Abstract
Background
Alterations in serotonin (5-HT) are suspected in the pathophysiology of irritable bowel syndrome (IBS). Tryptophan hydroxylase (TPH) is the rate-limiting enzyme in the biosynthesis of serotonin and has two isoforms, TPH1 and TPH2. Genetic variants in both genes have been studied in various disorders related to serotonin dysregulation. The aim of this study was to examine whether TPH gene variants were associated with IBS and IBS-related gastrointestinal (GI) symptoms.
Methods
Five single nucleotide polymorphisms (SNPs) from the TPH1 and one SNP from the TPH2 were genotyped in 199 IBS patients and 79 healthy controls. All subjects were Caucasian women of European origin. IBS patients filled in a daily diary with five GI symptoms and stool characteristics for 28 days.
Key Results
The TPH1 SNPs showed no association with the diagnosis of IBS. However among IBS patients, all five TPH1 SNPs showed some association with diarrhea and loose type of stool consistency, with p-values rating from 0.01 to 0.20. The TPH2 SNP showed a trend towards a reduced risk of IBS as well as possible associations with stool characteristics, both hard and loose stools. However, no p-values were less than the conservative multiple-comparison-adjusted threshold of 0.001 and hence these results must be interpreted cautiously.
Conclusions & Inferences
This study is the first to assess associations of TPH gene variants with IBS-related GI symptoms and stool characteristics. The possible association of TPH gene variants with diarrhea needs to be verified in an independent sample.
Keywords: IBS, TPH1, TPH2, polymorphisms, Genetic association
INTRODUCTION
Irritable bowel syndrome (IBS) is a common functional gastrointestinal (GI) disorder characterized by recurrent abdominal pain or discomfort associated with altered bowel habits.(1) Although the underlying pathophysiology of IBS is still not clearly understood, serotonin (5-hydroxy tryptophan [5-HT]) is known to be an important signaling molecule in the regulation of GI motility, sensation and secretion.(2) Approximately 95% of 5-HT in the body is found in the GI tract with 90% in enterochromaffin (EC) cells and 10% in serotonergic neurons of the myenteric plexus.(3) The therapeutic success observed with serotonin-modulating agents in patients with IBS supports the pivotal role of 5-HT in IBS pathophysiology.(4-6) Tryptophan hydroxylase (TPH) is the first and rate-limiting enzyme of 5-HT biosynthesis, catalyzing the oxygenation of tryptophan.(7) There are two isoforms of TPH, TPH1 and TPH2, with overall 71% identity in amino acid sequence in humans.(8, 9) The gene encoding TPH1 is located on chromosome 11p15.3-p14 with a size of 29 kilobases (kb) and composed of 11 exons.(10) TPH2 is encoded by a 93.6 kb gene on chromosome 12q21.1 which is also composed of 11 exons.(11) TPH1 and TPH2 have different expression patterns: TPH1 is expressed in the pineal gland, pituitary gland and peripheral organs, mostly EC cells in the gut, while TPH2 is mainly expressed in the CNS and peripheral serotonergic neurons.(8, 9, 12)
Polymorphisms in the TPH genes have been studied intensively in psychiatric or behavioral disorders whose underlying pathophysiology is related to 5-HT. However, there is as yet no published data related to TPH gene polymorphisms and GI symptoms or IBS. Therefore, the purpose of this study was to examine a possible association of TPH gene polymorphisms with IBS in women as well as their relationship to severity of GI symptoms.
METERIALS AND METHODS
Subjects
This study used DNA samples and survey data from three case-control studies of IBS carried out in western Washington State. Extensive description of the study samples can be found in previous reports.(13-15) IBS patients with a prior diagnosis who currently met the Rome III criteria(1) and healthy controls without history of functional GI disorders were recruited through community advertisement. IBS patients or healthy control subjects were excluded if they 1) had a history of co-existing GI pathology (e.g., inflammatory bowel disease) or surgery, renal or reproductive pathology (e.g., endometriosis), severe fibromyalgia, severe cardiovascular disease; or 2) were currently taking the following medications more than 3 day a week: antibiotics, anticholinergics, cholestyramine, narcotics, colchicines, docusate, an enema preparation, iron supplements, or laxatives. The protocols for this study and the three parent studies were approved by the University of Washington’s institutional review board and all the patients gave written informed consent. For this study, we restricted the sample to women who identified themselves as Caucasian to avoid population stratification bias. This resulted in an analysis set of 199 IBS subjects and 79 controls. Of the women with IBS, 20% (n = 41) met criteria for constipation-predominant, 44% (n = 88) met criteria for diarrhea-predominant and 26% (n = 52) met criteria for mixed IBS (Table 4).
Table 4.
TPH polymorphisms and IBS subgroups by Rome III criteria
| IBS Subgroups |
||||||
|---|---|---|---|---|---|---|
| Gene SNP no. |
SNP ID | Genotype | IBS-C (n = 41) |
IBS-D (n = 88) |
IBS-M (n = 52) |
IBS-U (n = 17) |
| TPH1 | rs4537731 | AA | 15 (37%) | 32 (36%) | 20 (38%) | 8 (47%) |
| 1 | AG | 20 (49%) | 42 (48%) | 27 (52%) | 7 (41%) | |
| GG | 6 (15%) | 14 (16%) | 5 (10%) | 2 (12%) | ||
|
| ||||||
| 2 | rs684302 | CC | 14 (34%) | 31 (35%) | 15 (29%) | 3 (18%) |
| CT | 18 (44%) | 46 (52%) | 23 (44%) | 10 (59%) | ||
| TT | 9 (22%) | 11 (13%) | 14 (27%) | 4 (24%) | ||
|
| ||||||
| 3 | rs211105 | TT | 28 (68%) | 51 (58%) | 31 (60%) | 13 (77%) |
| GT | 12 (29%) | 31 (35%) | 19 (37%) | 4 (24%) | ||
| GG | 1 ( 2%) | 6 ( 7%) | 2 ( 4%) | 0 | ||
|
| ||||||
| 4 | rs1800532 | CC | 16 (39%) | 35 (40%) | 17 (33%) | 4 (24%) |
| AC | 16 (39%) | 42 (48%) | 21 (40%) | 10 (59%) | ||
| AA | 9 (22%) | 11 (13%) | 14 (27%) | 3 (18%) | ||
|
| ||||||
| TPH2 | rs4570625 | GG | 27 (66%) | 57 (65%) | 34 (65%) | 12 (71%) |
| 6 | GT | 12 (29%) | 30 (34%) | 15 (29%) | 5 (30%) | |
| TT | 2 ( 5%) | 1 ( 1%) | 3 ( 6%) | 0 | ||
TPH, tryptophan hydroxylase; IBS, irritable bowel syndrome; SNP, single nucleotide polymorphism; IBS-C, IBS with constipation; IBS-D, IBS with diarrhea; IBS-M, Mixed IBS; IBS-U, Unsubtyped IBS
SNP selection and genotyping
Based on previously reported association studies we selected five single nucleotide polymorphisms (SNPs) in the TPH1 gene and one SNP in the TPH2 gene for genotyping. TPH1 SNPs are spanning the TPH1 gene from the upstream region (−6.5 kilo-basepairs (kbp); rs4537731, also known as −6526A/G), through intron 2 (rs684302) and intron 3 (rs21105) to intron 7 (rs1800532, also known as 218A/C and rs1799913, also known as 779A/C). The TPH2 SNP is located in the promoter region (−703 kbp; rs4570625, also known as −709G/T). For convenience, five TPH1 SNPs were numbered by their location in the gene and the TPH2 SNP was marked as SNP6 (Table 1).
Table 1.
Characteristics of TPH polymorphisms and their association with IBS in women
| Polymorphism |
Frequency, n (%) |
Odds ratioa |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene SNP no. |
SNP ID | Location | MAF | Genotype | IBS (n = 199) |
Control (n = 79) |
OR | 95%CI | p-valueb |
| TPH1 | rs4537731 | Promoter | 0.38 | AA | 76 (38%) | 29 (37%) | 1 | 0.84 | |
| 1 | region | AG | 96 (48%) | 41 (52%) | 0.86 | 0.49, 1.5 | |||
| GG | 27 (14%) | 9 (11%) | 1.03 | 0.43, 2.5 | |||||
|
| |||||||||
| 2 | rs684302 | Intron 2 | 0.44 | CC | 63 (32%) | 27 (34%) | 1 | 0.84 | |
| CT | 97 (48%) | 37 (47%) | 1.18 | 0.65, 2.1 | |||||
| TT | 39 (20%) | 15 (19%) | 1.21 | 0.57, 2.6 | |||||
|
| |||||||||
| 3 | rs211105 | Intron 3 | 0.21 | TT | 124 (62%) | 48 (60%) | 1 | 0.82 | |
| GT | 66 (33%) | 28 (35%) | 0.86 | 0.49, 1.5 | |||||
| GG | 9 ( 5%) | 3 ( 4%) | 1.20 | 0.31, 4.7 | |||||
|
| |||||||||
| 4 | rs1800532 | Intron 7 | 0.42 | CC | 72 (36%) | 28 (35%) | 1 | 0.97 | |
| AC | 89 (45%) | 36 (46%) | 0.96 | 0.53, 1.7 | |||||
| AA | 38 (19%) | 15 (19%) | 1.04 | 0.49, 2.2 | |||||
|
| |||||||||
| TPH2 | rs4570625 | Promoter | 0.21 | GG | 131 (66%) | 46 (58%) | 1 | 0.05 | |
| 6 | region | GT | 62 (31%) | 25 (32%) | 0.92 | 0.51, 1.6 | |||
| TT | 6 ( 3%) | 8 (10%) | 0.25 | 0.08, 0.8 | |||||
SNP, single nucleotide polymorphism; MAF, minor allele frequency; IBS, irritable bowel syndrome; OR, odds ratio; CI, confidence interval; TPH, tryptophan hydroxylase
Logistic regression adjusted for age
Overall p-value
Genomic DNA was extracted from fresh whole blood or frozen isolated white blood cells (WBCs) by buffy coat preparation(16) using Puregene DNA Purification kits (Gentra Systems Inc., Minneapolis, MN). Genotyping was done using TaqMan custom genotyping assays and an ABI 7300 Real-time PCR System (Applied Biosystems Inc., Foster City, CA). PCR reactions containing 100ng genomic DNA, 1x Genotyping Master Mix, 900 nM of each primer and 200 nM of each probe were performed in 96-well plates using the standard protocol for TaqMan MGB probes in a total volume of 25ul. After the cycling, end-point fluorescence was measured and genotype calling was carried out by the allelic discrimination analysis module. To assure quality control, 5% of genotyping was performed in duplicate which showed 100% reproducibility of our results.
Measures
IBS-related GI symptom score
IBS subjects filled in a daily diary for 28 days, recoding five items related to GI symptoms: abdominal pain or discomfort, bloating, constipation, diarrhea and intestinal gas. These were rated on a scale of 0 (not present), 1 (mild), 2 (moderate), 3 (severe) to 4 (very severe). The severity of each symptom was summarized as percent of days with moderate to very severe symptoms.(13) The consistency of each stool was also recorded in the diary with a scale of 1 (very hard), 2 (hard), 3 (formed), 4 (loose) to 5 (watery). Stool consistency over 28 days was summarized as percent of days with either ‘very hard’ or ‘watery’ stool.
Statistical analysis
Hardy-Weinberg equilibrium was tested using Chi-square tests. To test genotypic association between TPH SNPs and IBS, odds ratios (OR) and 95% confidence intervals were estimated using logistic regression, controlling for age. The homozygous genotype of the more common allele of each SNP served as the reference category. Associations with GI symptom scores of individuals SNPs were analyzed by the analysis of covariance (ANCOVA) with age as a covariate. Associations of SNPs with IBS predominant bowel pattern subgroups (Rome III criteria) were tested with Chi-square tests. To adjust for multiple comparisons with correct type I error, we used a very conservative method, the Bonferroni adjustment. We tested 9 hypotheses and 5 SNPs, hence we considered a p-value of less than 0.05/45 = 0.001 to be significant. Unadjusted p-values are presented and compared to the threshold of 0.001. Results are also discussed in terms of the overall pattern of results.
As a further exploration of the primary analyses described above, the Haploview 4.0 program (http://www.broad.mit.edu.offcampus.lib.washington.edu/mpg/haploview) was used to explore the haplotype structure of the TPH1 gene and determine linkage disequilibrium (D’ and R2) between SNP pairs.(17) Haplotype analysis was performed with Hapstat (http://www.bios.unc.edu/~lin/hapstat/) using a sliding window approach with three consecutive markers throughout the gene.
RESULTS
One hundred ninety nine women with IBS and 79 healthy women completed the study. The control group was younger than the IBS group (IBS: 40 ± 14; controls: 36 ± 13 years of age, p=0.03). To reduce ethnic variation and stratification effects, only unrelated Caucasian women were included in the analysis.
Genotype distributions
Genotype distributions for all SNPs were in Hardy-Weinberg Equilibrium in both control and patient groups (data not shown). Genotypes for SNP4 and SNP5 in the TPH1gene were identical in all cases and controls, indicating complete LD between SNPs (D’=1, r2=1.0). Therefore, we excluded SNP5 from further analysis.
Association of TPH gene variants with IBS diagnosis
Table 1 shows the logistic regression analysis of the association between TPH gene variants and IBS. There was no significant association between TPH1 gene SNPs and IBS. Although homozygosity for the rare allele (T) of TPH2 gene SNP6 was less common in IBS patients (3%) than in healthy controls (10%), the difference is based on only a few individuals and the unadjusted p-value was 0.05, not less than the threshold of 0.001.
The association between TPH1 variants and IBS was also examined at the level of haplotypes. The LD pattern of TPH1 variants was similar between our sample and the HapMap CEU data set with one block of strong LD and limited haplotype diversity with only five common haplotypes (Figure 1, Table 2). These five haplotypes accounted for 99% of the segregating haplotypes in this study population. No evidence was found for an association between any of the TPH1 haplotypes and IBS.
Figure 1.
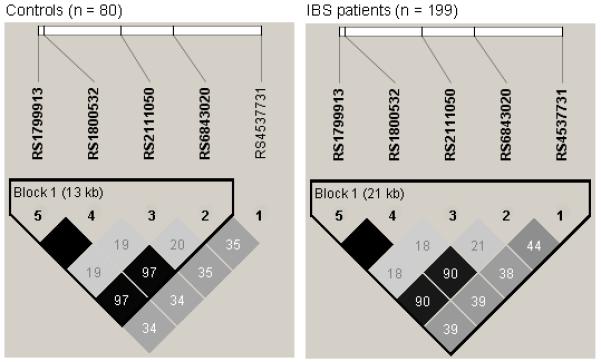
LD plot of the TPH1 SNPs genotyped in this study. Each diamond in the LD plot represents the strength of pairwise LD, with dark color indicating strong LD. The pairwise R2 values are written in the boxes. Haplotype blocks are indicated with a surrounding black line using the method of Gabriel et al (2002).(39)
Table 2.
TPH1 Haplotype Frequencies in controls and IBS.
| Haplotype |
Frequency |
Chi square | p-value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP1 | SNP2 | SNP3 | SNP4 | SNP5 | IBS | Control | |||
| a | A | T | T | A | A | 0.408 | 0.402 | 0.009 | 0.926 |
| b | G | C | G | C | C | 0.202 | 0.199 | 0.001 | 0.971 |
| c | A | C | T | C | C | 0.180 | 0.203 | 0.329 | 0.566 |
| d | G | C | T | C | C | 0.169 | 0.158 | 0.074 | 0.786 |
| e | A | T | T | C | C | 0.025 | 0.006 | 2.061 | 0.151 |
| f | A | C | G | C | C | 0.009 | 0.016 | 0.310 | 0.578 |
TPH, tryptophan hydroxylase; IBS, irritable bowel syndrome
Association of TPH gene variants with the severity of GI symptoms in women with IBS
We also investigated possible associations of TPH SNPs and IBS-related GI symptoms (Table 3). None of these associations were significant at the multiple-comparison-adjusted threshold of p < 0.001. However, an interesting pattern does appear. Two SNPs in the TPH1 gene showed associations with diarrhea symptoms. IBS patients homozygous for the minor allele (G) of TPH1 SNP1 reported higher severity of diarrhea symptoms than the other two genotype groups (AA and AG genotypes, p = 0.02), and also showed a trend towards more days with watery stools (p = 0.08). For TPH1 SNP3, patients carrying the minor allele (GT and GG genotypes) reported higher severity of diarrhea symptoms (p = 0.02) and more days with watery stools than patients homozygous for the major allele (TT genotype, p = 0.01) (table 3). Minor allele (G) carriers at SNP1 also reported more severe bloating symptoms (p = 0.02).
Table 3.
Association between TPH polymorphisms and the severity of GI symptoms in women with IBS
| Symptom Scorea, mean (sd) |
p-valueb | |||
|---|---|---|---|---|
| SNP1 (rs4537731) | AA (n = 70) | AG (n = 93) | GG (n = 25) | |
| GI symptom | ||||
| Abdominal pain | 27.9 (23.5) | 35.6 (25.2) | 34.8 (25.7) | 0.15 |
| Bloating | 21.5 (22.5) | 33.9 (30.9) | 36.4 (32.7) | 0.02 |
| Intestinal gas | 32.7 (24.4) | 38.1 (30.4) | 40.2 (34.0) | 0.61 |
| Constipation | 16.9 (21.6) | 18.9 (21.1) | 24.7 (24.4) | 0.33 |
| Diarrhea | 9.8 (15.4) | 12.3 (15.9) | 20.5 (30.3) | 0.05 |
| Stool consistency | ||||
| Very hard stool | 13.1 (19.2) | 11.1 (14.7) | 12.7 (17.3) | 0.73 |
| Watery stool | 7.2 ( 9.1) | 10.3 (16.5) | 14.4 (20.3) | 0.08 |
| SNP2 (rs684302) | CC (n = 59) | CT (n = 93) | TT (n = 36) | |
|
| ||||
| GI symptom | ||||
| Abdominal pain | 32.8 (21.8) | 34.6 (27.2) | 27.6 (22.6) | 0.38 |
| Bloating | 34.9 (30.1) | 28.3 (29.1) | 24.2 (25.3) | 0.22 |
| Intestinal gas | 37.7 (30.1) | 36.2 (30.9) | 34.5 (26.9) | 0.96 |
| Constipation | 19.4 (20.8) | 18.4 (23.1) | 19.7 (20.2) | 0.93 |
| Diarrhea | 15.2 (23.7) | 12.7 (16.0) | 7.2 (13.2) | 0.13 |
| Stool consistency | ||||
| Very hard stool | 10.4 (15.3) | 11.8 (15.5) | 15.5 (21.7) | 0.36 |
| Watery stool | 11.0 (15.5) | 10.7 (16.7) | 5.1 ( 6.0) | 0.11 |
| SNP3 ( rs211105) | TT (n = 116) | GT (n = 63) | GG (n = 9) | |
|
| ||||
| GI symptom | ||||
| Abdominal pain | 31.1 (24.6) | 36.8 (25.6) | 23.2 (17.1) | 0.20 |
| Bloating | 27.0 (26.9) | 35.0 (31.7) | 26.0 (29.5) | 0.27 |
| Intestinal gas | 36.2 (27.8) | 38.6 (33.3) | 24.0 (28.9) | 0.54 |
| Constipation | 19.0 (22.3) | 19.1 (21.9) | 17.4 (12.2) | 0.98 |
| Diarrhea | 9.5 (14.2) | 17.5 (24.0) | 15.0 (16.1) | 0.02 |
| Stool consistency | ||||
| Very hard stool | 13.8 (18.3) | 9.5 (14.2) | 7.3 (10.1) | 0.18 |
| Watery stool | 7.2 (10.0) | 14.0 (20.5) | 12.1 (16.5) | 0.01 |
| SNP4 (rs1800532) | CC (n = 68) | AC (n = 85) | AA (n = 35) | |
|
| ||||
| GI symptom | ||||
| Abdominal pain | 35.3 (23.7) | 32.4 (26.4) | 28.3 (22.5) | 0.40 |
| Bloating | 34.7 (30.5) | 27.7 (28.5) | 24.5 (25.6) | 0.14 |
| Intestinal gas | 38.0 (30.2) | 35.4 (31.0) | 35.5 (26.6) | 0.73 |
| Constipation | 19.3 (21.9) | 18.2 (22.4) | 20.0 (20.5) | 0.88 |
| Diarrhea | 14.3 (22.7) | 13.0 (16.2) | 7.4 (13.3) | 0.20 |
| Stool consistency | ||||
| Very hard stool | 11.3 (16.9) | 11.1 (14.0) | 15.8 (21.9) | 0.35 |
| Watery stool | 10.9 (15.3) | 10.5 (16.9) | 5.2 ( 6.0) | 0.13 |
| SNP6 (rs4570625) | GG (n = 126) | GT (n = 57) | TT (n = 5) | |
|
| ||||
| GI symptom | ||||
| Abdominal pain | 32.0 (24.1) | 33.8 (25.2) | 35.0 (40.4) | 0.84 |
| Bloating | 30.8 (29.3) | 27.1 (28.2) | 26.9 (29.9) | 0.83 |
| Intestinal gas | 37.9 (30.0) | 33.3 (29.6) | 32.8 (31.8) | 0.76 |
| Constipation | 19.2 (21.8) | 17.0 (18.7) | 34.3 (44.9) | 0.25 |
| Diarrhea | 11.8 (17.8) | 12.1 (16.1) | 30.7 (45.1) | 0.08 |
| Stool consistency | ||||
| Very hard stool | 12.2 (18.0) | 9.9 (11.6) | 32.7 (23.9) | 0.01 |
| Watery stool | 8.3 (12.5) | 11.5 (17.7) | 22.5 (29.4) | 0.07 |
TPH, tryptophan hydroxylase; GI, gastrointestinal; SNP, single nucleotide polymorphism
Each symptom score was calculated by the percentage of days with moderate-to-very-severe symptoms
Analysis of covariate controlling for age
Patients who were homozygous for the minor allele (T) of TPH2 SNP6 reported more days with both very hard and watery stools compared to other genotype groups (GG and GT genotypes). However, this result was due to two IBS patients, homozygous for the minor allele, who are very high on both hard and watery stool types.
IBS patients in the diarrhea-predominant subgroup showed a trend towards increased frequency of the SNP3 minor allele (GT and GG genotypes), but overall the genotype distribution of SNP3 did not significantly differ between subgroups (Table 4). None of the other GI symptoms such as abdominal pain or constipation was associated with any of the TPH gene variants.
DISCUSSION
Our study is the first to show possible associations of TPH gene variants with IBS and IBS-related GI symptoms in unrelated Caucasian women. While SNPs in the TPH1 gene were not associated with a diagnosis of IBS, they showed some association with daily reporting of GI symptoms including diarrhea, bloating and loose stools in women with IBS. A SNP in the TPH2 gene promoter region showed a trend of association with a reduced risk of IBS as well as stool consistency with both hard and loose stools in IBS patients. While these results were not statistically significant after accounting for multiple comparisons, they are intriguing and an attempt at replication with a larger independent data set would be warranted.
Alterations in 5-HT synthesis and metabolism has been considered as a possible underlying mechanism of IBS, due to the importance of 5-HT in the regulation of gut motility and secretion. Although findings have been inconsistent, various studies have been reported changes in 5-HT content, TPH1 expression, serotonin reuptake transporter (SERT) expression, and postprandial plasma serotonin levels in patients with IBS.(18, 19) TPH1 is predominantly expressed in EC cells in the gut.(9) Several studies have reported altered mucosal TPH1 transcript levels of RNA in patients with functional GI disorders. One study reported significantly reduced TPH1 mRNA levels in the colonic mucosa in both diarrhea-predominant and constipation-predominant IBS patients compared to healthy controls.(18) Another study showed higher mucosal TPH1 mRNA levels in patients with chronic constipation compared to healthy controls.(20) Several studies have shown increased postprandial plasma 5-HT levels in diarrhea-predominant IBS (21-24), and decreased plasma 5-HT levels in constipation-predominant IBS (22, 25).
Although we found no association between individual SNPs or haplotypes of the TPH1 gene and a diagnosis of IBS, SNP1 and SNP3 showed associations with daily diarrhea symptoms and loose stools in women with IBS, though these were non-significant after multiple comparison adjustment. TPH is the rate-limiting enzyme of 5-HT synthesis, therefore TPH gene variants could be candidates to influence concentrations of 5-hydroxyindoleacetic acid (5-HIAA, the major metabolite of 5-HT). One study reported that the AA genotype of TPH1 SNP1 was not associated with 5-HIAA concentrations in cerebrospinal fluid (CSF) although this genotype was associated with suicide attempts among patients with major depressive disorder.(26) However, a recent study showed that the major (A) allele of TPH1 SNP1 was associated with decreased CSF 5-HIAA concentrations (p = 0.0028) in healthy Caucasians.(27) This discrepancy may be due to different ethnicities of the study populations or different health conditions (major depressive patients versus healthy controls).
The TPH1 gene variants we investigated here have been shown to be associated with psychiatric disorders such as major depression, schizophrenia, and suicidal behavior.(26, 28) These findings provide indirect evidence that they are either functionally important in themselves or in LD with other functional genetic variants. Therefore, additional studies with larger sample sizes are needed to verify that the TPH1 gene represents a diarrhea susceptibility factor in IBS. In addition functional studies are needed to explain the biological mechanisms where by these SNPs contribute to IBS phenotype.
The SNP6 of TPH2 gene (also known as −703 G/T) is located in the promoter region and has been reported to modify serotonin availability by influencing gene expression.(29, 30) In addition, several genetic studies reported that TPH2 polymorphisms which are high LD with SNP6 are associated with CSF 5-HIAA levels.(31, 32) The TT genotype of SNP6 has been reported to be associated with higher amygdala response to emotional stimuli.(33, 34) Several genetic association studies demonstrated an association of the T allele with increased startle response(35), and higher risk for personality disorder (36) as well as attention-deficit/hyperactivity disorder (ADHD).(37)
Our result showed that being homozygous for the minor allele (T) of the TPH2 SNP6 was marginally associated with a reduced risk of IBS, without correction for multiple testing. This finding may be supported by the observation that the frequency of the minor allele of SNP6 in our controls was similar to the frequencies in the HapMap CEU data (21%) and healthy controls of German European origin (20%) in other genetic studies.(35, 38) However, we could not exclude the possibility that this is only by chance. Therefore, additional studies with larger sample sizes are needed to confirm this association as well.
Regarding GI symptoms, women with IBS who were homozygous for the minor allele (T) of SNP6 were more likely to have stools that were characterized as both ‘hard’ and ‘loose’. Among the five IBS patients with the TT genotype of SNP6, two subjects reported very high percentages of days with hard stool and also very high percentages of days with watery stools. These two subjects also reported high abdominal pain, intestinal gas and fatigue. At this time, we cannot determine whether these subjects were more likely to experience abdominal discomfort as a result of their genotype or whether they had a tendency to over-endorse GI symptoms as part of a more generalized tendency to report heightened levels of distress. Given the contradictory results of SNP6 in relation to presence of IBS versus the severity of symptoms, the fact that they are based on a very small number of patients with the TT genotype, and the concern about multiple comparisons, these results for SNP6 should not be interpreted too strongly.
A strength of this study lies in its use of a well-characterized cohort of women with IBS, who logged their GI symptoms by daily diary for 28 days. However there are several limitations to this study. First, our study samples originated from three different studies which might have introduced sampling bias. However, patients and controls of all three studies were recruited in the same way, from the same geographic region, and with the same inclusion and exclusion criteria. Second, the relatively small sample size and the exploration of a number of SNPs and outcome measures means the power of this study was low after accounting for multiple comparisons. The suggestive findings need to be verified in a larger independent sample.
In conclusion, we reported a preliminary finding of possible associations of TPH1 gene variants with IBS-related GI symptoms including diarrhea, bloating and loose stool in Caucasian women with IBS. Our findings also suggest a possible association of a TPH2 SNP with IBS diagnosis and stool characteristics.
Acknowledgments
ACKNOWLEGEMENTS AND DISCLOSURES: This study was supported by grants from National Institute of Nursing Research USA (NIH NR004142, NR001094 and NR 04913), the NIH Office of Research in Women’s Health (P30 NR04001), a research grand from Sigma-Theta-Tau International (STTI) and a Hester McLauw Nursing Scholarship Award from the University of Washington.
Footnotes
The authors of this manuscript declare that they have no conflict of interest.
REFERENCES
- 1.Drossman D, Corazziari E, Delvaux M, et al., editors. Rome III: The functional gastrointestinal disorders. 3rd ed Degnon Associates, Inc.; McLean,VA: 2006. [Google Scholar]
- 2.Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology. 2007;132:397–414. doi: 10.1053/j.gastro.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Gershon MD, Drakontides AB, Ross LL. Serotonin: Synthesis and Release from the Myenteric Plexus of the Mouse Intestine. Science. 1965;149:197–9. doi: 10.1126/science.149.3680.197. [DOI] [PubMed] [Google Scholar]
- 4.Camilleri M, Northcutt AR, Kong S, Dukes GE, McSorley D, Mangel AW. Efficacy and safety of alosetron in women with irritable bowel syndrome: a randomised, placebo-controlled trial. Lancet. 2000;355:1035–40. doi: 10.1016/S0140-6736(00)02033-X. [DOI] [PubMed] [Google Scholar]
- 5.Greenwood-Van Meerveld B, Venkova K, Hicks G, Dennis E, Crowell MD. Activation of peripheral 5-HT receptors attenuates colonic sensitivity to intraluminal distension. Neurogastroenterol Motil. 2006;18:76–86. doi: 10.1111/j.1365-2982.2005.00723.x. [DOI] [PubMed] [Google Scholar]
- 6.Prather CM, Camilleri M, Zinsmeister AR, McKinzie S, Thomforde G. Tegaserod accelerates orocecal transit in patients with constipation-predominant irritable bowel syndrome. Gastroenterology. 2000;118:463–8. doi: 10.1016/s0016-5085(00)70251-4. [DOI] [PubMed] [Google Scholar]
- 7.Lovenberg W, Jequier E, Sjoerdsma A. Tryptophan hydroxylation: measurement in pineal gland, brainstem, and carcinoid tumor. Science. 1967;155:217–9. doi: 10.1126/science.155.3759.217. [DOI] [PubMed] [Google Scholar]
- 8.Walther DJ, Bader M. A unique central tryptophan hydroxylase isoform. Biochem Pharmacol. 2003;66:1673–80. doi: 10.1016/s0006-2952(03)00556-2. [DOI] [PubMed] [Google Scholar]
- 9.Walther DJ, Peter JU, Bashammakh S, et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- 10.Paoloni-Giacobino A, Mouthon D, Lambercy C, et al. Identification and analysis of new sequence variants in the human tryptophan hydroxylase (TpH) gene. Mol Psychiatry. 2000;5:49–55. doi: 10.1038/sj.mp.4000647. [DOI] [PubMed] [Google Scholar]
- 11.Zill P, Baghai TC, Zwanzger P, et al. SNP and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene provide evidence for association with major depression. Mol Psychiatry. 2004;9:1030–6. doi: 10.1038/sj.mp.4001525. [DOI] [PubMed] [Google Scholar]
- 12.Zill P, Buttner A, Eisenmenger W, Moller HJ, Ackenheil M, Bondy B. Analysis of tryptophan hydroxylase I and II mRNA expression in the human brain: a post-mortem study. J Psychiatr Res. 2007;41:168–73. doi: 10.1016/j.jpsychires.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Jarrett ME, Cain KC, Burr RL, Hertig VL, Rosen SN, Heitkemper MM. Comprehensive self-management for irritable bowel syndrome: randomized trial of in-person vs. combined in-person and telephone sessions. Am J Gastroenterol. 2009;104:3004–14. doi: 10.1038/ajg.2009.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motzer SA, Jarrett M, Heitkemper MM, Tsuji J. Natural killer cell function and psychological distress in women with and without irritable bowel syndrome. Biol Res Nurs. 2002;4:31–42. doi: 10.1177/1099800402004001005. [DOI] [PubMed] [Google Scholar]
- 15.Jarrett ME, Kohen R, Cain KC, et al. Relationship of SERT polymorphisms to depressive and anxiety symptoms in irritable bowel syndrome. Biol Res Nurs. 2007;9:161–9. doi: 10.1177/1099800407307822. [DOI] [PubMed] [Google Scholar]
- 16.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic acids research. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 18.Coates MD, Mahoney CR, Linden DR, et al. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology. 2004;126:1657–64. doi: 10.1053/j.gastro.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Mawe GM, Coates MD, Moses PL. Review article: intestinal serotonin signalling in irritable bowel syndrome. Aliment Pharmacol Ther. 2006;23:1067–76. doi: 10.1111/j.1365-2036.2006.02858.x. [DOI] [PubMed] [Google Scholar]
- 20.Costedio MM, Coates MD, Brooks EM, et al. Mucosal serotonin signaling is altered in chronic constipation but not in opiate-induced constipation. Am J Gastroenterol. 2010;105:1173–80. doi: 10.1038/ajg.2009.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bearcroft CP, Perrett D, Farthing MJ. Postprandial plasma 5-hydroxytryptamine in diarrhoea predominant irritable bowel syndrome: a pilot study. Gut. 1998;42:42–6. doi: 10.1136/gut.42.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atkinson W, Lockhart S, Whorwell PJ, Keevil B, Houghton LA. Altered 5-hydroxytryptamine signaling in patients with constipation- and diarrhea-predominant irritable bowel syndrome. Gastroenterology. 2006;130:34–43. doi: 10.1053/j.gastro.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 23.Houghton LA, Atkinson W, Whitaker RP, Whorwell PJ, Rimmer MJ. Increased platelet depleted plasma 5-hydroxytryptamine concentration following meal ingestion in symptomatic female subjects with diarrhoea predominant irritable bowel syndrome. Gut. 2003;52:663–70. doi: 10.1136/gut.52.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuo XL, Li YQ, Yang XZ, et al. Plasma and gastric mucosal 5-hydroxytryptamine concentrations following cold water intake in patients with diarrhea-predominant irritable bowel syndrome. J Gastroenterol Hepatol. 2007;22:2330–7. doi: 10.1111/j.1440-1746.2006.04772.x. [DOI] [PubMed] [Google Scholar]
- 25.Dunlop SP, Coleman NS, Blackshaw E, et al. Abnormalities of 5-hydroxytryptamine metabolism in irritable bowel syndrome. Clin Gastroenterol Hepatol. 2005;3:349–57. doi: 10.1016/s1542-3565(04)00726-8. [DOI] [PubMed] [Google Scholar]
- 26.Galfalvy H, Huang YY, Oquendo MA, Currier D, Mann JJ. Increased risk of suicide attempt in mood disorders and TPH1 genotype. J Affect Disord. 2009;115:331–8. doi: 10.1016/j.jad.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreou D, Saetre P, Werge T, et al. Tryptophan hydroxylase gene 1 (TPH1) variants associated with cerebrospinal fluid 5-hydroxyindole acetic acid and homovanillic acid concentrations in healthy volunteers. Psychiatry Res. doi: 10.1016/j.psychres.2009.11.018. doi:10.1016/j.psychres.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Saetre P, Lundmark P, Wang A, et al. The tryptophan hydroxylase 1 (TPH1) gene, schizophrenia susceptibility, and suicidal behavior: a multi-centre case-control study and meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:387–96. doi: 10.1002/ajmg.b.30991. [DOI] [PubMed] [Google Scholar]
- 29.Chen GL, Vallender EJ, Miller GM. Functional characterization of the human TPH2 5′ regulatory region: untranslated region and polymorphisms modulate gene expression in vitro. Hum Genet. 2008;122:645–57. doi: 10.1007/s00439-007-0443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin YM, Chao SC, Chen TM, Lai TJ, Chen JS, Sun HS. Association of functional polymorphisms of the human tryptophan hydroxylase 2 gene with risk for bipolar disorder in Han Chinese. Arch Gen Psychiatry. 2007;64:1015–24. doi: 10.1001/archpsyc.64.9.1015. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Z, Roy A, Lipsky R, et al. Haplotype-based linkage of tryptophan hydroxylase 2 to suicide attempt, major depression, and cerebrospinal fluid 5-hydroxyindoleacetic acid in 4 populations. Arch Gen Psychiatry. 2005;62:1109–18. doi: 10.1001/archpsyc.62.10.1109. [DOI] [PubMed] [Google Scholar]
- 32.Lim JE, Pinsonneault J, Sadee W, Saffen D. Tryptophan hydroxylase 2 (TPH2) haplotypes predict levels of TPH2 mRNA expression in human pons. Mol Psychiatry. 2007;12:491–501. doi: 10.1038/sj.mp.4001923. [DOI] [PubMed] [Google Scholar]
- 33.Brown SM, Peet E, Manuck SB, et al. A regulatory variant of the human tryptophan hydroxylase-2 gene biases amygdala reactivity. Mol Psychiatry. 2005;10:884–8. doi: 10.1038/sj.mp.4001716. 05. [DOI] [PubMed] [Google Scholar]
- 34.Canli T, Congdon E, Gutknecht L, Constable RT, Lesch KP. Amygdala responsiveness is modulated by tryptophan hydroxylase-2 gene variation. J Neural Transm. 2005;112:1479–85. doi: 10.1007/s00702-005-0391-4. [DOI] [PubMed] [Google Scholar]
- 35.Armbruster D, Mueller A, Strobel A, Kirschbaum C, Lesch KP, Brocke B. Influence of functional tryptophan hydroxylase 2 gene variation and sex on the startle response in children, young adults, and older adults. Biol Psychol. 2010;83:214–21. doi: 10.1016/j.biopsycho.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 36.Jacob CP, Nguyen TT, Dempfle A, et al. A gene-environment investigation on personality traits in two independent clinical sets of adult patients with personality disorder and attention deficit/hyperactive disorder. Eur Arch Psychiatry Clin Neurosci. 2010;260:317–26. doi: 10.1007/s00406-009-0079-0. [DOI] [PubMed] [Google Scholar]
- 37.Walitza S, Renner TJ, Dempfle A, et al. Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase-2 gene in attention-deficit/hyperactivity disorder. Mol Psychiatry. 2005;10:1126–32. doi: 10.1038/sj.mp.4001734. [DOI] [PubMed] [Google Scholar]
- 38.Marziniak M, Kienzler C, Kuhlenbaumer G, Sommer C, Mossner R. Functional gene variants of the serotonin-synthesizing enzyme tryptophan hydroxylase 2 in migraine. J Neural Transm. 2009;116:815–9. doi: 10.1007/s00702-009-0236-7. [DOI] [PubMed] [Google Scholar]
- 39.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]


