Abstract
Molecular-genetic imaging is advancing from a valuable preclinical tool to guiding patient management. The strategy involves pairing an imaging reporter gene with a complementary imaging agent in a system that can be used to measure gene expression, protein interaction or track gene-tagged cells in vivo. Tissue-specific promoters can be used to delineate gene expression in certain tissues, particularly when coupled with an appropriate amplification mechanism. Here we show that the progression elevated gene-3 promoter (PEG-Prom), derived from a rodent gene mediating the malignant phenotype, can be used to drive imaging reporters selectively to enable detection of micrometastatic disease in murine models of human melanoma and breast cancer using bioluminescence and radionuclide-based molecular imaging techniques. Because of its strong promoter, tumor specificity and capacity for clinical translation, PEG-Prom-driven gene expression may represent a practical, new system for facilitating cancer imaging and therapy.
INTRODUCTION
Targeted imaging of cancer remains an important but elusive goal. Such imaging could provide early diagnosis, aid treatment planning and benefit therapeutic monitoring. By leveraging the expanding list of specific molecular characteristics of tumors and their microenvironment, molecular imaging has the potential to generate tumor-specific reagents. But many efforts at tumor-specific imaging are fraught by nonspecific localization of the putative targeted agents, eliciting unacceptably high background noise. While investigators use many strategies to provide tumor-specific imaging agents – largely in the service of maintaining high target-to-background ratios – they fall into two general categories, namely direct and indirect methods1. Direct methods employ an agent that reports directly on a specific parameter, such as a receptor, transporter or enzyme concentration, usually by binding directly to the target protein. Indirect methods use a reporter transgene strategy, in analogy to the use of green fluorescent protein (GFP) in vitro, to provide a read-out on cellular processes occurring in vivo by use of an external imaging device. Molecular-genetic imaging employs an indirect technique that has enabled the visualization and quantification of the activity of a variety of gene promoters, transcription factors and key enzymes involved in disease processes and therapeutics in vivo including Gli2, E2F13, telomerase4,5, and several kinases, including one that has proved useful in human gene therapy trials6,7.
A minimal promoter region of progression elevated gene-3 (Peg-3), a rodent gene, was identified by us (P.B.F.) for its association with malignant transformation and tumor progression using subtraction hybridization8. PEG-Prom drives downstream gene expression in a tumor-specific manner and has been tested in cancer cell lines of various tissues such as brain, prostate, breast and pancreas9-11, as well as in metastatic melanoma12. Transcription factors AP-1 and E1AF/PEA3 (ETS-1) are known to mediate the cancer-specific activity of PEG-Prom8,9,13. Previous studies have demonstrated the utility of PEG-Prom for cancer gene therapy through intratumoral delivery9-12,14. Here we describe a novel method for imaging a variety of metastatic cancers through systemic delivery of PEG-Prom. We hypothesize that the systemic delivery of PEG-Prom-driven imaging constructs will enable tumor-specific expression of reporter genes, not only within primary tumor, but also in associated metastases in a manner broadly applicable to tumors of different tissue origin or subtype.
RESULTS
Cancer-specificity of PEG-Prom via bioluminescence imaging
To test the specificity of PEG-Prom for tumor imaging in vivo, we used two different reporters, firefly luciferase (Luc) and the herpes simplex virus 1 thymidine kinase (HSV1-TK). Luc is often used with bioluminescence imaging (BLI) to establish proof-of-principle for imaging specific gene expression or gene-tagged cells in preclinical models, while HSV1-TK, also often used preclinically, has been translated to clinical studies. Accordingly, we generated two plasmid constructs, pPEG-Luc and pPEG-HSV1tk (Supplementary Fig. 1a,b). We chose to image the experimental metastasis models of two different tissues: human melanoma and breast cancer. As a gene delivery vehicle we used in vivo-jetPEI™, which is based on linear polyethylenimine (l-PEI), one of the most widely used cationic polymers for gene delivery. We chose that inert (nonviral) vehicle rather than a viral delivery system to avoid biased systemic delivery, as can be seen with viral vectors, which have a tendency to localize to liver upon intravenous (IV) administration15,16.
After confirmation of the presence of metastatic nodules in the lung by computed tomography (CT) at 4–6 weeks after IV administration of the human malignant melanoma cell line MeWo, or the human metastatic breast cancer cell line MDA-MB-231, animals received an IV dose of pPEG-Luc/PEI polyplex (Supplementary Fig. 1a). Twenty four and 48 h after plasmid DNA (pDNA) delivery, PEG-Prom-driven gene expression was assessed by BLI. The same pDNA delivery and imaging protocols were applied to a group of healthy animals as a negative control. Expression of Luc driven by PEG-Prom was observed only in the melanoma metastasis model (Mel) and not in control animals (Fig. 1a,b). Control animals demonstrated nearly background levels of BLI output at the 24 h time point that disappeared by the 48 h imaging session (Supplementary Fig. 2a). Quantification of the BLI signal intensity from the thoracic cavity, which represents Luc expression mainly in lung, shows significantly higher PEG-Prom activity in the Mel group compared to controls at both time points after pPEG-Luc administration (Fig. 1c), and more so at 48 h. Similar results were observed in the model of breast cancer metastasis (BCa) (Fig. 1f–h). Pseudo-color images of another representative animal from the same control group shown in Fig. 1a were readjusted for the BCa model such that the control and BCa groups are scaled to the same threshold values (Fig. 1f). As with the Mel model, quantified bioluminescence intensity from the thoracic cavity shows higher PEG-Prom activity in the BCa group compared to controls, and more markedly so 48 h after pPEG-Luc delivery (Fig. 1h). It took longer for the BCa group harboring MDA-MB-231 metastases than for the Mel group with MeWo metastases to provide a significant increase in BLI signal over background, likely resulting from the lower efficiency of gene delivery in the BCa model, as discussed below. BLI images of all of the animals in each group, Mel and BCa, as well as controls, at the same pseudo-color threshold values can be found in Supplementary Figs. 2 and 3, respectively.
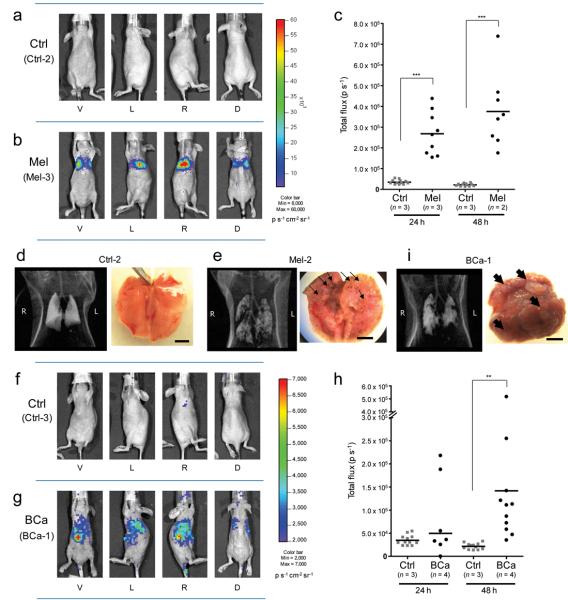
Figure 1.
Cancer-specific PEG-Prom activity shown by bioluminescence imaging (BLI) in experimental metastasis models of human melanoma (Mel) and breast cancer (BCa). (a) BLI of a representative healthy control animal (Ctrl-2). (b) BLI showing firefly luciferase (Luc) expression observed in a representative Mel model (Mel-3). Each animal was imaged from four directions (V, ventral; L, left side; R, right side; D, dorsal views) in order to cover the entire body. Pseudo-color images from the two groups were adjusted to the same threshold. (c) Quantification of BLI signal intensity in the control group (Ctrl) and Mel group at 24 and 48 h after injection of pPEG-Luc/PEI polyplex. Quantified values are shown in Total flux (photons per second, p s−1). *** P < 0.0001. (d,e) CT scans and gross anatomical views of lung from one representative animal from the control group (d) and the Mel group (e). (f,g) BLI of one representative animal from the control group (f, Ctrl-3) and the experimental breast cancer metastasis group (g, BCa-1). The pseudo-color images were adjusted to the same threshold. (h) Quantification of bioluminescent signal intensity in the Ctrl and BCa groups at 24 and 48 h after injection of pPEG-Luc/PEI polyplex. ** P = 0.0066. (i) A CT image and a macroscopic view of lung from a representative BCa animal. Displayed bioluminescent images (a,b,f,g) were obtained at 48 h after the systemic delivery of pPEG-Luc/PEI polyplex. Black arrows (e,i) indicate metastatic nodules observed in the lung. Scale bars, 5 mm.
On average an approximately three-fold higher level of Luc expression was observed from the Mel group compared to the BCa group at 48 h. CT scans and gross anatomical views revealed very different patterns of metastatic nodule formation in the lung of those two models. While MeWo cells formed small nodules uniformly scattered throughout the lungs (Fig. 1e, black arrows), MDA-MB-231 cells tended to form isolated large nodules (Fig. 1i, black arrows). Histological analysis demonstrated that metastases derived from MeWo cells in the Mel model were better vascularized (Supplementary Fig. 4a, white arrows), while necrotic centers were observed in the nodules formed in the lungs of BCa animals harboring metastases derived from MDA-MB-231 cells (Supplementary Fig. 4b, white arrows). In addition to decreasing the efficiency of gene delivery, the poor vascularization and consequent central necrosis of the BCa tumors may limit access of D-luciferin and oxygen to the tumor, which are necessary concomitants for productive BLI signal.
In order to exclude the possibility that tumor-specific expression of Luc by BLI might have resulted from the difference in transfection efficiency between normal and malignant mouse lung tissues, we quantified the amount of pDNA delivered to the lung of each animal. We performed quantitative real time PCR using a primer set designed to amplify a region of the Luc-encoding gene in the pPEG-Luc plasmid. Total DNA extracted from the lung tissues was used as a template. The difference in transfection efficiency between the control group and the Mel group was not significant (Supplementary Fig. 5). On the other hand, the BCa group had significantly lower transfection efficiency compared to the control. That result confirmed that the tumor-specific expression of Luc observed in these models was due to the tumor-selective activity of PEG-Prom rather than differential transfection efficiency between normal and malignant lungs. Poor vascularization and segregated large nodules most likely contributed to lower transfection efficiency observed in the lung of the BCa model (Supplementary Fig. 4). We also imaged constitutive cytomegalovirus (CMV) promoter activity in the lungs of the healthy control and Mel groups as an additional check on the tumor specificity of PEG-Prom imaging (Supplementary Fig. 6). BLI showed no significant difference in the CMV promoter-driven Luc expression level between the control and Mel groups at any time up to 45 h after the systemic delivery of pCMV-Tri/PEI polyplex. This result indicates further that the specificity of PEG-Prom imaging results from tumor-specific promoter activity and not from tumor microenvironmental effects, such as increased tumor vascularity or enhanced tumor vascular permeability, causing greater plasmid delivery or nonspecific effects on transgene expression.
BLI with systemically administered pPEG-Luc also enabled imaging of small metastatic deposits, i.e., micrometastases, outside of the lung parenchyma in both the Mel and BCa models. That was confirmed through harvesting regions producing BLI signal above background and performing correlative histological analysis (Figs. 2 and 3). Specifically, histological analysis on the tissue sections from a representative Mel model, Mel-2 (Fig. 2a), confirmed that Luc expression was associated with the metastatic sites formed in the lung, adrenal glands, the chest cavity adjacent to the sternum and abdominal inguinal adipose tissues adjoining the bladder (Fig. 2b,c,d,e, respectively). Similarly, correlation between metastatic sites and PEG-Prom activity was observed in a representative BCa model, BCa-3 (Fig. 3a), inside the lung, the peripancreatic area, the thoracic wall adjacent to the sternum, a lymph node located in the adipose connective tissues surrounding the bladder and the rib cage in the form of thin rows of micrometastatic deposits (Fig. 3b,c,d,e,f, respectively).
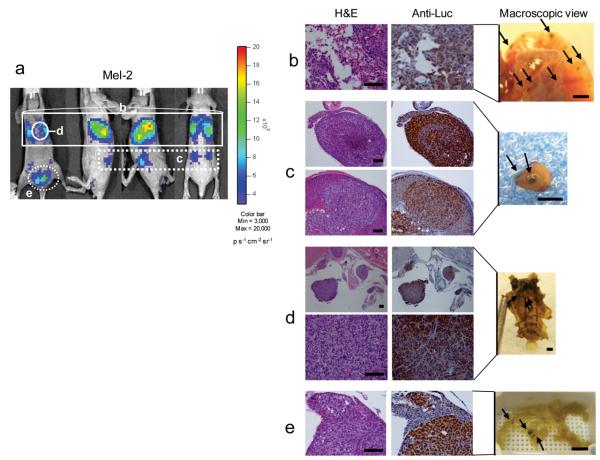
Figure 2.
Correlation between PEG-Prom-driven Luc expression and microscopic metastatic sites shown by histopathological analysis in a mouse model of melanoma metastasis (Mel). (a) BLI of a representative animal, Mel-2 from Supplementary Fig. 2b. These images were acquired at 48 h after the delivery of pPEG-Luc/PEI polyplex. (From left to right: ventral, left-sided, right-sided and dorsal views) (b–e) Hematoxylin and eosin (H&E) and firefly luciferase (Luc) staining of the formalin-fixed paraffin-embedded (FFPE) tissues collected from Mel-2. According to the imaging result (a, white solid or dotted rectangles and circles), organs tentatively associated with Luc expression were harvested for histological analysis. H&E staining confirms metastatic foci of melanoma cells in the lung (b, white solid rectangle in a), in the adrenal glands (c, white dotted rectangle in a), inside the thoracic cage adjacent to the sternum (d, white solid circle in a) and in abdominal inguinal adipose tissues adjacent to the urinary bladder (e, white dotted circle in a). Immunostaining of the consecutive sections with rabbit Luc-specific antibody (anti-Luc) shows precise correlation between the localization of microscopic metastasis and Luc expression (brown staining). Melanoma-associated melanin pigmentation was also observed in these organs (black arrows in ‘Macroscopic View’, b,c,d,e). Scale bars, 100 μm (H&E) and 2 mm (macroscopic views).
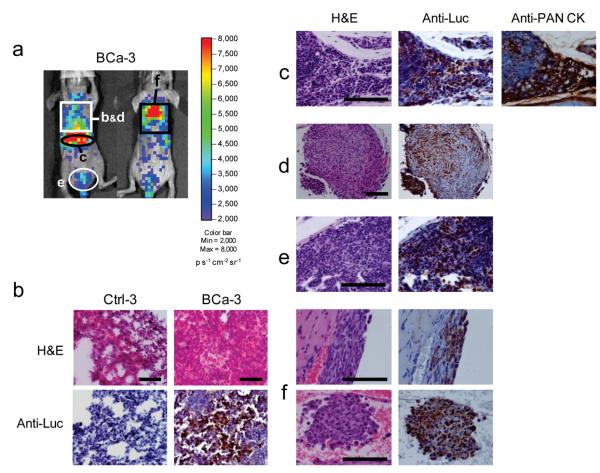
Figure 3.
Histological confirmation of the microscopic metastatic sites detected by the PEG-Prom-driven BLI system in a human breast cancer metastasis model (BCa). (a) BLI of a representative animal, BCa-3 from Supplementary Fig. 3b, 24 h after the systemic delivery of pPEG-Luc/PEI polyplex (from left to right: ventral and dorsal views). The organs associated with the expression of Luc from a (black or white circles and rectangles), were collected for histological correlation. (b) H&E and Luc staining on cryosections of the lung from BCa-3, which correlates with bioluminescent light output shown in the white rectangle in a. Stained lung cryosections of a control animal (Ctrl-3) from Supplementary Fig. 3a are shown for comparison. (c–f) H&E and Luc staining of the FFPE-tissue sections collected from BCa-3. In the peripancreatic area (c, black circle in a), inside the thoracic wall adjacent to the sternum (d, white rectangle in a) and in the lymph node located in the abdominal inguinal adipose tissues adjacent to the bladder (e, white circle in a), H&E staining confirmed the presence of metastatic lesions. Thin layers of breast cancer cells were found inside the rib cage (f, black rectangle in a): between 1st–3rd ribs (f, upper panel) and 4th–7th ribs (f, lower panel). Luc staining on the consecutive sections demonstrated the co-localization of the metastatic sites and Luc expression. Another consecutive section of peripancreatic tissues was stained for human pan-cytokeratin (PAN CK) to ensure the precise co-localization of Luc expression and MDA-MB-231 cells (c). Scale bars, 100 μm.
PEG-Prom-mediated cancer detection via radionuclide imaging
Although both malignant lung lesions and extrathoracic micrometastases could be detected with BLI, this technique is limited to preclinical studies. That is due to several factors, including the need to administer luciferase substrate, insufficient depth of penetration of BLI light output and difficulty in generating quantitative, tomographic BLI-based images. Accordingly, we generated a more clinically relevant PEG-Prom-driven gene expression imaging system, pPEG-HSV1tk (Supplementary Fig. 1b), which can be detected using radionuclide-based techniques, namely, single photon emission computed tomography (SPECT) or positron emission tomography (PET), upon administration of a suitably radiolabeled nucleoside analog.
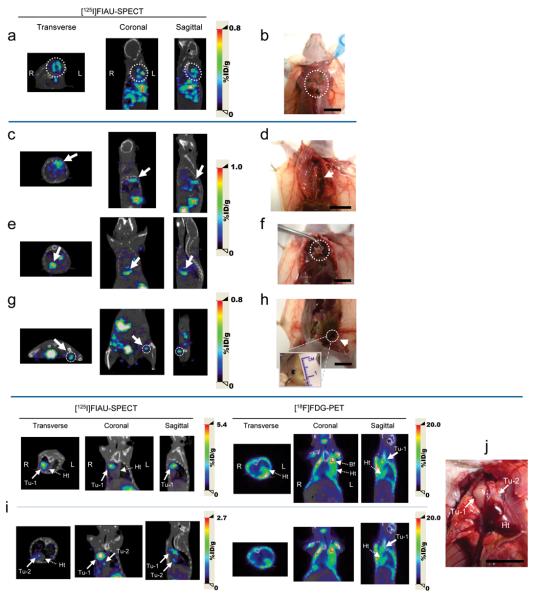
We used the Mel experimental metastasis model to demonstrate tumor-targeted imaging with SPECT-CT. Approximately 7 weeks after receiving MeWo cells as above, the Mel group and corresponding controls received pPEG-HSV1tk/PEI polyplex by IV injection. 46 h after pDNA delivery, the animals were injected with 2′-fluoro-2′-deoxy-β-D-5-[125I]iodouracil-arabinofuranoside ([125I]FIAU) and imaged at 24 and 48 h after receiving the radiotracer (Fig. 4a,b, Supplementary Videos 1 and 2). Quantification of radioactivity demonstrates a 31-fold higher accumulation of [125I]FIAU in the lungs of the Mel model compared to controls, indicating the tumor-specific expression of HSV1-TK under the control of PEG-Prom (Fig. 4c). We further confirmed tumor presence in presumptive extrathoracic metastatic sites through gross histological analysis after the 48 h imaging session (Fig. 5). Detected on the whole body SPECT-CT images (Fig. 5a, Supplementary Video 1) were multiple metastatic lesions in the dorsal neck of Mel-2 that corresponded to the intact histological specimen (Fig. 5b). Metastatic sites, such as one to the left of the spinal cord, another immediately above the diaphragm and the other in the left inguinal lymph node, similarly correlated in Mel-3 (Fig. 5c with d, Fig. 5e with f and Fig. 5g with h, respectively). In order to evaluate the accuracy of detection and translational potential of PEG-Prom-mediated imaging, we compared the ability of the PEG-Prom system to detect lesions to that of [18F]fluorodeoxyglucose (FDG), the clinical standard, in both Mel and BCa models. The same animals were imaged using each method (Fig. 5i,j, Supplementary Fig. 7) and in most instances there was good correlation between the metastatic nodules detected by these two radionuclide-based techniques. However, the PEG-Prom-based system was better able to detect nodules adjacent to the heart and brown fat tissues, areas known to sequester FDG and cause high regional background17,18. That finding is particularly significant in light of the fact that SPECT is inherently at least one order of magnitude less sensitive than PET, and accumulation of [125I]FIAU is limited to the tumor cells transfected with pPEG-HSV1tk/PEI that express the HSV1TK reporter, while FDG is taken up essentially by all viable malignant cells (Supplementary Fig. 7a).
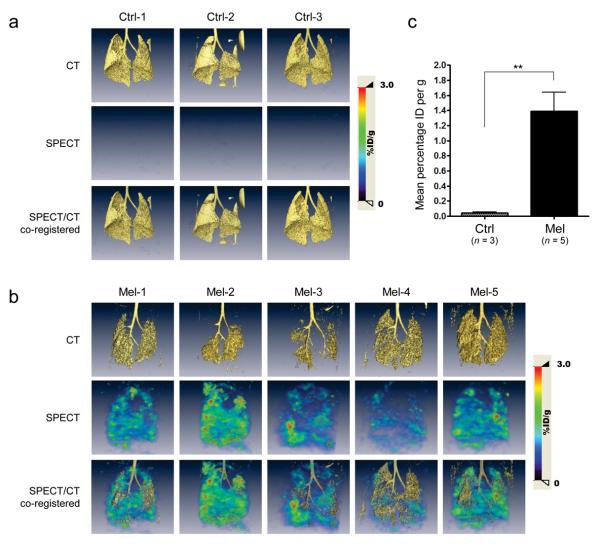
Figure 4.
Cancer-specific expression of HSV1-TK driven by PEG-Prom shown by SPECT-CT imaging in an experimental model of human melanoma metastasis (Mel). (a,b) CT, SPECT and co-registered [125I]FIAU SPECT-CT images of lungs of the healthy control group (a, n = 3; Ctrl-1–3) and of the metastasis model of melanoma (b, n = 5; Mel-1–5). Images were acquired at 48 h after IV injection of [125I]FIAU, which was 94 h after IV administration of pPEG-HSV1tk/PEI polyplex. Color scales are expressed as %ID/g (percentage injected dose per gram of tissue). (c) Quantification of lung SPECT images in a and b. ROIs of the same size and shape were drawn in the right lobes of the lung of each animal. Quantified radioactivity was expressed as Mean percentage ID per g (mean percentage injected dose per gram of tissue). ** P = 0.0070.
Figure 5.
Detection and localization of metastatic masses by SPECT-CT imaging after the systemic administration of pPEG-HSV1tk. (a,c,e,g) Transverse, coronal and sagittal views of co-registered SPECT-CT images of Mel-2 (a) and Mel-3 (c,e,g) from Fig. 4b. All images were obtained at 24 h after [125I]FIAU injection. (b,d,f,h) Gross anatomical details of the metastatic masses that were located based on the SPECT-CT images (a,c,e,g). Multiple metastatic sites were detected by imaging in Mel-2 (a, dotted circle). Necropsy of the corresponding area revealed melanoma masses under the brown adipose tissue in the upper dorsal area (b, dotted circle). (c) Accumulated radioactivity was detected adjacent to the thoracic mid-spine (arrow), which corresponded to a tumor at this location (d, arrow). Additional metastatic sites demonstrated by SPECT-CT imaging (e,g, arrow and dotted circle) correlated with melanoma masses uncovered immediately above the diaphragm (f, dotted circle) and in the left inguinal lymph node (h, arrow), respectively. (i,j) Cross-comparison of the PEG-Prom-mediated imaging and FDG-PET in a breast cancer metastasis model, BCa-1. Two nodules (Tu-1 and Tu-2) were detected by [125I]FIAU-SPECT near the heart (i) and were confirmed by necropsy (j). While Tu-1 was also detected by [18F]FDG-PET, Tu-2, a smaller nodule attached to the heart, was not obvious in the PET image. SPECT images were acquired 48 h post-injection of [125I]FIAU. The PET and SPECT images were acquired on the same day (i). (Tu, tumor; Ht, heart; Bf, brown fat) Color scales are expressed as %ID/g (percentage injected dose per gram of tissue). Scale bars, 10 mm.
DISCUSSION
Our goal was to develop a systemically deliverable construct that would enable molecular-genetic imaging of cancer. Necessary elements to provide such a construct include a sufficiently strong promoter with cancer specificity, potential for clinical translation and capacity to be linked to gene therapy. Promoters derived from human telomerase reverse transcriptase (hTERT)4, survivin19 and carcinoembryonic antigen (CEA)20 promoters and enhancer elements have been used in molecular-genetic imaging to provide tumor-specific reporter expression. However, because those studies employed adenoviral vectors, delivery was limited to local administration, or systemic administration resulted in expression only within the liver. By contrast here we could delineate metastases with PEG-Prom after systemic delivery using a nonviral vector. Often promoter activity must be amplified to drive the downstream gene for purposes of imaging or therapy. One such strategy for doing so involves the two-step transcriptional amplification (TSTA) system21,22 using the GAL4-VP16 fusion protein and GAL4 response elements19,20,23-25. However, PEG-Prom did not require amplification to achieve high-sensitivity imaging. SPECT-CT imaging demonstrated a metastatic to normal lung signal ratio of 31 out to four days after administration of pPEG-HSV1tk (Fig. 4c). PEG-Prom activity is comparable to the constitutively active SV40 promoter (data not shown). In keeping with previously reported in vitro results9, we demonstrate here that PEG-Prom proved to be tumor-specific in vivo using both imaging modalities and in both tumor models tested, with the potential for further generalization to other modalities and tumors. We further chose pPEG-HSV1tk because of its capacity to be translated clinically. Clinical molecular-genetic imaging and gene therapy have been accomplished using HSV1tk and radiolabeled nucleoside analogs7,26 and ganciclovir6,27-29, respectively. By using the l-PEI polyplex delivery vehicle we avoid the attendant problems of viral vectors in gene delivery, including immune reactions30 and oncogenesis. Using pDNA vectors the integration rate of the extrachromosomal gene into the host genome in vivo was negligible31-34. We also estimated the potential of in vivo-jetPEI™ as a pPEG-HSV1tk delivery vehicle for detecting metastases in bone and brain, organs considered to be relatively difficult to access through systemic administration. Although lower than within lung, quantitative real time PCR demonstrated delivery of significant amounts of pDNA to each of those tissues (Supplementary Fig. 8). Since poorly vascularized necrotic areas within tumors contain less viable cells and are also difficult to reach through systemic delivery of imaging agents, molecular-genetic imaging techniques are expected to detect more accurately well-vascularized as opposed to predominantly necrotic lesions.
Here we show how PEG-Prom can be used as an imaging agent for melanoma and breast cancer metastases in vivo and propose this promoter as potentially universal for this purpose. Such an agent might be used to detect tumors before their tissue of origin or subtype is identified, without concern for nonspecific expression in normal tissues. As with other imaging agents, PEG-Prom can be used not just for tumor detection, but also for preoperative planning, intraoperative management and therapeutic monitoring. The PEG-Prom imaging system can also be fashioned into a theranostic agent, through use of an internal ribosome entry site or other strategy enabling tandem gene expression. Promoters such as PSA (prostate-specific antigen) promoter23,24 for prostate cancer, mucin-1 promoter25,35 for breast cancer, and mesothelin promoter36 for ovarian cancer have been used to delineate primary tumors and lymph node metastasis through molecular-genetic imaging. However, the activities of those promoters have not been tested more broadly, which would be needed to image a wide variety of tumors. Similarly, although hTERT, survivin and CEA promoters were reported to be of a less tissue- and more cancer-specific nature, their activity relies on the transcription level of the marker genes. Rather, PEG-Prom is responsive directly to transcription factors unique to tumor cells. The PEG-3 gene is a truncated mutant form of the rat growth arrest- and DNA damage-inducible gene, Gadd34, which occurs uniquely during murine tumorigenesis and may function as a dominant-negative of Gadd34 promoting the malignant phenotype37. No homolog to PEG-Prom is found in the human genome including the promoter/enhancer region of the human GADD homolog, which makes the use of PEG-Prom in human subjects likely to produce only minimal background signal9,37.
In conclusion, these studies suggest that PEG-Prom may possess all of the necessary elements to provide a practical strategy for imaging and potentially image-guided therapy of a variety of cancers. Determination of the sensitivity and specificity of this system in vivo using radionuclide-based techniques in a variety of tissues, malignant and otherwise, is under way. Further optimization of the delivery vehicle and assessment of its effects on normal tissue are also being vigorously pursued.
METHODS
Plasmids
pPEG-Luc was constructed as described previously9. The firefly luciferase (Luc)-encoding gene in pPEG-Luc was replaced by the HSV1-TK-encoding sequence from pORF-HSVtk plasmid (InvivoGen) to generate pPEG-HSV1tk. We prepared pDNA with the EndoFree Plasmid Kit (Qiagen). Endotoxin level was ensured as < 2.5 endotoxin unit per mg pDNA.
Systemic DNA delivery
Low molecular weight l-PEI-based cationic polymer, in vivo-jetPEI™, (Polyplus-transfection) provided the gene delivery vehicle. DNA-polyplex was formed according to the Manufacturer's Instructions. 30 μg of pDNA and 3.6 μl of 150 mM in vivo-jetPEI™ were diluted in endotoxin-free 5% glucose separately and then mixed together to give an N:P ratio of 6:1 in a total volume of 400 μl. 400 μl were injected into the lateral tail vein of an animal as two 200 μl-injections with a 5 min-interval.
Generation of experimental metastasis models
Animal studies were undertaken in accordance with the rules and regulations of the Johns Hopkins Animal Care and Use Committee. BLI studies employed experimental metastasis models of human melanoma (Mel) and breast cancer (BCa). 5–6 week-old female NCR nu/nu mice (NCI-Frederick and Charles River Laboratories) received whole body irradiation (5 Gy) to ensure suppression of the residual immune system in nude mice. Within 24 h after irradiation, animals were randomly divided into three groups. One group was injected with 5 × 106 cells of the human malignant melanoma cell line MeWo (ATCC) intravenously to generate Mel. Another group of mice received IV injection of 2 × 106 cells of the human breast cancer cell line MDA-MB-231 for BCa. Another group was maintained as a control. We confirmed the metastatic nodule formation in the lung by CT at 4–7 weeks after cell injection. For the SPECT-CT and PET-CT studies we generated the Mel and BCa models as described above except that whole body irradiation was omitted. MeWo and MDA-MB-231 cell lines were maintained in MEM and RPMI-1640 media, respectively, supplemented with 10% FBS and 1% penicillin/streptomycin.
In vivo bioluminescence imaging
At 24 and 48 h after gene delivery, we imaged animals with the IVIS Spectrum (Caliper/Xenogen). For each imaging session mice were injected intraperitoneally with D-luciferin (150 mg kg−1) under anesthesia using 1.5–2.5% isoflurane/oxygen mixture. Images were acquired between 5–35 min after injection of D-luciferin. In order to compensate for the limitation of 2D image acquisiton, we imaged most animals in four different positions: ventral, left- and right-sided, dorsal. ROIs of the same size and shape, covering the entire thoracic cavity, were applied to every image acquired from all four orientations to account for intragroup variations in metastatic site localization. Total flux (p s−1) in the ROIs was measured. Image acquisition and BLI data analysis were done using Living Image software (Caliper Life Sciences).
SPECT-CT imaging and data analysis
At 46 h after injection of pPEG-HSV1tk/PEI polyplex, animals were injected IV with 51.8 mBq (1.4 mCi) of [125I]FIAU. 24 and 48 h after radiotracer injection image data were acquired with the X-SPECT small-animal SPECT-CT system (Gamma Medica-Ideas, Inc.) using the low-energy single pinhole collimator (1.0 mm aperture). Focused lung imaging was acquired with a radius of rotation (ROR) of 3.35 cm and whole body imaging with a ROR of 6.75 cm. At 24 h after injection, animals were imaged in 64 projections and 30 s of acquisition per projection, and at 48 h after injection with 60 s per projection. SPECT images were co-registered with the corresponding 512-slice CT images. Tomographic image datasets were reconstructed with the 2D ordered subsets-expectation maximum (OS-EM) algorithm with two iterations and four subsets, and AMIDE38 and Amira (Visage Imaging) software were used for analysis.
PET-CT imaging and data analysis
At 1 h after 9.25 mBq (0.25 mCi) of IV administration of FDG, whole body images were acquired with the eXplore Vista small animal PET scanner (GE Healthcare) using the 250–700 keV energy window. Acquisition time was 30 min (2 bed positions; 15 min per bed position). Animals were fasted for 6–12 h prior to receiving FDG and were kept warm on the heating pad in order to minimize radiotracer accumulation in non-tumor tissues. PET images were co-registered with the corresponding 512-slice CT images. Tomographic image datasets were reconstructed with the 3D OS-EM algorithm with three iterations and twelve subsets and analyzed with AMIDE38 software.
Immunohistochemistry
After the BLI data acquisition at 48 h after the pPEG-Luc/PEI polyplex delivery, each organ demonstrating expression of Luc was harvested and fixed in 10% neutral buffered formalin. Paraffin-embedded tissue slices and lung cryosections were stained with rabbit Luc-specific polyclonal antibody (Fitzgerald Industries International, Inc.). Horseradish peroxidase (HRP)-conjugated polyclonal goat anti-rabbit antibody was used as a secondary antibody. HRP activity was detected with 3,3′-diaminobenzidine substrate-chromogen (EnVision™+ Kit, Dako).
Statistical analysis
Error bars in graphical data represent means ± s.e.m. The two-tailed Student's t test was performed, with P < 0.05 considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the technical support provided by S. Dhara, S. Nimmagadda, M. Goggins and M. Griffith. We are grateful to J. Fox and G. Green for providing [125I]FIAU and assisting in the SPECT-CT and PET-CT imaging studies. We also thank C. Endres, B. Tsui, J. Yu and J. Fox for help with SPECT and PET data processing. The MDA-MB-231 cell line and pCMV-Tri construct were generous gifts from Z. Bhujwalla (Johns Hopkins University) and S. Gambhir (Stanford University), respectively. Funding was provided by National Institutes of Health U24 CA92871 (to M.G.P.), by the Predoctoral Molecular Imaging Scholar Program from the Society of Nuclear Medicine and the Korea Science and Engineering Foundation Fellowship Program (to H.C.B.), and by US National Institutes of Health P01 CA104177 and the US National Foundation for Cancer Research (to P.B.F.). P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research at the VCU Massey Cancer Center.
Footnotes
AUTHORS' CONTRIBUTIONS STATEMENT
H.C.B. designed and performed the experiments, analyzed data and prepared the manuscript. K.L.G. provided technical support in histopathology. J.L., K.L.G. and P.B.F. gave conceptual advice and edited the manuscript. M.G.P. and P.B.F. conceived the project. M.G.P. supervised the project and prepared the manuscript.
COMPETING INTEREST STATEMENT
The authors have no competing financial interests.
Supplementary Information accompanies the paper on www.nature.com/naturemedicine.
References
- 1.Blasberg RG, Tjuvajev JG. Molecular-genetic imaging: current and future perspectives. J Clin Invest. 2003;111:1620–1629. doi: 10.1172/JCI18855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, et al. ABCG2/BCRP expression modulates D-Luciferin based bioluminescence imaging. Cancer Res. 2007;67:9389–9397. doi: 10.1158/0008-5472.CAN-07-0944. [DOI] [PubMed] [Google Scholar]
- 3.Uhrbom L, Nerio E, Holland EC. Dissecting tumor maintenance requirements using bioluminescence imaging of cell proliferation in a mouse glioma model. Nat Med. 2004;10:1257–1260. doi: 10.1038/nm1120. [DOI] [PubMed] [Google Scholar]
- 4.Kishimoto H, et al. In vivo imaging of lymph node metastasis with telomerase-specific replication-selective adenovirus. Nat Med. 2006;12:1213–1219. doi: 10.1038/nm1404. [DOI] [PubMed] [Google Scholar]
- 5.Padmanabhan P, et al. Visualization of telomerase reverse transcriptase (hTERT) promoter activity using a trimodality fusion reporter construct. J Nucl Med. 2006;47:270–277. [PMC free article] [PubMed] [Google Scholar]
- 6.Freytag SO, et al. Phase I trial of replication-competent adenovirus-mediated suicide gene therapy combined with IMRT for prostate cancer. Mol Ther. 2007;15:1016–1023. doi: 10.1038/mt.sj.6300120. [DOI] [PubMed] [Google Scholar]
- 7.Yaghoubi SS, et al. Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Nat Clin Pract Oncol. 2009;6:53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su ZZ, Shi Y, Fisher PB. Subtraction hybridization identifies a transformation progression-associated gene PEG-3 with sequence homology to a growth arrest and DNA damage-inducible gene. Proc Natl Acad Sci U S A. 1997;94:9125–9130. doi: 10.1073/pnas.94.17.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su ZZ, et al. Targeting gene expression selectively in cancer cells by using the progression-elevated gene-3 promoter. Proc Natl Acad Sci U S A. 2005;102:1059–1064. doi: 10.1073/pnas.0409141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar D, et al. Eradication of therapy-resistant human prostate tumors using a cancer terminator virus. Cancer Res. 2007;67:5434–5442. doi: 10.1158/0008-5472.CAN-07-0195. [DOI] [PubMed] [Google Scholar]
- 11.Sarkar D, et al. Targeted virus replication plus immunotherapy eradicates primary and distant pancreatic tumors in nude mice. Cancer Res. 2005;65:9056–9063. doi: 10.1158/0008-5472.CAN-05-1261. [DOI] [PubMed] [Google Scholar]
- 12.Sarkar D, et al. A cancer terminator virus eradicates both primary and distant human melanomas. Cancer Gene Ther. 2008;15:293–302. doi: 10.1038/cgt.2008.14. [DOI] [PubMed] [Google Scholar]
- 13.Su Z, Shi Y, Fisher PB. Cooperation between AP1 and PEA3 sites within the progression elevated gene-3 (PEG-3) promoter regulate basal and differential expression of PEG-3 during progression of the oncogenic phenotype in transformed rat embryo cells. Oncogene. 2000;19:3411–3421. doi: 10.1038/sj.onc.1203666. [DOI] [PubMed] [Google Scholar]
- 14.Sarkar D, et al. Dual cancer-specific targeting strategy cures primary and distant breast carcinomas in nude mice. Proc Natl Acad Sci U S A. 2005;102:14034–14039. doi: 10.1073/pnas.0506837102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood M, et al. Biodistribution of an adenoviral vector carrying the luciferase reporter gene following intravesical or intravenous administration to a mouse. Cancer Gene Ther. 1999;6:367–372. doi: 10.1038/sj.cgt.7700090. [DOI] [PubMed] [Google Scholar]
- 16.Peng KW, et al. Organ distribution of gene expression after intravenous infusion of targeted and untargeted lentiviral vectors. Gene Ther. 2001;8:1456–1463. doi: 10.1038/sj.gt.3301552. [DOI] [PubMed] [Google Scholar]
- 17.Evans KD, Tulloss TA, Hall N. 18FDG uptake in brown fat: potential for false positives. Radiol Technol. 2007;78:361–366. [PubMed] [Google Scholar]
- 18.Shreve PD, Anzai Y, Wahl RL. Pitfalls in oncologic diagnosis with FDG PET imaging: physiologic and benign variants. Radiographics. 1999;19:61–77. doi: 10.1148/radiographics.19.1.g99ja0761. quiz 150-151. [DOI] [PubMed] [Google Scholar]
- 19.Ray S, et al. Noninvasive imaging of therapeutic gene expression using a bidirectional transcriptional amplification strategy. Mol Ther. 2008;16:1848–1856. doi: 10.1038/mt.2008.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao J, et al. Tumor-specific transcriptional targeting of suicide gene therapy. Gene Ther. 2002;9:168–175. doi: 10.1038/sj.gt.3301618. [DOI] [PubMed] [Google Scholar]
- 21.Iyer M, et al. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proc Natl Acad Sci U S A. 2001;98:14595–14600. doi: 10.1073/pnas.251551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 23.Burton JB, et al. Adenovirus-mediated gene expression imaging to directly detect sentinel lymph node metastasis of prostate cancer. Nat Med. 2008;14:882–888. doi: 10.1038/nm.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer M, et al. Noninvasive imaging of enhanced prostate-specific gene expression using a two-step transcriptional amplification-based lentivirus vector. Mol Ther. 2004;10:545–552. doi: 10.1016/j.ymthe.2004.06.118. [DOI] [PubMed] [Google Scholar]
- 25.Huyn ST, et al. A potent, imaging adenoviral vector driven by the cancer-selective mucin-1 promoter that targets breast cancer metastasis. Clin Cancer Res. 2009;15:3126–3134. doi: 10.1158/1078-0432.CCR-08-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs A, et al. Positron-emission tomography of vector-mediated gene expression in gene therapy for gliomas. Lancet. 2001;358:727–729. doi: 10.1016/s0140-6736(01)05904-9. [DOI] [PubMed] [Google Scholar]
- 27.Immonen A, et al. AdvHSV-tk gene therapy with intravenous ganciclovir improves survival in human malignant glioma: a randomised, controlled study. Mol Ther. 2004;10:967–972. doi: 10.1016/j.ymthe.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Klatzmann D, et al. A phase I/II study of herpes simplex virus type 1 thymidine kinase “suicide” gene therapy for recurrent glioblastoma. Study Group on Gene Therapy for Glioblastoma. Hum Gene Ther. 1998;9:2595–2604. doi: 10.1089/hum.1998.9.17-2595. [DOI] [PubMed] [Google Scholar]
- 29.Trask TW, et al. Phase I study of adenoviral delivery of the HSV-tk gene and ganciclovir administration in patients with current malignant brain tumors. Mol Ther. 2000;1:195–203. doi: 10.1006/mthe.2000.0030. [DOI] [PubMed] [Google Scholar]
- 30.Bonnet ME, Erbacher P, Bolcato-Bellemin AL. Systemic delivery of DNA or siRNA mediated by linear polyethylenimine (L-PEI) does not induce an inflammatory response. Pharm Res. 2008;25:2972–2982. doi: 10.1007/s11095-008-9693-1. [DOI] [PubMed] [Google Scholar]
- 31.Coelho-Castelo AA, et al. Tissue distribution of a plasmid DNA encoding Hsp65 gene is dependent on the dose administered through intramuscular delivery. Genet Vaccines Ther. 2006;4:1. doi: 10.1186/1479-0556-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang KK, et al. Safety evaluation of GX-12, a new HIV therapeutic vaccine: investigation of integration into the host genome and expression in the reproductive organs. Intervirology. 2003;46:270–276. doi: 10.1159/000073206. [DOI] [PubMed] [Google Scholar]
- 33.Manam S, et al. Plasmid DNA vaccines: tissue distribution and effects of DNA sequence, adjuvants and delivery method on integration into host DNA. Intervirology. 2000;43:273–281. doi: 10.1159/000053994. [DOI] [PubMed] [Google Scholar]
- 34.Ramirez K, et al. Preclinical safety and biodistribution of Sindbis virus measles DNA vaccines administered as a single dose or followed by live attenuated measles vaccine in a heterologous prime-boost regimen. Hum Gene Ther. 2008;19:522–531. doi: 10.1089/hum.2007.172. [DOI] [PubMed] [Google Scholar]
- 35.Dwyer RM, Bergert ER, O'Connor M,K, Gendler SJ, Morris JC. In vivo radioiodide imaging and treatment of breast cancer xenografts after MUC1-driven expression of the sodium iodide symporter. Clin Cancer Res. 2005;11:1483–1489. doi: 10.1158/1078-0432.CCR-04-1636. [DOI] [PubMed] [Google Scholar]
- 36.Tsuruta Y, et al. A fiber-modified mesothelin promoter-based conditionally replicating adenovirus for treatment of ovarian cancer. Clin Cancer Res. 2008;14:3582–3588. doi: 10.1158/1078-0432.CCR-07-5053. [DOI] [PubMed] [Google Scholar]
- 37.Su ZZ, et al. Potential molecular mechanism for rodent tumorigenesis: mutational generation of Progression Elevated Gene-3 (PEG-3) Oncogene. 2005;24:2247–2255. doi: 10.1038/sj.onc.1208420. [DOI] [PubMed] [Google Scholar]
- 38.Loening AM, Gambhir SS. AMIDE: a free software tool for multimodality medical image analysis. Mol Imaging. 2003;2:131–137. doi: 10.1162/15353500200303133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.