Abstract
On the basis of hyf-lacZ fusion studies, the hyf operon of Escherichia coli, noted for encoding the fourth hydrogenase isoenzyme (HYD4), is not expressed at a significant level in a wild-type strain. However, mutant FhlA proteins (constitutive activators of the hyc-encoded hydrogenase 3 isoenzyme) activated hyf-lacZ. HyfR, an FhlA homolog encoded by the hyfR gene present at the end of the hyf operon, also activated transcription of hyf-lacZ but did so only when hyfR was expressed from a heterologous promoter. The HYD4 isoenzyme did not substitute for HYD3 in H2 production. Optimum expression of hyf-lacZ required the presence of cyclic AMP receptor protein-cyclic AMP complex and anaerobic conditions when HyfR was the activator.
Three hydrogenase isoenzymes have been identified, purified, and characterized from Escherichia coli (6, 10, 16, 27, 37). The structural subunits and accessory proteins needed for these three isoenzymes are encoded by the hya, hyb, hyc, and hyp operons (9-11, 26, 29, 30, 34). The hya operon, hyaABCDEF (30), encodes the hydrogenase 1 (HYD1) isoenzyme and other accessory proteins required for processing of these subunits into the active form. This operon is induced under anaerobic conditions in the presence of formate or fumarate, repressed in the presence of nitrate, and requires acidic pH, ArcA, and AppY for optimal expression (12, 21, 32). However, hya mutants have no detectable phenotype (31). The hyb operon, hybABCDEFG (29), encodes the structural subunits of HYD2 as well as the needed accessory proteins (9). Based on genetic and physiological studies, HYD2 is responsible for uptake of hydrogen as an electron donor during anaerobic respiration, with fumarate serving as an electron acceptor (24, 29, 45).
The hyc operon encodes the structural subunits and necessary enzyme components to link HYD3 (36) to a unique formate dehydrogenase isoenzyme (FDH-H, encoded by fdhF) (46) to produce active formate hydrogenlyase complex (FHL) (10). This protein complex catalyzes the cleavage of formate to dihydrogen and carbon dioxide. Transcription of the hyc operon and fdhF requires the FhlA protein, a formate-dependent transcriptional activator (28, 38). In addition to FhlA-formate, molybdate is also required for transcription of the hyc operon, and this requirement is in part due to the need for the ModE-molybdate complex as a secondary activator (40). ModE, initially characterized as a molybdate-dependent repressor of the modABC operon carrying high-affinity molybdate transport genes (18), has subsequently been shown to act as a positive transcriptional regulator of the hyc operon (HYD3) as well as of the narXL operon (40), encoding a nitrate-responsive two-component regulatory system which activates transcription of narGHJI (respiratory nitrate reductase) (17). Additionally, optimal expression of hyc also requires the catalytic product of MoeA, a protein implicated in the activation of Mo during Mo-cofactor biosynthesis (19, 20). Mutated forms of FhlA that are independent of formate and/or molybdate have been described previously (42). These mutations are localized in the unique N-terminal region of the FhlA protein (23, 42). Deletion of the N-terminal 350 amino acids also produced an effector-independent transcriptional activator (FhlA165) (25, 41).
The E. coli genome sequence (8) revealed a 10-gene cluster (hyfABCDEFGHIJ), which is recognized as being the hyf operon (the fourth hydrogenase) based on similarity to corresponding Hyc proteins (2, 3). The proteins encoded by the hyf operon are proposed to constitute a proton-translocating formate hydrogenlyase (2). In support of this proposal, Bagramyan et al. (4, 5) reported an H+-K+ exchange reaction in osmotically stressed E. coli cells which was absent in a hyf mutant. On the basis of these and other studies, these investigators proposed that Hyf catalyzes dihydrogen production and ion transport when the cells are grown at a starting pH of 7.5. Skibinski et al. (43) reported that hyf-lac was expressed in wild-type E. coli in a formate-dependent manner, with FhlA serving as the activator. However, the maximum level of β-galactosidase activity produced by hyfA-lac was less than 100 U. Only when HyfR was produced from a multicopy plasmid was hyfA-lac expressed at a high level. It has been observed that mutant strains lacking all three known hydrogenases failed to produce hydrogenase activity assayed either by viologen reduction or by a more sensitive tritium exchange assay (J. C. Wendt and K. T. Shanmugam, unpublished data). These results suggest that the fourth hydrogenase encoded by the hyf operon is not produced in E. coli and that the hyf operon is silent in this organism. In this communication, we report that hyf-lacZ is not expressed to significant levels in wild-type E. coli, and this fact is independent of medium and growth conditions. We further report that hyfA-lacZ can be activated in the presence of effector-independent mutated forms of FhlA (FhlA132 and FhlA165) or native HyfR produced from a heterologous promoter, even when the gene is at single-copy level. In the presence of these activators, hyf expression is dioxygen sensitive and subject to catabolite repression.
Bacterial strains.
The bacterial strains, phages, and plasmids used in this study are listed in Table 1. All strains are derivatives of E. coli K-12.
TABLE 1.
E. coli strains, phages, and plasmids used in this study
| Strain, phage, or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Bacterial strains | ||
| BN4020 | fur-1::Tn5 | CGSC no. 7540 |
| BW25113 | lacIqrrnBT14 ΔlacZW116hsdR514 araBADAH33 ΔrhaBADLD78 | B. Wanner |
| RK4353 | Δ(argF-lac) U169 rpsL150 | Laboratory collection |
| YMC18 | endA thi hsdR Δlac rpoN::Tn10 | B. Magasanik |
| JW138 | Δhya Δhyb Φ(hycB′-lacZ) | Laboratory collection |
| SE1174 | fhlA102::Tn10 | Laboratory collection |
| SE1931 | fnr zcj::Tn10 | Laboratory collection |
| SE1265 | pfl-1 zba::Tn10 | Laboratory collection |
| SE1910 | Δ(modE-Km)2 | 18 |
| SE1989 | Δcya-Km CRP* | Laboratory collection |
| SE2147 | moeA113 zbi::Tn10 | 19 |
| VJS720 | modB247::Tn10 | V. Stewart |
| WS219 | RK4353 Δ(hyfB to hyfG)-Cm | This study |
| WS127 | Δ(srl-fhlA) Δlac λWS1-(hycA-lac) | 41 |
| Φ(hyfA′-′lacZ) derivatives | ||
| WS222 | RK4353 λWS4 | This study |
| WS228 | WS222 fhlA::Tn10 | WS222 × (P1)SE1174 |
| WS229 | WS222 rpoN::Tn10 | WS222 × (P1)YMC18 |
| WS230 | WS222 fnr zcj::Tn10 | WS222 × (P1)SE1931 |
| WS231 | WS222 moeA113 | WS222 × (P1)SE2147 |
| WS232 | WS222 Δ(hyfB to hyfG)-Cm | WS222 × (P1)WS219 |
| WS233 | WS222 pfl-1 zba::Tn10 | WS222 × (P1)SE1265 |
| WS235 | WS222 modB247::Tn10 | WS222 × (P1)VJS720 |
| WS236 | WS222 Δ(modE-Km)2 | WS222 × (P1)SE1910 |
| WS266 | RK4353 λWS10 | This study |
| WS267 | WS266 modB247::Tn10 | WS222 × (P1)VJS720 |
| WS268 | WS266 pfl-1 zba::Tn10 | WS266 × (P1)SE1265 |
| WS269 | WS266 rpoN::Tn10 | WS266 × (P1)YMC18 |
| WS270 | WS266 fhlA::Tn10 | WS266 × (P1)SE1174 |
| WS271 | WS266 Δ(modE-Km)2 | WS266 × (P1)SE1910 |
| WS272 | WS266 Δcya-Km | WS266 × (P1)SE1989 |
| WS273 | WS266 fnr zcj::Tn10 | WS266 × (P1)SE1931 |
| WS274 | WS266 moeA113 | WS266 × (P1)SE2147 |
| WS275 | WS266 Δ(hyfB to hyfG)-Cm | WS266 × (P1)WS219 |
| WS280 | WS266 fur::Tn5 | WS266 × (P1)BN4020 |
| AH266 | WS222 Δ(hyfA to hyfJ), Km | This study |
| AH267 | WS222 Δ(hyfR), Km | This study |
| Phages | ||
| P1 | Tn9 Cmrclr-100 | Laboratory collection |
| λRZ5 | λ′bla ′lacZ lacY+ | Laboratory collection |
| λWS4 | λbla+ Φ(hyfA′-lacZ) lacY+ | This study |
| λWS10 | λbla+laclqhyfR+ Φ(hyfA′-lacZ) lacY+ | This study |
| Plasmids | ||
| PKD4 | FRT-kan+-FRT bla+ | B. Wanner |
| pWS2 | pACYC184-fhlA+ | This study |
| pWS132 | pACYC184-fhlA132 | This study |
| pWS165 | pACYC184-fhlA165 | This study |
| pWS42 | pBR322 bcp hyfABCDEFGHIR′ | This study |
| pWS43 | pBR322 nlpB dapA gcvR bcp hyfA′ | This study |
| pWS44 | pBR322 nlpB dapA gcvR bcp hyfA′-lacZ | This study |
| pWTS3 | pWS42 Δhyf-Cm | This study |
| pWTS36 | pBR322-lacIqhyfR+bcp hyfA′-lacZ | This study |
| pWTS37 | pUC19-hyf promoter region | This study |
| pZCam | pZ1918-Cmr | This study |
Media, growth conditions, and materials.
Media used for bacterial growth were previously described (33). Luria broth (LB) (1.0% tryptone, 0.5% yeast extract, 0.5% NaCl), which served as rich medium, was supplemented with glucose (0.3%), sodium formate (15 mM), or sodium molybdate (1 mM) as needed. Glucose-minimal medium included 44 mM Na2HPO4, 5.5 mM KH2PO4, 34 mM NaCl, 41 μM Na2MoO4, 36 μM FeSO4, 7.5 mM (NH4)2SO4, 0.8 mM MgSO4, and 83 mM glucose. Antibiotics, when included, were used at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 30 μg/ml; chloramphenicol, 50 μg/ml (plates) and 10 μg/ml (liquid); and kanamycin, 50 μg/ml.
Transduction with phages P1 and λ was performed as previously described (33). Genetic and molecular biological experiments were carried out essentially as previously described (40). Biochemicals were purchased from Sigma Chemical Co. Other organic and inorganic chemicals came from Fisher Scientific and were of analytical grade. Restriction endonucleases and DNA-modifying enzymes were purchased from New England BioLabs and Promega.
Enzyme assays.
β-Galactosidase activity assays were carried out using cells in late exponential phase with cells that were permeabilized with sodium dodecyl sulfate and chloroform as previously described (33, 40). Units are expressed as nanomoles · minute−1 · (milligram of cell protein)−1. Under our experimental conditions, a Δlac mutant of E. coli was assayed at high cell density and produced enough o-nitrophenol to account for about 20 to 50 U of β-galactosidase activity. Due to this extremely low level of o-nitrophenyl-β-d-galactopyranoside hydrolysis, we used a value of 50 U of β-galactosidase activity as the basal level. Specific activity values represent the average of at least three independent experiments and varied by less than 15%. FHL activity of the cultures was determined by using whole cells to minimize dioxygen inactivation of FHL, with formate used as the electron donor (24). The amount of formate-dependent dihydrogen produced was determined by gas chromatographic methods (Varian gas chromatography with thermal conductivity detector and a 5-Å molecular sieve column).
Construction of Φ(hyfA-lacZ).
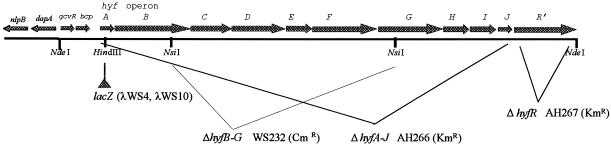
In order to construct a lacZ operon fusion for transcriptional analysis of the hyf operon, a 4.3-kb EcoRI-HindIII fragment from Kohara λ clone no. 424 (22), which carries the hyfA′ gene and 3.7 kb of upstream DNA, was cloned into plasmid pBR322 within the unique EcoRI and HindIII sites. The resulting plasmid, pWS43, was modified by inserting a 3.2-kb HindIII fragment from plasmid pZ1918 (39), which carries a promoterless lacZ gene, into the HindIII site. The resulting plasmid, pWS44, carries a hyfA′-lacZ fusion which is adjacent and opposite in orientation to the bla gene. In this plasmid, the lac fusion is located 296 bp downstream of the hyfA translation start site. This hyfA′-lacZ fusion was recombined in vivo with λRZ5 as previously described (33) in order to yield λWS4 (Fig. 1).
FIG. 1.
The hyf DNA from E. coli. Individual genes and direction of transcription are indicated above the line. Promoterless lacZ was inserted into the HindIII site in the hyfA gene in the construction of λWS4 and λWS10. The extent of deletion in each of the deletions and the corresponding strain are indicated.
Construction of Φ(hyfA-lacZ) hyfR+.
For the construction of λWS10, which carries the hyfA′-lacZ operon as well as the hyfR+ gene, the hyfR gene was amplified from plasmid pLC32-45 (14) by using two primers, 5′-ACTGTCCATGGCTATGTCAGACGAG-3′ and 5′-AAAAGAAGCTTACAACACCTCGCGA-3′. This PCR product was engineered to incorporate an NcoI site into the start codon (ATG) of the hyfR gene and a HindIII site past the translation stop codon. After amplification by Vent polymerase (New England Biolabs) and hydrolysis by NcoI and HindIII, the PCR product was ligated into the NcoI-HindIII sites of vector pTrc99A (1). The resulting plasmid, pWTS35, which also carries lacIq, expressed the hyfR gene from the trc promoter at low levels even in the absence of inducers of the lac operon (13). The lacIq and hyfR genes were removed from plasmid pWTS35 as an NsiI-ScaI fragment (3.5 kb) and ligated to an NsiI-ScaI fragment from plasmid pWS44, which carries the hyfA′-lacZ DNA. This construct, plasmid pWTS36, contains the hyfR+ gene and hyfA′-lacZ as well as 3.7 kb of hyf upstream DNA. In this construct, hyfR is still expressed from the trc promoter in the absence of isopropyl-β-d-thiogalactopyranoside (IPTG). The E. coli DNA in plasmid pWTS36 was recombined in vivo with λRZ5 as described previously (33) in order to produce λWS10 (Fig. 1).
For the construction of a plasmid which expresses hyfR and is also chloramphenicol resistant (pWTS34), a Cmr cartridge from plasmid pZCam was removed as a 988-bp HincII fragment and cloned into the FspI site of pTrc99A, thus creating pTrc99A-Cm. As per the construction of plasmid pWTS35, the PCR-amplified hyfR gene was cloned into the NcoI-HindIII sites of plasmid pTrc99A-Cm, resulting in plasmid pWTS34.
Construction of Δhyf and ΔhyfR.
Two different deletions of the hyf operon were constructed. The first, with an internal deletion between hyfB and hyfG, was constructed by starting with a 12-kb NdeI fragment from Kohara λ clone no. 424 (22), which was cloned into the NdeI site in plasmid vector pBR322. This plasmid, pWS42, which carries hyfABCDEFGHIJR′, was hydrolyzed with restriction enzyme NsiI so as to release a 5.6-kb internal fragment between the hyfB and hyfG genes (Fig. 1). This fragment was replaced with a 1.0-kb PstI fragment from pZCam carrying a Cmr gene cartridge. The resultant plasmid, pWTS3, carries the gene for chloramphenicol resistance in an orientation opposite to that of the hyf operon transcription between the hyfB and hyfG genes. The Cmr gene cartridge was expected to have a polar effect on the expression of downstream hyf (hyfHIJ and possibly hyfR) genes. In order to replace the wild-type hyf DNA in the chromosome with Δ(hyfB to hyfG)-Cm DNA, an 8.0-kb NdeI fragment from pWTS3 containing the Cmr gene and the neighboring hyf genes was removed and self-ligated by using T4 DNA ligase. This circular DNA lacks the bla gene and the origin of replication. Approximately 1 μg of the self-ligated 8.0-kb NdeI fragment was transformed into strain RK4353, and Cmr transformants were selected. One stable Cmr clone, strain WS219, was used in further studies. Cotransduction of the Cmr gene with a narQ::Tn10 mutation by P1 phage transduction confirmed that the gene for Cmr had recombined into the hyf operon.
The second deletion, which removed the entire hyf operon, was constructed as described previously (15). Hybrid primers that are complementary to E. coli hyfA and hyfJ and to the kanamycin gene in plasmid pKD4 (Hyf1, 5′-CGCTTTGTGGTGGCCGAACCACTGTGGTGTACAGGATGTAATACGTGTAGGCTGGAGCTGCTTC-3′, and Hyf2, 5-GGTCAACAGGGCGGTGTGGCTGGCGTCAATAACAATCTCACCAACATATGAATATCCTCCTTAG-3′) were obtained from Sigma-Genosys. Plasmid pKD4 was used as the template for PCR amplifications. About 1 μg of PCR product was electroporated into E. coli strain BW25113 with plasmid pKD46 pregrown in arabinose in order to induce the red recombinase. The resulting deletion (of hyfA to hyfJ) was confirmed by PCR. This mutation was transduced into strain WS222 for further studies (AH266).
A deletion which removed the entire hyfR gene was constructed by using the same method described above based on the procedures described by Datsenko and Wanner (15). The two primers used for PCR amplification of DNA and deletion of the hyfR gene were HyfR1 (5′-AAAAATTGCGTGAGAAGGATTTCTCATTAATAAGGACTGTTGATGGTGTAGGCTGGAGCTGCTTC-3′) and HyfR2 (5′-CCATTGGTTTCTCGCAATACCTGAACAATGCGCTGACGTTCTTCCATATGAATATCCTCCTTAG-3′). Upon construction, the ΔhyfR was transduced into strain WS222 (strain AH267).
The hyf operon is not expressed to significant levels in wild-type E. coli.
Based on genomic analysis, Andrews et al. (2) proposed that the Hyf hydrogenase, together with the FDH-H, couples formate oxidation to proton translocation. Recently, Bagramyan et al. (4, 5) reported that E. coli produced dihydrogen from formate which was Hyf dependent and inhibited by N,N′-dicyclohexylcarbodiimide. Production of dihydrogen, catalyzed by the fourth hydrogenase, required both growth of the culture at a starting pH of 7.5 and exposure to hyperosmotic stress before the assay. This hydrogenase activity was also proposed to be responsible for H+-K+ exchange. These results suggest that the hyf operon is expressed and that HYD4 isoenzyme is produced by E. coli during anaerobic growth at an alkaline starting pH.
Although the Hyf proteins are similar to the Hyc proteins, hyc and fhlA mutants are defective in dihydrogen production (28, 35, 38). E. coli mutants lacking all three known hydrogenase isoenzymes did not produce hydrogenase activity as determined by either dihydrogen-dependent dye reduction or by a more sensitive tritium exchange assay (Wendt and Shanmugam, unpublished). In the present study, E. coli mutants carrying a deletion within the hyf operon (AH266 and WS232) were cultured at a constant pH of 7.5 or 6.5, and the level of FHL activity in the cells was determined (Table 2). The parent and the deletion strains produced comparable levels of FHL activity when grown at pH 7.5. Although the FHL activity of cultures grown at a constant pH of 6.5 was higher, again, no significant difference in the levels of FHL activity between the parent and deletion strains could be observed. These results clearly show that the FHL activity observed in the pH 7.5 culture (constant pH) was derived from the HYD3 isoenzyme. In this regard, the hyf mutant is similar to the hya mutant, which also lacks a detectable phenotype (31). However, the HYD1 produced by the hya operon has been purified and characterized (16, 37), while a hydrogenase corresponding to Hyf was not detected in E. coli cells or extracts.
TABLE 2.
Formate hydrogenlyase activities of E. coli cultures grown in a pH-stata
| Strain | Relevant genotype | Growth medium | FHL activity (nmol · min−1 · mg of cell protein−1)
|
|
|---|---|---|---|---|
| pH 7.5 | pH 6.5 | |||
| WS222b | Wild type | LB + Glu (0.3%) | 15 | 48 |
| AH266 | Δ(hyfA to hyfJ) | 23 | 56 | |
| AH267 | Δ(hyfR) | 15 | 24 | |
| WS222 | Wild type | LB + Glu (0.1%) + formate (15 mM) | 40 | 148 |
| AH266 | Δ(hyfA to hyfJ) | 50 | 129 | |
| WS232 | Δ(hyfB to hyfG) | 66 | 136 | |
| AH267 | Δ(hyfR) | 83 | 103 | |
Cultures, in the indicated media, were grown anaerobically under argon gas phase at the indicated pH in a pH-stat. pH was maintained by the addition of 2 N KOH. FHL activity was determined as formate-dependent dihydrogen production by using a gas chromatograph. β-Galactosidase activities of all these cultures were below the detection limit of 50 nmol·min−1·(mg of cell protein)−1.
Strain WS222 carries Φ(hyfA-lacZ) via λWS4.
In order to evaluate the possibility that the fourth hydrogenase is produced in E. coli when cultured under specific medium composition (5), a hyc mutant (strain WS127) was grown in rich or minimal medium with or without glucose at a starting pH value of 6.5, 7.0, 7.5, or 8.0 (±0.1 M phosphate buffer) without pH control under a gas phase N2 atmosphere. These and other cultures grown with NaCl (0.2 or 0.3 M) at pH 7.5 or 8.0 did not produce any detectable dihydrogen measured as H2 by gas chromatography (data not presented). Strain JW138, lacking the three known hydrogenase isoenzyme genes (hya, hyb, and hyc), grown under similar conditions (initial medium pH value of 6.5, 7.0, 7.5, or 8.0 [±0.1 M phosphate buffer and ±0.3 M NaCl]) also did not produce detectable H2.
Maturation of the three known hydrogenases requires chaperone-like proteins, and the three proteins are interconnected at this level (9). It is possible that the inability to detect the fourth hydrogenase activity in a mutant lacking the other three hydrogenases is related to a need for such a chaperone-like protein or a specific protease produced by either the hya, hyb, or hyc operon for processing the fourth hydrogenase precursor protein to become the active enzyme. In previous studies, we have observed that the transcription of hycA-lac is unaffected by deleting the entire srl-fhlA region of the chromosome, which includes the hyp and hyc operons (41). By analogy, transcription of the hyf operon is expected to be independent of the ability of the coded proteins to function in the cell and thus should permit analysis of hyf expression as β-galactosidase activity by using a hyf-lacZ derivative.
In order to evaluate the level of transcription of the hyf operon, a λ phage carrying the hyfA′-lac fusion was constructed and inserted into the E. coli chromosome (strain WS222). Strain WS222 was cultured in a pH-stat at either pH 7.5 or 6.5, and irrespective of culture pH, β-galactosidase activity was not detected in these cells (data not presented). StrainWS222 did not produce detectable levels of β-galactosidase activity when cultured in either rich medium or minimal medium under a variety of anaerobic and aerobic growth conditions, including growth at pH 8.0 and in the presence of 0.3 M NaCl—conditions that, according to Bagramyan et al. (5), support Hyf-dependent activity (data not shown). Likewise, plasmid pWS44 (hyfA′-lacZ construct) used in the construction of this λ phage also did not support production of β-galactosidase activity either in the wild type or in various E. coli mutant strains tested (modE, moeA, fur, fnr, and crp mutants; data not shown). Attempts to isolate point mutations within the putative promoter region of hyf, which allowed expression of hyf-lacZ in a wild-type background, were also unsuccessful. The lack of expression of hyfA-lacZ suggests that this operon is not expressed under the physiological conditions tested and is apparently a silent operon. Skibinski et al. (43) reported that E. coli with a hyf-lacZ fusion produced about 15 U [nanomoles · minute−1 · (mg of protein)−1] of β-galactosidase activity, which was increased to about 50 U in the presence of formate. Under our experimental conditions, strain WS222 produced less β-galactosidase activity (Table 3) than did a lac deletion mutant without the λ phage carrying the fusion. These results show that the hyf operon is not expressed to significant levels in wild-type E. coli, and thus this operon should be considered a silent operon. However, the possibility that the hyf operon is expressed in the presence of an effector(s) which is not present in the cytoplasm when E. coli is cultured in the laboratory cannot be ruled out.
TABLE 3.
Regulation of expression of Φ(hyfA-lacZ) in the presence of various fhlA alleles and HyfR
| Mutated genea | β-Galactosidase activityb
|
||
|---|---|---|---|
| FhlA+ | FhlA165 | HyfR | |
| None (wild type) | <50 | 2,500 | 8,200 |
| fhlA | <50 | 2,500 | 9,900 |
| rpoN | <50 | <50 | <50 |
| hyf | NDc | ND | 7,200 |
| pfl | <50 (<50)d | 2,700 (2,700)d | 7,000 (7,500)d |
| modB | <50 (<50)e | 3,400 (3,400)e | 9,800 (11,000)e |
| modE | <50 | 3,300 | 12,500 |
| moeA | <50 | 1,800 | 9,000 |
| fur | <50 | ND | 11,000 |
| fnr | <50 | 2,600 | 22,000 |
All strains with FhlA plasmids are derivatives of E. coli strain WS222, which carries Φ(hyfA-lacZ) via λWS4, and all are listed in Table 1. For experiments with HyfR, mutant derivatives of strain WS266, which carries Φ(hyfA-lacZ) hyfR+ via λWS10, were used (Table 1). Cultures were grown in LBG (LB with glucose) medium under anaerobic conditions.
β-Galactosidase activity is expressed as nanomoles·minute−1·(milligram of cell protein)−1.
ND, not determined.
Formate was added to the growth medium at an initial concentration of 15 mM (values in parentheses).
Molybdate was present in the medium at an initial concentration of 1 mM (values in parentheses).
FhlA132 and FhlA165 proteins activate expression of the hyf operon.
The lack of expression of the hyf operon in E. coli could be due to the absence of an appropriate activator protein. A gene coding for a putative transcriptional activator, HyfR, is located at the end of the hyf operon (2). HyfR is similar to the FhlA protein (44% identical and 54% similar), which is the formate- and molybdenum-dependent activator of the hyc operon. HyfR, a protein with 663 amino acids, is missing the amino acids corresponding to the first 43 amino acids of the FhlA protein, which contains the region similar to the ABC-ATPases (41). Except for a stretch of about 60 amino acids (139 to 195 in HyfR and 179 to 234 in FhlA) in which the two proteins are 56% identical, HyfR and FhlA are dissimilar in their unique N-terminal regions. This N-terminal segment of the FhlA protein was proposed to be essential for formate binding in vivo (23, 41, 42). These differences in the N-terminal domain of the two proteins may be responsible for the inability of FhlA to activate hyfA-lacZ since the fhlA gene is constitutively expressed in anaerobic E. coli. Even when the copy number of fhlA+ was increased by introducing a plasmid carrying the fhlA+ gene (plasmid pWS2), the hyfA-lacZ expression was below the detection limit. It is apparent that the FhlA protein, either with or without formate and molybdate, is not an activator for the hyf operon. Although Skibinski et al. (43) reported that the FhlA activated hyfA-lac, the level of β-galactosidase activity produced by these cultures was only about 20 nmol · min−1 · (mg of protein)−1, and this was increased to about 50 U of activity in the presence of formate in the growth medium.
Both point mutations and deletions in the N-terminal domain of the FhlA protein were found to be effector independent, and some of the deletion derivatives activated hyc to a higher level than did the native protein (23, 25, 41, 42). Furthermore, the N-terminal domain of FhlA has also been reported to inhibit hyc activation by the deletion derivatives of FhlA (25). Since the central and C-terminal domains of FhlA and HyfR are more than 60% identical (70% similar), it is possible that the effector-independent forms of the FhlA protein would activate hyfA-lacZ. In the presence of FhlA132, which carries two point mutations (42), hyf-lac was expressed, and the level of β-galactosidase activity produced by the strain WS222(pWS132) was 330 U. The FhlA165 protein, which lacks the unique N-terminal region (amino acids 5 to 374) (41), increased activation of hyf-lac by about eight times, to about 2,500 U of β-galactosidase activity (Table 3). Although FhlA132 and FhlA165 activated hyc-lac expression at comparable levels (2,900 and 3,500 U of β-galactosidase activity, respectively) (41, 42), FhlA132 is only minimally effective with the hyf operon. This may be a consequence of the N-terminal domain (although carrying point mutations) significantly affecting the activation of transcription of hyf by the C-terminal domain of FhlA.
Activation of hyf by HyfR.
In a separate experiment, the hyfR gene was cloned and expressed from a heterologous promoter to determine whether HyfR, once produced within the cell, would activate the expression of hyfA-lacZ. In this experiment, the hyfR gene was cloned into phage λ, which also carries the hyfA-lacZ fusion (λWS10), in order to minimize the copy number effect. The HyfR protein, produced independent of its native control system, activated the hyf operon, and the level of β-galactosidase activity produced by strain WS266 (with λWS10) was about 8,000 U (Table 3). This level of expression is more than threefold higher than the value obtained with a strain carrying multiple copies of plasmid pWS165 coding for FhlA165. These results also confirm that the lack of transcription of hyf in wild-type E. coli is due to the absence of HyfR, the activator protein. Once produced in the cell, HyfR is an effective activator of the hyf operon (Table 3). However, HyfR failed to activate the hyc operon coding for the HYD3 isoenzyme, as evidenced by the lack of β-galactosidase activity from hyc-lacZ (strain WS127) or by dihydrogen production by a fhlA mutant (strain SE1174) carrying a plasmid expressing the hyfR+ gene (plasmid pWTS35) (data not presented). This difference is apparently due to the differences in the unique N-terminal domains of the two proteins. These results are in agreement with those of Skibinski et al. (43).
Even upon activation by either the FhlA165 or the HyfR protein, a hyc mutant that is hyf+ failed to produce detectable dihydrogen under any of the growth conditions tested. These results show that the Hyf proteins, although similar to the Hyc proteins, could not substitute for the HYD3 isoenzyme and other proteins of the FHL complex.
Formate and molybdate are not needed for hyf expression.
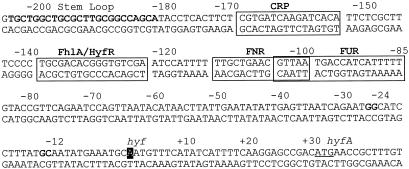
As expected, in the presence of FhlA165 as the activator, hyf-lac expression was not significantly affected by the presence or absence of either formate or molybdate (Table 3). Similar results were also obtained with HyfR, indicating that the hyf operon expression was not affected in strains carrying mutations in the production of formate (pfl) or molybdate transport (modB). A slight increase in hyf-lac expression in a modB mutant (8,200 versus 9,800 U of β-galactosidase activity) and a further increase when molybdate was added to the medium (9,800 to 11,000 U) both suggest that the observed effect is physiological. If molybdate is required for hyf-lac expression, the level is expected to be lower in a modB mutant and restored by molybdate addition, as was seen with other operons such as hyc-lac (40-42). A modest increase in the level of expression of hyf-lac occurred in a modE mutant compared to that of the wild-type parent, suggesting a potential repression by ModE. However, the upstream region of the hyf operon lacks a ModE consensus sequence (Fig. 2), and the observed effect is apparently physiological. A mutation in moeA had a minimal effect on hyf-lac expression with FhlA165 and no effect with HyfR as the activator. These results suggest that molybdenum apparently had a minimal indirect effect on hyf expression (Table 3). Although a consensus sequence for the iron-dependent control protein Fur can be found upstream of the transcription start site, a fur mutation had only a minimal effect on the level of expression of hyf-lac (Table 3). However, a mutation in fnr increased the level of β-galactosidase activity produced by the culture about threefold with HyfR as the activator (Table 3) but had no significant effect on expression activated by the mutant FhlA protein. A putative FNR consensus sequence (44) in the hyf upstream DNA can be detected between positions −111 and −98 (Fig. 2). It is possible that binding of FNR at this site may not have an impact on binding of the smaller constitutive activator FhlA165 but may minimize the ability of the larger HyfR protein to bind at the target sequence, which is only about 25 bases upstream of the predicted FNR site. This possibility will be tested in future experiments by using purified HyfR, FhlA, and FhlA165 proteins.
FIG. 2.
The hyf operon upstream DNA. The putative consensus sequences for the CRP, FNR, Fur, and FhlA/HyfR proteins are enclosed by rectangles. A possible stem-loop between the stop codon of the upstream bcp gene and hyf promoter is shown in boldface type. The transcription start site is in reverse type. The −12 and −24 positions are also in shown in boldface type. A putative start codon for hyfA is both underlined and enclosed within a rectangle.
CRP-cAMP is required for activation of hyf operon.
A cyclic AMP receptor protein-cyclic AMP complex (CRP-cAMP) consensus sequence is also present in the hyf upstream sequence centered at position −160.5 (Fig. 2), which is 30 bp upstream of the HyfR/FhlA consensus. In order to evaluate the significance of this sequence, the level of expression of hyf was determined in wild-type and cya mutant strains in the absence of added glucose to evaluate the role of CRP-cAMP with HyfR as the activator (Table 4). When the culture was grown in LB without glucose, the level of β-galactosidase activity increased 3.5-fold to about 28,000 U from a value of 8,200 U of activity in the presence of glucose (Table 4). In the presence of the cya mutation, the level of β-galactosidase activity produced by the LB culture of strain WS272 decreased ninefold to about 3,000 U, and the addition of 3 mM cAMP restored hyf-lac expression to a level higher than that observed with the wild type grown in the same medium. These results show that hyf expression is subject to catabolite repression. It should be noted that although FhlA132 and FhlA165 activated hyf-lac expression and apparently bind to the hyf upstream DNA at the same location as does HyfR, the cya mutation had a slightly positive effect on hyf-lacZ-dependent β-galactosidase activity produced in the presence of FhlA165 (data not presented). This difference in the responses between FhlA165 and HyfR to CRP-cAMP is probably related to the absence of N-terminal domain in the smaller protein, FhlA165.
TABLE 4.
Regulation of HyfR-dependent expression of Φ(hyfA-lacZ) requires cAMP for maximal activation and is repressed in aerobically grown cells
| Strain | Relevant genotypea | β-Galactosidase activityb
|
||
|---|---|---|---|---|
| LBG | LB | LB + cAMP | ||
| WS266 | Wild type | 8,200 | 27,800 | 26,200 |
| WS272 | cya | 5,100 | 3,260 | 39,800 |
| WS266 (aerobic) | Wild type | ND | 180 | ND |
All strains are derivatives of WS266, which carries Φ(hyfA-lacZ) hyfR+ via λWS10. Cultures were grown under anaerobic conditions, except where indicated. cAMP, when present, was added at a final concentration of 3 mM.
β-Galactosidase activity is expressed as nanomoles·minute−1·(milligram of cell protein)−1. ND, not determined.
HyfR does not activate transcription of hyf-lacZ in aerobically grown cultures.
Although strain WS266, with HyfR as the activator, produced more than 25,000 U of β-galactosidase activity when grown anaerobically in LB medium, aerobic cultures produced less than 200 U of β-galactosidase activity (Table 4). However, FhlA132 and FhlA165 did activate transcription of hyf-lacZ under aerobic conditions to levels comparable to those of the anaerobically grown cultures (data not presented). These mutant proteins have previously been shown to activate hyc transcription aerobically, so their activation of hyf is not unexpected (41). The lack of expression of hyf-lac by HyfR when the cells were grown aerobically demonstrated that when activated by HyfR, expression of hyf is oxygen sensitive. In the unique N-terminal domain of HyfR, a cysteine-rich amino acid sequence can be detected (200-CSDLSASHCACLPRC-214). This segment of the protein may potentially play a role in redox-dependent regulation of the hyf operon, as has been shown previously for the well-studied FNR protein (7). Although FhlA165 successfully activated transcription of hyf-lacZ in an in vitro transcription-translation experiment, aerobically purified HyfR protein was unable to activate transcription in vitro (data not shown), a finding that was in agreement with the putative oxygen sensitivity of the protein. Biochemical experiments with HyfR protein purified under aerobic and anaerobic conditions will help identify the oxygen-sensitive nature of hyf expression.
Although the hyf operon is apparently silent in wild-type E. coli, two mutant FhlA proteins (FhlA132 and FhlA165) and constitutively expressed HyfR protein were able to activate transcription of hyf. The ability of mutated forms of FhlA proteins to activate this operon represents a unique way to activate transcription of what seems to be a vestigial, unexpressed operon. Appropriate altered forms of known regulatory proteins may help activate corresponding silent genes or operons in E. coli or other organisms in order to elucidate the potential physiological role(s) of these proteins in the cell.
Acknowledgments
We thank P. Kiley, V. Stewart, and B. Magasanik for providing various strains used in this study. We thank Ken Rudd for providing the Kohara λ phages used in this study.
This work was supported by funds from the Florida Agricultural Experiment Station.
Footnotes
Florida Agricultural Experiment Station Journal Series no. R-09737.
REFERENCES
- 1.Amann, E., J. Brosius, and M. Ptashne. 1983. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene 25:167-178. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, S. C., B. C. Berks, J. McClay, A. Ambler, M. A. Quail, P. Golby, and J. R. Guest. 1997. A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenlyase system. Microbiology 143:3633-3647. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, S. C., P. M. Harrison, and J. R. Guest. 1991. A molecular analysis of the 53.3 minute region of the Escherichia coli linkage map. J. Gen. Microbiol. 137:361-367. [DOI] [PubMed] [Google Scholar]
- 4.Bagramyan, K., N. Mnatsakanyan, A. Poladian, A. Vassilian, and A. Trchounian. 2002. The roles of hydrogenases 3 and 4, and the F0F1-ATPase, in H2 production by Escherichia coli at alkaline and acidic pH. FEBS Lett. 516:172-178. [DOI] [PubMed] [Google Scholar]
- 5.Bagramyan, K., A. Vassilian, N. Mnatsakanyan, and A. Trchounian. 2001. Participation of hyf-encoded hydrogenase 4 in molecular hydrogen release coupled with proton-potassium exchange in Escherichia coli. Membr. Cell Biol. 14:749-763. [PubMed] [Google Scholar]
- 6.Ballantine, S. P., and D. H. Boxer. 1986. Isolation and characterisation of a soluble active fragment of hydrogenase isoenzyme 2 from the membranes of anaerobically grown Escherichia coli. Eur. J. Biochem. 156:277-284. [DOI] [PubMed] [Google Scholar]
- 7.Bates, D. M., B. A. Lazazzera, and P. J. Kiley. 1995. Characterization of FNR* mutant proteins indicates two distinct mechanisms for altering oxygen regulation of the Escherichia coli transcription factor FNR. J. Bacteriol. 177:3972-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner, F. R., G. Plunkett, 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 9.Blokesch, M., A. Magalon, and A. Bock. 2001. Interplay between the specific chaperone-like proteins HybG and HypC in maturation of hydrogenases 1, 2, and 3 from Escherichia coli. J. Bacteriol. 183:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bock, A., and G. Sawers. 1996. Fermentation, p. 262-282. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 11.Bohm, R., M. Sauter, and A. Bock. 1990. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol. Microbiol. 4:231-243. [DOI] [PubMed] [Google Scholar]
- 12.Brondsted, L., and T. Atlung. 1994. Anaerobic regulation of the hydrogenase 1 (hya) operon of Escherichia coli. J. Bacteriol. 176:5423-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brosius, J., M. Erfle, and J. Storella. 1985. Spacing of the −10 and −35 regions in the tac promoter. Effect on its in vivo activity. J. Biol. Chem. 260:3539-3541. [PubMed] [Google Scholar]
- 14.Clarke, L., and J. Carbon. 1976. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell 9:91-99. [DOI] [PubMed] [Google Scholar]
- 15.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis, K., P. Patel, J. C. Wendt, and K. T. Shanmugam. 1990. Purification and characterization of two forms of hydrogenase isoenzyme 1 from Escherichia coli. J. Bacteriol. 172:5750-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 18.Grunden, A. M., R. M. Ray, J. K. Rosentel, F. G. Healy, and K. T. Shanmugam. 1996. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J. Bacteriol. 178:735-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasona, A., R. M. Ray, and K. T. Shanmugam. 1998. Physiological and genetic analyses leading to identification of a biochemical role for the moeA (molybdate metabolism) gene product in Escherichia coli. J. Bacteriol. 180:1466-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasona, A., W. T. Self, R. M. Ray, and K. T. Shanmugam. 1998. Molybdate-dependent transcription of hyc and nar operons of Escherichia coli requires MoeA protein and ModE molybdate. FEMS Microbiol. Lett. 169:111-116. [DOI] [PubMed] [Google Scholar]
- 21.King, P. W., and A. E. Przybyla. 1999. Response of hya expression to external pH in Escherichia coli. J. Bacteriol. 181:5250-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohara, Y., K. Akiyama, and K. Isono. 1987. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50:495-508. [DOI] [PubMed] [Google Scholar]
- 23.Korsa, I., and A. Bock. 1997. Characterization of fhlA mutations resulting in ligand-independent transcriptional activation and ATP hydrolysis. J. Bacteriol. 179:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, J. H., P. Patel, P. Sankar, and K. T. Shanmugam. 1985. Isolation and characterization of mutant strains of Escherichia coli altered in H2 metabolism. J. Bacteriol. 162:344-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonhartsberger, S., A. Ehrenreich, and A. Bock. 2000. Analysis of the domain structure and the DNA binding site of the transcriptional activator FhlA. Eur. J. Biochem. 267:3672-3684. [DOI] [PubMed] [Google Scholar]
- 26.Lutz, S., A. Jacobi, V. Schlensog, R. Bohm, G. Sawers, and A. Bock. 1991. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol. Microbiol. 5:123-135. [DOI] [PubMed] [Google Scholar]
- 27.Maier, T., and A. Bock. 1996. Generation of active [NiFe] hydrogenase in vitro from a nickel-free precursor form. Biochemistry 35:10089-10093. [DOI] [PubMed] [Google Scholar]
- 28.Maupin, J. A., and K. T. Shanmugam. 1990. Genetic regulation of formate hydrogenlyase of Escherichia coli: role of the fhlA gene product as a transcriptional activator for a new regulatory gene, fhlB. J. Bacteriol. 172:4798-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menon, N. K., C. Y. Chatelus, M. Dervartanian, J. C. Wendt, K. T. Shanmugam, H. D. Peck, Jr., and A. E. Przybyla. 1994. Cloning, sequencing, and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J. Bacteriol. 176:4416-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menon, N. K., J. Robbins, H. D. Peck, Jr., C. Y. Chatelus, E. S. Choi, and A. E. Przybyla. 1990. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J. Bacteriol. 172:1969-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menon, N. K., J. Robbins, J. C. Wendt, K. T. Shanmugam, and A. E. Przybyla. 1991. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J. Bacteriol. 173:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richard, D. J., G. Sawers, F. Sargent, L. McWalter, and D. H. Boxer. 1999. Transcriptional regulation in response to oxygen and nitrate of the operons encoding the [NiFe] hydrogenases 1 and 2 of Escherichia coli. Microbiology 145:2903-2912. [DOI] [PubMed] [Google Scholar]
- 33.Rosentel, J. K., F. Healy, J. A. Maupin-Furlow, J. H. Lee, and K. T. Shanmugam. 1995. Molybdate and regulation of mod (molybdate transport), fdhF, and hyc (formate hydrogenlyase) operons in Escherichia coli. J. Bacteriol. 177:4857-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sankar, P., J. H. Lee, and K. T. Shanmugam. 1985. Cloning of hydrogenase genes and fine structure analysis of an operon essential for H2 metabolism in Escherichia coli. J. Bacteriol. 162:353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sankar, P., J. H. Lee, and K. T. Shanmugam. 1988. Gene-product relationships of fhlA and fdv genes of Escherichia coli. J. Bacteriol. 170:5440-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauter, M., R. Bohm, and A. Bock. 1992. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol. Microbiol. 6:1523-1532. [DOI] [PubMed] [Google Scholar]
- 37.Sawers, R. G., and D. H. Boxer. 1986. Purification and properties of membrane-bound hydrogenase isoenzyme 1 from anaerobically grown Escherichia coli K12. Eur. J. Biochem. 156:265-275. [DOI] [PubMed] [Google Scholar]
- 38.Schlensog, V., and A. Bock. 1990. Identification and sequence analysis of the gene encoding the transcriptional activator of the formate hydrogenlyase system of Escherichia coli. Mol. Microbiol. 4:1319-1327. [DOI] [PubMed] [Google Scholar]
- 39.Schweizer, H. P. 1993. Two plasmids, X1918 and Z1918, for easy recovery of the xylE and lacZ reporter genes. Gene 134:89-91. [DOI] [PubMed] [Google Scholar]
- 40.Self, W. T., A. M. Grunden, A. Hasona, and K. T. Shanmugam. 1999. Transcriptional regulation of molybdoenzyme synthesis in Escherichia coli in response to molybdenum: ModE-molybdate, a repressor of the modABCD (molybdate transport) operon is a secondary transcriptional activator for the hyc and nar operons. Microbiology 145:41-55. [DOI] [PubMed] [Google Scholar]
- 41.Self, W. T., A. Hasona, and K. T. Shanmugam. 2001. N-terminal truncations in the FhlA protein result in formate- and MoeA-independent expression of the hyc (formate hydrogenlyase) operon of Escherichia coli. Microbiology 147:3093-3104. [DOI] [PubMed] [Google Scholar]
- 42.Self, W. T., and K. T. Shanmugam. 2000. Isolation and characterization of mutated Fh1A proteins which activate transcription of the hyc operon (formate hydrogenlyase) of Escherichia coli in the absence of molybdate. FEMS Microbiol. Lett. 184:47-52. [DOI] [PubMed] [Google Scholar]
- 43.Skibinski, D. A., P. Golby, Y. S. Chang, F. Sargent, R. Hoffman, R. Harper, J. R. Guest, M. M. Attwood, B. C. Berks, and S. C. Andrews. 2002. Regulation of the hydrogenase-4 operon of Escherichia coli by the σ54-dependent transcriptional activators FhlA and HyfR. J. Bacteriol. 184:6642-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spiro, S., and J. R. Guest. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6:399-428. [DOI] [PubMed] [Google Scholar]
- 45.Wendt, J. C. 1989. Regulation of the hydrogen uptake (hup) gene of Escherichia coli. M.S. thesis. University of Florida, Gainesville.
- 46.Zinoni, F., A. Birkmann, T. C. Stadtman, and A. Bock. 1986. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc. Natl. Acad. Sci. USA 83:4650-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]