Abstract
Bacterial assemblages from subsurface (100 m depth), meso- (200–1000 m depth) and bathy-pelagic (below 1000 m depth) zones at 10 stations along a North Atlantic Ocean transect from 60°N to 5°S were characterized using massively parallel pyrotag sequencing of the V6 region of the 16S rRNA gene (V6 pyrotags). In a dataset of more than 830,000 pyrotags we identified 10,780 OTUs of which 52% were singletons. The singletons accounted for less than 2% of the OTU abundance, while the 100 and 1,000 most abundant OTUs represented 80% and 96%, respectively, of all recovered OTUs. Non-metric Multi-Dimensional Scaling and Canonical Correspondence Analysis of all the OTUs excluding the singletons revealed a clear clustering of the bacterial communities according to the water masses. More than 80% of the 1,000 most abundant OTUs corresponded to Proteobacteria of which 55% were Alphaproteobacteria, mostly composed of the SAR11 cluster. Gammaproteobacteria increased with depth and included a relatively large number of OTUs belonging to Alteromonadales and Oceanospirillales. The bathypelagic zone showed higher taxonomic evenness than the overlying waters, albeit bacterial diversity was remarkably variable. Both abundant and low-abundance OTUs were responsible for the distinct bacterial communities characterizing the major deep-water masses. Taken together, our results reveal that deep-water masses act as bio-oceanographic islands for bacterioplankton leading to water mass-specific bacterial communities in the deep waters of the Atlantic.
Keywords: bacterial diversity, massively parallel tag sequencing, North Atlantic Ocean, deep water masses, bacterial biogeography
INTRODUCTION
Prokaryotes represent an important component of the marine plankton, comprising up to 70% and 75% of the total biomass in surface (Fuhrman et al. 1989) and deep (Aristegui et al. 2009) waters, respectively. They serve a fundamental role in mediating a wide range of biogeochemical cycles (Azam et al. 1983; Karl 2002). The introduction of molecular tools has substantially increased our knowledge about marine microbial community structure and has shown that the vast majority of environmental microbes represents novel taxa that have yet to be cultivated (Handelsman 2004; Olsen et al. 1986). High-throughput sequencing methods and pyrosequencing allow for efficient deep molecular sampling efforts of the microbial populations (Gilbert et al. 2009; Huber et al. 2007; Huse et al. 2008; Sogin et al. 2006) and sidestep the need to clone individual DNA molecules (Margulies et al. 2005). Furthermore, data from these massively parallel sequencing approaches provide an exhaustive description of the taxonomic affiliation of microbial community. This information forms the basis for estimating both the richness and evenness of the microbial populations present in the environment which are, in turn, essential to refine our knowledge on the biogeography of marine microbes (Galand et al. 2009b; Galand et al. 2010; Martiny et al. 2006; Pommier et al. 2005) and to relate microbial diversity and ecosystem properties (Andersson et al. 2009).
By applying a high-throughput pyrosequencing strategy, Sogin et al. (2006) found a remarkably high bacterial diversity in the deep-water masses of the North Atlantic and in diffuse flow hydrothermal vents. The study highlighted the existence of thousands of low-abundance populations, coined the “rare biosphere”, which accounted for most of the observed phylogenetic bacterial diversity (Sogin et al. 2006). The rare phylotypes are assumed to be recruited by immigration and to have extremely low loss rates from grazing and viral lysis (Pedros-Alio 2006). Conventional molecular techniques fail to detect microbial phylotypes that make up the long tail of taxon rank-distribution curves because dominant populations (comprising > 1% of the total community) mask the detection of the highly diverse, low-abundance organisms. The massively parallel pyrotag sequencing approach, however, allows for deep sequencing that can capture information about these low-abundance populations (Galand et al. 2009a; Palacios et al. 2008; Sogin et al. 2006).
The spatial variability of microbial diversity across habitats has not been investigated extensively (Galand et al. 2010; Pommier et al. 2007). The availability of resources, selective loss factors (grazing and viral lysis), and physical parameters such as temperature and salinity can influence microbial population structure. This is in accordance with the deterministic theory “everything is everywhere, but, the environment selects” (Baas Becking 1934) which has been debated extensively recently (de Wit & Bouvier 2006; O'Malley 2008). In contrast, stochastic neutral models of biodiversity and biogeography (Hubbel 2001; Sloan et al. 2006) postulate that immigration, dispersal rates, size of the habitat (i.e., taxa-area relationships) (Woodcock et al. 2007) and ecological invariance among microbial phylotypes shape microbial community structure.
Distinct water masses characterize the hydrodynamic conditions of the ocean, most notably the major water masses driving the thermohaline ocean circulation (Tomczak & Godfrey 2003). Non-sinking free-living prokaryotes inhabiting oceanic deep waters might be trapped in these distinct water masses leading overall to water mass-specific prokaryotic community composition and activity (Agogué et al. 2008; Galand et al. 2009b; Galand et al. 2010; Varela et al. 2008a,b). Hence, dispersal and immigration of free-living microbes in the ocean might be more limited than generally assumed. These physical boundary conditions of oceanic water masses might constrain the applicability of some of the fundamental theories and models on the biogeographic distribution of microbes in the ocean.
This study aimed at describing the composition of bacterial assemblages in the North Atlantic Ocean throughout the water column by using massively parallel pyrotag sequencing to resolve the spatial distribution of bacterial richness and evenness in different water masses. We hypothesized that the bacterial communities exhibit a biogeography according to the water masses. Hence, we expected that bacterial communities collected several 1000 km apart from each other but originating from the same water mass are more similar than bacterial communities collected less than a few hundred meters apart but from different water masses. We used data from the hypervariable V6 region of the bacterial 16S rRNA gene from 45 samples collected from the main deep water masses along a 8000 km north-to-south transect in North Atlantic Ocean ranging from 60°N to 5°S.
MATERIAL AND METHODS
Study site and sampling
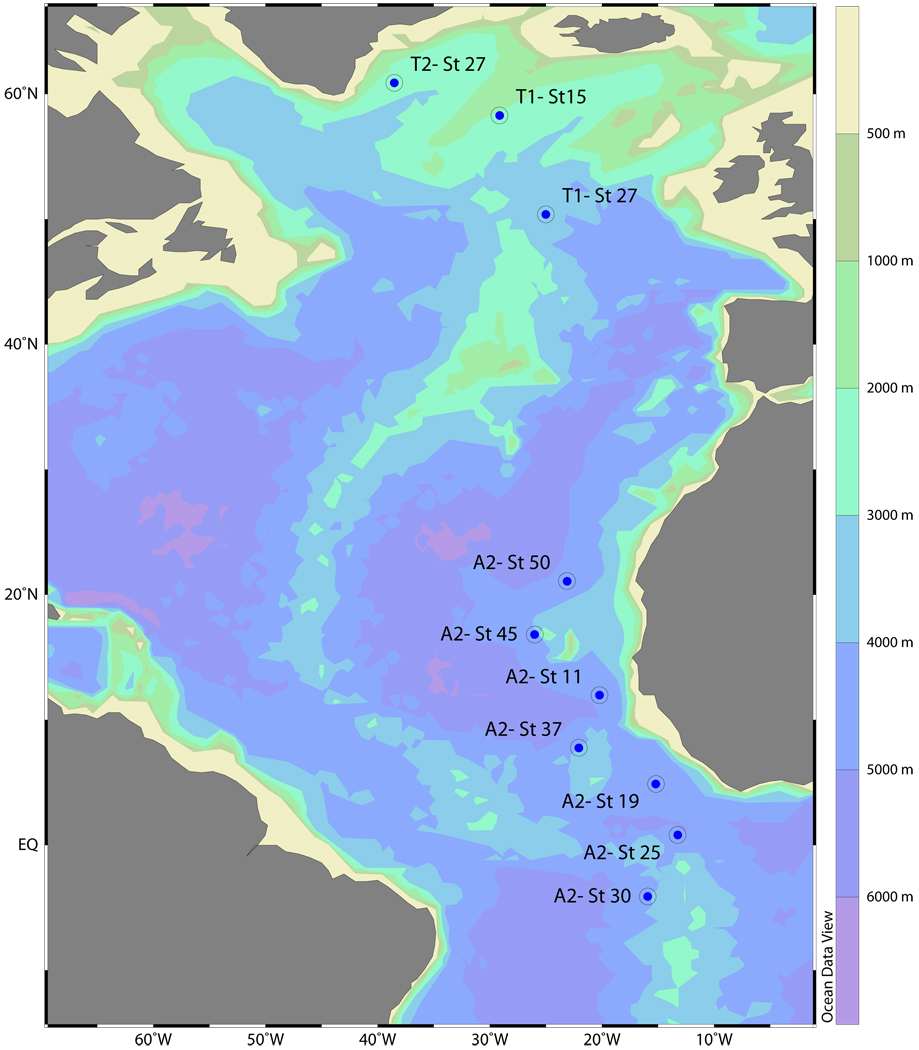
Sampling was conducted during the cruises TRANSAT-1 (September 2002) and -2 (May 2003) and ARCHIMEDES-2 (November/December 2006) on board R/V Pelagia following the North Atlantic Deep Water (NADW) from 60°N to 5°S in the eastern basin of the Atlantic Ocean (Fig. 1). Ten stations were occupied, and samples were taken from 6 to 10 sampling depths at each station from 100m to 4500m depth. In total, 45 samples were collected for massively parallel pyrotag sequencing (Table S1).
Figure 1.
Location of the stations where samples for pyrosequencing were taken during the Transat-1, Transat-2 and Archimedes-2 cruises in the North Atlantic. Characteristics of the samples are given in Table S1.
The water masses along the eastern North Atlantic section were identified by their distinct potential temperature and salinity characteristics (Table S2, Fig. S1) (van Aken 2000a, b). At 4000–5000m depth, the Lower Deep Water (LDW) has low salinity (34.9) and temperature (1.9 – 2.6°C) (Table S2, Fig. S1). The NEADW (North East Atlantic Deep Water) is characterized by a temperature between 2.5 and 4.1°C and a higher salinity than LDW (Table S2, Fig. S1). The core of NEADW was identifiable throughout the transect at around 2750m depth. Two types of mesopelagic waters were found in the (sub)equatorial region: the transitional and South Atlantic Central Water (tCW/SACW) exhibiting the same temperature and salinity characteristics, and the Antarctic Intermediate Water (AAIW) characterized by lower salinity and temperature than tCW/SACW (Table S2, Fig. S1). In the northernmost region of the transect, the Northern Intermediate Water (NIW) was found at 500m depth, showing the same temperature but higher salinity than AAIW. Labrador Sea Water (LSW) was clearly identifiable at depths between 1200–2100m throughout the transect (Table S2, Fig. S1). Samples from the distinct water masses were collected with 10-L NOEX (no oxygen exchanges) bottles mounted in a CTD (conductivity, temperature, depth) frame. Samples were collected from: (i) subsurface waters (lower euphotic layer, 100–150m layer), (ii) mesopelagic waters including tCW and SACW (250 – 500m layer), AAIW (750 – 900m layer) and NIW (500m), and (iii) bathypelagic waters including LSW (1200 – 2100m layer), NEADW (1750 – 4000m layer) and LDW (4000 – 5000m layer) (Table S1).
DNA extraction, pyrosequencing and identification of the bacterial phylotypes
DNA extraction
During the TRANSAT cruises, 1 L of seawater from each depth was filtered onto a 0.2 µm polycarbonate filter (Millipore) and the filters were subsequently stored at −80°C until further processing in the lab. During the ARCHIMEDES-2 cruise, 10 L of seawater from each depth was filtered through a 0.22 µm Sterivex filter GP unit (Millipore). Lysis buffer (40 mM EDTA, 50 mM Tris-HCl, 0.75 M sucrose) was then added into the Sterivex (1.8 mL) and the filters were subsequently stored at −80°C until analysis. Extraction of total DNA was performed using an UltraClean soil DNA and Mega soil DNA isolation kit (Mobio) for TRANSAT-1 & -2 and ARCHIMEDES-2 samples, respectively.
PCR amplicon library construction and pyrosequencing
The hypervariable V6 region of the 16S rRNA of bacteria was amplified from TRANSAT samples using primers 967F, 5’-gcctccctcgcgccatcag-CAACGCGAAGAACCTTACC-3’ and 1046R, 5’-gccttgccagcccgctcagCGACAGCCATGCANCACCT-3’ and pyrosequenced on a Roche Genome Sequencer 20 under conditions described in Sogin et al. (2006). A cocktail of five fused primers at the 5’ end of the V6 region (E. coli positions 967– 985) and four primers at the 3’ end (E. coli positions 1046–1028) that capture the full diversity of rRNA sequences represented in molecular databases (Huber et al. 2007; Sogin et al. 2006) amplified ARCHIMEDES-2 environmental DNA samples for pyrosequencing on a Roche Genome GS FLX system. For both the TRANSAT and ARCHIMEDES-2 we prepared amplicon libraries from at least three independent PCR cocktails to minimize the impact of potential early-round PCR errors. To minimize effects of sequencing errors, we employed a quality trimming procedure to remove low quality pyrotags and to eliminate sequences with multiple undetermined residues or mismatches to the PCR primers at the beginning of a read (Huse et al. 2007).
Clustering and assignment of the OTUs: identification of bacterial phylotypes
The clustering of V6 pyrotags into Operational Taxonomic Units (OTUs) was done with the new single-linkage preclustering (SLP) algorithm to smooth sequencing errors and reduce noise, followed by primary pairwise, average linkage clustering (PW-AL) described in Huse et al. (2010). The advantage of this new method is that it corrects for sequencing errors and minimizes the propagation of OTUs with sequencing effort. This method provides a comparable reduction in spurious OTUs as a previously published algorithm (e.g. PyroNoise, Quince et al. 2009), but requires less computational expense (Huse et al. 2010, see also the Discussion part on pyrosequencing errors). OTUs were created using clustering thresholds of 3% corresponding to 97% similarity.
We assigned taxonomic identifiers to OTUs by using the rRNA indexing algorithm Global Assignment of Sequence Taxonomy (GAST) (Sogin et al. 2006), which compares OTUs to known rRNA genes that have already been placed in a phylogenetic framework of more than 1,000,000 nearly full-length rRNA reference sequences (RefSSU) based on the SILVA database (Pruesse et al. 2007). GAST methodology is freely available through the VAMPS (Visualisation and Analysis of Microbial Population Structure) website (http://vamps.mbl.edu/resources/faq.php#gasting). The V6 reference database (V6RefDB: high-quality, full-length 16S rRNA sequences) is publically available at: http://vamps.mbl.edu.
Accession numbers and data availability
The OTU sequences and supporting data have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/home/). Run IDs are SRR029056–SRR029102 inclusive. One run corresponds to one sample. The sequences obtained from each run are downloadable from the SRA (sequence read archive) web site of NCBI. In addition, the VAMPS site http://vamps.mbl.edu provides individual and edited sequences and analytical functions for interrogating the data.
Diversity estimation and statistical analysis
Diversity indices
The non-parametric ACE and the Chao1 richness index were calculated with the CatchAll software program (Bunge et al. 2010).
The Gini index of evenness (Wittebolle et al. 2009) was calculated on the relative abundance of all OTUs except the singletons using the ineq function in the ineq package of the software package R for subsurface, meso- and bathypelagic samples. The higher the Gini index is, the more unevenly distributed are the OTUs. The Gini index was used as it has been shown to be more robust to differences in sample size than other evenness indices (Peet 1974).
Cluster analysis
Non-metric multidimensional scaling (MDS) (Kruskal 1964a, b) was used to determine the similarity between samples. This data-reduction method shows the differences (or similarities) between samples by reducing the comparisons between samples from a multidimensional space to fewer dimensions, preferably 2 or 3. Differences between samples were calculated based on the relative abundance of (i) all the OTUs, (ii) all the OTUs except the singletons and (iii) the 1,000 most abundant OTUs. MDS analysis was also applied to (i) all the pyrotags except the singletons, (ii) the abundant pyrotags (frequency > 1% within a sample) and (iii) the rare pyrotags (frequency < 0.01% within a sample). The similarities are presented in a multidimensional space by plotting more similar samples closer together (Kruskal 1964a, b).
Analysis of similarity (ANOSIM) was used to verify the significance of the MDS clustering by testing the hypothesis that bacterial communities from the same cluster were more similar to each other than to communities in different clusters. A Bray–Curtis similarity matrix computed from the relative abundance of all OTUs except the singletons was used to generate one-way ANOSIM statistics with 999 permutations.
SIMPER analysis
Similarity percentage (SIMPER) (Clarke & Warwick 2001) was used to determine which individual sequence contributed most to the dissimilarity between water masses (Dataset S1). The SIMPER analysis was also used to determine the percentage of similarity (i) between each station and the northernmost station of the transect (station 27, Transat-2 cruise) (Fig. 1, Table S1) within specific water layers and (ii) between water layers, based on the relative abundance of all OTUs except the singletons. The data of the NEADW from Sogin et al. (2006) were included in these analyses to allow full comparison of all the stations occupied from 60°N to 5°S, i.e., a stretch of 8000 km.
Mantel analysis
The Mantel test was used to analyze the phylogenetic composition of the 1,000 most abundant OTUs and the singletons among all the samples. Mantel analysis was also used to determine the relationships between (i) bacterial assemblage structure, (ii) environmental factors and (iii) bacteria-related parameters. The three similarity matrices (Euclidian distance, n = 45) included the following variables: (i) the relative abundance of all OTUs except the singletons, (ii) temperature, salinity, concentration of inorganic nutrients (nitrite, nitrate, ammonia, silicate and phosphate), dissolved organic carbon (DOC) and nitrogen (DON), apparent oxygen utilization (AOU), latitude, longitude and depth and (iii) bacterial-related variables comprising bacterial abundance and production, percentage of high nucleic acid cells (HNA), potential respiration (measured as activity of the electron transport system), alpha- and beta-glucosidase, leucine aminopeptidase and alkaline phosphatase activity and its enzyme kinetics.
CCA analysis
A canonical correspondence analysis (CCA) was used to investigate the variations in the relative abundance of the 1,000 most abundant OTUs under the constraint of our set of environmental variables. We assumed a unimodal response of OTUs to environmental variations. Generally, nonlinear models are required for analysis of ecological data collected over a large range of habitats (Ter Braak & Verdonschot 1995). When a linear response is assumed, the percentage of explained variation is lower (redundancy analysis not shown). The null hypothesis that the bacterial assemblage is independent of the environmental parameters was tested using constrained ordination with a Monte Carlo permutation test (499 permutations). The parameters were selected to obtain significant canonical axes (p < 0.05) and to maximize the percentage of variance explained. The cluster, the SIMPER and the Mantel analyses were performed with PRIMER 6.1.7 (Primer-E, Ltd) and XLSTAT Pro (2006) software. The CCA analysis was performed with the Canoco version 4.5 software (Ter Braak 1989).
Variability of the 100 most abundant OTUs per water mass
The deviation (in %) from the mean relative abundance in all the samples was calculated for the 100 most abundant OTUs for the different water masses. This deviation was calculated for each water mass and for each of the 100 most abundant OTUs as follows:
where X̄WM is the mean relative abundance of a OTU in a specific water mass and X̄T is the mean relative abundance of this OTU in all the samples.
RESULTS
The sequencing effort yielded on average 18,111 ± 14,869 reads per sample, and ranged from 2,083 to 62,100 pyrotags among the 45 samples (Table 1). For all samples combined, we identified a total of 49,517 unique pyrotag sequences. The pyrotag length averaged 62.51 ± 3.47 bp, varying from 51 to 165 bp. On average, the unique V6 pyrotags (i.e., present in only one out of the 45 samples analyzed at an abundance >1) accounted for 14 ± 3% of the total number of pyrotags in each sample. On average each sample had 2,249 ± 1,529 unique bacterial sequences representing nearly 835 ± 421 OTUs (Operational Taxonomic Units) at the 3% difference level. The non-parametric Chao1 and ACE estimate predicted an average number per sample of 1,416 ± 787 and 1,733 ± 1,220 OTUs, respectively. The rarefaction curves (Fig. S2) indicate that, despite obtaining on average more than 18,000 pyrotags and 10,700 OTUs identified as Bacteria, our sampling of bacterial richness was not complete.
Table 1.
Sequencing information and diversity estimates for the 45 samples of the North Atlantic Ocean obtained by pyrosequencing.
| Average per sample | Standard deviation | Range | |
|---|---|---|---|
| Pyrotag length (bp) | 62.51 | ± 3.47 | 51 – 165 |
| Total number of pyrotags | 18111 | ± 14869 | 2083 – 62100 |
| Total unique pyrotags | 2249 | ± 1529 | 479 – 7241 |
| % of total unique pyrotags | 14 | ± 3 | 6 – 23 |
| Total OTUs at 3% difference | 835 | ± 421 | 245 – 2063 |
| Chao1 estimator of richness at 3% difference | 1416 | ± 787 | 431 – 4032 |
| ACE estimator of richness at 3% difference | 1733 | ± 1220 | 552 – 7453 |
To examine another aspect of diversity, the Gini’s index of evenness was calculated for each OTU except the singletons (Fig. S3). Most of the OTUs were highly unevenly distributed (Gini > 0.5) comprising both abundant and rare OTUs. The OTUs from subsurface samples exhibited a higher evenness than the OTUs from meso- and bathypelagic samples, indicating that OTUs from subsurface were more equally distributed among samples than OTUs from deeper layers.
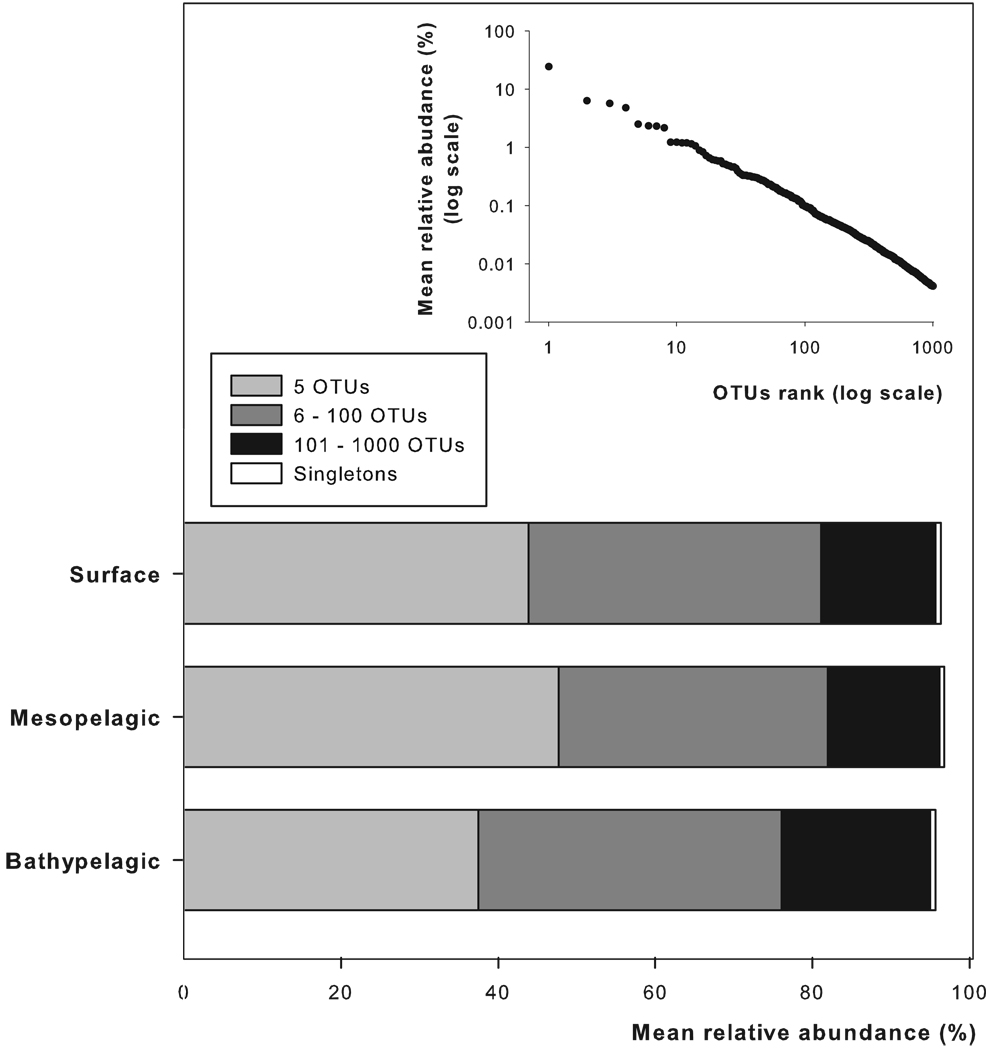
The 100 and 1,000 most abundant OTUs represented on average 80 ± 3% and 96 ± 1%, respectively, of the total OTU abundance in the individual samples with no significant difference among the different depth layers or water masses (Fig. 2). Significant differences between depth layers were only observed in the relative contribution of the 5 most abundant OTUs (Kruskal-Wallis test, H = 7.01, p = 0.03; Fig. 2) representing between 37% and 47% of total OTU abundance. The 5 most abundant OTUs and the 6 – 100 (34–39%) most abundant OTUs contributed roughly equally to the total OTU abundance (Fig. 2). The rank-frequency distribution of the 1,000 most abundant OTUs indicates that only a few OTUs are very abundant with a long tail of low-abundance OTUs (inset, Fig. 2). The rank-frequency distribution of meso- and bathypelagic communities exhibited a steeper slope (slope of 0.58 ± 0.01 and 0.57 ± 0.01, respectively) than that of subsurface (slope of 0.32 ± 0.01) OTUs (slope comparison, Student-t test, p < 0.001 for all comparisons). The rank-frequency distributions of OTUs of meso- and bathypelagic waters were not significantly different (Student-t test, p > 0.05).
Figure 2.
Depth distribution of the 5, 6 – 100 and 101 – 1,000 most abundant OTUs and the singletons. Inset: Rank-frequency distribution of the 1,000 most abundant OTUs.
Singleton OTUs (i.e., present in only one sample at an absolute abundance of 1) comprised 5,567 OTUs out of the 10,780 distinct OTUs in our analysis. They represented half (52%) of the total number of OTUs. However, they accounted for less than 2% of the total OTUs abundance (Fig. 2) and they were equally distributed among the water layers.
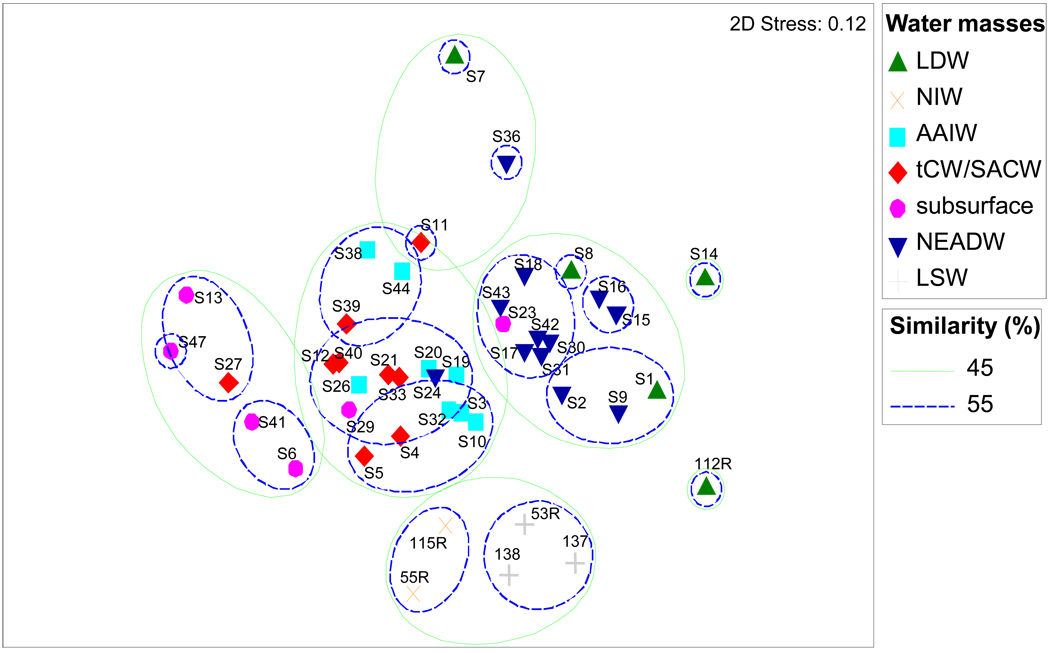
Non-metric Multidimensional Scaling (MDS) based on the relative abundance of all OTUs except singletons was used to discriminate bacterial community composition in the different water masses. Cluster analysis showed that bacterial community composition clustered according to the water masses (Fig. 3). The samples separated into one cluster containing bacterial communities of the subsurface zone, one cluster of the mesopelagic waters (AAIW and tCW/SACW) and two deep-water clusters (NEADW and LDW) at 45% of similarity. Two clusters of deep LSW and mesopelagic NIW bacterial communities were identified at the 55% similarity level. The bacterial communities of bathypelagic waters were less similar to each other than samples from subsurface and intermediate waters (Fig. 3). Bacterial communities from the same water mass but separated by thousands of kilometers (S6 from St.A2-11 and S41 from St. A2-45 belonging to subsurface; S12 from St. A2-19 and S40 from St. A2-45 belonging to tCW/SACW; S42 from St. A2-50 and S17 from St. A2-25 belonging to NEADW) were more similar to each other than communities separated by only a few hundred meters at individual sites (S14 and S21 from St. A2-25; S7 and S10 from St. A2-19) but originating from different water masses (Fig. 3, Table S1). The ANOSIM test showed that the differences between the water mass clusters were significant (p < 0.05) except for AAIW and tCW/SACW (p = 0.062) and for LSW and NIW (p = 0.1; Table S3). For this latter pairwise comparison, the R-value was still high (R = 1), and the insignificant difference might be due to the low number of samples for each water mass (n = 3 for LSW and n = 2 for NIW) allowing only 10 permutations (Table S3). A MDS of the 1,000 most abundant OTUs versus all OTUs excluding the singletons produced the same clustering (data not shown). Including singletons in the MDS analysis, however, resulted in a lack of water mass-specificity of bacterial communities (data not shown). SIMPER analysis indicated that the differences in bacterial community composition between water masses are explained by the combination of abundant and rare OTUs (Dataset S1 in Suppl. Information).
Figure 3.
Non-metric multidimensional analysis based on relative abundance of all OTUs except the singletons. Individual samples were affiliated to their respective water-mass. Superimposed circles represent clusters of samples at similarity values of 45 and 55% (Bray-Curtis similarity). Characteristics of the samples are indicated in Table S1. The final solution was based on 25 iterations with a final stress of 0.12.
The clustering of all pyrotags except the singletons was comparable to the clustering of all OTUs except the singletons (Fig S4a). The clustering of the rare sequences (frequency < 0.01% of total pyrotag abundance within a sample, Fig S4c) was similar to the clustering of the abundant sequences (frequency > 1% within a sample, Fig. S4b), albeit with a generally lower percentage of similarity. The differences in the rare bacterial sequences between the water mass clusters were significant (ANOSIM, global R = 0.65, p < 0.001) and the matrices of the abundant and rare pyrotags were significantly related (Mantel test, r = 0.68, p < 0.001).
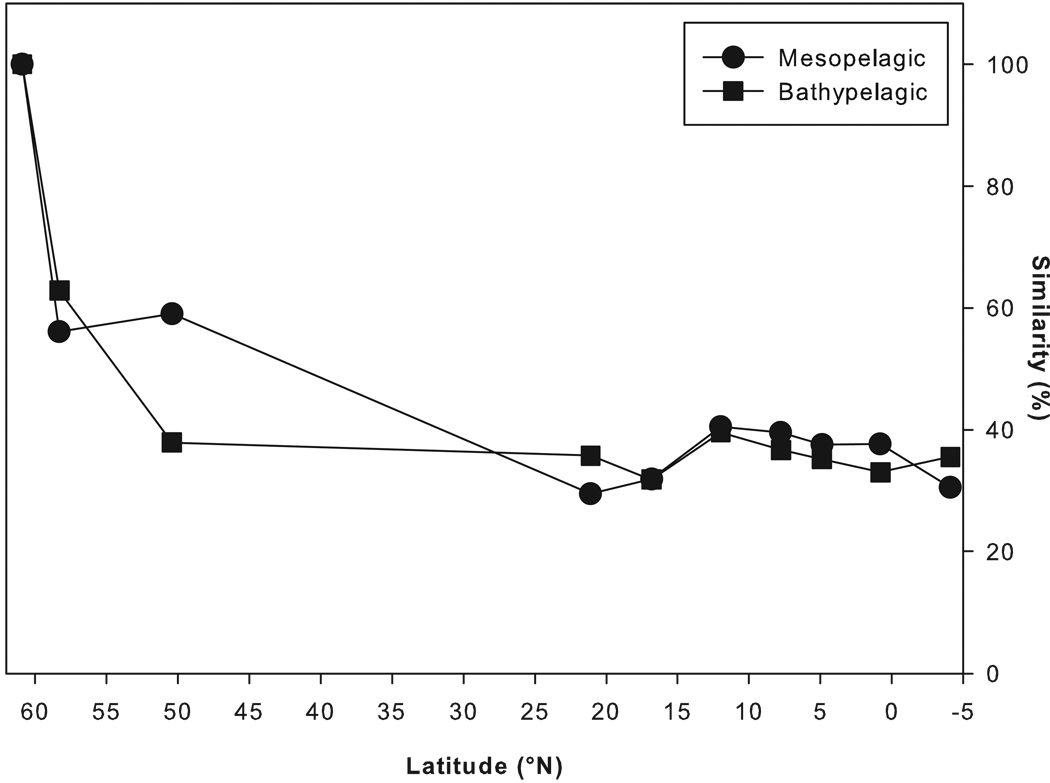
The similarity of the bacterial communities of the bathy- and mesopelagic waters decreased rapidly from the northernmost station at 60°N (station 27, Transat-2 cruise) to ≈ 50°N and remained fairly constant thereafter towards the equator (Fig. 4). Besides this pronounced latitudinal trend in similarity in the northern part of the North Atlantic, a pronounced stratification of the bacterial communities was detected (Fig. S5). In the northern part of the North Atlantic, the meso- and bathypelagic bacterial communities were more similar (47.4%) than in the southern part of the North Atlantic (37.7%) indicating an increasing stratification of bacterial communities with decreasing latitude (Fig. S5).
Figure 4.
Similarity in relative abundance of all OTUs except the singletons (in %, calculated through SIMPER analysis) between each station and the northernmost station of the transect (station 27, Transat-2) within bathy- and mesopelagic water layers.
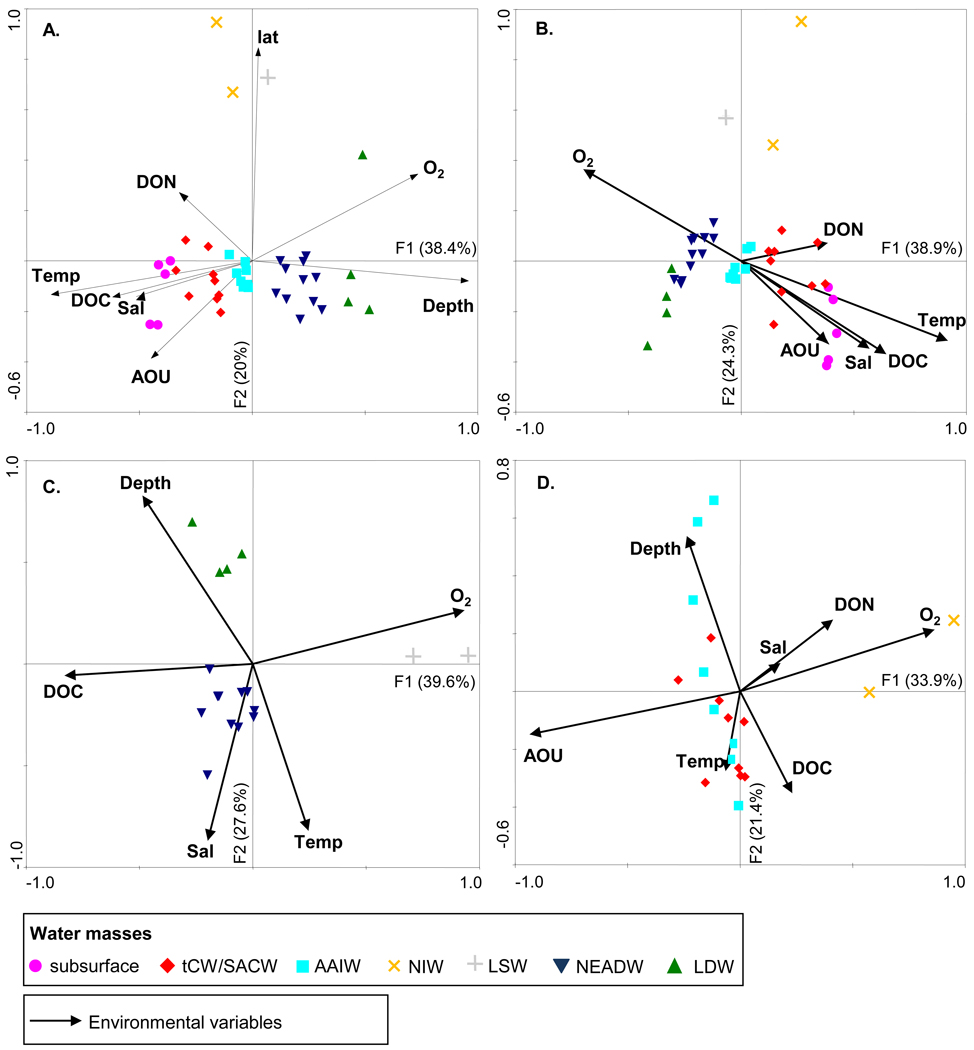
The relationship between bacterial assemblage structure (based on the relative abundance of all OTUs), environmental factors and the bacterial activity parameters was assessed by correlating the three distance matrices with a Mantel test. The bacterial assemblage structure correlated with the environmental factors (r = 0.57, p = 0.0001) but not with bacterial activity parameters (r = 0.06, p = 0.072). The potential link of environmental factors with bacterial community structure was analyzed by the ordination technique of canonical correspondence analysis (CCA). The CCA indicated that samples clearly clustered according to the water masses (Fig. 5a). Depth and latitude emerged as highly significant explanatory variables, separating samples along the first and the second axes, respectively (Fig. 5a). When depth and latitude were removed from the analysis (Fig. 5b), O2 concentration, temperature and DOC clearly separated subsurface and bathypelagic samples along the first axis. DON concentration and salinity appeared to be key factors for determining subsurface bacterial assemblage structure. When CCA was applied to bathypelagic samples (Fig. 5c), depth and temperature appeared to be significant explanatory variables, separating bathypelagic samples along the first axis, while DOC and O2 concentrations separated samples along the second axis. The combination of depth, temperature, salinity, DOC and O2 concentrations explained 67% of the total variance in the relative abundance of the 1,000 most abundant OTUs in bathypelagic samples. When potential density was used in the analysis instead of depth, the distribution of the bathypelagic samples in the CCA analysis was similar. When CCA was applied to mesopelagic samples (Fig. 5d), the O2 concentration and AOU (apparent oxygen utilization) separated samples along the first axis, while depth and DOC concentration separated samples along the second axis. The combination of these parameters explained about 55% of the total variance in the relative abundance of the 1,000 most abundant OTUs in mesopelagic samples.
Figure 5.
Canonical correspondence analysis of the relative abundance of the 1,000 most abundant OTUs (a) for all samples, (b) for all samples without depth and latitude as explanatory variables, (c) for bathypelagic samples and (d) for mesopelagic samples. Monte Carlo permutation tests for the first and all axes were highly significant for all the four CCA analyses (p < 0.002). Abbreviations of the environmental and bacteria-related variables: lat, latitude; O2, oxygen concentration; AOU, apparent oxygen utilization; DOC, dissolved organic carbon; DON, dissolved organic nitrogen; Sal, salinity; NH4, ammonium concentration.
Phylogenetic affiliation of North Atlantic deep-water bacterial communities
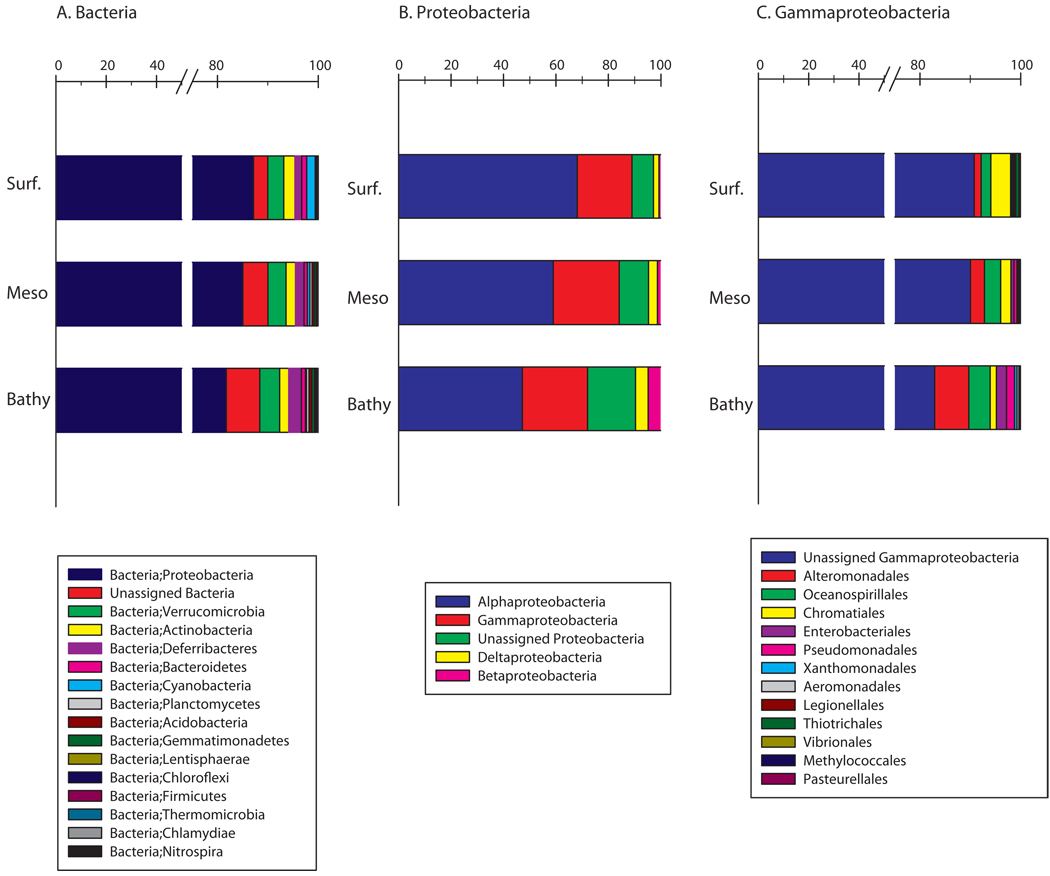
Among the 1,000 most abundant OTUs, Proteobacteria were, overall, the most abundant phylum (Fig. 6a), representing 84 ± 8% of the 1,000 most abundant OTUs. The bathypelagic zone exhibited the highest proportion of unassigned bacteria and a higher evenness than the overlying waters. Deferribacteres and, to a lesser extent, Verrucomicrobia increased in relative abundance with depth, and contributed 2.5% and 3.9%, respectively, to the bacterial abundance in the bathypelagic zone. Chloroflexi contributed to total bacterial abundance 5-fold more in bathypelagic zone (0.25%) than in subsurface (0.05%). In contrast, Cyanobacteria decreased with depth from 1.7% of total bacterial abundance in the subsurface layer to 0.2% in the bathypelagic zone (Fig. 6a).
Figure 6.
Relative abundance and affiliation of the 1000 most abundant bacterial OTUs of each zone of the Atlantic Ocean (subsurface n=6, mesopelagic n=20 and bathypelagic n=19). (a) on the phylum level, (b) the class level within Proteobacteria, (c) the order level within Gammaproteobacteria.
Within Proteobacteria, Alphaproteobacteria, mostly composed of the SAR11 cluster (data not shown), was the most abundant class and accounted for 47% and 68% of the 1,000 most abundant proteobacterial OTUs in the bathypelagic and subsurface waters, respectively (Fig. 6b). As at the phylum level, the evenness increased with depth at the class level, and was higher in the bathypelagic zone than in the subsurface and mesopelagic zones. Gamma-, Delta- and Betaproteobacteria increased in relative abundance with depth, and represented 25%, 5% and 5%, respectively, of Proteobacteria in the bathypelagic waters.
Within Gammaproteobacteria, unassigned Gammaproteobacteria comprised the major fraction amounting to 83% of the total Gammaproteobacteria in the bathypelagic and to 91% in the subsurface waters (Fig. 6c). Also at the order level, evenness increased with depth and some groups were found to be specific to a specific water layer. For example, higher proportions of Alteromonadales (7%) and, to a lesser extent, Oceanospirillales (4%), Enterobacteriales (2%) and Pseudomonadales (1.6%) were found in the bathypelagic than in the subsurface zone (Fig. 6c). In contrast, Chromatiales was more restricted to subsurface (4%) than deep layers (1%).
In total, 1262 OTUs were present in all water layers, representing 24% of the OTUs without counting the singletons. Among these OTUs, 56 of them remain essentially at the same relative abundance (± 0.01), representing 1% of all the OTUs except the singletons.
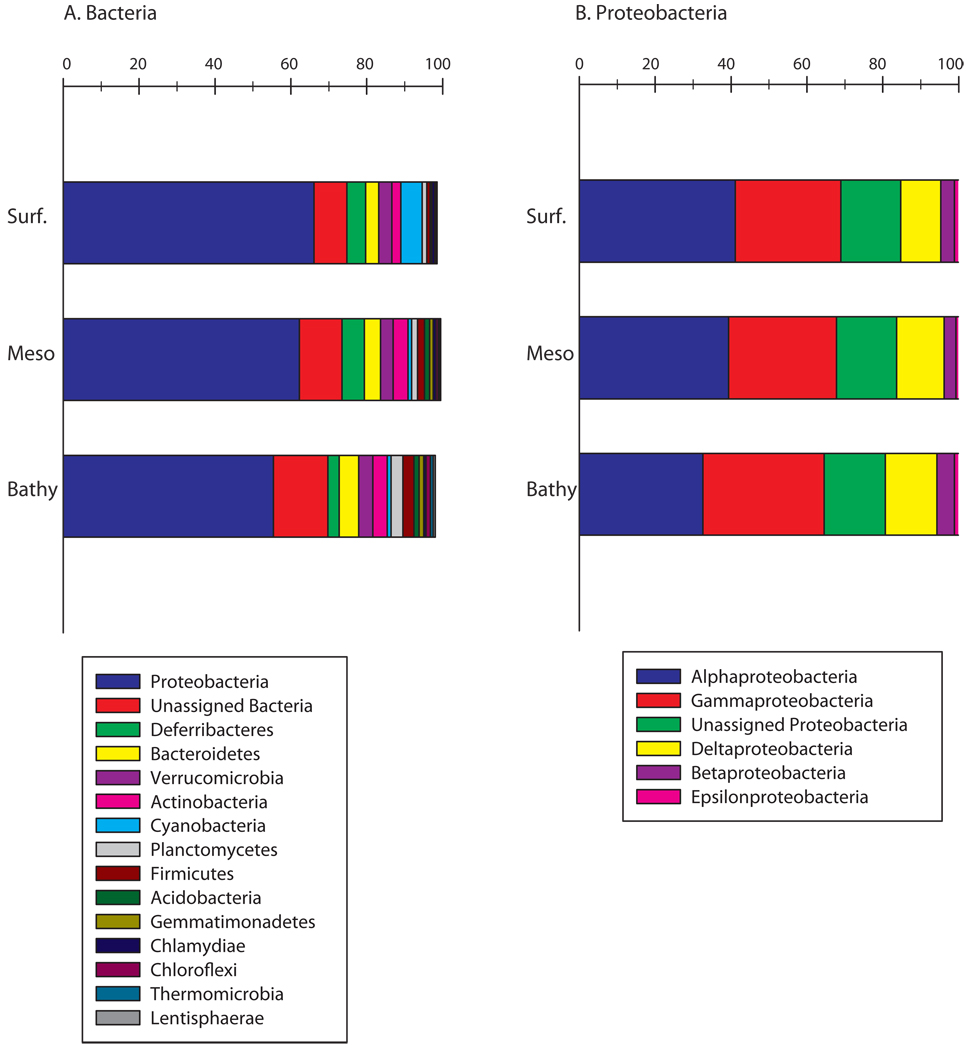
Analysis of the taxonomic composition of the singletons revealed that they were as diverse as the abundant OTUs (Fig. 7). Most of the singletons belonged to the Proteobacteria (61%) but 11% were unassigned (Fig. 7a). Within Proteobacteria, Alpha- (38%) and Gammaproteobacteria (29%) dominated and 16% of the gammaproteobacterial OTUs remained unassigned (Fig. 7b). Overall, the phylogenetic composition of the singleton community was similar among all three water layers and similar to the bacterial assemblage structure of the 1,000 most abundant OTUs at the phylum (Mantel test, r = 0.56, p = 0.001) and the class level within the Proteobacteria (Mantel test, r = 0.33, p = 0.001).
Figure 7.
Relative abundance and affiliation of the singletons of each zone of the Atlantic Ocean (subsurface n=6, mesopelagic n=20 and bathypelagic n=19). (a) on the phylum level, and (b) the class level within Proteobacteria.
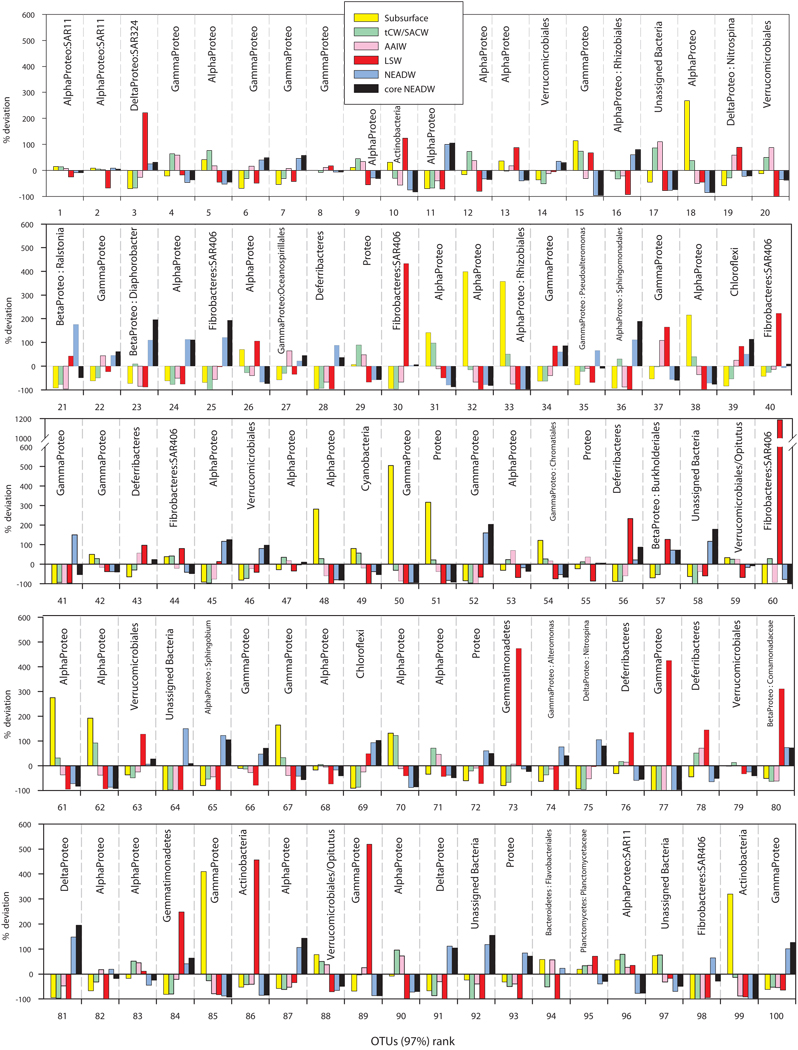
The deviation (in %) from the mean relative abundance in all the samples of the 100 most abundant OTUs (accounting for 79 ± 8% of the OTUs abundance over all the water masses) was calculated for each water mass (Fig. 8). Lower abundance OTUs exhibit higher water mass specificity than the more abundant OTUs. OTUs specific for LSW included members of the SAR324 clade (belonging to the Deltaproteobacteria; OTU #3, Fig. 8 and Table S4) which accounted for 7% of the 100 most abundant OTUs, members of the SAR406 clade (belonging to Fibrobacteres, OTUs #30, 40 and 60) which showed the greatest variability observed among the 100 most abundant OTUs, members of the Gammaproteobacteria (OTUs #77, 89), Gemmatimonadetes (OTU #73) and Actinobacteria (OTU #86). OTUs specific for the NEADW included members of the Alphaproteobacteria (OTUs #11, 16, 24, 65 and 87), the Betaproteobacteria (OTU #23), the Gammaproteobacteria (OTUs #35, 41, 52, 66, 74 and 100), the Deltaproteobacteria (OTUs #75, 81, 91), the Fibrobacteres (members of the SAR406 clade, OTUs #25 and 98) and the Verrucomicrobiales (OTU #46) (Fig. 8 and Table S4). Members of the Chloroflexi-type SAR202 cluster (OTUs #39, 69) were generally specific for deep waters (NEADW and LSW). A large number of OTUs specific for mesopelagic waters were affiliated to unassigned Alphaproteobacteria (OTUs #12, 53, 71, 83 and 90) and one OTU to the Verrucomicrobiales phylum (OTU #20) (Fig. 8 and Table S4). Surface-specific OTUs were mainly members of unassigned Alphaproteobacteria (OTUs #18, 32, 38, 48, 61, 70) and Gammaproteobacteria (OTUs # 50, 67, 85) and one OTU from the Actinobacteria (OTU #99). Members of the SAR11 clade (OTUs #1, 2, and 96) exhibited a ubiquitous distribution, albeit they were relatively less abundant in deep-water masses (NEADW, core NEADW and LSW) than in subsurface and mesopelagic waters (250 – 500m and AAIW) (Fig. 8 and Table S4). The SAR11 clade contributed about 37% to the relative abundance of the 100 most abundant OTUs.
Figure 8.
Deviation (in %) from the mean relative abundance of the 100 most abundant OTUs for all the samples for the different water masses.
DISCUSSION
The pyrosequencing approach allowed a deep sampling of bacterial communities in North Atlantic waters. The analyses of 45 samples, originating from 10 stations along a 8,000 km north-to-south transect provide an exhaustive description of the distribution of bacterial communities in the major deep-water masses of the North Atlantic.
We found water mass-specific clustering of bacterial communities throughout the major North Atlantic water masses. These differences in the phylogenetic composition of the bacterial community between water masses are not only due to the most abundant OTUs, but the less abundant OTUs including the rare ones (but not the singletons) are also responsible for the water mass-specificity of the bacterial community composition. This implies that the less abundant bacterial populations also exhibit biogeographical traits as the most abundant ones. Previous studies suggested that the rare biosphere should exhibit a high rate of dispersal (Fenchel & Finlay 2004; Finlay 2002) and might act as a “seed-bank” of micro-organisms which might become abundant in case of environmental changes (Palacios et al. 2008; Pedros-Alio 2006). Kirchman et al. (2010), however, argued against the “seed-bank” hypothesis as rare phylotypes of the winter bacterial community remained rare also in the summer community in the Arctic Ocean. The distribution patterns observed for rare OTUs in the North Atlantic Ocean indicate that the rare bacterial biosphere contributes to the water mass-specificity of bacterial communities. Hence, the rare biosphere is most likely not maintained by stochastic immigration but appears to be a water mass property as also shown for the Arctic Ocean (Galand et al. 2010). A ubiquitous distribution of members of the rare bacterial biosphere, as previously suggested (Pedros-Alio 2006), was not found in the deep-waters of the Atlantic challenging the idea that “everything is everywhere” (Baas Becking 1934).
The singletons, i.e., the OTUs present only once in all the samples, are included in the rare bacterial biosphere (Sogin et al. 2006). Before speculating on the potential ecological role of the rare bacterial biosphere and its origin, we need to address the question whether the presence of singletons generated by the pyrosequencing approach are fact or artifact. Random sequencing errors and miscalled bases could potentially be an explanation for the high number of singleton sequences (Gomez-Alvarez et al. 2009; Quince et al. 2009; Reeder & Knight 2009) and could inflate actual diversity estimates (Kunin et al. 2009). Increased sampling effort leads to a greater probability that OTUs from a given template will include variant sequences and therefore inflate the number of observed singletons. However, the frequency of variant sequences will be a function of the relative abundance of a unique starting template and the number of generated OTUs for the entire community. Sequences that correspond to high frequency templates are more likely to produce variants. A systematic and stringent trimming procedure was used in this study to remove low quality sequences and reads that did not represent the targeted region. This procedure results in a per-base error rate of 0.2–0.4% (Huse et al. 2007). Moreover, the newly developed clustering method applied in this study removes spurious OTUs produced by pyrosequencing errors and minimizes the inflation of OTUs with sequencing effort (Huse et al. 2010). In our study, the singletons represent only 2% of the total OTU abundance in each depth zone of the North Atlantic. No clear clustering according to water masses was obtained when the singletons were included in the MDS analysis, while the bacterial community structure using all the OTUs except the singletons exhibits a water-mass specific clustering. As the singletons have the same phylogenetic composition as the 1,000 most abundant OTUs, some of the singletons might be artifacts resulting from noise in high-throughput pyrosequencing of the most abundant OTUs (Reeder & Knight 2009).
Biogeography of bacterial communities in the thermohaline ocean circulation of the North Atlantic
Distance, latitudinal gradients and area have been shown to influence bacterial community composition in lakes and the ocean (Fuhrman et al. 2008; Hewson et al. 2006; Reche et al. 2005). Recent phylogenetic surveys revealed a pronounced stratification among specific groups of planktonic Bacteria (DeLong 2005; Galand et al. 2010; Hewson et al. 2006; Kirchman et al. 2010; Martin-Cuadrado et al. 2007; Suzuki et al. 2004; Treusch et al. 2009) and Archaea (Agogué et al. 2008; Galand et al. 2009b; Garcia-Martinez & Rodriguez-Valera 2000). In our study, the similarity in bacterial community composition decreases rapidly in the meso- and bathypelagic waters from 60°N to 50°N and remains fairly stable thereafter. Moreover, the similarity in bacterial community composition between meso- and bathypelagic waters was higher in the northern than in the southern part of the transect. This reflects the large scale deep-water formation in the northern North Atlantic leading to a more uniform deep-water bacterial community in the generally younger deep waters in the northern North Atlantic also exhibiting smaller differences in physico-chemical parameters between meso- and bathypelagic waters than further south towards the (sub)tropical Atlantic. Further south, distance was influencing bacterial community composition much less than water mass identity in the (sub)tropical Atlantic Ocean.
Analysis of the affiliation of the 100 most abundant OTUs revealed obvious water mass-specific signatures of bacterial community structure. Due to their distinct temperature and salinity characteristics, water masses might act as presumed dispersal barriers for bacterioplankton and hence, might limit immigration of bacterial phylotypes (Aristegui et al. 2009) from adjacent water masses. In the bathypelagic realm, depth and temperature were the main factors determining the composition of bacterial communities and discriminating samples from different water masses. Salinity, temperature and consequently density act as potential oceanographic barriers separating water masses and their inhabiting bacterial communities (Pommier et al. 2007) generating bio-oceanographic islands with specific signatures of bacterial community composition such as shown most pronouncedly for North East Atlantic Deep Water and Labrador Sea Water. Besides water mass identity and latitude, the specific environmental properties of the water masses, such as the concentrations of DOC and O2, and AOU determine the bacterial community composition in the North Atlantic Ocean.
In contrast to our current perception of the deep ocean as a rather low diversity environment, bacterial community richness and evenness in the bathypelagic waters of the North Atlantic are as high and as variable as subsurface and mesopelagic waters. This observation is consistent with previous reports on bacterial diversity of the North Pacific and Atlantic (Hewson et al. 2006) and in the eastern Mediterranean Sea (Moeseneder et al. 2001) using fingerprinting techniques. This reported heterogeneity in deep bacterial assemblages contrasts the very slow assemblage growth rates estimated to be 0.061 ± 0.008 d−1 in the deep Pacific (Aristegui et al. 2009) and deep-water movement (1.5 cm s−1) (van Aken 2007). The heterogeneity in deep-water bacterial community composition has been interpreted to result from episodic input of organic matter from surface waters and patchiness (Hewson et al. 2006). However, it appears that microbial life in the dark ocean is likely more dependent on slowly sinking or buoyant, laterally advected suspended particles than hitherto assumed (Bochdansky et al. 2010) which might generate the water mass-specific biogeochemical conditions leading ultimately to water mass-specific bacterial assemblages (Baltar et al. 2009).
Vertical distribution of bacterial phylotypes
As found in previous studies on deep-sea bacterial diversity (DeLong et al. 2006; Fuhrman & Davis 1997; Lopez-Garcia et al. 2001; Pham et al. 2008), Proteobacteria, mostly from the alpha subdivision, dominate the bacterial community. Members of the alphaproteobacterial clade SAR11, ubiquitously present in the ocean (DeLong et al. 2006; Field et al. 1997; Giovannoni et al. 1990; Morris et al. 2002), decreased in relative abundance with depth.
The relative abundance of Gammaproteobacteria, however, increased with depth as reported previously for the North Atlantic (Lauro & Bartlett 2008; Sogin et al. 2006) and Antarctic Polar Front (Lopez-Garcia et al. 2001). Within Gammaproteobacteria, members of the order Alteromonadales are abundant in bathypelagic waters. Several psychropiezophilic (cold and pressure-loving) bacterial isolates such as Moritella sp. PE36, Psychromonas sp. CNPT3 and Shewanella sp. KT99 are belonging to this order (DeLong et al. 1997; Lauro et al. 2007). We also found members of the Oceanospirillales more abundant in the bathypelagic North Atlantic waters than in the subsurface and mesopelagic layers. Members of this order are also known to be symbionts of the deep-sea worm Osedax sp. (Goffredi et al. 2007; Rouse et al. 2009). Betaproteobacteria also increased in relative abundance with depth. OTUs affiliated to this class are closely related to endosymbionts of bivalves found at hydrothermal vents (Kimura et al. 2003). Their relatively high abundance in the free-living mode in bathypelagic waters, particularly in the North East Atlantic Deep Water (NEADW) might indicate their spreading from hydrothermal vents and/or seafloor as the NEADW flows mainly along the eastern slope of the mid-Atlantic ridge. Free-living Betaproteobacteria are common in freshwaters (Kirchman et al. 2005) but not abundant in the bathypelagic realm. However, metagenomic analysis from deep-sea methane vents have reported Betaproteobacteria-related phylotypes (Pernthaler et al. 2008), suggesting a potential niche for these organisms within methane-rich deep marine environments. In the North Pacific Ocean, Brown et al. (2009) recently reported an increasing relative abundance of Betaproteobacteria V9 pyrotags at 800m and 4400m depth. The bathypelagic bacterial community of the North Atlantic was also enriched in Deltaproteobacteria, as previously shown for the deep Arctic Ocean as well (Galand et al. 2010) and for Station ALOHA in the North Pacific Subtropical Gyre (DeLong et al. 2006).
The comparison of the phylogenetic composition of the 1,000 most abundant OTUs reveals an increase of OTUs richness and evenness with depth. Thus, the bacterial community of the North Atlantic is more diverse in the bathypelagic realm than that at the base of the euphotic layer. The composition of the 100 most abundant OTUs indicates distinct water mass-specific OTUs, especially in deep waters. In meso- and bathypelagic waters, the Deltaproteobacteria were dominated by the SAR324 cluster (Wright et al. 1997), a typical deep-water clade (DeLong et al. 2006; Lopez-Garcia et al. 2001; Pham et al. 2008; Zaballos et al. 2006). The SAR406 cluster (Gordon & Giovannoni 1996) previously detected in various oceanic provinces (Gallagher et al. 2004; Pham et al. 2008) is another abundant clade of the deep Atlantic ocean, specific to the LSW. Chloroflexi-type SAR202 cluster, described as highly abundant in the bathypelagic subtropical North Atlantic (Morris et al. 2004; Varela et al. 2008a; Wright et al. 1997) was found to be a specific member of the NEADW.
Concluding remarks
By applying a high-throughput pyrosequencing strategy, we achieved a deep coverage of the bacterial populations in the deep-water masses of the North Atlantic. The bacterial assemblages clearly clustered according to the distinct water masses. The low abundance OTUs were as important for the observed water mass-specificity of bacterial community composition as the more abundant OTUs. Density differences separate water masses and their inhabiting bacterial communities, generating bio-oceanographic islands with specific signatures of bacterial community composition such as shown most pronouncedly for the deep waters. The deep bacterial assemblages exhibit a higher richness and evenness than bacterial assemblages at the base of the euphotic layer suggesting that the bathypelagic waters might offer a more heterogeneous environment for microbial life than hitherto assumed.
Supplementary Material
Acknowledgements
We acknowledge the help of H.M. van Aken for the characterization of the water masses, J. M. Arrieta for collecting and extracting the samples of the TRANSAT-1 cruise, M. Brink and D. de Corte for extracting the samples of ARCHIMEDES-2 cruise, F. Baltar for the dataset on ectoenzymatic activity, D.M Welch and S.M. Huse for the dataset on OTUs sequences and T. Reinthaler for providing the parameters datasets of Transat and Archimedes cruises. The captain and crew of RV Pelagia provided excellent service at sea.
Funding. This research was supported by two Marie Curie Fellowships of the European Community to D.L. and H.A., by the Alfred P. Sloan Foundation’s ICoMM field project, the W. M. Keck Foundation and a subcontract from the Woods Hole Center for Oceans and Human Health from the National Institutes of Health and the National Science Foundation (NIH_NIEHS 1 P50 ES012742-01 and NSF_OCE 0430724-J. Stegeman PI to M.L.S.). Shiptime was provided through grants of the Earth and Life Science Division of the Dutch Science Foundation (ALW-NWO) (TRANSAT and ARCHIMEDES projects) to G.J.H.
Abbreviations
- LDW
lower Deep Water
- NEADW
Northeast Atlantic Deep Water
- SACW
South Atlantic Central Water
- LSW
Labrador Sea Water
- NIW
Northern Intermediate Water
- tCW
transitional Central Water
- AAIW
Antarctic Intermediate Water
- DOC
dissolved organic carbon
- DON
dissolved organic nitrogen
- AOU
apparent oxygen utilization
- HNA
high nucleic acid
- MDS
non-metric multidimensional scaling
- CCA
canonical correspondence analysis
Footnotes
Author contribution. G.J.H. and M.S. sampling scheme and sample analyses, H.A., D.L. and P.N. analyzed the data, G.J.H., P.N. and M.S. contributed the reagents/materials/analysis tools, H.A., D.L. and G.J.H wrote the paper.
Competing interests. The authors have declared that no competing interests exist.
REFERENCES
- Agogué H, Brink M, Dinasquet J, Herndl GJ. Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature. 2008;456:788–791. doi: 10.1038/nature07535. [DOI] [PubMed] [Google Scholar]
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME Journal. 2009;4:171–181. doi: 10.1038/ismej.2009.108. [DOI] [PubMed] [Google Scholar]
- Aristegui J, Gasol JM, Duarte CM, Herndl GJ. Microbial oceanography of the dark ocean's pelagic realm. Limnology and Oceanography. 2009;54:1501–1529. [Google Scholar]
- Azam F, Fenchel T, Field JG, et al. The Ecological Role of Water-Column Microbes in the Sea. Marine Ecology Progress Series. 1983;10:257–263. [Google Scholar]
- Baas Becking LGM. Geobiologie of inleiding tot de milieukunde. The Hague, the Netherlands: 1934. [Google Scholar]
- Baltar F, Aristegui J, Gasol JM, Sintes E, Herndl GJ. Evidence of prokaryotic metabolism on suspended particulate organic matter in the dark waters of the subtropical North Atlantic. Limnology and Oceanography. 2009;54:182–193. [Google Scholar]
- Bochdansky AB, van Aken HM, Herndl GJ. Role of macroscopic particles in deep-sea oxygen consumption. Proceedings of the National Academy of Sciences, USA. 2010;107:8287–8291. doi: 10.1073/pnas.0913744107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MV, Philip GK, Bunge JA, et al. Microbial community structure in the North Pacific ocean. ISME Journal. 2009;3:1374–1386. doi: 10.1038/ismej.2009.86. [DOI] [PubMed] [Google Scholar]
- Bunge JA, Woodard L, Connolly S. CatchAll: Parametric and nonparametric estimation of species richness and population size. 2010 Manuscript in preparation. [Google Scholar]
- Clarke KR, Warwick RM. PRIMER-E Ldt. Plymouth, UK: 2001. Change in marine communities: an approach to statistical analysis and interpretation. [Google Scholar]
- DeLong EF. Microbial community genomics in the ocean. Nature Reviews: Microbiology. 2005;3:459–469. doi: 10.1038/nrmicro1158. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Franks DG, Yayanos AA. Evolutionary Relationships of Cultivated Psychrophilic and Barophilic Deep-Sea Bacteria. Applied and Environmental Microbiology. 1997;63:2105–2108. doi: 10.1128/aem.63.5.2105-2108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Preston CM, Mincer T, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- Fenchel T, Finlay BJ. The ubiquity of small species: Patterns of local and global diversity. BioScience. 2004;54 [Google Scholar]
- Field KG, Gordon D, Wright T, et al. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Applied and Environmental Microbiology. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eucaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Davis AA. Widespread archaea and novel bacteria from the deep sea as shown by 16S rRNA gene sequences. Marine Ecology Progress Series. 1997;150:275–285. [Google Scholar]
- Fuhrman JA, Sleeter TD, Carlson CA, Proctor LM. Dominance of bacterial biomass in the Sargasso Sea and its ecological implications. Marine Ecology Progress Series. 1989;57:207–217. [Google Scholar]
- Fuhrman JA, Steele JA, Hewson I, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proceedings of the National Academy of Sciences. 2008;105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proceedings of the National Academy of Sciences, USA. 2009a;106:22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME Journal. 2009b;3:860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- Galand PE, Potvin M, Casamayor EO, Lovejoy C. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME Journal. 2010;4:564–576. doi: 10.1038/ismej.2009.134. [DOI] [PubMed] [Google Scholar]
- Gallagher JM, Carton MW, Eardly DF, Patching JW. Spatio-temporal variability and diversity of water column prokaryotic communities in the eastern North Atlantic. FEMS Microbiology Ecology. 2004;47:249–262. doi: 10.1016/S0168-6496(03)00281-2. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J, Rodriguez-Valera F. Microdiversity in uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of Group I. Molecular Ecology. 2000;9:935–948. doi: 10.1046/j.1365-294x.2000.00953.x. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, et al. The seasonal structure of microbial communities in the Western English Channel. Environmental Microbiology. 2009;11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic Diversity in Sargasso Sea Bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Goffredi SK, Johnson SB, Vrijenhoek RC. Genetic Diversity and Potential Function of Microbial Symbionts Associated with Newly Discovered Species of Osedax Polychaete Worms. Applied and Environmental Microbiology. 2007;73:2314–2323. doi: 10.1128/AEM.01986-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME Journal. 2009;11:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Gordon DA, Giovannoni SJ. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific Oceans. Applied and Environmental Microbiology. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: Application of Genomics to Uncultured Microorganisms. Microbiology and Molecular Biology Reviews. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson I, Steele JA, Capone DG, Fuhrman JA. Remarkable heterogeneity in meso- and bathypelagic bacterioplankton assemblage composition. Limnology and Oceanography. 2006;51:1274–1283. [Google Scholar]
- Huber JA, Mark Welch D, Morrison HG, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, et al. Exploring Microbial Diversity and Taxonomy Using SSU rRNA Hypervariable Tag Sequencing. PLoS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000255. e10000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Mark Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biology. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environmental Microbiology. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl DM. Nutrient dynamics in the deep blue sea. Trends in Microbiology. 2002;10:410–418. doi: 10.1016/s0966-842x(02)02430-7. [DOI] [PubMed] [Google Scholar]
- Kimura H, Higashide Y, Naganuma T. Endosymbiotic microflora of the vestimentiferan tubeworm (Lamellibrachia sp.) from a bathyal cold seep. Marine Biotechnology. 2003;5:593–603. doi: 10.1007/s10126-002-0117-7. [DOI] [PubMed] [Google Scholar]
- Kirchman DL, Cottrell MT, Lovejoy C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environmental Microbiology. 2010;12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- Kirchman DL, Dittel AI, Malmstrom RR, Cottrell MT. Biogeography of major bacterial groups in the Delaware Estuary. Limnology and Oceanography. 2005;50:1697–1706. [Google Scholar]
- Kruskal JB. Multidimensional-Scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika. 1964;29:1–27. [Google Scholar]
- Kruskal JB. Nonmetric Multidimensional Scaling - a numerical method. Psychometrika. 1964b;29:115–129. [Google Scholar]
- Kunin V, Engelbrekton A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors lead to artificial inflation of diversity estimates. Environmental Microbiology. 2009;1:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lauro FM, Bartlett DH. Prokaryotic lifestyles in deep sea habitats. Extremophiles. 2008;12:15–25. doi: 10.1007/s00792-006-0059-5. [DOI] [PubMed] [Google Scholar]
- Lauro FM, Chastain RA, Blankenship LE, Yayanos AA, Bartlett DH. The Unique 16S rRNA Genes of Piezophiles Reflect both Phylogeny and Adaptation. Applied and Environmental Microbiology. 2007;73:838–845. doi: 10.1128/AEM.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia P, Lopez-Lopez A, Moreira D, Rodriguez-Valera F. Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front. FEMS Microbiology Ecology. 2001;36:193–202. doi: 10.1016/s0168-6496(01)00133-7. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado A-B, Lopez-Garcia P, Alba J-C, et al. Metagenomics of the Deep Mediterranean, a Warm Bathypelagic Habitat. PLoS ONE. 2007;2:e914. doi: 10.1371/journal.pone.0000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, et al. Microbial biogeography: putting microorganisms on the map. Nature Reviews: Microbiology. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Moeseneder MM, Winter C, Arrieta JM, Herndl GJ. Terminal-restriction fragment length polymorphism (T-RFLP) screening of a marine archaeal clone library to determine the different phylotypes. Journal of Microbiol Methods. 2001;44:159–172. doi: 10.1016/s0167-7012(00)00247-5. [DOI] [PubMed] [Google Scholar]
- Morris RM, Rappe MS, Connon SA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Morris RM, Rappe MS, Urbach E, Connon SA, Giovannoni SJ. Prevalence of the Chloroflexi-Related SAR202 Bacterioplankton Cluster throughout the Mesopelagic Zone and Deep Ocean. Applied and Environmental Microbiology. 2004;70:2836–2842. doi: 10.1128/AEM.70.5.2836-2842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial Ecology and Evolution: A Ribosomal RNA Approach. Annual Review of Microbiology. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Palacios C, Zettler E, Amils R, Amaral-Zettler L. Contrasting microbial community assembly hypotheses: a reconciling tale from the Rio Tinto. PLoS One. 2008;3:e3853. doi: 10.1371/journal.pone.0003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedros-Alio C. Marine microbial diversity: can it be determined? Trends in Microbiology. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Peet RK. The measurements of species diversity. Annual Review of Ecology and Systematics. 1974;5:285–307. [Google Scholar]
- Pernthaler A, Dekas AE, Brown CT, et al. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proceedings of the National Academy of Sciences, USA. 2008;105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VD, Konstantinidis KT, Palden T, DeLong EF. Phylogenetic analyses of ribosomal DNA-containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre. Environmental Microbiology. 2008;10:2313–2330. doi: 10.1111/j.1462-2920.2008.01657.x. [DOI] [PubMed] [Google Scholar]
- Pommier T, Canback B, Riemann L, et al. Global patterns of diversity and community structure in marine bacterioplankton. Molecular Ecology. 2007;16:867–880. doi: 10.1111/j.1365-294X.2006.03189.x. [DOI] [PubMed] [Google Scholar]
- Pommier T, Pinhassi J, Hagstrom A. Biogeographic analysis of ribosomal RNA clusters from marine bacterioplankton. Aquatic Microbial Ecology. 2005;41:79–89. [Google Scholar]
- Quince C, Lanzen A, Curtis TP, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nature Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Reche I, Pulido-Villena E, Morales-Baquero R, Casamayor EO. Does ecosystem size determine aquatic bacterial richness? Ecology. 2005;86:1715–1722. [Google Scholar]
- Reeder J, Knight R. The 'rare biosphere': a reality check. Nature Methods. 2009;6:636–637. doi: 10.1038/nmeth0909-636. [DOI] [PubMed] [Google Scholar]
- Rouse G, Wilson N, Goffredi S, et al. Spawning and development in Osedax boneworms (Siboglinidae, Annelida) Marine Biology. 2009;156:395–405. [Google Scholar]
- Sloan WT, Lunn M, Woodcock S, et al. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environmental Microbiology. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, et al. Microbial diversity in the deep sea and the underexplored "rare biosphere". Proceedings of the National Academy of Sciences, USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Preston CM, Beja O, et al. Phylogenetic screening of ribosomal RNA gene-containing clones in bacterial artificial chromosome (BAC) libraries from different depths in Monterey Bay. Microbial Ecology. 2004;48:473–488. doi: 10.1007/s00248-004-0213-5. [DOI] [PubMed] [Google Scholar]
- Ter Braak CJ, Verdonschot PF. Canonical correspondence analysis and related multivariate methods in aquatic ecology. Aquatic Sciences. 1995;57:255–289. [Google Scholar]
- Ter Braak CJF. CANOCO - an extension of DECORANA to analyze species-environment relationships. Hydrobiologia. 1989;184:169–170. [Google Scholar]
- Tomczak M, Godfrey JS. Regional Oceanography: An Introduction. 2nd edn. Delhi: Daya Publishing House; 2003. [Google Scholar]
- Treusch AH, Vergin KL, Finlay LA, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME Journal. 2009;3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- van Aken HM. The hydrography of the mid-latitude northeast Atlantic Ocean: I: The deep water masses. Deep Sea Research Part I. 2000a;47:757–788. [Google Scholar]
- van Aken HM. The hydrography of the mid-latitude Northeast Atlantic Ocean: II: The intermediate water masses. Deep Sea Research Part I. 2000b;47:789–824. [Google Scholar]
- van Aken HM. The oceanic thermohaline circulation. New York: Springer; 2007. [Google Scholar]
- Varela MM, van Aken HM, Herndl GJ. Abundance and activity of Chloroflexi-type SAR202 bacterioplankton in the meso- and bathypelagic waters of the (sub)tropical Atlantic. Environmental Microbiology. 2008a;10:1903–1911. doi: 10.1111/j.1462-2920.2008.01627.x. [DOI] [PubMed] [Google Scholar]
- Varela MM, van Aken HM, Sintes E, Herndl GJ. Latitudinal trends of Crenarchaeota and Bacteria in the meso- and bathypelagic water masses of the Eastern North Atlantic. Environmental Microbiology. 2008b;10:110–124. doi: 10.1111/j.1462-2920.2007.01437.x. [DOI] [PubMed] [Google Scholar]
- Wittebolle L, Marzorati M, Clement L, et al. Initial community evenness favours functionality under selective stress. Nature. 2009;458:623–626. doi: 10.1038/nature07840. [DOI] [PubMed] [Google Scholar]
- Wright TD, Vergin KL, Boyd PW, Giovannoni SJ. A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Applied and Environmental Microbiology. 1997;63:1441–1448. doi: 10.1128/aem.63.4.1441-1448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaballos M, Lopez-Lopez A, Ovreas L, et al. Comparison of prokaryotic diversity at offshore oceanic locations reveals a different microbiota in the Mediterranean Sea. FEMS Microbiology Ecology. 2006;56:389–405. doi: 10.1111/j.1574-6941.2006.00060.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.