Abstract
Poly(A) polymerases (PAPs) are found in most living organisms and have important roles in RNA function and metabolism. Here we report the crystal structure of human PAPD1, a noncanonical PAP that can polyadenylate RNAs in the mitochondria (also known as mtPAP) and oligouridylate histone mRNAs (TUTase1). The overall structure of the palm and fingers domains is similar to that in the canonical PAPs. The active site is located at the interface between the two domains, with a large pocket that can accommodate the substrates. The structure reveals the presence of a previously unrecognized domain in the N-terminal region of PAPD1, with a backbone-fold that is similar to that of RNP-type RNA binding domains. This domain (named the RL domain), together with a β-arm insertion in the palm domain, contributes to dimerization of PAPD1. Surprisingly, our mutagenesis and biochemical studies show that dimerization is required for the catalytic activity of PAPD1.
Introduction
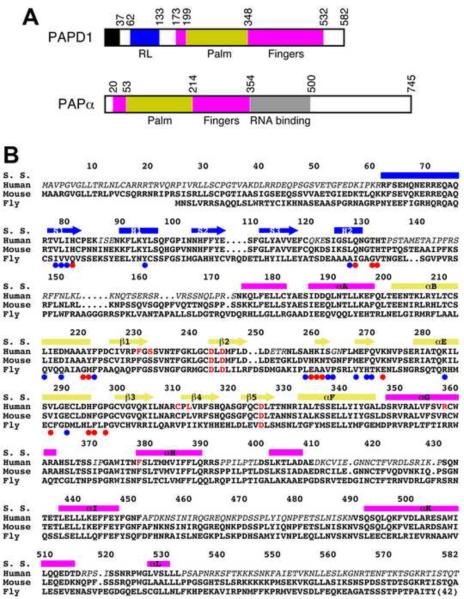
Poly(A) polymerases (PAPs) are template-independent polymerases belonging to the nucleotidyl transferase superfamily (Aravind and Koonin, 1999). They are found in the nucleus, cytoplasm, and mitochondrion in eukaryotes as well as in bacteria, underscoring their functional importance. For example, almost all eukaryotic mRNAs, with the exception of histone mRNAs, are polyadenylated during maturation in the nucleus, which is important for their stability, transport to the cytoplasm, and translation (Colgan and Manley, 1997; Edmonds, 2002; Mandel et al., 2008; Proudfoot and O'Sullivan, 2002; Vinciguerra and Stutz, 2004; Zhao et al., 1999). A canonical PAP, PAPα in humans, catalyzes the polyadenylation of these mRNA precursors. The enzyme contains three domains: an N-terminal (palm) domain, a middle (fingers) domain, and a C-terminal (RNA binding) domain (Fig. 1A) (Bard et al., 2000; Martin et al., 2000). The active site is located in the cleft between the palm domain and RNA binding domain (RBD) (Balbo and Bohm, 2007; Martin et al., 2004). Three highly conserved Asp residues in the palm domain coordinate two divalent metal ions (Mg2+ or Mn2+), which are essential for catalytic activity.
Figure 1.
Sequence conservation of PAPD1. (A). Schematic drawing of the domain organization of human PAPD1 and human PAPα. The various domains are labeled. The mitochondrial targeting sequence of PAPD1 is shown in black. (B). Sequence alignment of human, murine and Drosophila melanogaster (Fly, CG11418) PAPD1. The secondary structure elements (S. S.) are labeled and colored according to the domains. Residues shown in red are in the active site. The colored dots highlight those residues with >50 Å2 (red) or >20 Å2 (blue) buried surface area in the dimer interface. Residues missing in the current structure of human PAPD1 are shown in italic. The sequence conservation between human and Drosophila PAPD1 for residues in the RL domain (25% identity) is lower compared to that for the palm and fingers domains (39% identity).
A class of noncanonical PAPs has also been identified, including seven such enzymes in humans (Kwak and Wickens, 2007; Martin and Keller, 2007; Mullen and Marzluff, 2008; Stevenson and Norbury, 2006). For example, Star-PAP is regulated by PIP2 and targets select mRNAs in the nucleus (Mellman et al., 2008). GLD2 controls the length of poly(A) tails and regulates mRNA translation in the cytoplasm (Barnard et al., 2004). It also mono-adenylates microRNA-122, leading to its stabilization (Katoh et al., 2009). Compared to the canonical PAPs, the noncanonical enzymes lack the RNA binding domain that follows directly after the fingers domain (Fig. 1A), and it has been shown that GLD2 functions as a heterodimer with the RNA binding protein GLD3 (Kwak et al., 2004; Wang et al., 2002). Moreover, some of these noncanonical PAPs can add poly- or oligo-uridylates to their substrate RNAs, and hence are also known as poly(U) polymerases (PUPs) or terminal uridylyltransferases (TUTases or TUTs) (Kwak and Wickens, 2007; Martin and Keller, 2007; Mullen and Marzluff, 2008; Rissland et al., 2007). This modification also has important roles for RNA function and stability. Star-PAP is required for uridylation of U6 snRNA (Trippe et al., 2006), and TUTase4 regulates the biogenesis of the let-7 microRNA (Heo et al., 2009).
PAPD1 (also known as mtPAP, TUTase1) is a noncanonical PAP that polyadenylates mitochondrial RNAs (Nagaike et al., 2005; Nagaike et al., 2008; Tomecki et al., 2004). This polyadenylation is required to complete the stop codon (UAA) for some mitochondrial mRNAs, and the poly(A) tail can probably regulate RNA stability as well. PAPD1 may also be important for the maturation of mitochondrial tRNAs. In contrast to GLD2, PAPD1 appears to be active on its own and does not require a separate RNA binding protein (Nagaike et al., 2005; Tomecki et al., 2004). Consistent with its role in the mitochondrion, PAPD1 is the only PAP in humans that contains a mitochondrial targeting sequence at the N terminus (residues 1–37) (Fig. 1A). However, recent studies show that this enzyme, as well as TUTase3, can oligo-uridylate histone mRNAs for their degradation, suggesting that PAPD1 can also function as a TUTase in the cytoplasm (Mullen and Marzluff, 2008). Three single-nucleotide polymorphisms (SNPs) in PAPD1 have been linked to extreme obesity in cattle, one of which leads to a missense mutation, D39G (human PAPD1 numbering), just after the mitochondrial targeting sequence (Xiao et al., 2006).
While the canonical PAPs have been studied extensively at the structural level (Balbo and Bohm, 2007; Bard et al., 2000; Martin et al., 2000; Martin et al., 2004), currently no structural information is available on the noncanonical PAPs. In addition, the sequence conservation between the two classes of enzymes is very low, around 10–15%, despite their sharing the palm and fingers domains. We report here the crystal structure of a noncanonical PAP, human PAPD1. The structure defines the active site architecture of this enzyme, and reveals a domain (RL domain) in the N-terminal region that contributes to its dimerization and has a backbone-fold similar to RNP-type RNA binding domains. Our mutagenesis and biochemical studies unexpectedly show that this dimerization is required for the catalytic activity of PAPD1.
Results and Discussion
Structure determination
We set out to identify a derivative of human PAPD1 that would produce crystals suitable for structural studies. After screening a large number of bacterial expression constructs containing various regions of PAPD1, we were able to obtain crystals for a derivative consisting of residues 44–538. However, the quality of these crystals was poor, and the best diffraction that was observed extended to about 3.5 Å resolution. We attempted to improve the diffraction quality by crystallizing the enzyme in complex with substrates (MgATP and/or oligonucleotides), and mutated one of the conserved Asp residues in the active site, D325A, equivalent to the D154A mutation in yeast PAP that was used to obtain a structure of that enzyme in complex with MgATP and oligo(A) (Balbo and Bohm, 2007). Interestingly, the mutant protein gave better quality crystals than the wild-type enzyme, even though no nucleotides were bound, and we were able to determine its structure at 3.1 Å resolution by the selenomethionyl anomalous diffraction method (Supplementary Table 1) (Hendrickson, 1991).
The refined structure has good agreement with the crystallographic data and the expected bond lengths, bond angles, and other geometric parameters (Table 1). The two molecules of PAPD1 in the asymmetric unit have similar overall structures, with rms distance of 0.45 Å between equivalent Cα atoms, and similar temperature factor values (average B of 48 and 51 Å2 for the two molecules). The somewhat higher R factor values are due to the many poorly ordered and disordered segments in the structure (Fig. 1B). A few of the poorly ordered segments were modeled as poly-alanines (Fig. 2A), due to insufficient electron density for the side chains, while the others were too discontinuous to be modeled. This would reduce the agreement between the structure model and the crystallographic data. This problem appears to be inherent to this protein, as we refined the model against a different reflection data set, collected on another crystal at a different synchrotron source, and obtained similar R factor values.
Table 1.
Summary of crystallographic information
| Maximum resolution (Å) | 3.1 |
| Number of observations | 73,621 |
| Rmerge (%)1 | 8.0 (28.9) |
| I/σI | 9.0 (2.3) |
| Redundancy2 | 2.4 (2.0) |
| Resolution range used for refinement | 30–3.1 |
| Number of reflections2 | 28,985 |
| Completeness (%) | 92 (75) |
| R factor (%) | 24.7 (32.6) |
| Free R factor (%) | 32.8 (45.6) |
| rms deviation in bond lengths (Å) | 0.010 |
| rms deviation in bond angles (°) | 1.4 |
| Average temperature factor value (Å2) | 50 |
| Number of protein atoms | 5,353 |
| Number of solvent atoms | 0 |
The numbers in parentheses are for the highest resolution shell. See also Table S1.
The Friedel pairs are kept as separate reflections.
Figure 2.
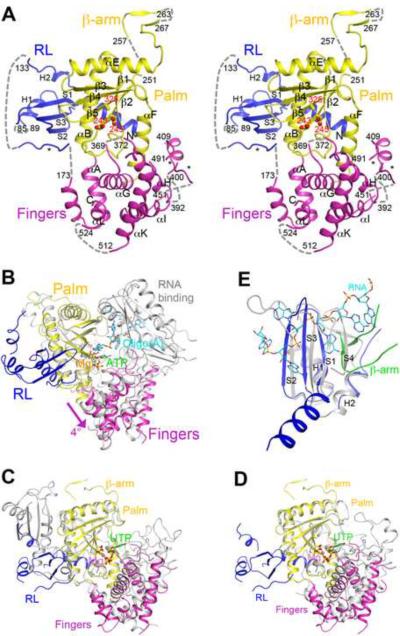
Structure of human PAPD1 monomer. (A). Stereo drawing of the structure of human PAPD1 monomer. The two catalytic Asp residues and the D325A mutation in the active site are shown in stick models and labeled in red. Three segments modeled as poly-alanines are indicated with the stars. The missing segments of the structure are indicated with the dashed lines (gray). (B). Overlay of the structure of human PAPD1 (in color) with that of yeast PAP (in gray) in complex with Mg2+ (pink sphere), ATP (green) and oligo(A) (cyan) (Balbo and Bohm, 2007). The fingers domain of PAPD1 is in a slightly more open conformation compared to that in PAP, indicated with the arrow (magenta). (C). Overlay of the structure of human PAPD1 (in color) and T. brucei TUTase2 (in gray) (Deng et al., 2005). The inserted domain in the palm domain of TUTase2 is in a different location compared to the RL domain of PAPD1. (D). Overlay of the structure of human PAPD1 (in color) and T. brucei TUTase4 (in gray) (Stagno et al., 2007a; Stagno et al., 2007b). For stereo versions of panels C and D, please see Supplementary Fig. 3. (E). Overlay of the RL domain of human PAPD1 (in blue) with the first RRM of PABP (in gray) in complex with oligo(A) (cyan) (Deo et al., 1999). The β-arm of the second monomer of PAPD1 is shown in green, and it clashes with the fourth strand of the RRM. All the structure figures were produced with PyMOL (www.pymol.org). See also Figs. S1–3.
Overall structure of PAPD1 monomer
The structure of PAPD1 contains three domains: the canonical palm and fingers domains and a previously unrecognized domain in the N-terminal region (Fig. 2A). Residues 199–347 form the palm domain, and the three catalytic aspartic acid residues (Asp243, Asp245, and Asp325, with the Asp325 residue mutated to Ala in the current structure) are located on the surface of the five-stranded β-sheet in this domain (Fig. 2A). A simulated-annealing omit difference electron density map for this region is shown in Supplementary Fig. 1. The structure reveals that residues 253–273 form a β-hairpin that protrudes far away from the rest of the monomer. This structural feature is involved in dimerization of PAPD1 (see below), and we have named it the β-arm (Fig. 2A).
Residues 173–198 and 348–532 form the fingers domain (Fig. 2A). While the helices in this domain have good electron density, many of the connecting loops are poorly ordered or disordered in the crystal, even though their sequences are quite conserved among PAPD1 orthologs (Fig. 1B). These disordered loops may be important for the function of PAPD1. Two of these loops were modeled as poly-alanines (Fig. 2A), guided by the structure of yeast PAP (Balbo and Bohm, 2007). Residues 533–538 at the C-terminus of this derivative were disordered in the crystal. In fact, secondary structure predictions suggest that the entire C-terminal segment of PAPD1, residues 533–582, is unstructured, and PAPD1 does not have an RNA binding domain directly following the fingers domain (Fig. 1A).
The overall structure of the palm and fingers domains of PAPD1 is similar to that in the canonical PAPs, despite their low sequence conservation (Supplementary Fig. 2). Compared to the structure of yeast PAP in complex with MgATP and oligo(A) (Balbo and Bohm, 2007), the two domains are somewhat more open relative to each other, corresponding to a rotation of approximately 4° (Fig. 2B). Substrate binding is expected to lead to a more closed conformation for PAPD1, as has been observed for yeast PAP (see below). Canonical PAP in the absence of substrates assumes a much more open conformation (Bard et al., 2000; Martin et al., 2000), and undergoes large conformational changes upon substrate binding (Balbo and Bohm, 2007). In contrast, PAPD1 assumes a mostly closed conformation even in the absence of substrates, and may have a smaller conformational change upon substrate binding.
The overall structure of the palm and fingers domains of PAPD1 is also similar to that of the RNA editing TUTase2 (Fig. 2C, Supplementary Fig. 3) (Deng et al., 2005) and the minimal TUTase4 (Fig. 2D, Supplementary Fig. 3) (Stagno et al., 2007a; Stagno et al., 2007b) from T. brucei, even though these proteins also share less than 20% sequence identity with PAPD1 (Supplementary Fig. 2). Like PAPD1, these two TUTases do not contain an RNA binding domain following the fingers domain.
A separate domain in the N-terminal region of PAPD1
Unexpectedly, the structure of PAPD1 shows that residues 62–133 in the N-terminal region forms a separate domain (Fig. 2A) with a backbone fold that is similar to RNP-type RNA binding domains (RBDs, or RNA recognition motifs, RRMs), including that in canonical PAPs, as indicated by a search through the Protein Data Bank with the program DaliLite (Holm et al., 2008). The sequence conservation among these proteins is very low, 10–20% amino acid identity. The closest structural homolog is the poly(A) binding protein (PABP) (Deo et al., 1999), with a Z score of 5.5 (Fig. 2E). This domain is connected to the rest of PAPD1 by a 40-residue linker (Fig. 1B), a segment of which is modeled as poly-alanines in one monomer (Fig. 2A), partially stabilized by crystal packing. In the other monomer, this linker is disordered.
Residues 62–133 of PAPD1 forms a 3-stranded anti-parallel β-sheet with two α-helices on one face, and the β-arm from the other monomer of the PAPD1 dimer adds two more strands, making a five-stranded β-sheet for this domain (Fig. 2E, see below). Most RBDs contain a 4-stranded β-sheet with two helices on one face, which roughly match the first four strands of the β-sheet and the two helices in PAPD1 (Fig. 2E). However, the fourth strand in PAPD1 (from the β-arm of the other monomer) runs parallel to the first strand, while it runs anti-parallel to the first strand in the RBDs (Fig. 2E). A 5-stranded β-sheet has been observed in a few of the other RBDs, for example the RRM2 and RRM3 of polypyrimidine tract binding protein (PTB) (Conte et al., 2000; Oberstrass et al., 2005). However, the fifth strand is located at the other edge of the β-sheet in these two RRMs, next to the second strand, and so the domain in PAPD1 is quite different from these RRMs.
While this domain in PAPD1 has structural similarity to an RBD, it is not known whether it can actually bind RNA. In its current conformation in the structure, the domain is positioned about 40 Å from the active site, on the opposite face of the palm domain (Fig. 2A), and therefore cannot directly contribute to RNA binding in the active site. Moreover, the open face of the β-sheet is generally involved in RNA binding in related domains, such as PABP (Fig. 2E) (Deo et al., 1999), U1A (Oubridge et al., 1994), the AU-rich element binding protein Hrp1 (Perez-Canadillas, 2006), and the RRM in CstF-64 (Canadillas and Varani, 2003). In PAPD1, however, this face and an extra α-helix at the N-terminus of the domain are in tight contact with the palm domain (Fig. 2A), mediated predominantly by hydrophobic residues. The amino acid sequences of the first and third strands do not contain the RNP1 and RNP2 sequence motifs for RNA binding (Burd and Dreyfuss, 1994; Kielkopf et al., 2004; Query et al., 1989). In yeast PAP, on the other hand, the helices of the domain contribute to binding the oligo(A) RNA, while the open face of the β-sheet is not involved in binding (Fig. 2B) (Balbo and Bohm, 2007). These helices are exposed to the solvent in PAPD1 (Fig. 2A), although they do not have strongly electro-positive surface patches that could mediate interactions with the RNA phosphate backbone.
The structural analysis therefore suggests that this separate domain in PAPD1 may be primarily involved in protein-protein interactions rather than substrate binding and catalysis, and we have named it the RL (RBD-like) domain. Similar observations have been made on other RBDs, such as that in the splicing factor U2AF (Kielkopf et al., 2004; Kielkopf et al., 2001) and the 68 kD subunit of the pre-mRNA 3'-end processing factor CF-Im (Brown and Gilmartin, 2003; Dettwiler et al., 2004). Due to the long linker between this domain and the rest of the structure, we cannot exclude the possibility of a large conformational change that could bring this domain close to the active site. However, such a change will disrupt the dimer (see below) as well as expose a large hydrophobic surface patch on the palm domain.
In the structure of T. brucei TUTase2, the palm domain carries an inserted domain with a backbone fold similar to RBDs, though the function of this domain is not known (Deng et al., 2005). The location of this domain in TUTase2 is different from the RBD in PAPD1 (Fig. 2C, Supplementary Fig. 3).
The active site of PAPD1
The active site of PAPD1 is located in a large pocket at the interface between the palm and fingers domains (Fig. 3A). As our attempts to obtain the structure of a substrate complex were not successful, we modeled the binding modes of MgATP and the last nucleotide (the −1 nucleotide) of the RNA substrate based on the observations in yeast PAP (Balbo and Bohm, 2007). Both nucleotides can be readily accommodated in the pocket in this model (Fig. 3A), suggesting that the active site is formed properly in PAPD1. This is consistent with biochemical studies showing that PAPD1 can carry out catalysis on its own, in the absence of another RNA binding protein (Nagaike et al., 2005). The RBD of yeast PAP interacts with the other nucleotides in the RNA substrate (Fig. 2B), and is not involved in the recognition of MgATP and the −1 nucleotide of the RNA (Balbo and Bohm, 2007).
Figure 3.
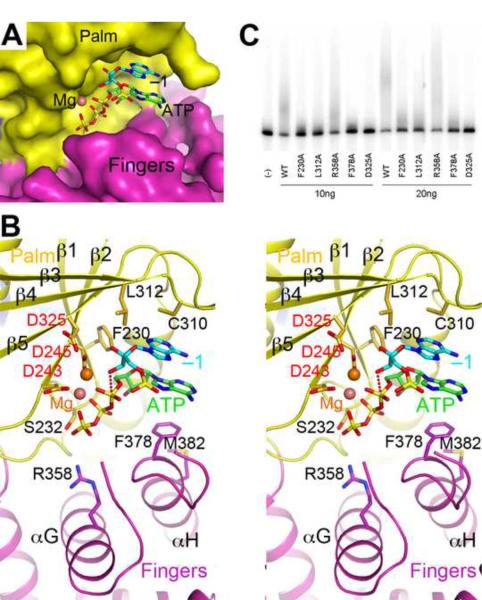
Model of the active site of human PAPD1 with substrates. (A). Molecular surface of the active site region of PAPD1. The bound positions of Mg2+ (pink sphere), ATP (green) and the last nucleotide of the RNA substrate (cyan, labeled −1) are modeled from the structure of yeast PAP (Balbo and Bohm, 2007). (B). Stereo drawing of the active site model of human PAPD1. Side chains with potentially important roles in catalysis and/or substrate binding are shown as stick models and labeled. The side chain of Asp325 and the two Mg2+ ions are modeled. The inline nucleophilic attack of the 3' hydroxyl group of the last nucleotide of the RNA substrate (the −1 nucleotide) on the α-phosphate of ATP, modeled based on the structure of yeast PAP (Balbo and Bohm, 2007), is indicated by the dashed line in red. (C). Poly(A) polymerase assays for active site mutants of PAPD1. 10 or 20 ng of each enzyme was used in the assay, and the D325A mutant was included as a control.
Residues that have important roles in catalysis and/or interactions with the substrates are highly conserved among PAPD1 orthologs (Fig. 1B). Many of these residues are also conserved in yeast PAP (Balbo and Bohm, 2007) as well as the T. brucei TUTases (Deng et al., 2005; Stagno et al., 2007a; Stagno et al., 2007b) (Supplementary Fig. 2). The three catalytic Asp residues in the palm domain coordinate two metal ions and activate the 3'-hydroxyl group of the −1 nucleotide, which is in the correct position for the inline attack on the α-phosphate of ATP to initiate the reaction (Fig. 3B). The model suggests that the phosphate groups of ATP may also interact with the main-chain amide and side-chain hydroxyl groups of Ser232 (just after β1 of palm domain) and the side chain of Arg358 (αG of fingers domain). The −1 nucleotide is also located near the side chains of Phe230 and Leu312 (Fig. 3B).
The adenine base of ATP is π-stacked with that of the −1 nucleotide (Fig. 3B). The other face of this adenine is located near Phe378 (just prior to αH of fingers domain) in the model. It is expected that with a more closed conformation for the fingers domain upon substrate binding, the adenine base of ATP and the Phe side chain could be in π-stacking interactions, as was observed in the TUTases from T. brucei, where a Tyr residue is conserved at this position (Deng et al., 2005; Stagno et al., 2007a; Stagno et al., 2007b). Yeast PAP has a Val residue at this position, and its side chain has van der Waals interactions with the adenine base.
To assess the importance of these residues for the catalytic activity of PAPD1, we created the F230A, L312A, R358A and F378A mutants and characterized their poly(A) polymerase activity. The results of the assays demonstrated that the F230A, L312A and F378A mutants had greatly reduced PAP activity (Fig. 3C), confirming their functional importance and the relevance of our model for the active site of PAPD1. On the other hand, the R358A mutant had little effect on activity, indicating that this side chain is not important for catalysis.
PAPD1 can use all four nucleotides as substrates in vitro
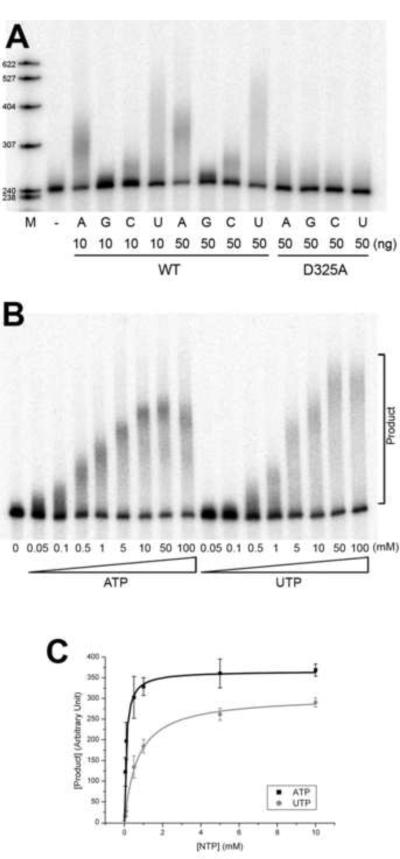
The adenine base of ATP does not appear to be specifically recognized by PAPD1. In the current structure, none of the residues of the enzyme are located within hydrogen-bonding distances of the base. Our in vitro studies in fact suggest that PAPD1 can utilize all four nucleotides as substrates, although it is more active with ATP or UTP (Fig. 4A). Therefore, PAPD1 does not have a strict substrate specificity towards the nucleotide, which is consistent with the structural observation. The lowest activity was observed with GTP, and PAPD1 added only a short oligo(G) tail to the RNA in these assays (Fig. 4A). The in vitro data are also consistent with observations in vivo on the composition of mitochondrial mRNA tails, which indicated a predominance of A with rare occurrences of G and C (Tomecki et al., 2004).
Figure 4.
PAPD1 has activity towards all four nucleotides in vitro. (A). Catalytic activity of PAPD1 towards the four nucleotides, ATP, GTP, CTP and UTP. 10 and 50 ng of the wild-type (WT) enzyme were used, and 50 ng of the D325A mutant were also assayed as a control. (B). Concentration titration of the activity of PAPD1 towards ATP and UTP. The concentrations used for the nucleotides are indicated. (C). The poly(A) and poly(U) polymerase reaction of PAPD1 follows Michaelis-Menten kinetics. The error bars represent ± 1SD.
To characterize further the catalytic activity of PAPD1 towards ATP and UTP, we determined the kinetic parameters for these substrates from a concentration titration (Fig. 4B). The reactions followed Michaelis-Menten kinetics, and Km values for ATP and UTP were calculated to be 0.1 and 0.7 mM, respectively (Fig. 4C). The Vmax values for ATP and UTP were 370 and 310 (arbitrary unit), respectively. Overall, the kinetic analysis suggests that the activity towards ATP is nine-fold higher than UTP (based on Vmax/Km values) in this in vitro assay. The reaction contained 20 nM PAPD1 and approximately the same concentration of the RNA substrate.
Studies with the T. brucei TUTases suggest that water-mediated hydrogen-bonds may be important for substrate selectivity (Stagno et al., 2007a; Stagno et al., 2007b). Studies with yeast PAP and other polymerases show that an induced-fit behavior of the enzymes may also be important for substrate selectivity (Balbo and Bohm, 2007; Balbo et al., 2005; Balbo et al., 2007; Doublie et al., 1999). On the other hand, PAPD1 appears to have both PAP and TUTase activity (Mullen and Marzluff, 2008; Nagaike et al., 2005; Tomecki et al., 2004) in vivo. It remains to be seen how the enzyme is selective for ATP in the mitochondrion but UTP in the cytoplasm.
A dimer of PAPD1
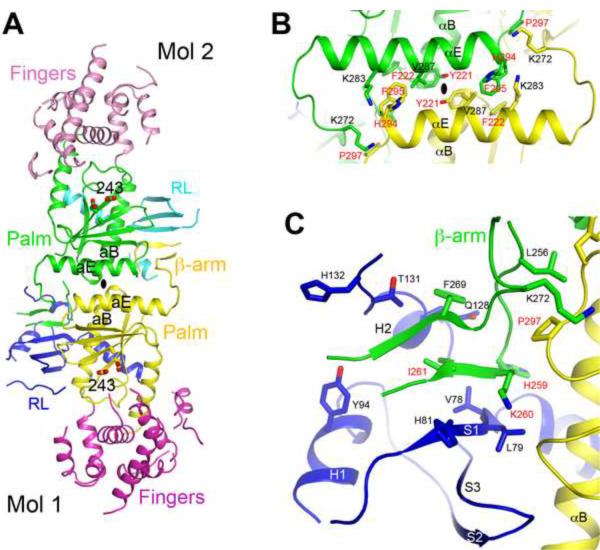
The crystal structure of human PAPD1 reveals a dimer, formed by the two molecules in the asymmetric unit (Fig. 5A). This is the only tight association among PAPD1 molecules in the crystal. Dimerization of PAPD1 was originally suggested by results from gel-filtration chromatography, during purification of the recombinant protein. After determination of the structure, we further confirmed this dimerization using solution light scattering experiments (Supplementary Fig. 4). This appears to be a unique property of PAPD1, as the canonical PAPs as well as the T. brucei TUTases are monomeric in solution. A different nucleotidyl transferase, the A. fulgidus tRNA CCA-adding enzyme, is also a dimer (Tomita et al., 2006; Xiong and Steitz, 2004), though it bears no similarity to the dimer of PAPD1. The active site of PAPD1 is located approximately 25 Å from the dimer interface, with no contributions from residues in the other monomer of the dimer (Fig. 5A).
Figure 5.
The dimer of human PAPD1. (A). Schematic drawing of the human PAPD1 dimer. The domains in the two monomers are given different colors and labeled, and Mol 1 is the one shown in Fig. 2A. The catalytic Asp residues in the active sites are shown in stick models. The two-fold axis of the dimer is indicated with the oval in black. (B). Detailed interactions between helices αE and αB in one contact area of the dimer interface. Residues labeled in red have been selected for mutagenesis experiments. (C). Detailed interactions between the RL domain of one monomer and the β-arm of the other monomer in the other contact area of the dimer interface. See also Fig. S4.
The PAPD1 dimer buries approximately 1,900 Å2 of the surface area of each monomer, suggesting that the dimer is likely stable, consistent with our studies in solution. Residues in the dimer interface are generally conserved among PAPD1 orthologs (Fig. 1B), suggesting that they may also be dimers. The dimer is formed through two major areas of contact between the monomers. In one area, the αE helix in the palm domain of one monomer is located next to its equivalent in the other monomer, and the C-terminal ends of the αB helices of the two monomers are positioned close together as well (Fig. 5A). This area of contact consists primarily of van der Waals interactions. Residues His294 (with 80 Å2 of surface area burial), Phe295 (100 Å2) and Pro297 (80 Å2) near the end of helix αE of one monomer are located near residues Lys272 (110 Å2) and Lys283 (40 Å2) near the beginning of helix αE and residue Phe222 (20 Å2) near the end of helix αB of the other monomer (Fig. 5B). In addition, residue Tyr221 (60 Å2) interacts with its symmetry-mate across the two-fold axis of the dimer.
In the other area of contact, the β-arm from one monomer is placed next to the RL domain of the other monomer, forming a five-stranded β-sheet (Fig. 5C). Besides the main-chain hydrogen-bonding interactions in the formation of this larger β-sheet, van der Waals interactions for several side chains are also important for this part of the dimer interface. Residues His259 (150 Å2), Lys260 (100 Å2) and Ile261 (80 Å2) in the first strand of the β-arm contribute the most to the buried surface area in this region (Fig. 1B).
Dimerization is required for PAPD1 activity
To assess the structural observations on the PAPD1 dimer, we introduced mutations in the interface and characterized their effects on dimerization. Three groups of site-specific mutations, Y221A/F222A (helix αB), H294A/F295A/P297A (αE), and H259A/K260A/I261A (β-arm), were selected based on the structural information. Gel-filtration experiments showed that the Y221A/F222A and H294A/F295A/P297A mutants existed in a monomer-dimer equilibrium (data not shown), suggesting that the mutations have destabilized the dimer. In comparison, the H259A/K260A/I261A mutant remained dimeric, consistent with the structural observation that the main-chain atoms of these residues in the β-arm also make important contributions to the dimer interface (Fig. 5C).
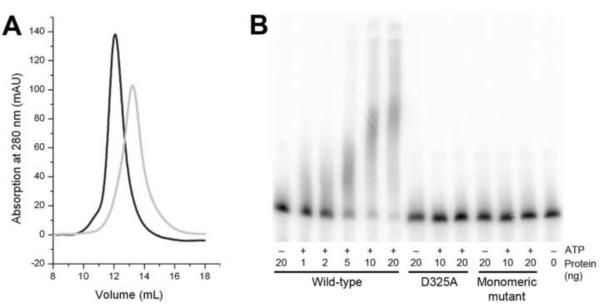
We next created combinations of the three groups of mutations and found that the H259A/K260A/I261A/H294A/F295A/P297A sextuple mutant, which simultaneously disrupted both areas of contact in the dimer interface, was a stable monomer in solution (Fig. 6A), suggesting that the mutation did not disrupt the overall structure of the protein. In comparison, the other combinations produced mutants that were unstable in solution, indicated by the presence of a large amount of aggregates in gel-filtration chromatography (data not shown). Similarly, deletion mutants lacking the entire N-terminal RL domain or the β-arm were also found to be unstable in solution. Overall, the mutagenesis experiments confirmed the structural observations and the stability of the PAPD1 dimer.
Figure 6.
Dimerization is required for the catalytic activity of PAPD1. (A). Gel-filtration data showing that a sextuple mutation in the dimer interface, H259A/K260A/I261A/H294A/F295A/P297A, produces a stable monomer in solution. The profile for wild-type PAPD1 is shown in black, and that for the mutant in gray. The void volume on this column is at 7.8 ml, aldolase (158 kD) at 12.1 ml, and albumin (67 kD) at 13.4 ml. (B). Poly(A) polymerase assay for wild-type PAPD1 (residues 44–538), the D325A active-site mutant, and the sextuple mutant H259A/K260A/I261A/H294A/F295A/P297A (labeled monomeric mutant).
To assess the functional importance of PAPD1 dimerization, we carried out poly(A) polymerase assays. Wild-type PAPD1 (residues 44–538) showed robust PAP activity in the presence of Mg2+ and especially Mn2+ cations (Fig. 6B). Mutation of one of the conserved Asp residues in the active site, D325A, abolished activity (Fig. 6B). Surprisingly, the sextuple mutant, H259A/K260A/I261A/H294A/F295A/P297A, which forms a stable monomer in solution, also failed to show any PAP activity (Fig. 6B), indicating that dimerization is essential for PAPD1 function. It is also unlikely that the mutant can have activity towards the UTP substrate.
As the active site of PAPD1 is located far from the dimer interface (Fig. 5A), it is not clear why dimerization is required for its PAP activity. One possibility is that dimerization could be important for substrate binding. Electrophoretic mobility shift assays were not able to detect interactions between wild-type or monomeric mutant PAPD1 and the RNA substrate at concentrations used in the poly(A) polymerase assay (data not shown). Binding was observed at much higher concentrations of the wild-type and mutant enzymes, but this was probably non-specific and not catalytically productive. On the other hand, some of the residues in the dimer interface are located in the catalytic palm domain (Fig. 5A). Dimerization could indirectly stabilize the conformation of the active site region, while monomers may have detrimental changes in the active site.
In summary, we have produced the structure of a noncanonical poly(A) polymerase, human PAPD1. The structure revealed the enzyme active site as well as a RBD-like (RL) domain in the N-terminal region that contributes to dimerization of the enzyme. Mutagenesis and kinetic studies showed unexpectedly that dimerization is essential for PAP activity. Our observation of a properly formed active site in PAPD1 also has implications for GLD2, Star-PAP and other noncanonical PAPs. In addition, most of these noncanonical enzymes contain additional sequences in the N-terminal region, prior to the palm and fingers domains. In fact, Star-PAP has an authentic RBD in this region, which is likely involved in RNA binding. Divergent RBD-like domains in this region in the other enzymes could mediate dimerization, as observed for PAPD1, or other protein-protein interactions, for example recruitment of a separate RNA binding protein.
Experimental procedure
Protein expression and purification
Residues 44–538 of human PAPD1 were subcloned into the pET28a vector (Novagen). The expression construct contained an N-terminal hexa-histidine tag, which was not removed for crystallization. The D325A mutant was created with the QuikChange kit (Stratagene). The native protein (wild-type and D325A mutant) was over-expressed overnight in E. coli BL21 Rosetta (DE3) cells (Novagen) at 20°C in the presence of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Gold Biotechnology, Inc). The soluble protein was eluted from nickel affinity beads (Qiagen) by a buffer containing 50 mM Tris (pH 8.5), 300 mM NaCl and 250 mM imidazole. The eluted protein was further purified by gel-filtration chromatography and then concentrated to 15 mg/mL in a buffer containing 50 mM Tris (pH 8.5), 300 mM NaCl and 5 % (v/v) glycerol. Purified protein samples were flash frozen with liquid nitrogen and then stored at −80°C.
The selenomethionyl (SeMet) protein was produced in E. coli BL21 Rosetta (DE3) cells grown in defined M9 media supplemented with selenomethionine and specific amino acids to block endogenous methionine biosynthesis (Doublie et al., 1996). The SeMet protein was purified with the same protocol as the native protein, and was concentrated to 35 mg/mL.
The oligomerization state of wild-type PAPD1 was analyzed by size exclusion chromatography followed by static multi-angle light scattering experiments. Gel filtration using a Superose-12 column (GE Healthcare) was also used to analyze the solution behavior of the protein samples.
Protein crystallization
Crystals of the SeMet D325A mutant were obtained at 20°C by the microbatch method under paraffin oil (Hampton Research). The precipitant solution contained 100 mM MES (pH 6.0), 6% (v/v) PEG4000, 35 mM NaCl, and 5 mM MgSO4. The protein was at 20 mg/mL concentration, and 0.5 mM MgATP was included as an additive. For cryo-protection, the concentration of PEG4000 in the precipitant solution was raised to 30%, and the crystals were flash frozen in liquid nitrogen for diffraction analysis and data collection at 100 K.
Data collection and processing
A selenomethionyl multi-wavelength anomalous diffraction (MAD) data set to 3.5 Å resolution and a selenomethionyl single-wavelength anomalous diffraction (SAD) data set to 3.1 Å resolution were collected on an ADSC Quantum4 CCD at the X4A beamline of the National Synchrontron Light Source (NSLS) at Brookhaven National Laboratory. The diffraction images were processed using the HKL package (Otwinowski and Minor, 1997). The crystal belongs to space group P21, with unit cell parameters of a=68.7 Å, b=76.9 Å, c=87.6 Å, and β=103.4°. There are two molecules of PAPD1 in the crystallographic asymmetric unit. The data processing statistics are summarized in Table 1 for the SAD data and Supplementary Table 1 for the MAD data.
Structure determination and refinement
The structure of PAPD1 was determined by the multi-wavelength anomalous diffraction method using the anomalous signal of Se (Hendrickson, 1991). Twelve out of the fourteen expected Se sites in the asymmetric unit were located using the program SOLVE and the reflection phases were calculated and improved with the program SOLVE/RESOLVE (Terwilliger, 2003). The overall figure of merit from the MAD phasing was 0.39 (Supplementary Table 1). The initial model for one molecule was built with the program O (Jones et al., 1991) and Coot (Emsley and Cowtan, 2004). The locations of the Se sites, as well as comparison to the structure of yeast PAP (Balbo and Bohm, 2007), guided the tracing of electron density. Residue Met138 is disordered in both monomers, explaining the two missing Se sites. The second molecule in the asymmetric unit was located by molecular replacement using the program COMO (Jogl et al., 2001). The structure refinement was carried out with the program CNS (Brunger et al., 1998) and Phenix (Adams et al., 2002). We also tried the Refmac refinement program (Murshudov et al., 1997), but could not reduce the R factor further. The statistics on the structure refinement are summarized in Table 1. No solvent molecules were included in the atomic model.
Several segments of the structure were modeled as poly-alanines (Fig. 2A), as there was insufficient side chain electron density for the residues. For the segments in the fingers domain, the location of the residues was guided by the yeast PAP structure (Balbo and Bohm, 2007). For the linker between the N-terminal domain and the palm domain, electron density was recognized in only one of the two molecules (Fig. 2A). These residues were assigned arbitrary residue numbers of greater than 1000, to distinguish them from the natural sequence. The average temperature factor value (50 Å2) for these poly-alanine residues was comparable to that for the rest of the protein. If these residues were removed from the atomic model, both the R and free R factor values would increase by 1%.
Polyadenylation assay
The plasmid used was derived from pG3SVL-A, containing the SV40 late polyadenylation site. Pre-cleaved mRNA substrate uniformly labeled with α-32P-UTP was prepared by in vitro transcription using SP6 RNA polymerase (Promega) from the plasmid linearized by HpaI (Ryan et al., 2004; Takagaki et al., 1988). Polyadenylation reaction was carried out at 30°C for 30 minutes. Each reaction contained 1–20 ng of wild-type or D325A mutant protein in 15 μL of reaction buffer, which was composed of ~100 count/second labeled pre-mRNA substrate, 50 mM Tris (pH 8.0), 40 mM KCl, 1 mM DTT, 0.1 mM ATP, 0.5 mM MgCl2, 0.1 mM MnCl2, and 2 units of RNase inhibitor (Promega). RNA products were fractionated on 6% polyacrylamide gel in 8.3 M urea and visualized by PhosphorImager.
Electrophoretic mobility shift assay (EMSA)
Various amounts of wild-type and mutant PAPD1 were mixed with body-labeled pre-mRNA substrate (~100 count/second), incubated at room temperature for 15 min, and then fractionated on 6% native polyacrylamide gel. The data were visualized by PhosphorImager.
Supplementary Material
Acknowledgements
We thank Farhad Forouhar and Jayaraman Seetharaman for help with data collection at the synchrotron source; Randy Abramowitz and John Schwanof for access to the X4A beamline at the NSLS; Dafne Campigli Di Giammartino for help with the polyadenylation assays; Kehui Xiang, Takashi Nagaike and Andrew Bohm for helpful discussions. This research is supported in part by grants from the NIH to LT (GM077175) and JLM (GM028983).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data deposition: The atomic coordinates and structure factors have been deposited at the Protein Data Bank, with the accession code 3PQ1.
References
- Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building a new software for automated crystallographic structure determination. Acta Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. DNA polymerase b-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucl Acid Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo PB, Bohm A. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure. 2007;15:1117–1131. doi: 10.1016/j.str.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo PB, Meinke G, Bohm A. Kinetic studies of yeast polyA polymerase indicate an induced fit mechanism for nucleotide specificity. Biochem. 2005;44:7777–7786. doi: 10.1021/bi050089r. [DOI] [PubMed] [Google Scholar]
- Balbo PB, Toth J, Bohm A. X-ray crystallographic and steady state fluorescence characterization of the protein dynamics of yeast polyadenylate polymerase. J Mol Biol. 2007;366:1401–1415. doi: 10.1016/j.jmb.2006.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard J, Zhelkovsky AM, Helmling S, Earnest TN, Moore CL, Bohm A. Structure of yeast poly(A) polymerase alone and in complex with 3'-dATP. Science. 2000;289:1346–1349. doi: 10.1126/science.289.5483.1346. [DOI] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3' processing by human cleavage factor Im. Mol Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Canadillas JMP, Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003;22:2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Develop. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Conte MR, Grune T, Ghuman J, Kelly G, Ladas A, Matthews S, Curry S. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 2000;19:3132–3141. doi: 10.1093/emboj/19.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Ernst NL, Turley S, Stuart KD, Hol WGJ. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SML. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- Doublie S, Kapp U, Aberg A, Brown K, Strub K, Cusack S. Crystallization and preliminary X-ray analysis of the 9 kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 1996;384:219–221. doi: 10.1016/0014-5793(96)00316-x. [DOI] [PubMed] [Google Scholar]
- Doublie S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure. 1999;7:R31–R35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- Edmonds M. A history of poly A sequences: from formation to factors to function. Prog Nucl Acid Res Mol Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan KD. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim Y-K, Ha M, Yoon M-J, Cho J, Yeom K-H, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogl G, Tao X, Xu Y, Tong L. COMO: A program for combined molecular replacement. Acta Cryst. 2001;D57:1127–1134. doi: 10.1107/s0907444901006783. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S-I, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Develop. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Develop. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci USA. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:830–837. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3'-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W. RNA-specific ribonuleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W, Doublie S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Moglich A, Keller W, Doublie S. Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J Mol Biol. 2004;341:911–925. doi: 10.1016/j.jmb.2004.06.047. [DOI] [PubMed] [Google Scholar]
- Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'. Genes Develop. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Ueda T. Polyadenylation in mammalian mitochondria: insights from recent studies. Biochim Biophys Acta. 2008;1779:266–269. doi: 10.1016/j.bbagrm.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Perez-Canadillas J-M. Grabbing the message: structural basis of mRNA 3' UTR recognition by Hrp1. EMBO J. 2006;25:3167–3178. doi: 10.1038/sj.emboj.7601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, O'Sullivan J. Polyadenylation: a tail of two complexes. Curr Biol. 2002;12:R855–R857. doi: 10.1016/s0960-9822(02)01353-2. [DOI] [PubMed] [Google Scholar]
- Query CC, Bentley RC, Keene JD. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3' processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proc Natl Acad Sci USA. 2007a;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and apo structures of a minimal RNA uridylyltransferase. J Mol Biol. 2007b;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson AL, Norbury CJ. The CidI family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a Poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. SOLVE and RESOLVE: Automated structure solution and density modification. Meth Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucl Acid Res. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Ishitani R, Fukai S, Nureki O. Complete crystallographic analysis of the dynamics of CCA sequence addition. Nature. 2006;443:956–960. doi: 10.1038/nature05204. [DOI] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke B-J. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P, Stutz F. mRNA export: an assembly line from genes to nuclear pores. Curr Opin Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Xiao Q, Wu X-L, Michal JJ, Reeves JJ, Busboom JR, Thorgaard GH, Jiang Z. A novel nuclear-encoded mitochondrial poly(A) polymerase PAPD1 is a potential candidate gene for the extreme obesity related phenotypes in mammals. Int J Biol Sci. 2006;2:171–178. doi: 10.7150/ijbs.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature. 2004;430:640–645. doi: 10.1038/nature02711. [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore CL. Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.