Abstract
Posttranscriptional gene regulation frequently occurs through elements in mRNA 3′ untranslated regions (UTRs)1,2. Although crucial roles for 3′UTR-mediated gene regulation have been found in Caenorhabditis elegans3,4,5, most C. elegans genes have lacked annotated 3′UTRs6,7. Here we describe a high-throughput method to reliably identify polyadenylated RNA termini, and we apply this method, called poly(A)-position profiling by sequencing (3P-Seq), to determine C. elegans 3′UTRs. Compared to standard methods also recently applied to C. elegans UTRs8, 3P-Seq identified 8,581 additional UTRs while excluding thousands of shorter UTR isoforms that do not appear to be authentic. Analysis of this expanded and corrected dataset suggested that the high A/U content of C. elegans 3′UTRs facilitated genome compaction, since the elements specifying cleavage and polyadenylation, which are A/U-rich, can more readily emerge in A/U rich regions. Indeed, 30% of the protein-coding genes have mRNAs with alternative, partially overlapping end regions that generate another 10,498 cleavage and polyadenylation sites that had gone largely unnoticed and represent potential evolutionary intermediates of progressive UTR shortening. Moreover, a third of the convergently transcribed genes utilize palindromic arrangements of bidirectional elements to specify UTRs with convergent overlap, which also contributes to genome compaction by eliminating regions between genes. Although nematode 3′UTRs have median length only one-sixth that of mammalian 3′UTRs, they have twice the density of conserved microRNA sites, in part because additional types of seed-complementary sites are preferentially conserved. These findings reveal the influence of cleavage and polyadenylation on the evolution of genome architecture and provide resources for studying posttranscriptional gene regulation.
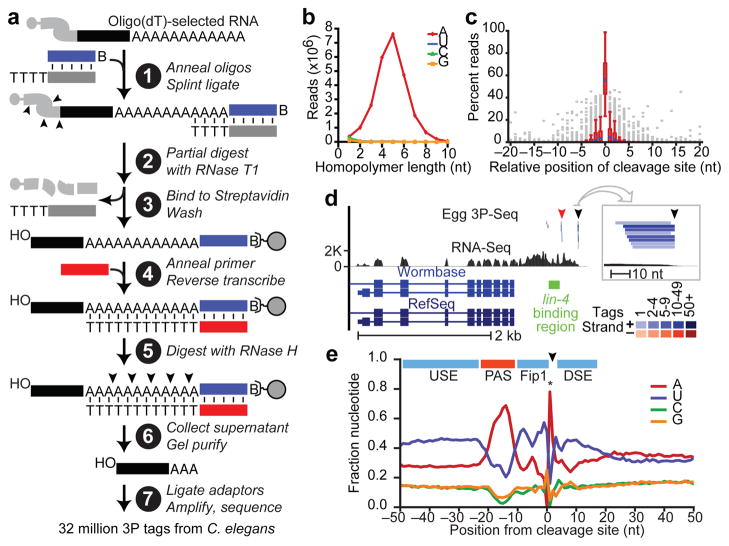
We developed a high-throughput method to reliably identify 3′ ends of mRNAs and other polyadenylated transcripts (Fig. 1a). This method, called poly(A)-position profiling by sequencing (3P-Seq), begins with a splint-ligation that favors ends of poly(A) tails when appending a biotinylated primer-binding site (Fig. 1a, step 1). After partial digestion with T1 nuclease (which cuts after Gs; step 2), the polyadenylated ends are captured (step 3), and the poly(A) tail is reverse transcribed with dTTP as the only deoxynucleoside triphosphate (step 4). Digestion with RNaseH releases the polyadenylated ends (step 5), which are purified (step 6) and prepared for high-throughput sequencing (step 7).
Figure 1. Identification of C. elegans 3′ UTRs.
a, Schematic of the 3P-Seq protocol. See text for description. b, Sequence composition of homopolymer runs that were found at 3′ termini of candidate 3P tags and included ≥1 untemplated nucleotide. c, Cleavage heterogeneity surrounding the most abundant cleavage site (position 0). Box plots show results for 380 cleavage sites that were both between two non-A residues (which enabled precise mapping) and within the top quintile of 3P-tag abundance. d, The lin-14 3′UTRs. 3P tags from egg were mapped relative to RNA-Seq data10, prior mRNA annotations from the indicated databases6,11, and the proposed lin-4-binding region 4. Distal and proximal cleavage sites are indicated (black and red arrowheads, respectively). A 50-nucleotide region containing the distal 3P cluster is enlarged (box). Each tag sequence with a unique genome match is depicted as a bar, colored by tag frequency (key). e, Nucleotide sequence composition at mRNA end regions. Shown above are elements implicated in cleavage and polyadenylation (Supplementary Fig. 3c)30, with colors reflecting their nucleotide composition (A-rich, red; U-rich, blue). The sharp adenosine peak at position +1 (*) was due only partly to cleavage prior to an A. Also contributing to this peak (and to both depletion of A at position −1 and blurring of sequence composition at other positions) was cleavage after an A, for which the templated A was assigned to the poly(A) tail, resulting in a 1 nucleotide offset from the cleavage-site register.
3P-Seq was designed to identify the 3′ ends of polyadenylated RNAs without recourse to oligo(dT) priming. Oligo(dT) priming can prime on internal A-rich regions of transcripts, thereby yielding artifacts difficult to distinguish from authentic polyadenylated transcripts because the artifacts also have untemplated A’s9. Although untemplated adenylates at the ends of 3P tags could not have arisen from internal-priming artifacts, in principle, such nucleotides could have arisen from polymerase/sequencing errors. Countering this possibility was the observation that homopolymeric runs containing untemplated nucleotides at the ends of candidate 3P tags were overwhelmingly A’s (Fig. 1b). Thus, non-genomic terminal adenylates at the ends of 3P tags [a beneficial consequence of incomplete RNase H digestion near duplex termini (Fig. 1a)] provided compelling evidence that they derived from distal ends of bona fide polyadenylated transcripts.
To ensure proper assignment to polyadenylated transcripts, we considered as 3P tags only reads that both mapped uniquely to the genome and possessed at least two 3′-terminal A’s, of which at least one was untemplated. Nearly 32 million reads from C. elegans met these criteria, including millions from each major developmental stage (embryo, L1, L2, L3, L4, adult) as well as dauer L3 worms and germline-deficient glp-4(bn2) mutant adults (Supplementary Table 1).
Microheterogeneity at the cleavage and polyadenylation sites (hereafter called cleavage sites) often produced clusters of related 3P tags (Fig. 1c,d). All tags ending within 10 nucleotides of the most frequently implicated cleavage site were consolidated into a cluster, with this candidate cleavage site representing that of the cluster (Supplementary Dataset 1). Candidate sites were classified as mRNA cleavage sites if they were bridged by RNA-Seq reads10 to the stop codons of RefSeq mRNAs6,11, as illustrated for lin-14 (Fig. 1d).
3P-Seq identified 24,036 distinct 3′UTRs, including at least one UTR for 16,261 (83%) of the RefSeq mRNAs (Supplementary Dataset 2). For 5,331 mRNAs, we revised the longest-isoform annotation by >10 nucleotides (usually by extending it), and for 5,852 mRNAs without 3′UTR annotations, we identified a UTR (Supplementary Fig. 1a). A parallel effort within the modENCODE project used oligo(dT)-based methods to also generate a greatly expanded dataset of C. elegans 3′UTRs8; 8,581 of the 24,036 UTRs identified by 3P-Seq were not identified in that study (Supplementary Table 2). Our data were shared with the modENCODE consortium, thereby enabling them to annotate 8,758 novel UTRs that the oligo(dT)-based methods missed (L. Hillier and R. Waterston, personal communication; Supplementary Table 3). Of the 3,280 RefSeq mRNAs not assigned 3′UTRs using 3P-Seq, most were from predicted genes without evidence of expression (Supplementary Fig. 1b). Of the remainder, most were expressed at extremely low levels (Supplementary Fig. 1c). We estimated that only 124 ± 56 (95% confidence interval) sites were missed by requiring that tags have an untemplated A (Supplementary Fig. 1d). Most histone mRNAs were assigned 3′UTRs, consistent with oligo(dT)-based results8, but the polyadenylated forms of these mRNAs did not accumulate to levels detectable on RNA blots (Supplementary Fig. 2).
Apart from the A-rich segment corresponding to the polyadenylation signal (PAS) AAUAAA and its close variants (Supplementary Fig. 3a,b, Supplementary Table 4), the mRNA end regions were U-rich, presumably a feature of the binding sites of factors that enhance cleavage and polyadenylation (Fig. 1e, Supplementary Fig. 3c). Indeed, end regions that lacked a common PAS had exaggerated U-rich features surrounding an A-rich segment located where the PAS normally occurs (Supplementary Fig. 3d), which suggests that appropriate U-rich context can compensate for lack of a strong PAS12.
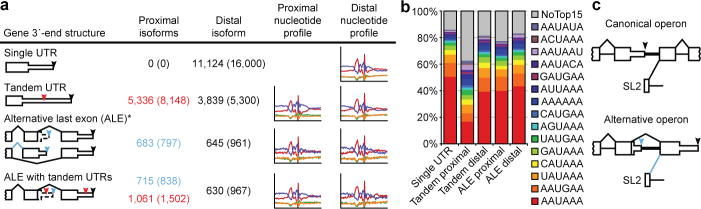
3P-Seq was particularly useful for reliably identifying alternative UTR isoforms. Genes with tandem 3′UTRs possess proximal cleavage sites that, when utilized, create a shorter UTR that is a subfragment of longer versions (Fig. 2a). When identifying these shorter isoforms, we required that 1) the proximal site be represented by ≥ 2 independent 3P tags, 2) that these tags constitute ≥ 1% of the tags mapping between the distal site and the stop codon, and 3) the site be in an end region nonoverlapping with that of a more distal site (i.e., that the two cleavage sites be ≥ 40 nucleotides apart). These criteria identified 7,798 shorter isoforms, which corresponded to 31% of the Entrez genes with 3P-supported UTRs (Fig. 2a). As expected for sites sometimes bypassed by the cleavage and polyadenylation machinery to allow production of longer isoforms, a larger fraction lacked a common PAS (Fig. 2b). Although less conserved than PASs for distal-most sites, PASs for proximal sites were more conserved than expected by chance (Supplementary Fig. 4b). Proximal isoforms had lengths typical of C. elegans UTRs, whereas distal isoforms were longer than typical UTRs (Supplementary Fig. 4a; P <10−300, Wilcoxon rank-sum test), hinting at even more elaborate UTR-mediated regulation.
Figure 2. Alternative 3′ UTRs in C. elegans.
a, Distribution of the 24,036 3P-Seq-supported UTRs among the types of alternative isoforms. For genes with ALEs that have tandem isoforms (bottom), the ALE tally indicates the number of distal isoforms of proximal ALEs (blue) and the tandem tally indicates the proximal tandem isoforms of all ALEs (red). In all cases, the distal isoform is the 3′-most cleavage site for each gene (black arrowhead). Also depicted are proximal tandem sites and proximal ALE sites (red and blue arrowheads, respectively). Listed (in parenthesis) is the number of cleavage sites associated with each isoform type for the 34,525 3P-Seq-supported cleavage sites (which exceeded the number of unique UTRs because OERs produced multiple cleavage sites for the same UTR). The nucleotide composition near proximal and distal sites is shown (right). b, Frequency of PAS motifs for isoform types indicated. c, Schematics of canonical and alternative operons.
Mangone et al. report a large class of proximal isoforms that lack PASs and instead have A-rich regions immediately following the cleavage sites8. 3P-Seq, which avoids oligo(dT) priming, provided no evidence for this novel class of isoforms, suggesting that it is composed of false-positives that arose from internal priming on A-rich UTR regions, as illustrated for the ubc-18 3′UTR and confirmed by an RNase-protection experiment (Supplementary Fig. 5a,b). Of the 5,728 cleavage sites not supported by 3P-Seq, 3,900 were sites of putative proximal isoforms (Supplementary Table 3), of which ~70% appear to have resulted from internal-priming artifacts (Supplementary Fig. 5c).
Genes with alternative last exons (ALEs) generate messages with completely different UTRs (Fig. 2a). We identified 1,398 ALEs distributed across 1,277 Entrez genes. Previous methods identified <25% of these ALEs (Supplementary Fig. 5d), presumably because data acquisition or analyses had focused on regions downstream of annotated stop codons8, which illustrates advantages of 3P-Seq for identifying unanticipated UTRs. The PAS motifs and nucleotide composition associated with proximal ALE ends were comparable to those at distal ends (Fig. 2a,b), and the distal isoforms tended to be longer than both proximal isoforms and single UTRs (Supplementary Fig. 4c, P <10−5 and <10−14, respectively, Wilcoxon rank-sum test).
Our analyses also identified a novel gene architecture, called the “alternative operon.” C. elegans operons are each arrays of genes transcribed from a single promoter and split into separate mRNAs through the biochemically coupled processes of 3′-end formation and trans-splicing to splicing leader 2 (SL2)13. Hypothesizing that this coupling could result in SL2 trans-splicing to 3′-splice sites downstream of ALEs, we searched for a gene structure that differed from the canonical operon by a splice junction bridging exons from different genes of an operon (Fig. 2c). This search identified 12 alternative operons, including the smg-6 locus (Supplementary Fig. 6; Supplementary Table 5).
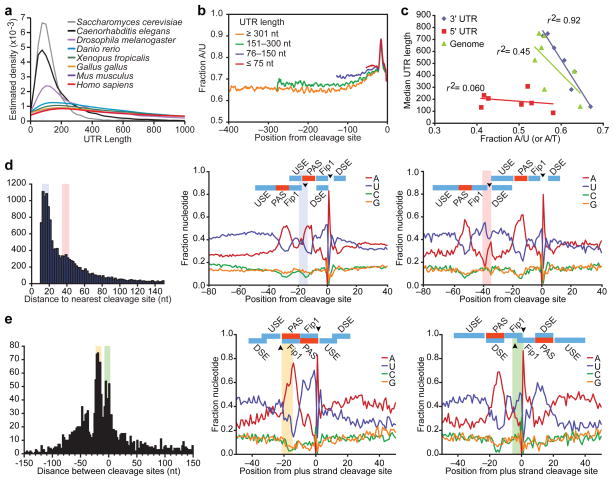
Among representative metazoans, C. elegans had the shortest 3′UTRs, with a length distribution approaching that of S. cerevisiae (Fig. 3a) and a median length of 130 nucleotides, only one sixth that of human. C. elegans 3′UTRs were also the most A/U rich. Shorter UTRs tended to be the most A/U rich (Fig. 3b), and even after masking the UTR end regions, which are exceptionally U/A rich, a cross-species comparison revealed a significant inverse correlation between 3′UTR length and 3′UTR A/U content (P = 0.0003, r2 = 0.92, Pearson correlation), whereas correlations between either 5′UTR length and A/U content or 3′UTR length and genomic A/T content were less significant (P = 0.30 and 0.05, respectively; Fig. 3c). We speculate that this strong inverse correlation is causal; i.e., higher A/U content favors the emergence of A/U-rich motifs that create proximal mRNA ends within existing 3′UTRs, thereby generating progressively shorter UTRs.
Figure 3. Evolution and topology of 3′-end formation.
a, 3′UTR length distributions for the indicated species, considering the most distal annotated isoform for each gene. b, A/U content for C. elegans 3′UTRs of the indicted lengths. c, Relationship between 3′UTR length and 3′UTR A/U content (disregarding content of the last 40 UTR nucleotides), 3′UTR length and genomic A/T content, and 5′UTR length and 5′UTR A/U content for the metazoan species in (a) (r2, Pearson correlation coefficients). d, OERs. Distances between neighboring cleavage sites are plotted (left). For peaks in the distribution at 15–20 and 35–40 nucleotides (shaded), nucleotide compositions of OERs are shown (middle and right, respectively), with proposed RNA-recognition elements colored as in Fig. 1e. Arrowheads indicate cleavage sites, with shading also indicating positions of upstream cleavage. e, Convergent UTR overlap. Distances between convergent 3′ ends are plotted (left), with negative values indicating overlap. For peaks at 15–22 and (−2) 8 nucleotides of overlap (shaded), nucleotide compositions are shown (middle and right, respectively) as in (d), with shading indicating positions of minus-strand cleavage.
Also potentially related to progressive UTR shortening were the 7,117 UTRs with with ≥ 2 closely spaced alternative cleavage sites. We did not classify these as tandem UTRs because the cleavage sites were very close to each other (<40 nucleotides, usually 12–22 nucleotides), implying overlapping end regions (OERs). This overlap tended to be phased, such that U-rich cis-acting elements could serve dual functions, binding alternative factors, depending on which cleavage site was being recognized (Fig. 3d). Although previous studies do not distinguish these isoforms from the heterogeneity normally found at UTR ends, proximal and distal OER isoforms were distinct in that each tended to have their own A-rich PASs (Fig. 3d). The 10,498 additional cleavage sites from OERs thus represented the largest class of alternative mRNA isoforms in C. elegans (Fig. 2a, compare UTR tallies with cleavage-site tallies).
The few additional nucleotides of distal OER isoforms presumably are dedicated to end recognition and processing (Fig. 3d), leaving little space for regulatory sites that could impart differential regulation. Thus, the importance of the OER isoforms might pertain instead to UTR evolution. The potential of the U-rich regions to serve dual functions would favor the emergence of new cleavage sites with OERs. Moreover, the higher A/U content of C. elegans UTRs compared to that of intergenic regions would favor the emergence of more upstream sites than downstream sites, which in turn could lead to progressive UTR shortening as the original signals acquire mutations rendering them less able to compete for factors (Supplementary Fig. 7a). If nematode UTRs had a propensity to drift towards a minimum UTR length, longer UTRs, which have avoided this shortening, might display more evidence of cleavage-site retention. Indeed, the PASs of long UTRs were more frequently conserved than were those of shorter UTRs (Supplementary Fig. 7b, P < 10−15, Kolmogorov-Smirnov test).
The alternating U- and A-rich elements defining UTR end regions provided opportunity for motifs to also serve double duty on opposite strands. Indeed, overlap of convergent UTRs occurred with a tri-modal distribution peaking at 5, 20, and 40 nucleotides, in which the A-rich PASs of each strand reciprocally served as U-rich motifs of the other strand (Fig. 3e). The bidirectionality of these composite sites was often selectively maintained (Supplementary Fig. 8a). Previously, a single peak in the distribution was observed at ~20 nucleotides of overlap, which was attributed to selective pressure to avoid RNAi8. Our data indicated a more complex overlap distribution that is better explained by preferential emergence of end regions where end elements of a convergent gene already provide some of the alternating A- and U-rich segments needed for end recognition. Although more extensive overlap can act to enforce mutually exclusive transcriptional regulation14, expression of the overlapping gene pairs were no less correlated than were random pairs (Supplementary Fig. 8b). Hence, gene topology utilizing palindromic arrangement of bidirectional elements provides a mechanism for genome compaction, effectively minimizing intergenic space downstream of 2,448 genes (a sixth of all genes with 3P-Seq–identified ends) without significantly impacting their regulatory autonomy.
Before considering targeting of the newly annotated 3′UTRs by microRNAs (miRNAs), we updated the set of confidently identified miRNAs using ~23 million genome-matching small-RNA sequences15. Methods shown to identify miRNAs reliably in mammals16 provided confident support for 145 annotated genes and 12 additional genes (Supplementary Table 6; Supplementary Fig. 9; Supplementary Text). Five of the newly identified miRNAs derived from mirtrons (Supplementary Table 6), which are spliced and debranched introns that fold into pre-miRNA hairpins, thereby bypassing Drosha processing17. Although mirtrons are typically thought to be spliced from pre-mRNAs, three newly identified mirtrons and two pre-miRNAs reclassified as mirtrons (mir-255 and mir-2220) derived from host transcripts that did not appear to be protein coding (Supplementary Fig. 10; Supplementary Table 6). We also generated developmental expression profiles for the 159 confidently annotated genes (Supplementary Fig. 11; Supplementary Tables 6, 7; Supplementary Text).
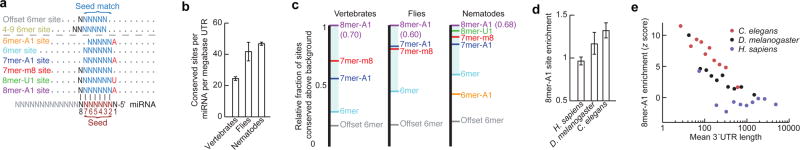
Methods used previously to detect miRNA site conservation in vertebrate genome alignments18 found six types of preferentially conserved sites that matched the miRNA seed region (Fig. 4a), including two types (8mer-U1 and 6mer-A1) not observed in vertebrates (Supplementary Figs. 12a-c, 13; Supplementary text). Efficacy of these six types was confirmed using two large-scale experimental datasets: mRNA fragments crosslinked to C. elegans miRNA silencing complexes19 and mRNA changes in C. elegans miR-124 mutants20 (Supplementary Fig. 12d; Supplementary Table 8).
Figure 4. MicroRNA targeting.
a, Expanded repertoire of seed-matched sites preferentially conserved in nematode 3′UTRs. Sites conserved only marginally above chance are above the dashed line. Watson-Crick-matched residues, blue or black; residues independent of the miRNA sequence, red. b, Density of miRNA sites conserved above background, combining all site types at the maximally sensitive cutoff. Error bars, one standard deviation (calculated by repeating the analysis for each site type 50 times, each time using a different cohort of control sequences that matched the properties of the miRNA sequences18). c, Relative strength of miRNA site types across clades. Within each clade, two species of comparable divergence were selected. For each miRNA site type, the fraction of sites conserved above background in the two species was normalized to that of the 8mer-A1 (shown in parentheses). d, Enrichment of 8mer-A1 3′UTR sites above expectation based on dinucleotide content. Error bars, one standard deviation, derived as in (b). e, Relationship between 3′UTR length and site enrichment. Site enrichment is ploted for 3′UTRs of the indicated species sorted by length into ten equally sized bins.
Summing results for all six site types indicted that C. elegans UTRs have at least 9,166 ± 197 (95% confidence interval) selectively maintained miRNA sites, and that at least 27.4% ± 4.7% of the C. elegans 3′UTRs have been under selective pressure to retain miRNA targeting (Supplementary Fig. 12b,c). This percentage was nearly three-fold greater than that detected previously in nematodes21 and about half that observed for human UTRs18, despite substantially shorter lengths of nematode UTRs, fewer nematode genomes available, and fewer conserved miRNA families in nematodes (62, compared to 87 in vertebrates). As in vertebrates18, few preferentially conserved sites had mismatches or wobbles to the seed nucleotides (Supplementary Fig. 14). Indeed, the three most compelling sites with seed mismatches (two let-7 sites in lin-41 and one let-7 site in hbl-1 ; Supplementary Table 9) had all been implicated by earlier genetic studies22,23. The updated miRNA target predictions will be presented in TargetScanWorm, release 5.2 (targetscan.org).
Compared to human 3′UTRs, C. elegans 3′UTRs had twice the density of selectively conserved miRNA sites (Fig. 4b, Supplementary Fig. 15d). This difference was attributed partly to the two additional site types conserved in nematodes and partly to the higher fractions of 6mer and 7mer sites preferentially conserved in nematodes (Fig. 4c). Drosophila, which has intermediate 3′UTR lengths (median 224 nucleotides), had intermediate fractions of sites conserved (Fig. 4c). Because the relative conservation of site types correlates well with their efficacy18, species with shorter 3′UTRs presumably have increased relative efficacy of site types that impart marginal repression in vertebrates, such as most 6mer sites. With this increased miRNA targeting promiscuity, C. elegans could cope with shorter UTRs without sacrificing as much miRNA-mediated regulation.
MicroRNA sites were enriched in C. elegans 3′UTRs irrespective of conservation (P <10−3, binomial test; Fig. 4d). This enrichment was not observed in other regions of C. elegans mRNAs nor for human miRNA sites in any region of human mRNAs (Supplementary Fig. 15a). Perhaps in humans, the evolutionary depletion of detrimental miRNA sites balances the selective retention of beneficial sites, whereas in C. elegans, with its short UTRs, the depletion is not sufficient to balance the selective retention of beneficial sites (Supplementary text). In this model, miRNA site enrichment would be a property of short 3′UTRs in any context. Indeed, enrichment of miRNA sites inversely correlated with mean 3′UTR length in both interspecies and intraspecies comparisons (Fig. 4d,e). Increased miRNA site density and increased efficacy of marginal site types in the context of short 3′UTRs are both likely to generalize to other cis-regulatory elements. Indeed, in C. elegans, the ~6,400 tandem UTR events occurred at one event per 560 nucleotides, a density over five times that reported in human UTRs24,25.
3P-Seq provided a more comprehensive and reliable view of C. elegans 3′UTRs and the basis for insights into their formation, evolution, and regulation. The method should provide analogous results when applied to other eukaryotes with poorly annotated 3′UTRs, i.e., most sequenced eukaryotes. 3P-Seq should also be informative for human studies, where it could shed light on shorter UTR isoforms, including those associated with cell proliferation and oncogenic transformation26,27,28.
Methods Summary
Nematodes were grown and RNA isolated as described29. 3P-Seq was performed as outlined in Fig. 1a. Reads that both mapped to a single locus in the genome and possessed ≥ 2–3′-terminal A’s (≥1 untemplated) were carried forward as 3P tags. Tags were iteratively clustered into representative cleavage sites and bridged to transcript models with RNA-Seq data (accession SRA003622.7)10. Poly(A) signals were identified as hexamers with position-dependent enrichment similar to AAUAAA. Cleavage sites 5′ of terminal exons indicated ALEs. For each last exon, cleavage sites mapping between the stop codon and the 3′-most 3P-Seq – supported cleavage site indicated tandem isoforms. Conservation analysis was as described18, except five UTR conservation bins were used for D. melanogaster and four were used for C. elegans, in order to compensate for the smaller total sequence space of 3′UTRs in these species. For comparisons between mammals, flies and nematodes, pairs of species analyzed were H. sapiens and M. domestica, D. melanogaster and D. willistoni, and C. elegans and C. briggsae, each ofwhich had a 3′UTR nucleotide-level divergence rate of ~0.55.
Methods
3P-Seq Libraries
Nematodes were grown and RNA isolated as described29. For each library, 30 μg total RNA was enriched for polyadenylated mRNA [Dynabeads Oligo(dT)25, Invitrogen]. Enriched RNAs were ligated to a 5′-phosphorylated, 3′-biotinylated oligonucleotide adaptor (p-agcguguagggcaccauGCACATAC-Biotin; lowercase, RNA; uppercase, DNA) using the splint DNA oligonucleotide (ATGGTGCCCTACACGCTTTTTTTTT) and 1 U Rnl II RNA ligase (New England Biolabs) in a 20 μl reaction for 16 hours, according the manufacturer’s instructions. After phenol extraction and precipitation, the RNA was partially digested with 3 U RNase T1 (Biochemistry grade, Ambion) in a 100 μl reaction for 20 minutes at 22°C, and 3′ fragments were captured with 100 μl streptavidin-coated beads (Dynabeads M-280 streptavidin, Invitrogen) in 400 μl B buffer (5 mM Tris-Cl, pH 7.5, 0.5 mM EDTA, 1 M NaCl) for 15 minutes rotating at room temperature (~22°C). After one wash in B buffer, beads were washed twice in 400 μl W buffer (10 mM Tris-Cl, pH 7.5, 1 mM EDTA, 50 mM NaCl) at 50°C, then equilibrated in reverse transcription (RT) buffer (Invitrogen). The RT primer (GTATGTGCATGGTGCCCTACACGCT) was annealed and then extended with dTTP as the only deoxynucleoside triphosphate, using 1 U reverse transcriptase (Superscript III, Invitrogen) in 25 μl for 20 minutes at 48°C. Polyadenylated RNA fragments were released into solution by adding 1 U RNase H (Invitrogen) and digesting for 25 minutes at 37°C. After precipitation, fragments were ligated to a pre-adenylated 3′ adaptor (AppAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT-C3spacer) with 10 U T4 RNL1 (NEB) in a 10 μl reaction for 2 hours at 22°C, and ligation products were gel purified (excising 75–300 nucleotide RNAs) and prepared for Illumina sequencing with a protocol used for strand-specific mRNA-Seq31. Because sequencing started at the residues corresponding to the 3′ ends of the RNA fragments, which for the 3P tags were all adenylates, cluster definition was somewhat compromised, which lowered the yield of 3P tags. In experiments performed after those described here, we obtained higher yields of 3P tags when defining the clusters from images in the middle of the run by starting the Illumina basecalling with the middle images and then reading the images from the first part of the run after the clusters have been defined. A detailed 3P-Seq protocol is available at web.wi.mit.edu/bartel/pub/protocols.html.
Distal mRNA Cleavage Sites
The reverse complements of the sequencing reads were considered candidate 3P tags. These candidate tags were aligned to the C. elegans genome (WS190) with Bowtie32, using alignment parameters “-q --solexa-quals -5–3 -l 25 -n 1 -e 240 -m 1” to allow for the presence of untemplated nucleotides at their 3′ termini. Sequences that both mapped to a single genomic locus and possessed ≥2–3′-terminal adenylates, of which at least one was untemplated, were carried forward as 3P-Seq tags. The most 3′-terminal non-adenosine base of each tag was considered a candidate cleavage site. Genomic loci were then marked off, using a set of RefSeq transcripts with non-redundant 3′ UTRs, with each locus corresponding to the region between the annotated 5′ terminus of the transcript and the annotated 5′ terminus of the downstream gene on the same genomic strand. Candidate sites mapping to each locus were sorted from most abundant to least, with equally abundant sites ordered 3′-most first. Clusters were then built from all sites within a 21 nt window centered on the site with the most tags (combining data from all libraries). This process was iterated until all 3P tags were assigned to clusters (with some clusters containing only one tag). The central site of each cluster was then evaluated as a potential mRNA cleavage site using RNA-Seq data (accession SRA003622.7)10. The number of RNA-Seq reads covering each base of the transcript was tallied, and a 50-nucleotide window was slid from the stop codon to the candidate terminus, after masking bases contained within annotated introns. A candidate site was assigned to an upstream protein-coding region if the median per-base RNA-Seq coverage in all windows was above 0. These sites were filtered further, requiring that the median per-base RNA-Seq coverage in the implied UTR was ≥5% that of the corresponding protein-coding region and that the maximum per-base RNA-Seq coverage in the UTR did not exceed ≥5 times that of the coding-region maximum. Among the sites that passed these filters, the distal cleavage site of a gene within the locus was the site of the 3′-most cluster that contributed ≥1% of the 3P tags from the locus.
Poly(A) Signals
Genes with single UTRs were used to search for position-dependent enrichment of hexamer motifs near the cleavage site. To establish the region where PASs were expected to occur, AAUAAA enrichment was analyzed at each position within the 50 nucleotides upstream of cleavage sites. At each position, significance was determined by the binomial test against the first-order Markov expectation for AAUAAA (Supplmentary Fig. 3). The region with significant AAUAAA enrichment (9–25 nucleotides upstream of the cleavage site) was analyzed for enrichment of other hexamers, after removing the UTRs with AAUAAA in the region. The most significantly enriched alternative hexamer was identified as above, and sequences containing this hexamer were removed and the process was iterated another 13 times. Enrichment analysis was also performed by a similar process, except the first-order Markov expectations were replaced with the hexamer frequency in an equally wide control window starting 50 nt upstream of the cleavage site, and significant enrichment was determined by the Fisher’s exact test. PASs were assigned to each cleavage site by searching the region 9–25 nt upstream of the site, considering the 15 most significantly enriched hexamers (Supplementary Figure 3; Supplementary Table 4) and in cases of matches to more than one hexamer, giving preference to the one most significantly enriched in the global analysis that calculated enrichment using upstream control sequences.
Proximal Alternative Sites
For each Entrez gene, candidate cleavage sites (as defined above) were considered as proximal alternative cleavage sites if they 1) mapped between the 5′-most end and the 3′-most 3P-supported cleavage site of the gene, 2) were from clusters containing ≥1% of the tags from the gene, and 3) were from clusters containing two independent 3P-tags. Tags were considered independent if they either 1) were sequenced in independent libraries, 2) mapped to different cleavage sites, or 3) mapped to the same cleavage site but had different numbers of terminal adenylates. For each RefSeq transcript, proximal alternative sites 3′ of the stop codon were classified as proximal tandem sites. Candidate ALEs were identified by proximal alternative sites that mapped internally to genes, excluding the exons 3′ of the distal-most stop codon for each Entrez gene annotation33. Identification of ALEs was particularly challenging in C. elegans for two reasons. First, many gene annotations had limited experimental validation. Second, C. elegans has a high density of genes34, at least 15% of which are organized as operons35. Identification of ALEs thereby depended on experimental validation of the exons as alternative. ALEs were required to have the support of one of the following: (1) an EST spanning the ALE and aligning to both an upstream and downstream exon relative to the ALE; (2) 3P tags mapping to exons downstream of the ALE; (3) an RNA-Seq read spanning the exon junction between the upstream and downstream exons. In addition, ALEs were required to have an in-frame stop codon before the cleavage site. If no appropriate stop codon was annotated for novel ALEs, the nearest upstream exon was extended to the ALE cleavage site and the first in-frame stop codon was used.
Experimental evaluation of cleavage sites
Probes for RNase-protection experiments were designed to span proximal and distal cleavage sites of genes identified as having tandem UTRs either by both 3P-Seq and oligo(dT)-based methods (rpl-12, kin-19) or by only oligo(dT)-based methods (ubc-18)8. Templates for T7 transcription were amplified from N2 bristol genomic DNA using the following primer pairs: GAACAGCCCAATCCGTTGG, CAACACCAGTGTCTTTCGATAC (rpl-12); CTCTTTTGGCTCCAAATGCC, AGGGTGTTACGGGAAATAGC (kin-19); GGAGCACACTCGAAAGCACG, CCGTGTTGTTATCGGCAACATC (ubc-18). Amplicons were cloned into pGEM-T easy and screened for inserts antisense with respect to the vector T7 promoter. Probes were bodylabeled during in vitro transcription (Maxiscript, Ambion) and gel purified on denaturing 5% acrylamide gels. RPAs were performed with 105 CPM probe and 15 μg total RNA, hybridized overnight at 42°C and digested for 45 minutes in a 1:50 dilution of RNase A/T1 at 22oC (RPA III, Ambion). Products were resolved on denaturing 5% acrylamide gels and visualized by phosphorimaging.
Conservation Analyses
3′UTR alignments were extracted from Multi-Z alignments, (6-way for nematodes, 15-way for Drosophila) from the UCSC genome browser36, starting with D. melanogaster RefSeq annotations for Drosophila UTRs. Conservation analyses were as described18, except that five UTR conservation bins were used for D. melanogaster and four were used for C. elegans, in order to compensate for the smaller total sequence space of 3′UTRs in these species. Analyses of miRNA sites considered 62 C. elegans miRNA families with nucleotides 2–8 conserved throughout the Caenorhabditis clade (Supplementary Table 7) and 51 Drosophila miRNA families with nucleotides 2–8 conserved to D. pseudoobscura (Supplementary Table 10). For imperfect site types, only the position of the bulged or mismatched nucleotide needed to be conserved, not the nucleotide itself. For comparisons between mammals, flies and nematodes (Fig. 4; Supplementary Fig. 15) pairs of species analyzed were H. sapiens and M. domestica, D. melanogaster and D. willistoni, and C. elegans and C. briggsae, each of which had a 3′UTR nucleotide-level divergence rate of ~0.55.
k-mer Enrichment
For each type of miRNA site, 1,000 cohorts of control k-mers were chosen to match the site length, number of G+C nucleotides, and number of CpG dinucleotides. Enrichment was calculated by comparing the number of site occurrences in the region of interest to the mean number of occurrences for the controls. The P value was the fraction of control cohorts with ratios of observed-to-expected occurrences (based on a first-order Markov model) more extreme than that of the sites. When analyzing enrichment in Drosophila and human 3′UTRs (Fig. 4; Supplementary Fig. 15), only RefSeq annotations with “validated” status were used.
Supplementary Material
Acknowledgments
We thank Chris Burge and members of the Bartel lab for helpful discussions and the WIBR Genome Technology Core for sequencing. This work was supported by NIH grant GM067031 (D.P.B.), a National Science Foundation predoctoral fellowship (C.H.J.) and a Krell Institute/Department of Energy Computational Sciences Graduate Fellowship (R.C.F.).
Footnotes
Author Contributions C.H.J. performed the experiments and computational analyses of 3P-Seq data. R.C.F. performed the computational analyses of miRNA targeting and motif conservation. J.G.R. performed the computational analyses of miRNAs. All authors contributed to study design and manuscript preparation.
Author information 3P-Seq reads and 3P tags were deposited at the GEO as fastq and BED files, respectively (GSEXXXXX). MicroRNA genes were deposited at miRBase. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
References
- 1.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 2.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahringer J, Kimble J. Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3' untranslated region. Nature. 1991;349:346–348. doi: 10.1038/349346a0. [DOI] [PubMed] [Google Scholar]
- 4.Wightman B, Burglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3'-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991;5:1813–1824. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- 5.Merritt C, Rasoloson D, Ko D, Seydoux G. 3' UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers A, et al. WormBase 2007. Nucleic Acids Res. 2008;36:D612–617. doi: 10.1093/nar/gkm975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangone M, Macmenamin P, Zegar C, Piano F, Gunsalus KC. UTRome.org: a platform for 3'UTR biology in C. elegans. Nucleic Acids Res. 2008;36:D57–62. doi: 10.1093/nar/gkm946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangone M, et al. The Landscape of C. elegans 3'UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nam DK, et al. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci U S A. 2002;99:6152–6156. doi: 10.1073/pnas.092140899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillier LW, et al. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res. 2009;19:657–666. doi: 10.1101/gr.088112.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J. 2010;29:1523–1536. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans D, et al. A complex containing CstF-64 and the SL2 snRNP connects mRNA 3' end formation and trans-splicing in C. elegans operons. Genes Dev. 2001;15:2562–2571. doi: 10.1101/gad.920501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci U S A. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batista PJ, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark AM, et al. The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lall S, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 22.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 23.Abrahante JE, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 24.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3' untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 30.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3'-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10 :R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium, C. e. S. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 35.Blumenthal T. Trans-splicing and operons. WormBook. 2005:1–9. doi: 10.1895/wormbook.1.5.1. [DOI] [PubMed] [Google Scholar]
- 36.Karolchik D, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.