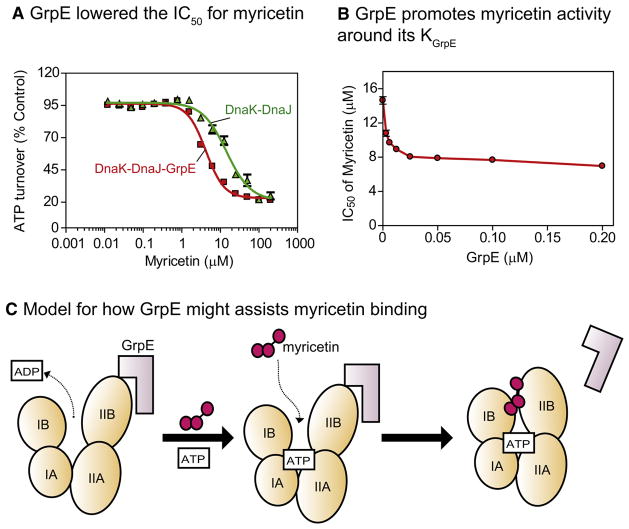
Because the binding sites for GrpE and myricetin are both in the IB and IIB subdomains, we were interested in how this co-chaperone might influence compound binding. We had already found that myricetin did not suppress the stimulatory activity of GrpE, but we were also interested in whether the co-chaperone might impact myricetin-binding affinity. Towards that question, we tested the activity of myricetin in the presence of both DnaJ and GrpE. At saturating concentration of GrpE, we found that the apparent affinity of myricetin was significantly enhanced (Figure 5A). By altering the concentration of GrpE and measuring the IC50 value of myricetin, we confirmed that the co-chaperone potentiated the activity of the compound (Figure 5B). This result suggests that GrpE, by opening the nucleotide-binding cleft of DnaK, may promote binding to myricetin; thereby increase affinity during ATP cycling (Figure 5C).
Figure 5. GrpE enhanced myricetin activity on DnaK-DnaJ.
(A) GrpE (2 μM) decreased the IC50 of myricetin for the DnaK-DnaJ complex. (B) Dose dependence of GrpE action on the IC50 value of myricetin. DnaK = 0.6 μM and DnaJ = 1 μM. Results are the average of triplicates and the error bars represent standard error of the mean. (C) Model for how GrpE might impact myricetin binding. Only the NBD is shown for clarity. Myricetin does not block GrpE-mediated stimulation, so it either dissociates prior to the next cycle or otherwise does not interfere with GrpE function.
Myricetin reduces the ability of DnaJ to stimulate the substrate binding activity of DnaK
Because myricetin appeared to block the ability of DnaJ to accelerate ATP turnover in DnaK, we hypothesized that it might also interrupt substrate binding by reducing conversion to the high affinity, ADP-bound state. To test this hypothesis, we measured DnaK’s binding to a model substrate, luciferase, using an ELISA-based approach. Briefly, we partially digested luciferase to increase the exposed hydrophobic regions and then absorbed this substrate into 96-well microtiter plates. Using an anti-DnaK antibody to measure bound chaperone, we first confirmed that DnaJ is able to stimulate DnaK’s binding to immobilized luciferase (Figure 6A) (Laufen et al., 1999). Maximal stimulation was observed at 60 nM of DnaJ and, at higher levels, DnaK’s binding gradually diminished, likely because of competition between DnaJ and DnaK for shared sites on luciferase. We found that 20 μM myricetin inhibited DnaJ-mediated stimulation of binding, with the amount of DnaJ required to promote binding increased from 62 nM to 1 μM and the maximal amount of bound DnaK also decreased by approximately 40% (Figure 6A).
Figure 6. Myricetin blocked DnaJ-mediated enhancement of DnaK’s binding to substrate and interfered with the DnaK-DnaJ interaction.
(A) Myricetin inhibited the DnaJ-mediated stimulation of DnaK binding to partially-digested firefly luciferase. Each data point is the average of triplicates and the error bars represent the standard error of the mean. (B) Labeled DnaJ was titrated into fluorescent DnaK and the apparent binding affinity (Kapp) measured by FRET. Myricetin, but not the control compound (catechin), partially blocked binding. The results are the average of triplicate and the error bars represent standard error of the mean. Further results are shown in Figure S5. (C) Model for the allosteric mechanism of myricetin. By impacting the clam-like motions of the subdomains, myricetin might impact DnaJ binding at a distal site. The hydrophobic cleft between subdomains IA and IIA is shown in green.
Myricetin inhibits binding of DnaJ to DnaK
Binding of myricetin to DnaK might block the ability of DnaJ to act on DnaK through at least two different mechanisms. In theory, myricetin might either (a) inhibit the physical interactions between DnaJ and DnaK or (b) block the allosteric activity of DnaJ on DnaK without directly impacting the strength of the protein-protein interaction. To begin to differentiate between these models, we directly measured the binding of DnaJ to DnaK by Förster resonance energy transfer (FRET) (Ruan et al., 2009). Briefly, DnaK was labeled with the fluorescent donor Alexa-488 (Em. 525 nm) and DnaJ with Black Hole Quencher-10 (BHQ-10), a FRET acceptor that absorb strongly around 507 nm. In the absence of myricetin, the apparent affinity (Kapp) between DnaJ and DnaK was estimated to be approximately 560 nM, consistent with previous studies (Suh et al., 1998). Addition of myricetin significantly reduced binding of DnaJ to DnaK (Figure 6B), with the Kapp weakened by 5.3-fold (at 25 μM myricetin). A control flavonoid, (±)-catechin, did not inhibit this interaction (Figure 6B) Together, these results suggest that myricetin inhibits DnaJ by preventing contacts between the chaperone and co-chaperone (Figure 6C).
DISCUSSION
Design of High Throughput Screens for the DnaK-DnaJ Combination: Methods for Targeting Multi-Protein Complexes
A key effort in chemical biology is to identify inhibitors of protein-protein interactions (Grant, 2009; May et al., 2007; Zhang et al., 2009). Further, allosteric modulators of these interactions are becoming an increasingly important goal in screening campaigns. With these concepts in mind, we targeted the DnaK-DnaJ combination. The key design criteria were (a) screen in the presence of high ATP concentrations (≫ Km) and (b) screen against the re-constituted combination of DnaK and DnaJ. The first design aspect (i.e. high ATP) was chosen to favor identification of allosteric, rather than competitive, modulators. For the DnaK system, this criterion is important because of the high cellular levels of ATP and its unusually tight binding to nucleotide (mid-nanomolar) (Massey, 2010). The second design aspect (i.e. inclusion of DnaJ) was selected because a majority of the measurable signal would be expected to emerge from stimulatory activities of the co-chaperone. Thus, compounds appearing as “actives” in the screen might be expected to preferentially influence the co-chaperone dependent activity (e.g. following the axiom, “you get what you screen for”). Together, we term this approach “gray box” screening because its goals are distinct from experiments that target purified proteins. Rather, it has more in common with “black box”, cell-based screens, but with fewer possible options for target deconvolution.
“Gray box” screens might be particularly appropriate for systems, such as Hsp70s, that are regulated by multiple stimulatory and inhibitory protein partners. Hsp70, being the hub of many cellular activities (Daugaard et al., 2007), is regulated by many co-chaperones, including J domain-containing proteins, nucleotide exchange factors (e.g. GrpE, BAGs, Hsp110 and HspBP1), and tetratricopeptide repeat (TPR) motif containing proteins (e.g. CHIP and HOP) (Bukau et al., 2006; Daugaard et al., 2007; Meimaridou et al., 2009). Interestingly, many of these interactions appear to converge on modulation of ATPase rate, and it might be a particularly attractive surrogate for emergent chaperone functions. In fact, we have used this method to identify chemical probes against the mammalian Hsp70 system that have revealed critical roles for ATP turnover in models of protein misfolding disease (Jinwal et al., 2009; Wisen et al., 2010). We anticipate that a variety of other, important multi-protein systems, such as chromatin remodeling complexes, the mTor complex, or the exocyst, might be similarly targeted Finally, it is also worth strongly emphasizing that the phenotypic complexity of other systems or technical issues may make a “gray box” approach cumbersome or undesirable.
Identification of an Unanticipated Allosteric Site on DnaK-DnaJ
In our natural product screens, we identified flavonoids as inhibitors of the DnaK-DnaJ complex. There are >9,000 naturally occurring flavonoids (Martens and Mithofer, 2005), and these compounds have been reported to have antioxidant, antitumor, antiviral, antifungal and antibacterial activities (Friedman, 2007; Khan and Mukhtar, 2008; Yang et al., 2009). These compounds carry out their diverse biological activities by targeting various enzymes, including receptor tyrosine kinases, cyclin-dependent kinases, and P-glycoprotein (Conseil et al., 1998; Friedman, 2007; Gamet-Payrastre et al., 1999; Teillet et al., 2008). The diversity of these targets suggests that flavonoids have relatively promiscuous binding profiles. Yet, plant-derived flavonoids have been productive and interesting starting points for drug discovery (Cuccioloni et al., 2009; Shapiro, 2004; Wiseman et al.). Interestingly, one common feature of flavonoid targets is that they often bind ATP (Gamet-Payrastre et al., 1999; Teillet et al., 2008); however, NMR, fluorescence and modeling studies suggested that the binding site of myricetin on DnaK is distinct from the ATP-binding pocket (see Figure 3). This finding is consistent with recent studies on other flavonoids (Wiseman et al., 2010). Despite this interesting mechanism, we found that the SAR for inhibition of DnaK-DnaJ is similar to that reported for flavonoids against other enzymes, such as receptor tyrosine kinase or protein kinase C (Gamet-Payrastre et al., 1999; Teillet et al., 2008). Thus, it is highly unlikely that myricetin or its close derivatives will bind selectively to DnaK-DnaJ in vivo. Rather, they might have been selected through evolution for their broad spectrum of targets (Khan and Mukhtar, 2008), an interesting alternative to the single-target, drug discovery model.
Although myrecitin is not a selective or particularly potent inhibitor of DnaK-DnaJ, these results suggest a new “druggable” site on the chaperone and they reveal a previously unanticipated allosteric network in DnaK. We found that myricetin, by binding to the upper nucleotide binding cleft between the IB and IIB subdomains, specifically interfered with binding to DnaJ and, subsequently, it interrupted co-chaperone stimulated activities. How can myricetin binding in the IB and IIB subdomain affect a remote site between the IA and IIA subdomains, where DnaJ binds to DnaK? Both NMR and modeling studies suggest that the IB and IIB subdomains move closer to each other in the “closed”, ATP-bound conformation (Bhattacharya et al., 2009; Woo et al., 2009). Simultaneously, the hydrophobic surface between the IA and IIA subdomains is expected to be more accessible in the ATP-bound form (Bhattacharya et al., 2009; Woo et al., 2009). Thus, it seems plausible that domains I and II undergo a clam-like movement during ATP hydrolysis, linking closure of the IB and IIB subdomains to the opening of IA and IIA. For that reason, if myricetin inserts between the IB and IIB subdomains, it might disrupt the DnaJ docking site at the distal IA and IIA region (see Figure 6C). This model clearly requires additional structural evaluation.
SIGNIFICANCE
Hsp70s are highly conserved molecular chaperones that play crucial roles in maintaining cellular proteostasis. They are also emerging drug targets for a range of diseases, including cancer, microbial infection and neurodegeneration. In this study, we identified myricetin as a new inhibitor of the E. coli Hsp70, DnaK. NMR and modeling studies suggested that this compound binds to a flexible site between the IB and IIB subdomains of DnaK and that it specifically blocks the activities of the co-chaperone DnaJ. This mechanism is unexpected because myricetin’s binding site is at least 20 Å from where DnaJ interacts with DnaK, in a region not previously implicated in DnaJ-mediated allostery. Thus, although myricetin, because of its poor selectivity and stability, is unlikely to be a suitable scaffold for drug development per se, this work identified an unanticipated allosteric site that might be suitable for targeting. Finally, this study provides a “gray box” strategy for identifying modulators of protein-protein interactions in reconstituted multiprotein complexes in vitro.
EXPERIMENTAL PROCEDURES
Materials and general protocols
Apigenin, baicalein, catechin, epigallocatechin-3-gallate, epigallocatechin, luteolin, morin, myricetin, quercetin, and rhamnetin were purchased from Sigma (St. Louis, MO). Taxifolin and kaempferol were bought from Fluka (Switzerland). Hieracin and epicatechin gallate were acquired from MicroSource (Gaylordsville, CT). The ATPase assays were performed using a previously established protocol (Chang et al., 2008a).
Plasmid construction and protein purification
DnaK, GrpE and DnaJ were purified, and the His-tags were cleaved as previously indicated (Chang et al., 2010). Nucleotide-free (apo) DnaK was purified by a slight modification of the previous protocol. Briefly, after the first low-salt washing step of the ATP column, apo-DnaK was eluted using 50 mM Tris buffer (pH 7.5) containing 1 M NaCl. His-tagged J-domain (residue 2–108) was first purified as previously mentioned (Chang et al., 2010), but an additional purification step using a Superdex 200 gel filtration column (GE Healthcare) was included. Purified DnaJ, J domain, GrpE and DnaK were dialyzed into 25 mM Tris buffer (pH 7.5) containing 10 mM KCl (for DnaJ and J domain, 150 mM KCl) and 5 mM MgCl2, and stored at −80 °C.
Construction of natural extract library and screening against DnaK/DnaJ complex
Approximately 300 mg of powdered natural product was extracted with ~500 μL of methanol. After vortexing and 15 minutes of sonication, the samples were centrifuged at 10,000 rpm for 5 minutes and the supernatant collected. After removing the organic solvent, the dried material was suspended in DMSO at 1 mg/mL. The resulting extract library was screened in the ATPase assay at 40 μg/ml final concentration, using a previously described protocol (Chang et al., 2008a). The concentrations of DnaJ and DnaK were 1.0 and 0.6 μM, respectively.
Identification of the active component from tea
White tea leaves were extracted with EtOAc, MeOH and acetone. The combined extracts were then concentrated in vacuo and the residue was partitioned between H2O and CH2Cl2. The organic phase was partitioned between 90% MeOH and n-hexanes and the aqueous MeOH fraction was further partitioned between 60% MeOH and CH2Cl2. Using ATPase assays for bioactivity-guided fractionation, we identified the 60% MeOH fraction as the sample with inhibitory activity against E. coli DnaK. This sample was then subjected to ODS flash chromatography (Supelco, Bellefonte, PA) with aqueous MeOH, yielding six fractions. The most active fraction was purified by reversed-phase HPLC (Beckman-coulter, Brea, CA) on Waters Spherisorb ODS-2 using a linear gradient elution of an aqueous MeOH system followed by reversed-phase HPLC with 20% aqueous MeOH. This procedure yielded epicatechin 3-gallate (ECG) as the major active component. The identification of this compound was confirmed by comparison of the 1H NMR spectrum and ESI-MS spectrum with those of an authentic sample purchased from MicroSource (see Figure S1).
Structure activity relationship studies
To identify available molecules structurally similar to ECG, we performed similarity searches for the core 2-phenyl benzopyran in the >100,000 compounds of the M-screen database (University of Michigan). From this analysis, 80 flavonoids were selected. Within the collection, 59 compounds were selected from the MS2000 library MicroSource), 10 from the Maybridge Hit finder library (Maybridge), 4 from the NIH Clinical Collection (BioFocus), 4 from the ChemDiv library, 1 from the Chembridge library, and 1 from the National Cancer Institute Collection. The compounds were “cherry-picked” and serially diluted (1:2) in ddH2O from 125 μM to 0.9 μM into white, 384-well low-volume plates (4 μL/well). Compounds were tested in duplicate, using a previously described protocol (Miyata et al., 2010a). Flavonoids that showed inhibition or had structure similar to active compounds were purchased and retested in 96-well malachite green-based ATPase assay against DnaK/J complex (at 0.6 and 1 μM) as described (Chang et al., 2008a).
NMR spectroscopy
The NMR experiments were carried out as previously described (Wisen et al., 2010). Briefly, 15N-labeled DnaK2-388 was purified and concentrated to approximately 0.5 mM, exchanged into NMR buffer (25 mM Tris, 10 mM MgCl2, 5 mM KCl, 10% 2H2O, 0.01% sodium azide, pH 7.1), and stored at −80 °C. Prior to NMR analysis, DnaK2-388 samples were thawed and supplemented with 5 mM ADP. After collecting reference spectra, myricetin was titrated to a final concentration of 1 mM from a 100 mM stock in DMSO. Control experiments showed that DMSO alone had had no detectable effects on the spectrum of DnaK2-388 at concentrations up to 1 %, and that it had minimal effects on the spectra of DnaK2-605 at the same concentration. 2D HSQC-TROSY NMR spectra were collected at 30 °C on a cryoprobe-equipped Varian Inova 800 MHz spectrometer, with data collection times of approximately 2 hours per spectrum. NMR data were processed using NMRPipe and analyzed with Sparky. Combined 1H and 15N chemical shift changes were measured using a weighted function and the assignments generated previously. Residues were selected as significant if the chemical shift was greater than 0.02 ppm (see Figure S3A).
Protein dynamic simulations
Coordinates of E. coli DnaK NBD were obtained from the PDB (PDB: 1DKG; (Harrison et al., 1997)) and the co-crystallized nucleotide-exchange factor GrpE was discarded. The missing side chains and short loops (≤ 5 amino acids) of NBD were introduced using Molecular Operating Environment (v2005.06) (Chemical Computing Group, Inc., Quebec, Canada) and PyMOL (DeLano Scientific, Palo Alto, CA). AMBER 10 (Case et al., 2008) package was used to perform unrestrained all-atom Langevin Dynamics (LD) simulations. The protein was modeled using the FF99SB force field (Hornak et al., 2006). Model II of a modified Generalized Born approach (Onufriev et al., 2004) was used to implicitly model aqueous solvation. Collision frequency of 1 ps−1 was used. Three states of DnaK were simulated using LD: apo-NDB, ATP-bound NDB, and ADP+Pi-bound NDB. Since 1DKG was crystallized without bound cofactors, positions for these cofactors were obtained by superimposing the crystal structures of the closely related homolog bovine Hsc70. ATP, Mg2+, and K+ ions were obtained from PDB: 1KAZ (O’Brien et al., 1996) and ADP, PO43−, Mg2+, and K+ ions were obtained from PDB: 1BUP (Sousa and McKay, 1998).
Parameters for Pi and Mg2+ were generated using the ANTECHAMBER (Wang et al. 2006) module of AMBER, and AM1-BCC charges (Jakalian et al., 2000) were applied. Nucleotide parameters developed by Meagher et al. (Meagher et al., 2003) were used for ATP and ADP. The SHAKE algorithm (Ryckaert et al., 1977) was used to restrain hydrogen atoms. Hydrogen atoms were first minimized, followed by residue side chains and finally an all-atom minimization. Five independent LD simulations for each of the three different NBD states were initiated with different random-number seeds. The non-bonded interactions cutoff was set to 999 Å. Default dielectric values were used: interior = 1 and exterior = 78.5. Heating and restrained equilibrations were followed. The system was heated gradually from 100 K to 300 K during the first two equilibrations, and the temperature remained at 300 K for the remaining equilibrations and production phase. Restraints were placed on all heavy atoms and gradually relaxed over the first four equilibrations using force constants from 2.0 to 0.1 kcal/mol·Å2. Restraints were maintained on backbone atoms in the fifth equilibration using a force constant of 0.1 kcal/mol·Å2. All restraints were removed in the sixth equilibration. The first three equilibrations were performed for 20 ps each, followed by 50 ps for the fourth and fifth equilibrations. The sixth, unrestrained equilibration was run for 740 ps and the production phase was run for 5 ns. A time step of 1 fs was used and snapshots were collected every 1 ps.
For the simulations of myricetin bound to DnaK, myricetin was docked into the initial NBD+ADP+Pi structure using AutoDock 4.01 (Morris et al., 2009). This was the same structure built from the crystal structures and used to initiate the LD simulations. A grid box of (90, 100, 94) points and 0.375 Å spacing was centered at the interface between IB/IIB subdomains and over the residues with myricetin-induced NMR chemical-shifts. The docking calculation involved a Lamarckian genetic algorithm (Morris et al., 1998); an initial population size of 200 and a maximum of 75 million energy evaluations were applied. The lowest-energy pose of myricetin, which was consistent with the NMR chemical-shifts, was added to the NBD. GAFF (Wang et al., 2004) with AM1-BCC charges was used to parameterize myricetin in ANTECHAMBER. Five independent LD simulations of the myricetin+ADP+Pi+NBD system were generated using protocol described above.
Tryptophan fluorescence
The tryptophan fluorescence experiments were carried out as previously reported with slight modification (Theyssen et al., 1996). To study whether myricetin blocked ATP binding, 6 μM apo-DnaK was incubated with 125 μM myricetin with total volume of 40 μL in a 96-well black plate (Corning) for one hour at 37 °C. Next, 10μL of 500 μM ATP or buffer (200 mM Tris, 40 mM KCl, 12 mM MgCl2, pH 7.4) was added to each well, and then the emission spectrum between 300 – 400 nm was recorded (excitation at 295 nm) using a SpectraMax M5 (Molecular Devices). Alternatively, the tryptophan fluorescence quench caused by myricetin was measured as follow. Myricetin, at concentration indicated in the figure, was added to 5 μM of DnaK in the presence or absence of 2 mM ATP with a total volume of 30 μL in a black 96-well plate and the fluorescence at 342 nm (excitation at 295 nm) was measured by SpectraMax M5 and the controls with every component except DnaK was subtracted from the final values. Experiments described in Figure 3E were carried out as above, but with slightly different DnaK and ATP concentrations (3 μM and 1 mM, respectively) in lower total volume (20 μL) and the readings were performed in black, 384-well round bottom plates (Corning).
Enzyme-linked Immunosorbant Assay
Binding of DnaK to denatured luciferase was measured as previously described (Miyata et al., 2010b).
DnaK-DnaJ Interaction by FRET
The assays were carried out as recently reported with slight modifications (Ruan et al., 2009). DnaK (10 mg/mL; 200 μL) was labeled with Alexa Fluor 488 5-TFP (Invitrogen, Carlsbad, CA) and DnaJ (10 mg/mL; 300 μL) was labeled using the N-hydroxysuccinimide ester of BHQ-10 Carboxylic Acid (Biosearch Technologies, Novato, CA). Both reactions were carried out at 10:1 molar ratio of dye:protein in bicarbonate buffer (100 mM NaHCO3, 5 mM MgCl2, 10 mM KCl, pH = 9.5). DnaK was labeled for 1.5 hours at room temperature while DnaJ was labeled for 1 hour at 30 °C. After the incubation, the unreacted dye was removed and the buffer was exchanged to 25 mM HEPES buffer (pH = 7.4) using Zeba™ Desalt Spin Columns (2 mL, MWCO = 7000 Da) (Thermo Scientific, Rockford, IL). The average extent of labeling for DnaK and DnaJ was then determined to be approximately 2.5 and 0.5 fluorophore/dye per protein, respectively, using the ε495 = 71,000 M−1cm−1 (Alexa Fluor 488) and ε507 = 30,000 M−1 cm−1 (BHQ-10). Labeled DnaK was diluted to 100 nM using 100 mM Tris-HCl buffer (20 mM KCl, 6 mM MgCl2, 0.01% Triton-X-100, pH = 7.45) containing compound (or solvent control) and 2 mM ATP. After pre-incubation at room temperature for 30 minutes, 10 μL of this diluted sample was added to 384-well, black round bottom plates (Corning). Next, 10 μL of a BHQ-10-labeled DnaJ solution was added to each well and the samples were incubated at 37 °C for 30 minutes. After the incubation, the fluorescence at 525 nm (Ex. 480, cut-off 515 nm) was measured using a SpectraMax M5 microplate reader. The result were analyzed by GraphPad Prism 4.0 using a hyperbolic fit with a non-zero intercept (ΔF = ΔFmax·[J]/(Kapp+[J])+b). ΔF = fluorescence change; ΔFmax = maximum fluorescence change; Kapp = apparent Kd; [J] = DnaJ concentration.
Supplementary Material
Acknowledgments
The authors thank Jack Dixon for helpful comments and Leah Makley for experimental assistance. This work was supported by grants MCB-0844512 and NS059690 to J.E.G.
Footnotes
Supplemental information includes five figures and a table.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ang D, Chandrasekhar GN, Zylicz M, Georgopoulos C. Escherichia coli grpE gene codes for heat shock protein B25.3, essential for both lambda DNA replication at all temperatures and host growth at high temperature. Journal of bacteriology. 1986;167:25–29. doi: 10.1128/jb.167.1.25-29.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Kurochkin AV, Yip GN, Zhang Y, Bertelsen EB, Zuiderweg ER. Allostery in Hsp70 chaperones is transduced by subdomain rotations. Journal of molecular biology. 2009;388:475–490. doi: 10.1016/j.jmb.2009.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Theyssen H, Schroder H, McCarty JS, Virgallita G, Milkereit P, Reinstein J, Bukau B. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. The Journal of biological chemistry. 1995;270:16903–16910. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Valencia A, McMacken R, Sander C, Bukau B. The chaperone function of DnaK requires the coupling of ATPase activity with substrate binding through residue E171. The EMBO journal. 1994;13:1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, et al. AMBER. Vol. 10. University of California; San Francisco: 2008. [Google Scholar]
- Chang L, Bertelsen EB, Wisen S, Larsen EM, Zuiderweg ER, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Analytical biochemistry. 2008a;372:167–176. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Chang L, Thompson AD, Ung P, Carlson HA, Gestwicki JE. Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (HSP70/DNAK) The Journal of biological chemistry. 2010;385:21282–21291. doi: 10.1074/jbc.M110.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, Sun YJ, Wang C, Hsiao CD. Crystal structures of the 70-kDa heat shock proteins in domain disjoining conformation. The Journal of biological chemistry. 2008b;283:15502–15511. doi: 10.1074/jbc.M708992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conseil G, Baubichon-Cortay H, Dayan G, Jault JM, Barron D, Di Pietro A. Flavonoids: a class of modulators with bifunctional interactions at vicinal ATP- and steroid-binding sites on mouse P-glycoprotein. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9831–9836. doi: 10.1073/pnas.95.17.9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccioloni M, Mozzicafreddo M, Bonfili L, Cecarini V, Eleuteri AM, Angeletti M. Natural occurring polyphenols as template for drug design. Focus on serine proteases. Chemical biology & drug design. 2009;74:1–15. doi: 10.1111/j.1747-0285.2009.00836.x. [DOI] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS letters. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Evans CG, Chang L, Gestwicki JE. Heat Shock Protein 70 (Hsp70) as an Emerging Drug Target. J Med Chem. 2010;53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman M. Overview of antibacterial, antitoxin, antiviral, and antifungal activities of tea flavonoids and teas. Molecular nutrition & food research. 2007;51:116–134. doi: 10.1002/mnfr.200600173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamet-Payrastre L, Manenti S, Gratacap MP, Tulliez J, Chap H, Payrastre B. Flavonoids and the inhibition of PKC and PI 3-kinase. General pharmacology. 1999;32:279–286. doi: 10.1016/s0306-3623(98)00220-1. [DOI] [PubMed] [Google Scholar]
- Gassler CS, Buchberger A, Laufen T, Mayer MP, Schroder H, Valencia A, Bukau B. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Molecular microbiology. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- Grant SK. Therapeutic protein kinase inhibitors. Cell Mol Life Sci. 2009;66:1163–1177. doi: 10.1007/s00018-008-8539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Christen P. Mutations in the interdomain linker region of DnaK abolish the chaperone action of the DnaK/DnaJ/GrpE system. FEBS letters. 2001;497:55–58. doi: 10.1016/s0014-5793(01)02435-8. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science (New York, NY. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horswill AR, Savinov SN, Benkovic SJ. A systematic method for identifying small-molecule modulators of protein-protein interactions. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15591–15596. doi: 10.1073/pnas.0406999101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakalian A, Bush BL, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method. J Comp Chem. 2000:132–146. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Structural basis of J cochaperone binding and regulation of Hsp70. Molecular cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinwal UK, Miyata Y, Koren J, 3rd, Jones JR, Trotter JH, Chang L, O’Leary J, Morgan D, Lee DC, Shults CL, et al. Chemical manipulation of hsp70 ATPase activity regulates tau stability. J Neurosci. 2009;29:12079–12088. doi: 10.1523/JNEUROSCI.3345-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Mukhtar H. Multitargeted therapy of cancer by green tea polyphenols. Cancer letters. 2008;269:269–280. doi: 10.1016/j.canlet.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science (New York, NY) 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. Journal of the American Chemical Society. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- Martens S, Mithofer A. Flavones and flavone synthases. Phytochemistry. 2005;66:2399–2407. doi: 10.1016/j.phytochem.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Massey AJ. ATPases as drug targets: insights from heat shock proteins 70 and 90. Journal of medicinal chemistry. 2010;53:7280–7286. doi: 10.1021/jm100342z. [DOI] [PubMed] [Google Scholar]
- May LT, Leach K, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors. Annual review of pharmacology and toxicology. 2007;47:1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- Meagher KL, Redman LT, Carlson HA. Development of polyphosphate parameters for use with the AMBER force field. Journal of computational chemistry. 2003;24:1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- Meimaridou E, Gooljar SB, Chapple JP. From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. Journal of molecular endocrinology. 2009;42:1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- Miyata Y, Chang L, Bainor A, McQuade TJ, Walczak CP, Zhang Y, Larsen MJ, Kirchhoff P, Gestwicki JE. High-Throughput Screen for Escherichia coli Heat Shock Protein 70 (Hsp70/DnaK): ATPase Assay in Low Volume by Exploiting Energy Transfer. J Biomol Screen. 2010a doi: 10.1177/1087057110380571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y, Chang L, Bainor A, McQuade TJ, Walczak CP, Zhang Y, Larsen MJ, Kirchhoff P, Gestwicki JE. High Throughput Screen for Escherichia coli Heat Shock Protein 70 (Hsp70/DnaK): ATPase Assay in Low Volume By Exploiting Energy Transfer. J Biomol Screen. 2010b doi: 10.1177/1087057110380571. (accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olsen AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of computational chemistry. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien MC, Flaherty KM, McKay DB. Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis. The Journal of biological chemistry. 1996;271:15874–15878. doi: 10.1074/jbc.271.27.15874. [DOI] [PubMed] [Google Scholar]
- Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annual review of biochemistry. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Ruan Q, Skinner JP, Tetin SY. Using nonfluorescent Forster resonance energy transfer acceptors in protein binding studies. Analytical biochemistry. 2009;393:196–204. doi: 10.1016/j.ab.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Russell R, Jordan R, McMacken R. Kinetic characterization of the ATPase cycle of the DnaK molecular chaperone. Biochemistry. 1998;37:596–607. doi: 10.1021/bi972025p. [DOI] [PubMed] [Google Scholar]
- Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-integration of Cartesian equations of motion of a system with constraints-molecular-dynamics of N-Alkanes. J Comp Phys. 1977:23. [Google Scholar]
- Schroder H, Langer T, Hartl FU, Bukau B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. The EMBO journal. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell SM, Eisen C, Ang D, Zylicz M, Georgopoulos C. Isolation and characterization of dnaJ null mutants of Escherichia coli. Journal of bacteriology. 1990;172:4827–4835. doi: 10.1128/jb.172.9.4827-4835.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro GI. Preclinical and clinical development of the cyclin-dependent kinase inhibitor flavopiridol. Clin Cancer Res. 2004;10:4270s–4275s. doi: 10.1158/1078-0432.CCR-040020. [DOI] [PubMed] [Google Scholar]
- Sousa MC, McKay DB. The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change. Biochemistry. 1998;37:15392–15399. doi: 10.1021/bi981510x. [DOI] [PubMed] [Google Scholar]
- Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Molecular cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teillet F, Boumendjel A, Boutonnat J, Ronot X. Flavonoids as RTK inhibitors and potential anticancer agents. Medicinal research reviews. 2008;28:715–745. doi: 10.1002/med.20122. [DOI] [PubMed] [Google Scholar]
- Theyssen H, Schuster HP, Packschies L, Bukau B, Reinstein J. The second step of ATP binding to DnaK induces peptide release. Journal of molecular biology. 1996;263:657–670. doi: 10.1006/jmbi.1996.0606. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science (New York, NY) 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case D. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Wiseman RL, Zhang Y, Lee KP, Harding HP, Haynes CM, Price J, Sicheri F, Ron D. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Molecular cell. 38:291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RL, Zhang Y, Lee KP, Harding HP, Haynes CM, Price J, Sicheri F, Ron D. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell. 2010;38:291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisen S, Bertelsen EB, Thompson AD, Patury S, Ung PM, Chang L, Evans CG, Walter GM, Wipf P, Carlson HA, et al. Binding of a Small Molecule at a Protein-Protein Interface Regulates the Chaperone Activity of Hsp70-Hsp40. ACS Chem Biol. 2010;5:611–622. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HJ, Jiang J, Lafer EM, Sousa R. ATP-induced conformational changes in Hsp70: molecular dynamics and experimental validation of an in silico predicted conformation. Biochemistry. 2009;48:11470–11477. doi: 10.1021/bi901256y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Wang X, Lu G, Picinich SC. Cancer prevention by tea: animal studies, molecular mechanisms and human relevance. Nature reviews. 2009 doi: 10.1038/nrc2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Hamilton AD. Strategies for targeting protein-protein interactions with synthetic agents. Angewandte Chemie (International ed) 2005;44:4130–4163. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nature reviews. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science (New York, NY) 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.