Abstract
Signals through the pre-B cell antigen receptor (pre-BCR) and IL-7 receptor (IL-7R) coordinate pre-B cell expansion with subsequent Igκ recombination. While many downstream effectors of each receptor are known, how they integrate to mediate development has remained unclear. Herein, we report that pre-BCR mediated activation of the Ras/MEK/ERK signaling pathway silences Ccnd3 transcription and coordinates cell cycle exit with the induction of E2A and the initiation of Igκ recombination. These activities are opposed by IL-7R mediated STAT 5 activation which promotes Ccnd3 expression and concomitantly inhibits Igκ transcription by binding to Eκi and inhibiting E2A recruitment. Our data reveal how pre-BCR signaling poises pre-B cells to undergo differentiation upon escape from IL-7R signaling.
INTRODUCTION
Following commitment to the B cell lineage, pro-B cells initiate recombination of the immunoglobulin heavy chain variable (V) to the diversity (D) and joining (J) gene segments. In-frame rearrangements allow the expression of Igµ which then assembles with Igα/Igβ, and the surrogate light chain complex (SLC, λ5 and VpreB), to form the pre-B cell antigen receptor (pre-BCR)1,2. Initial signals transmitted through the pre-BCR synergize with those elicited through the IL-7 receptor (IL-7R) to expand the B cell progenitor pool 3−5.
For subsequent development, pre-B cells must cease dividing and then initiate light chain recombination, primarily at the Igκ locus3,4. These two fundamental processes are, by necessity, sequential as concurrent replication and the introduction of double-strand breaks during recombination would compromise genomic integrity6. While it is clear that the pre-BCR and the IL-7R control proliferation and recombination, it is not known how receptor initiated signals integrate to affect this developmental transition. It has been hypothesized that proliferating pre-B cells exit cell cycle upon termination of SLC expression7,8. In this model, it is the absence of pre-BCR signals that mediate cell cycle exit. However, recent experiments in which SLC was constitutively expressed indicate that down-regulation of the pre-BCR is not required to either limit proliferation or to initiate Igκ rearrangement9. Furthermore, loss of downstream components of the pre-BCR signaling cascade, including SLP-65 and PLCγ2, arrests development at the cycling pre-BCR+ stage10−12. These observations indicate that the signaling mechanisms limiting proliferation are intrinsic to the pre-BCR.
Cessation of cell cycle in pre-B cells is not sufficient to induce Igκ recombination13. Rather, Igκ recombination appears to be directly regulated by the pre-BCR. Transgenic Igm expression increases Igκ locus accessibility in recombinase activating gene 2 (Rag2−/−) pro-B cells14 while targeted mutation of some pre-BCR signaling cascade components both enhances proliferation and attenuates Igκ recombination11,15. These latter observations implicate common proximal signaling molecules in Igκ recombination and cell cycle exit.
The signaling intermediate Ras has been implicated in B lymphopoiesis. Expression of dominant negative (DN) Ras results in a developmental block at the pro-B cell stage, before expression of the pre-BCR16. In contrast, expression of constitutively active (CA) Ras in Rag1 deficient mice induces a unique population of cells expressing markers of later B cell development17. These latter results indicate that downstream of the pre-BCR, Ras is an important mediator of developmental progression. Consistent with this conclusion, expression of CA-Ras in JH−/− mice induces extensive Igκ rearrangement18. The molecular mechanisms by which Ras contributes to Igκ rearrangement are not known. Furthermore, the relationships between Ras mediated Igκ recombination and cell cycle regulation have not been explored.
Downstream of the pre-BCR, the interferon regulatory factor family members, IRF4 and IRF8, are required in vivo for initiating light chain recombination, suppressing SLC and exiting cell cycle19,20. Culture of Irf4−/−Irf8−/− bone marrow with IL-7 yields populations of actively cycling pre-BCR+ cells while reintroduction of IRF4, in the presence of IL-7, silences SLC20 and induces Igκ recombination21,22. These and other observations23 demonstrate the importance of IRF4 in the generation of immature B cells.
While in vivo Irf4−/−Irf8−/− mice manifest a complete block in B lymphopoiesis, attenuation of IL-7R signaling in Irf4−/−Irf8−/− pre-B cell cultures induces cell cycle exit and Igκ recombination22. These observations suggest the existence of two molecular pathways that function synergistically to activate Igκ rearrangement: one that is independent of IRF-4 and requires attenuation of IL-7R signaling and another dependent on pre-BCR induced IRF-4 expression. The regulation of Igκ recombination by distinct signaling pathways is consistent with the known composite regulation of Igκ transcription. IRF-4 augments Igκ transcription through the Igκ 3’ enhancer (Eκ3’) while the IL-7R represses Igκ trancription through the intronic Igκ enhancer (Eκi)22.
In addition to directly regulating Igκ recombination, IRF-4 upregulates CXCR4 and promotes CXCL2 responsiveness of pre-B cells22. Given that there are discrete bone marrow niches of IL-7 and CXCL12 expression24 the in vivo phenotype of Irf4−/−Irf8−/− B cell progenitors may thus reflect, in part, an inability to migrate into IL-7 deficient microenvironments22.
Several other transcription factors have been implicated in the regulation of the Igκ locus25; however, expression of the transcription factor E2A with RAG1 and RAG2 is sufficient to induce Igκ recombination in a non-lymphoid cell line 26. Furthermore, gene-targeting studies have demonstrated that mutation of the E2A binding motifs in the Eκi reduces Igκ rearrangement similarly as ablation of the entire Eκi27. These, and other studies23,28, demonstrate that E2A is an important direct regulator of Igκ transcription. The activity of E2A proteins, and the regulation of E2A dependent developmental events, is modulated by expression of the inhibitors of DNA binding proteins, Id1–329.
The mechanisms regulating Igκ recombination have been intensely studied. In contrast, the important regulators controlling cell cycle progression in pre-B cells have only recently been identified13,30. Cyclin D3 expression is upregulated in pre-B cells and the pre-B cell proliferative expansion is ablated in Ccnd3−/− mice30. Furthermore, exit from cell cycle, and the initiation of Igκ recombination, is associated with termination of Ccnd3 transcription30. Therefore, to understand the relationships between the signaling elements controlling cell cycle exit and Igκ recombination, we first sought to identify the negative regulators of Ccnd3 transcription and relate these to those controlling E2A.
Herein, we demonstrate that pre-BCR mediated Ras/MEK/ERK activation couples cell cycle exit to Igκ recombination. Downstream of ERK, Aiolos silences Ccnd3 while reciprocal regulation of E2A and Id3 initiates Igκ transcription. The IL-7R antagonizes Igκ transcription through STAT 5, which binds to the Igκ intronic enhancer and prevents E2A recruitment. These findings identify a central signaling pathway controlling pre-BCR development and provide a molecular framework for understanding how the balance between pre-BCR and IL-7R signaling determines cell fate.
RESULTS
Ras-MEK activation is required for cell cycle exit and Igκ recombination in pre-B cells
To better understand how proliferation is regulated during B lymphopoiesis, we sought to identify the specific signaling pathways that suppressed Ccnd3 transcription in pre-B cells. For these initial studies, we used cultured Irf4−/−Irf8−/− pre-B cells as our model of differentiation. In the presence of IL-7, these pre-BCR+ cells continually proliferate. However, upon attenuation of IL-7 signaling they exit cell cycle and initiate Igκ recombination19,22.
In complete media alone, withdrawal of IL-7 was associated with diminished abundance of Ccnd3, Ccnd2 and Ccne encoding mRNAs (Fig. 1a) as well as the corresponding protein products (Fig. 1b). Initial pharmacological inhibitor studies failed to identify any negative regulators of Ccnd3 transcription30. However, subsequent studies demonstrated that inhibiting either MEK or ERK abrogated IL-7 withdrawal-induced cyclin downregulation (Fig. 1a, 1b). The observed modulation of cyclin E was proportional to that observed for the D-type cyclins, consistent with cyclin D mediated regulation of cyclin E31.
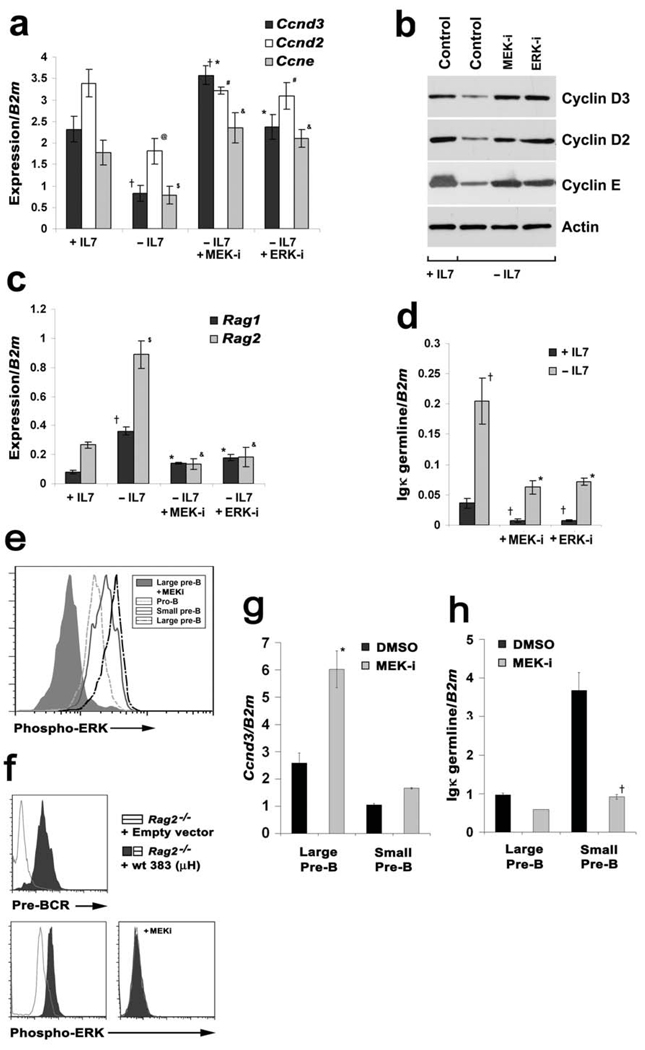
Figure 1. MEK/ERK regulate both cyclins and Igκ recombination machinery.
(a) Irf4−/−Irf8−/− pre-B cells were cultured in medium with (+ IL-7, 7.5ng/ml) or without (−IL-7, 0.1ng/ml) IL-7 for (a–b) 24 or (c–d) 48 hours. Cells were treated with either a MEK inhibitor PD98059 (MEK-i, 10µM) or an ERK inhibitor (ERK-i, 5µM) as indicated for the last six hours of culture. (a) Quantitative PCR (qPCR) for Ccnd3, Ccnd2 and Ccne expression; †P<0.001 as compared with +IL-7 Ccnd3 expression and *P<0.001 as compared with −IL-7 Ccnd3 expression; @P<0.001 as compared with +IL-7 Ccnd2 expression and #P<0.001 as compared with −IL-7 Ccnd2 expression; $P<0.001 as compared with +IL-7 Ccne expression and &P<0.001 as compared with −IL-7 Ccne expression. (b) Immunoblot for cyclin D3, cyclin D2 and cyclin E from Irf4−/−Irf8−/− pre-B total cell lysates. Cells were cultured according to the conditions described above. Actin immunoblot serves as an internal loading control. Analysis of three independent experiments indicated that expression of DN-Ras or DN-MEK increased cyclin D3 expression in −IL-7 cultures 4-fold as compared to untransfected controls (P<0.001, data not shown). (c) qPCR for Rag1 and Rag2 expression; †P<0.001 as compared with +IL-7 Rag1 expression and *P<0.001 as compared with −IL-7 Rag1 expression; $P<0.001 as compared with +IL-7 Rag2 expression and &P<0.001 as compared with −IL-7 Rag2 expression. (d) qPCR analysis for Igκ germline transcription; †P<0.001 as compared with +IL-7 Igκ germline transcription and *P<0.001 as compared with −IL-7 Igκ germline transcription expression. For all quantitative real time PCR analysis, beta 2 microglobulin (B2m) was used as the endogenous reference gene and data represent mean ± SD from four independent experiments. (e) Pro-B cell (B220+CD43+IgM−) or large and small pre-B cell (B220+CD43low/−IgM−) populations were isolated by FACS, permeabilized, stained with anti-phospho-ERK antibodies and analyzed by flow cytometry. Indicated cells were treated with PD98059 (10µM) for 2 hours prior to analysis. (f) Rag2−/− pro-B cells were infected with either control MIG1 retrovirus or virus encoding Igμ. After 48 hours, indicated cell populations were surface stained with anti-pre-BCR antibodies (SL156, upper panel) or permeabilized and stained with anti-phospho-ERK antibodies (lower panels). In lower right panel, Igμ expressing cells were analyzed after incubation for 2 hours with PD98059. (g and h) Large and small WT pre-B cells were isolated by flow cytometry, cultured in either PD98059 or vehicle for six hours and analyzed by qPCR for the expression of (g) Ccnd3 and (h) Igκ germline transcription. *P<0.001 as compared with Ccnd3 expression in large pre-B cells and †P<0.001 compared with κ germline transcription expression in small pre-B cells.
We next examined if the MEK/ERK pathway could regulate processes necessary for light chain recombination. Transcription of Rag1, Rag2 (Fig. 1c) and Igκ germline (Fig. 1d) was induced upon IL-7 withdrawal. Inhibition of either MEK or ERK attenuated both Rag and Igκ germline transcription following IL-7 withdrawal (Fig. 1c, 1d). MEK inhibition for 24 hours did not affect surface expression of the pre-BCR (data not shown). These data suggested that signaling through the MEK/ERK pathway directly impacts both cell cycle and the initiation of Igκ recombination.
To determine if the above observations were relevant to normal B lymphopoiesis, we examined ERK activation in different B cell progenitor populations. Pro-B (B220+CD43+IgM−), large pre-B (B220+CD43low/−IgM−) and small pre-B cells were isolated from wild-type (WT) bone marrow by flow cytometry, permeabilized and stained with anti-phospho-ERK antibodies. As can be seen in Fig. 1e, ERK activation was observed in all three populations with the highest levels detected in large pre-B cells and the lowest in pro-B cells.
The pattern of ERK activation paralleled that of pre-BCR expression which is highest in large pre-B cells and is then attenuated upon transition to the small pre-B cell stage9. To determine if expression of the pre-BCR augmented ERK activation, cultured Rag2−/− pro-B cells were infected with control retrovirus (MIGR1-IRES-GFP) or retrovirus encoding Igµ32 and GFP+ cells were assessed by flow cytometry. As can be seen in Fig. 1f, expression of Igµ led to the surface expression of a pre-BCR and this was associated with increased ERK activation.
To assess the function of MEK/ERK activation, large and small pre-B cells were isolated by fluorescence-activated cell sorting (FACS), treated for six hours in short-term cultures with MEK inhibitor and then assayed for Ccnd3 and germline Igκ transcription (Fig. 1g and 1h). As expected, large pre-B cells had relatively high levels of Ccnd3 transcripts and low levels of Igκ germline transcripts compared to small pre-B cells. In both cell populations, inhibition of MEK suppressed the abundance of Igκ germline transcripts and augmented those of Ccnd3. These results validate our initial observations in cultured Irf4−/−Irf8−/− pre-B cells and indicate that pre-BCR mediated MEK/ERK activation plays a central role in coordinating Ccnd3 suppression with the induction of Igκ germline transcription.
To further explore the contributions of the MEK/ERK signaling pathway to B lymphopoiesis, cultured Irf4−/−Irf8−/− pre-B cells were infected with control retrovirus or retrovirus encoding either a dominant negative (DN) mutant of Ras (DN-Ras, N17-H-Ras) or MEK (DN-MEK, MKK1-E8-K97M). GFP expressing cells were isolated by FACS, expanded in IL-7, and then cultured in either the presence or absence of IL-7. Both pharmacological inhibition and expression of DN-Ras and DN-MEK attenuated ERK activation (Fig. 2a). Interestingly, IL-7 withdrawal did not significantly modulate ERK activation in Irf4−/−Irf8−/− cells and ERK phosphorylation was relatively low in Rag2−/− pro-B cells irrespective of IL-7. These observations are consistent with the above results (Fig. 1e, f), and the findings of others, indicating that ERK activation primarily lies downstream of the pre-BCR33,34,35.
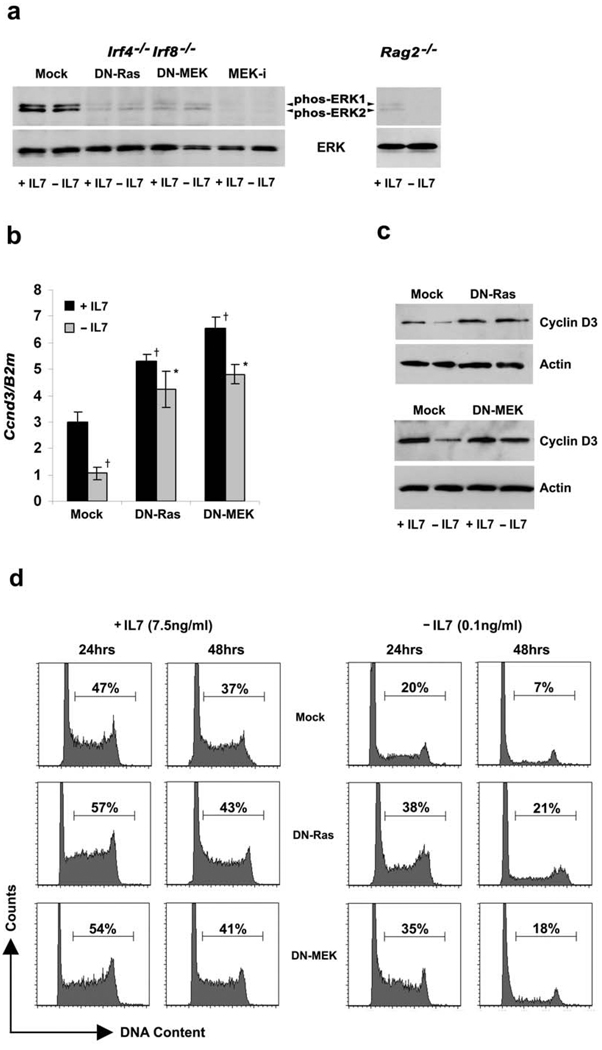
Figure 2. Downstream of pre-BCR, Ras-MEK activation required for cell cycle exit following IL-7 withdrawal.
Irf4−/−Irf8−/− pre-B cells expressing mock MIGR1 alone, DN-Ras or DN-MEK were cultured in medium with or without IL-7 for 24 or 48 hours as indicated or untransfected Irf4−/−Irf8−/− pre-B cells were cultured with or without MEK–i for the last six hours of culture. (a) Immunoblot for phospho-ERK and total ERK using total cell lysates from Irf4−/−Irf8−/− pre-B cells cultured according to the conditions described above and from Rag2−/− pro-B cells cultured in presence and absence of IL-7. Total-ERK immunoblot serves as an internal loading control. Data are representative of three independent experiments. (b) qPCR for Ccnd3 expression; †P<0.001 as compared with +IL-7 Ccnd3 expression and *P<0.001 as compared with −IL-7 Ccnd3 expression. B2m was used as the endogenous reference gene. Data represent mean ± SD from four independent experiments. (c) Immunoblot for cyclin D3 from Irf4−/−Irf8−/− pre-B total cell lysates. Cells were cultured according to the conditions described above. Actin immunoblot serves as an internal loading control. Data are representative of three independent experiments. (d) Cell cycle analysis of empty vector, DN-Ras or DN-MEK expressing Irf4−/−Irf8−/− pre-B cells cultured as described. Data are representative of three independent experiments.
Ectopic expression of either DN-Ras or DN-MEK increased the level of Ccnd3 mRNA in the presence of IL-7 and prevented the usual decrease associated with IL-7 withdrawal (Fig. 2b). Parallel changes in cyclin D3 protein expression were observed (Fig. 2c). Ectopic expression of both DN-Ras and DN-MEK similarly increased cyclin D2 and cyclin E expression (Supplementary Fig. 1a–1c). Ectopic expression of DN-Ras or DN-MEK did not significantly alter expression of p21Cip1 (Supplementary Fig. 1d) and only partially blunted the induction of p27Kip1 observed following IL-7 withdrawal (Supplementary Fig. 1e).
We next examined the role of Ras and MEK activation in cell cycle exit following IL-7 withdrawal. Irf4−/−Irf8−/− pre-B cells expressing either DN-Ras or DN-MEK were cultured in the presence or absence of IL-7 for up to 48 hours. Cell aliquots were permeabilized, stained with propidium iodide and analyzed by flow cytometry. In the presence of IL-7, both DN-Ras and DN-MEK modestly increased the fraction of cells in S/G2M (Fig. 2d). However, expression of either DN-Ras or DN-MEK significantly blunted the cell cycle exit associated with IL-7 withdrawal; 48 hours after IL-7 withdrawal when only 7% of control transfected cells were in S/G2M, 21% of those expressing DN-Ras and 18% of those expressing DN-MEK were in cell cycle. These data indicate that escape from IL-7 is not sufficient to terminate pre-B cell proliferation. In addition, activation through the Ras/MEK pathway is required to suppress Ccnd3 transcription and exit cell cycle.
We next examined the role of Ras/MEK/ERK activation in regulating the mechanisms of Igκ recombination. As demonstrated in Fig. 3, withdrawal of IL-7 from cultured Irf4−/−Irf8−/− pre-B cells induced the expression of Rag1 and Rag2 (Fig. 3a and 3b) and enhanced Igκ germline transcription (Fig. 3c). These responses were significantly attenuated by expression of either DN-Ras or DN-MEK.
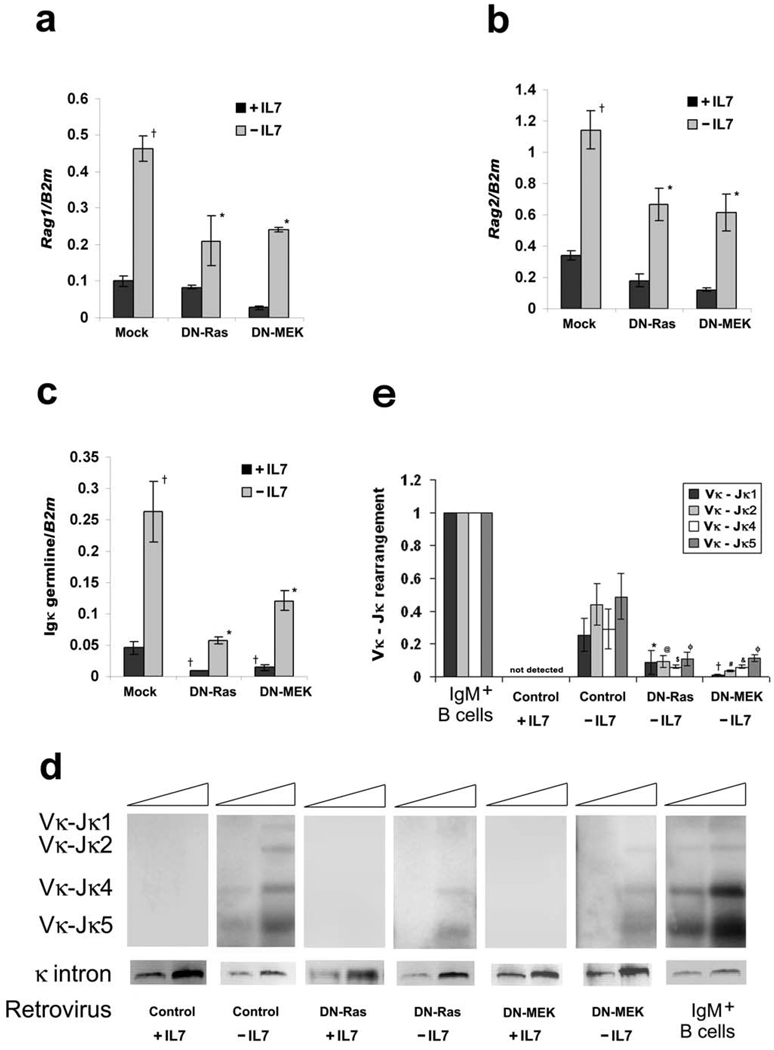
Figure 3. Ras-MEK activation is required for the induction of Igκ recombination following IL-7 withdrawal.
Irf4−/−Irf8−/− pre-B cells expressing mock MIGR1, DN-Ras or DN-MEK were cultured in medium with or without IL-7 for 48 hours. (a–c) qPCR for (a) Rag1, (b) Rag2 and (c) Igκ germline expression. †P<0.001 as compared with mock infected +IL-7 (a) Rag1, (b) Rag2 and (c) Igκ germline transcription and *P<0.001 as compared with −IL-7 (a) Rag1, (b) Rag2 and (c) Igκ germline transcription. B2m was used as the endogenous reference gene. Data represent mean ± SD from four independent experiments. (d) Semi-quantitative PCR analysis of Igκ rearrangement in Irf4−/−Irf8−/− pre-B cells cultured in medium as described for 48 hours. DNA from WT splenic IgM+ B cells was used as the positive control. PCR reactions employed degenerate Vκ primers and an Igκ intron primer (Supplementary Table 1) with 5 fold template dilutions. A region upstream of the Igκ intron was amplified to control for input genomic DNA. Amplified products were detected by hybridization with a k intron probe. Data are representative of three independent experiments. (e) Quantitative analaysis of Vκ-Jκ rearrangement (shown in d). The relative intensities for Vκ-Jκ1, Vκ-Jκ2, Vκ-Jκ4, Vκ-J5κ, and input genomic DNA (control) were calculated by ImageJ software (NIH); values from rearrangements were divided by the control and then depicted within a bar diagram, setting the values of normalized Vκ-Jκ1, Vκ-Jκ2, Vκ-Jκ4 and Vκ-J5κ rearrangement in IgM+ pre-B cells as 1. *P<0.05 and †P<0.01 as compared with −IL-7 control Vκ-Jκ1 expression; @P<0.01 and #P<0.001 as compared with −IL-7 control Vκ-Jκ2 expression; $P<0.05 and &P<0.01 as compared with −IL-7 control Vκ-Jκ4 expression and φP<0.01 as compared with −IL-7 control Vκ-Jκ5 expression. Data represent mean ± SD from three independent experiments.
To determine if the observed regulation of Rag and Igκ germline transcription affected Igκ recombination, genomic DNA was isolated from aliquots of cultured Irf4−/−Irf8−/− pre-B cells expressing MIGR1, DN-Ras or DN-MEK. Samples were then subjected to PCR with oligonucleotide primers complementary to Vκ and Jκ1–5 and the products visualized as described22. The results of a typical experiment are provided in Fig 3d. Fig. 3e provides a quantitative comparison of the relative band intensities corresponding to each V-Jκ1–5 recombination from three independent experiments. As can be seen, expression of either DN-Ras or DN-MEK significantly diminished Igκ recombination. From these observations, we conclude that the Ras/MEK/ERK signaling pathway coordinates both cell cycle exit and Igκ recombination in pre-B cells.
Ras-MEK mediated cyclin D3 suppression, and cell cycle exit, are dependent on Aiolos
We next asked if the Ras/MEK/ERK pathway suppressed proliferation and induced Igκ recombination through the same or different downstream signaling pathways. Aiolos is a lymphoid restricted transcription factor that is specifically upregulated in pre-B cells36 by pre-BCR dependent signaling pathways37,38. Furthermore, pre-B cell proliferation is increased in Aiolos−/− mice39. These findings prompted us to examine if Aiolos was playing a role in Ras/MEK mediated cell cycle exit.
We first characterized the expression of Aiolos, and the family member Ikaros40, in FACS isolated pro-B (CD19+CD43highIgM−) and pre-B (CD19+CD43low/−IgM−) cells from wild-type (WT) bone marrow. As expected37, Aiolos expression was increased in pre-B cells relative to pro-B cells, whereas the expression of Ikaros was similar in both populations (Fig. 4a). We next determined if the Ras/MEK/ERK signaling pathway could modulate Aiolos expression. Irf4−/−Irf8−/− pre-B cells ectopically expressing either DN-Ras or DN-MEK were cultured in the presence or absence of IL-7 and then assayed for the expression of Aiolos (Fig. 4b) and Ikaros (Fig. 4c). As demonstrated, blocking the Ras/MEK pathway attenuated Aiolos expression but had no significant effect on Ikaros expression.
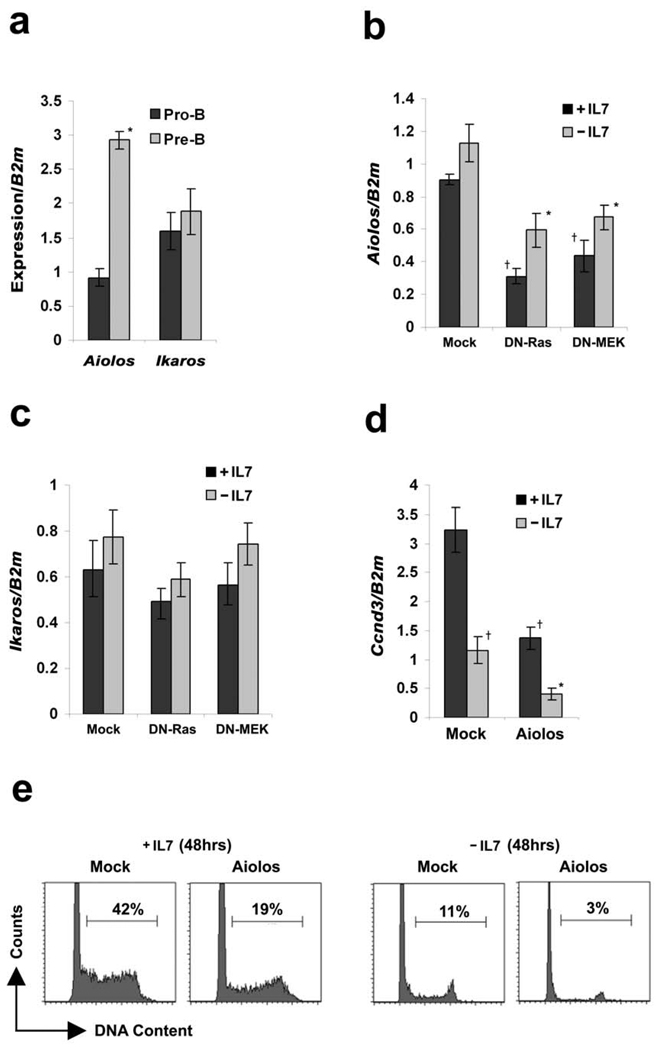
Figure 4. Aiolos is induced in pre-B cells and can suppress Ccnd3 transcription.
(a) WT pro-B cells (CD19+CD43highIgM−) and pre-B cells (CD19+CD43low/−IgM−) were isolated by flow sorting. The relative expression of Aiolos and Ikaros was analyzed by quantitative real time PCR. *P<0.001 as compared with Aiolos expression in pro-B cells. (b–c) qPCR analysis of (b) Aiolos and (c) Ikaros in Irf4−/−Irf8−/− pre-B cells expressing vector alone, DN-Ras or DN-MEK cultured in medium with or without IL-7 for 24 hours. †P<0.001 as compared with vector alone +IL-7 Aiolos expression and *P<0.001 as compared with −IL-7 Aiolos expression. (d) qPCR analysis for Ccnd3 expression in Irf4−/−Irf8−/− pre-B cells expressing either MIGR1 or Aiolos cultured in medium with or without IL-7 for 24 hours. †P<0.001 as compared with vector infected +IL-7 Ccnd3 expression and *P<0.001 as compared with −IL-7 Ccnd3 expression. B2m was used as the endogenous reference gene. Data represent mean ± SD from four independent experiments. (e) Cell cycle analysis of mock and Aiolos expressing Irf4−/−Irf8−/− pre-B cells cultured in medium with or without IL-7 for 48 hours. Data are representative of three independent experiments.
To determine if Aiolos could regulate Ccnd3, retroviral infection was used to establish cultured Irf4−/−Irf8−/− pre-B cell populations over-expressing murine Aiolos. In both the presence and absence of IL-7, ectopic expression of Aiolos downregulated Ccnd3 mRNA (Fig. 4d). Furthermore, irrespective of IL-7, this was associated with a marked decrease of cells in S/G2M (Fig. 4e). Expression of Aiolos suppressed Ccnd2 and Ccne expression but had no significant effect on the expression of p21Cip1 or p27Kip1 (Supplementary Figs. 2a–2b). Furthermore, Aiolos did not modulate Rag1 or Rag2 expression or enhance Igκ transcription (Supplementary Fig. 2c and 2d). These data suggest that, in the context of pre-B cell differentiation, Aiolos functions downstream of Ras/MEK to specifically suppress Ccnd3 expression and facilitate cell cycle exit.
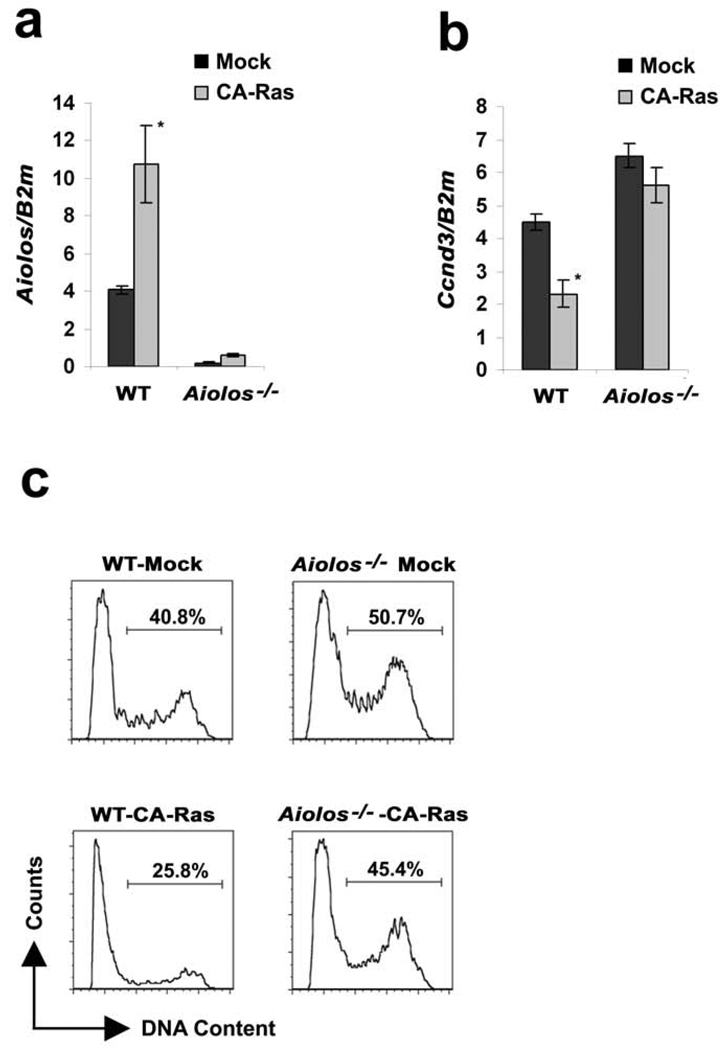
To examine if Aiolos was required for Ras-mediated cyclin D3 downregulation, pre-B cells (CD19+CD43lowIgM−) were isolated by FACS from WT and Aiolos−/− bone marrow and cultured cells were infected with retrovirus encoding CA-Ras. GFP expressing cells were then isolated by FACS and cultured in IL-7. Expression of CA-Ras induced Aiolos expression in WT pre-B cells but not Aiolos−/− pre-B cells (Fig. 5a). As expected, CA-Ras also downregulated Ccnd3 in WT cells. In contrast, in Aiolos−/− pre-B cells Ccnd3 mRNA was elevated and could not be suppressed by CA-Ras (Fig. 5b). Both Ccnd2 (Supplementary Fig. 3a) and Ccne (Supplementary Fig 3b) demonstrated a similar Aiolos dependent regulation by Ras. Cell cycle analysis revealed that ectopic expression of CA-Ras attenuated the fraction of cells in S/G2M in WT but not Aiolos−/− pre-B cells (Fig 5c). These data indicate that the induction of Aiolos by Ras/MEK/ERK mediates cell cycle exit by inhibiting Ccnd3 expression.
Figure 5. Downstream of Ras, Aiolos is required for suppressing Ccnd3 and cell cycle exit following IL-7 withdrawal.
Pre-B cells (CD19+CD43lowIgM−) were sorted from WT and Aiolos−/− mice and cultured for 24 hours in the presence of IL-7 (15ng/ml). Cells expressing either MIGR1 or CA-Ras were cultured in the presence of IL-7 (7.5ng/ml) for 48 hours. (a–b) The relative expression of (a) Aiolos and (b) Ccnd3 in WT and Aiolos−/− pre-B cells expressing either mock or CA-Ras was analyzed by qPCR. (a) *P<0.001 as compared with Aiolos expression in mock infected wild-type pre-B cells and (b) *P<0.001 as compared with Ccnd3 expression in mock infected wild-type pre-B cells. B2m was used as the endogenous reference gene. (c) Cell cycle analysis of mock and CA-Ras expressing WT and Aiolos−/− pre-B cells cultured with IL-7 for 48 hours.
The magnitude of Ras-mediated Aiolos induction in cultured WT pre-B cells (Fig. 5a) and in Irf4−/−Irf8−/− pre-B cells (Fig. 4b) was similar to the difference in expression observed between WT pro- and pre-B cells (Fig. 4a). However, overall Aiolos expression levels were lower in Irf4−/−Irf8−/− pre-B cells than WT pre-B cells. This later observation is consistent with previous findings that IRF4 contributes to Aiolos induction 20. However, our data indicate that sufficient Aiolos can be induced in the absence of IRF4 to mediate cell cycle exit.
Regulation of E2A and Id3 by the Ras/MEK/ERK signaling pathway enhances Igκ transcription
We next sought to identify which signaling intermediates downstream of Ras/MEK/ERK regulated Igκ recombination. We focused on Igκ transcription because it was almost entirely dependent upon Ras activation (Fig. 3c).
Igκ transcription is dependent upon E2A23,26,27, which in turn is negatively regulated by the Id proteins (Id1–3)41–43. Therefore, we examined if Ras/MEK/ERK activation could play a role in the coordinate regulation of E2A and Id3 during B lymphopoiesis.
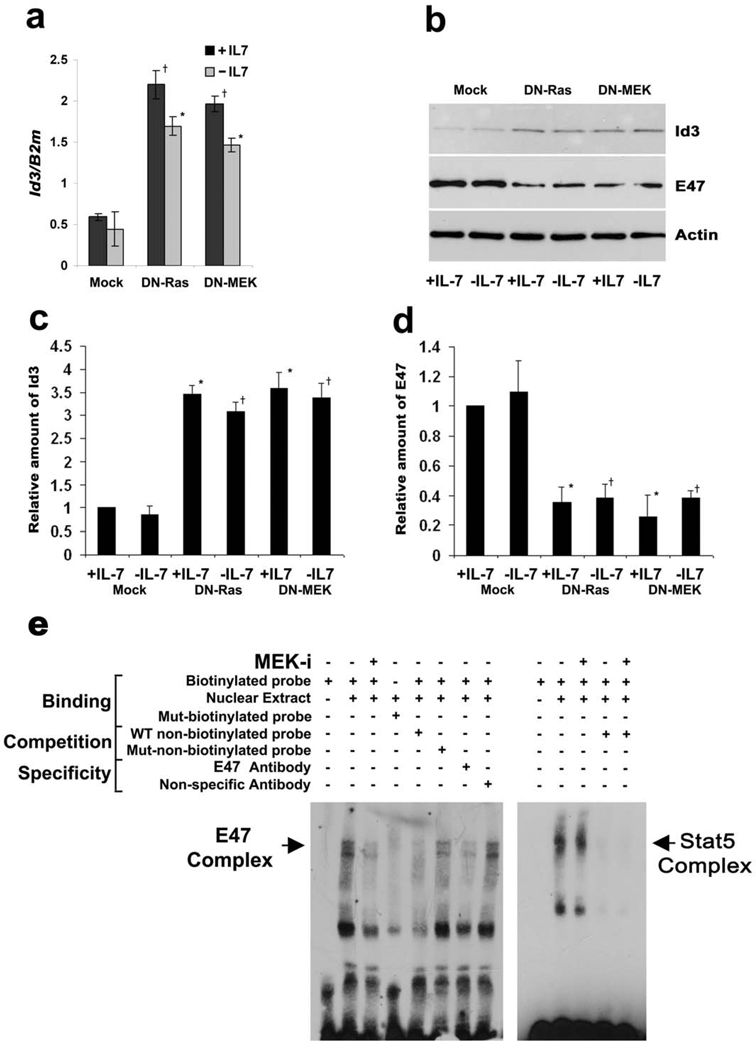
Irf4−/−Irf8−/− untransfected pre-B cells or cells expressing DN-Ras or DN-MEK were cultured in the presence or absence of IL-7 and assayed for expression of the Id proteins. As can be seen in Fig. 6a, inhibiting the Ras/MEK pathway significantly augmented the expression of Id3. In contrast, DN-Ras or DN-MEK had no effect on Id1 expression (Supplementary Fig. 4a) and moderately enhanced Id2 expression (Supplementary Fig. 4b). Immunoblotting of lysates from Irf4−/−Irf8−/− cells expressing either DN-Ras or DN-MEK confirmed that activation of the Ras/MEK/ERK pathway suppressed Id3 expression (Fig 6b). Immunblotting of these same lysates also demonstrated that Ras/MEK induced E47 expression. Expression of both Id3 and E47 was not significantly affected by IL-7.
Figure 6. Downregulation of Id3 and upregulation of E2A by Ras-MEK signaling increases E2A binding activity.
(a) qPCR analysis for expression of Id3. Irf4−/−Irf8−/− pre-B cells expressing MIGR1, DN-Ras or DN-MEK were cultured in medium with or without IL-7 for 24 hours. †P<0.001 as compared with mock infected +IL-7 Id3 and *P<0.001 as compared with −IL-7 Id3. B2m was used as the endogenous reference gene. Data represent mean ± SD from four independent experiments. (b) Western blot analysis of Id3 and the E2A protein E47 from indicated lysates of Irf4−/−Irf8−/− pre-B cells expressing empty vector, DN-Ras or DN-MEK cultured in medium with or without IL-7 for 30 hours. Actin immunoblot serves as an internal loading control. Data are representative of three independent experiments. (c–d) The relative amount of (c) Id3 and (d) E47 normalized with actin in arbitrary units (a.u.) were calculated from western blots (n=3) by ImageJ software (NIH). *P<0.001 as compared with mock infected +IL-7 Id3 and E47 expression and †P<0.001 as compared with −IL-7 Id3 and E47 expression. (e) In vitro binding of E47 was assayed in EMSAs. Nuclear extracts were prepared from Irf4−/−Irf8−/− pre-B cells cultured in medium with IL-7 for 30 hours and then treated with MEK inhibitor (MEK-i, 10µM of PD98059) for the last six hours of culture. An equal amount of nuclear proteins was used for each reaction. E2A binding was assayed with a probe corresponding to the E1 box of Eκi. E47/DNA complexes were verified using an E47 antibody. STAT 5 protein/DNA complexes were assessed as a control.
Quantitation of triplicate immunoblots indicated that inhibiting Ras or MEK led to an average 3.3-fold enhancement of Id3 expression (Fig. 6c) and a 2.9-fold decrease in E47 expression (Fig. 6d) irrespective of IL-7. While each observed change was moderate, the predicted change in the E47/Id3 ratio, and therefore of free E47, was substantial (9 to 10 fold). Therefore, we performed electrophoretic mobility shift assays (EMSAs) using nuclear extracts from Irf4−/−Irf8−/− cells and an oligonucleotide probe corresponding to E1 box of Eκi (Fig. 6d). In either the presence or absence of IL-7, an E47 containing complex was detected that bound the WT, but not mutant, labeled oligonucleotide probe. This complex was attenuated by addition of either anti-E47 antibodies or by excess unlabelled WT oligonucleotides. In contrast, the complex was not diminished by either addition of isotype-matched control antibodies or by unlabelled oligonucleotides containing mutations in the E2A binding motif. Inhibition of MEK greatly attenuated the formation of detectable E47 containing binding complexes. In contrast, inhibition of MEK had no detectable effect on the binding activity of STAT 5 to a known binding site in FcγRI44. Consistent with previous observations, IL-7 had no significant effect on nuclear E2A binding activity22 (Supplementary Fig. 5). These data indicate that MEK activation enhances E2A binding activity by coordinately regulating the expression of E2A and Id3.
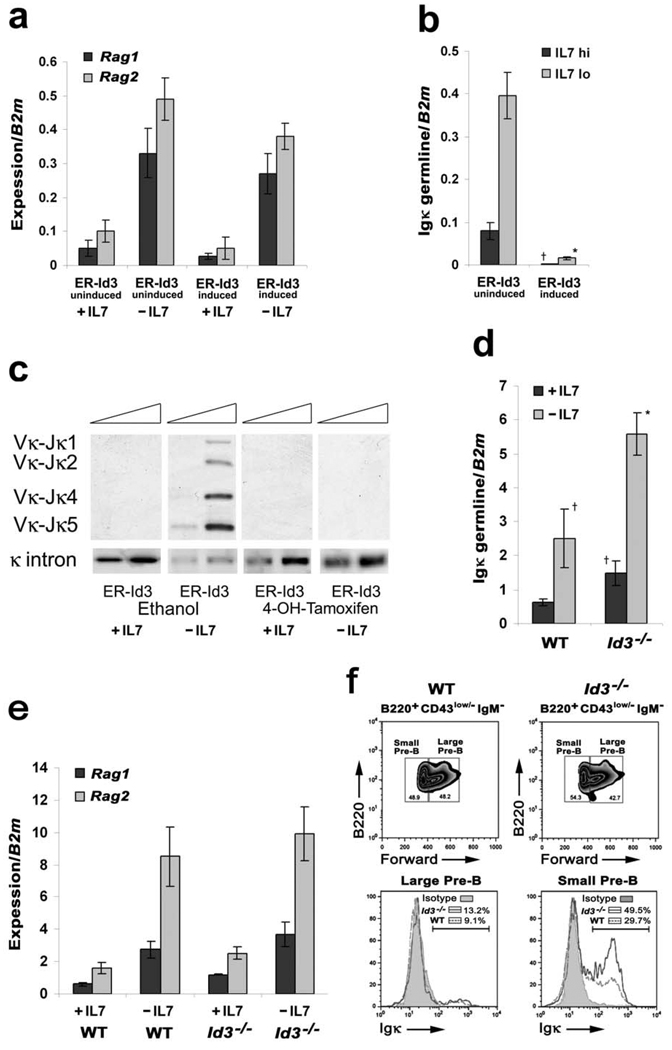
To examine the potential importance of Ras/MEK/ERK regulation of Id3, Irf4−/−Irf8−/− pre-B cells expressing a retrovirally encoded estrogen receptor (ER)-Id3 fusion protein were cultured with or without IL-7 in the presence or absence of 4-OH-tamoxifen. Although the expression of ER-Id3 and induction with tamoxifen had no significant effect on Rag1 and Rag2 expression (Fig. 7a), Id3 ablated detectable Igκ germline transcription (Fig. 7b). This was associated with complete suppression of detectable Vκ to Jκ recombination (Fig. 7c). Id3 induction had no effect on expression of the D- and E-type cyclins (Supplementary Fig. 4c) or on the cell cycle inhibitors p21Cip1 and p27Kip1 (Supplementary Fig. 4d). These data implicate Id3 as a specific regulator of Igκ transcription in pre-B cells.
Figure 7. Increased Igκ transcription and rearrangement in Id3−/− mice.
(a–b) qPCR analysis for (a) Rag1 and Rag2 and (b) Igκ germline expression in Irf4−/−Irf8−/− pre-B cells expressing inducible Id3 (ER-Id3-IRES-GFP) cultured in medium with or without IL-7 for 48 hours. Cells were either mock treated with ethanol (solvent of 4-OH-tamoxifen) or 4-OH-tamoxifen (1µM) to induce Id3 expression. †P<0.001 as compared with mock infected +IL-7 Igκ germline transcription and *P<0.001 as compared with −IL-7 Igκ germline transcription. B2m was used as the endogenous reference gene. Data represent mean ± SD from four independent experiments. (c) Semi-quantitative PCR analysis of Igκ rearrangements in Irf4−/−Irf8−/− pre-B cells expressing mock MIGR1 or ER-Id3 cultured as described above. DNA from WT splenic IgM+ B cells was used as the positive control. Data are representative of three independent experiments. (d–e) qPCR analysis for relative expression of (d) Igκ germline and (e) Rag1 and Rag2 transcription using sorted pre-B cells (CD19+CD43low/−IgM−) from WT and Id3−/− bone marrow and cultured in medium with or without IL-7 for 48hrs. †P<0.001 as compared with wild-type +IL-7 Igκ germline transcription and *P<0.001 as compared with −IL-7 Igκ germline transcription. B2m was used as the endogenous reference gene. Data represent mean ± SD from three independent experiments. (f) Large and small pre-B cells (CD19+CD43low/−IgD−) were sorted from WT and Id3−/− bone marrow and the surface expression of Igk was assayed by flow cytometry. Isotype control indicated. Data are representative of three independent experiments.
To determine if Id3 was required to suppress Igκ transcription in vivo, WT or Id3−/− pre-B cells were isolated by FACS, cultured in IL-7 and then subjected to IL-7 withdrawal. As can be seen in Fig. 7d, Igκ transcription was significantly enhanced in Id3−/− pre-B cells in both the presence and absence of IL-7. Consistent with results obtained from ectopic Id3 expression, the expression of Rag1 and Rag2 was not altered in Id3−/− pre-B cells as compared to WT cells (Fig. 7e); furthermore, Ccnd2, Ccnd3 and Ccne expression was comparable between WT and Id3−/− cells (Supplementary Fig. 6). To determine if Id3 regulates Igκ recombination in vivo, large and small pre-B cells from WT and Id3−/− mice were isolated, stained with anti-Igκ antibodies and analyzed by flow cytometry. As demonstrated in Fig. 7f, a greater fraction of Id3−/− large and small pre-B cells expressed Igκ on their cell surface, consistent with Igκ dysregulation. These data indicate that Ras/MEK/ERK enhances Igκ transcription through the coordinated regulation of E2A and Id3.
STAT 5 regulates accessibility of Eκi to E2A
For Ras/MEK/ERK activation to efficiently induce cell cycle exit and Igκ recombination, pre-B cells must escape the effects of IL-7. Our data predict that IL-7 inhibits differentiation by opposing the Ras/MEK/ERK signaling pathway. However, IL-7 did not significantly alter Aiolos expression (Fig. 4b) or E2A binding activity22 (Supplementary Fig. 5). This suggests that IL-7 modulates the activities of Ras/MEK/ERK downstream of Aiolos and E2A. To examine this possibility, we first sought to identify which IL-7R dependent signaling pathways enhance Ccnd3 transcription and inhibit Igκ transcription.
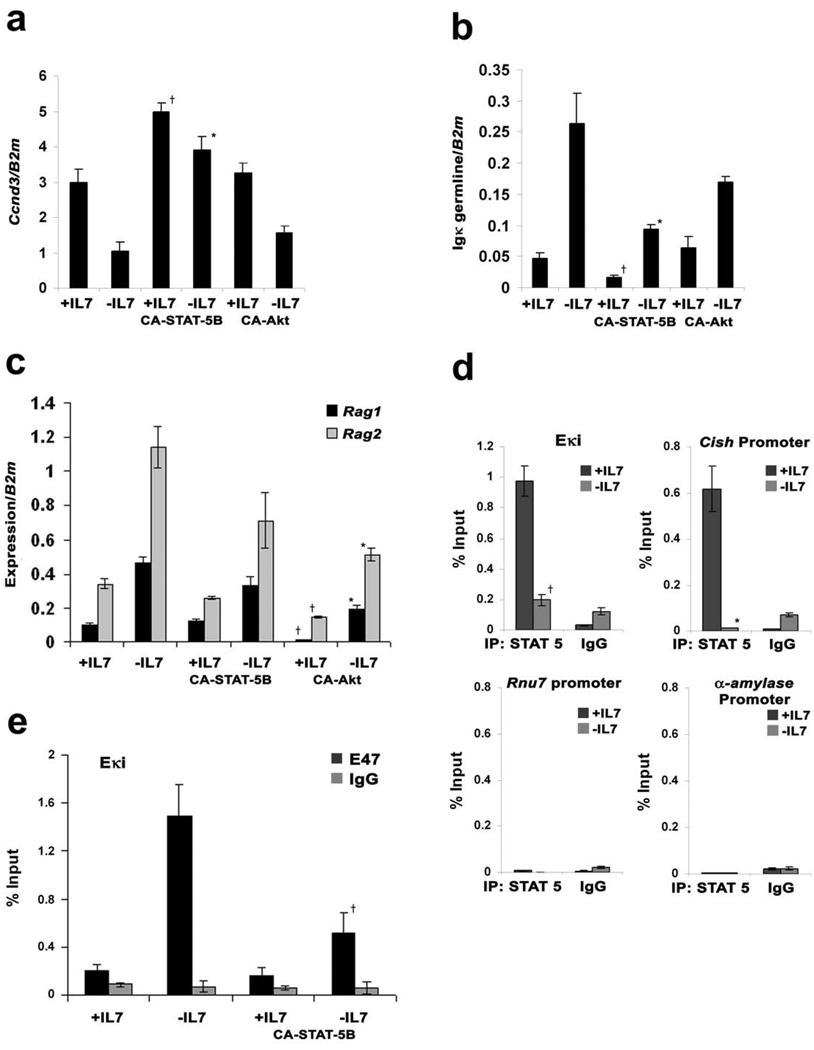
Downstream of the IL-7R, there are two primary signaling pathways: JAK3/STAT 5 and PI-3 kinase/Akt45. Therefore, we examined if retroviral mediated expression of either CA-STAT 5B46,47 or CA-Akt48 could supplant the biological activities of IL-7 in cultures of Irf4−/−Irf8−/− pre-B cells. As demonstrated in Fig. 8a, CA-STAT 5B enhanced Ccnd3 expression in IL-7 treated cells and prevented the attenuation associated with IL-7 withdrawal. CA-Akt did not significantly affect Ccnd3 expression. CA-STAT 5B also repressed Igκ germline transcription in cells cultured with or without IL-7 (Fig. 8b). In contrast, CA-Akt, and not CA-STAT 5B, repressed Rag1 and Rag2 induction following IL-7 withdrawal (Fig. 8c). From these data, we conclude that that STAT 5 is the main downstream signaling effector of the IL-7R regulating Ccnd3 and Igκ transcription 47.
Figure 8. STAT 5 regulates Igκ transcription and binds directly to Eκi.
Irf4−/−Irf8−/− pre-B cells expressing MIGR1, CA-STAT 5B or CA-Akt were cultured in medium with or without IL-7 for (a) 24 and (b–d) 48 hours. (a) qPCR analysis for Ccnd3 expression; †P<0.001 as compared with mock infected +IL-7 Ccnd3 expression and *P<0.001 as compared with mock infected −IL-7 Ccnd3 expression. B2m was used as the endogenous reference gene. Data represent mean ± SD from three independent experiments. (b) qPCR analysis for Igκ germline expression. †P<0.001 as compared with mock infected +IL-7 Igκ germline transcription and *P<0.001 as compared with mock infected −IL-7 Igκ germline transcription. B2m was used as the endogenous reference gene. Data represent mean ± SD from three independent experiments. (c) qPCR analysis shows Rag1 and Rag2 expression. †P<0.001 as compared with mock infected +IL-7 Rag1 (or Rag2) expression and *P<0.001 as compared with mock infected −IL-7 Rag1 (or Rag2) expression. B2m was used as the endogenous reference gene. Data represent mean ± SD from three independent experiments. (d) Irf4−/−Irf8−/− pre-B cells were cultured in medium with or without IL-7 for 48 hours and were subjected to ChIP assays with anti-STAT 5 antibodies. †P<0.001 as compared with binding of STAT 5 to Eκi with IL-7 and *P<0.001 as compared with binding of STAT 5 to Cish promoter. Data represent mean ± SD from three independent experiments. (e) Irf4−/−Irf8−/− pre-B cells expressing CA-STAT 5B or vector alone were cultured in medium with or without IL-7 for 24 hours and were subjected to ChIP assays with anti-E47 antibodies. †P<0.001 as compared with binding of E47 to Eki without IL-7.
IL-7-mediated Igκ repression is dependent upon the Eκi. Furthermore, repression is associated with inhibition of E2A binding to Eκi in vivo22. Therefore, we next examined if STAT 5 could modulate E2A binding to Eκi. We first determined if STAT 5 bound directly to Eκi in vivo. Aliquots of Irf4−/−Irf8−/− cells, cultured in the presence or absence of IL-7, were subjected to quantitative chromatin immunoprecipitation (ChIP) assays with anti-STAT 5B antibodies. As demonstrated in Fig. 8d, STAT 5B bound to Eκi in an IL-7 dependent manner. Binding was robust and comparable to that obtained for the Cish promoter, a validated STAT 5 target49. In contrast, there was little detectable binding to either the Rnu7 or α-amylase promoters.
We next determined if STAT 5B could attenuate E2A binding to Eκi. Irf4−/−Irf8−/− pre-B cells expressing CA-STAT 5B or control cells were cultured in the presence or absence of IL-7. Aliquots of each cell population were then assayed by quantitative ChIP for E47 binding to endogenous Eκi. As demonstrated in Fig. 8e, withdrawal of IL-7 was associated with enhanced binding of E47 to Eκi. This enhancement was attenuated by expression of CA-STAT 5B. These data are consistent with a model in which downstream of the IL-7R, STAT 5 inhibits Igκ transcription by directly limiting the accessibility of Eκi to E2A.
DISCUSSION
It has been proposed that differentiation of pre-B cells into immature B cells occurs when they escape pro-proliferative IL-7R signals22. Our findings demonstrate that this is not sufficient. In addition, pre-BCR mediated Ras/MEK/ERK activation is required to orchestrate the principal molecular events of this developmental transition: cell cycle exit and Igκ recombination (schematic model provided in Supplementary Fig. 7). ERK directs these outcomes by divergent mechanisms, utilizing lineage and developmentally restricted downstream effector molecules. The identification of a central, IL-7R modulated differentiation pathway provides a framework for understanding how signaling through the two principal receptors that dictate pre-B cell fate, the pre-BCR and IL-7R, coordinately regulate proliferation and differentiation. Furthermore, our findings provide an instructive example of how the molecular context of signal activation dictates cell fate.
A major function of the Ras/MEK/ERK pathway was to silence Ccnd3 and terminate proliferation. This result was surprising as Ras has been demonstrated to induce proliferation in a variety of cell types50. However, the unique anti-proliferative effect of Ras in pre-B cells was dependent upon Aiolos, a lineage and developmentally restricted transcription factor37, which coupled Ras activation to the silencing of Ccnd3. A central role for Aiolos in cell cycle termination is consistent with observations that Aiolos−/− mice have increased numbers of proliferating pre-B cells39. Furthermore, Aiolos is upregulated by expression of the pre-BCR36–38. In contrast to the necessity of Aiolos in regulating pre-B cell proliferation, the family member Ikaros36 appeared to have no substantial role. Aiolos also suppresses SLC expression37, indicating that it may silence a program of pre-B cell specific gene products.
While it is clear that Aiolos is required for Ras-mediated silencing of Ccnd3, it is likely that Ras is not the only inducer of Aiolos. Inhibiting Ras only partially attenuated Aiolos expression. The transcription factor IRF4 has also been shown to induce Aiolos20 in pre-B cells. Although compromised, Aiolos induction in Irf4−/−Irf8−/− pre-B cells is sufficient to allow cell cycle exit and Igκ recombination.
In addition to silencing Ccnd3, Ras/MEK/ERK augmented Igκ transcription by enhancing E2A expression and downregulating Id3. Ras/MEK/ERK only induced moderate changes in the level of each protein. However, this translated into a large increase in E2A binding activity. Among the Id proteins, Ras preferentially regulated Id3 and loss of Id3 was sufficient to dysregulate Igκ expression. These findings reveal that there is some functional specificity among the Id proteins expressed during B lymphopoiesis. Our results are in contrast to previous observations that Ras induces Id3 expression in double positive (DP) thymocytes and attenuates E2A activity51. However, the Egr transcription factors which upregulate Id3 in thymocytes, were only transiently suppressed following MEK inhibition in Irf4−/−Irf8−/− pre-B cells (data not shown). Therefore, it is not surprising that TCR dependent signals in DP thymocytes would regulate Id3 differently than signals through the pre-BCR.
Downstream of the Ras/MEK/ERK pathway, the specific molecular events of development were mediated by discrete and non-overlapping mechanisms. Aiolos repressed Ccnd3 transcription but had no effect on cell cycle inhibitors or on the mediators of Igκ recombination. Conversely, E2A enhanced Igκ transcription but had no significant effect on the cell cycle machinery or on Rag expression. This latter finding is consistent with prior observations that E2A is required for initial Rag expression in pro-B cells but not for subsequent receptor-induced expression52.
Proximal Ras/MEK/ERK activation coordinates a series of nuclear events required for development. This could potentially lead to a dangerous state as an aberration in one of the downstream effectors of ERK activation could uncouple cell cycle exit from Igκ recombination. However, RAG protein levels are tied to cell cycle progression as RAG and p27 are degraded through a common pathway53. It is likely that this, and potentially other mechanisms intrinsic to cell cycle progression54, ensure genomic integrity.
Our observations may help resolve several apparent discrepancies in the literature concerning the role of Ras in lymphocyte development. Inhibiting the Ras/MEK/ERK pathway, either by expressing DN-Ras16 or by lineage specific mutation of Erk1 and Erk2 induces a block at the pro-B cell stage55. In Erk1−/−Erk2−/− mice, this is associated with a proliferative defect in the remaining CD19+CD43+ cells and increased expression of SLC. In contrast, transgenic expression of CA-Ras throughout development induces extensive Igκ rearrangement18. We would argue that these two phenotypes arise from differential expression of Ras targets in pro and pre-B cells. In particular, the upregulation of Aiolos in pre-B cells is predicted to change the consequences of Ras activation. Other unexplored factors, such as differences in the amplitude or duration of Ras activation56 in pro- and pre-B cells, may determine specific functional responses.
Tracing the signaling pathways downstream of Ras/MEK/ERK has revealed one mechanism for how the IL-7R suppresses pre-BCR-mediated differentiation. Ras activation greatly increases nuclear E2A binding activity. However, this is not sufficient to induce Igκ transcription as E2A cannot bind Eκi unless IL-7R signaling is attenuated. This suppressive function of the IL-7R is mediated by STAT 5 and is associated with the direct binding of STAT 5 to Eκi. It is not clear how STAT 5 limits Eκi accessibility. However, STAT 5 has been demonstrated to be capable of both repressing57 and activating58 transcription. In addition to repressing Igκ transcription, STAT 5 coordinately enhanced Ccnd3 transcription irrespective of pre-BCR signaling. The dominance of STAT 5 at the Igκ and Ccnd3 loci provides a molecular explanation for why proliferation prevails over Igκ recombination when both the IL-7R and the pre-BCR are transmitting signals.
It is likely that the Ras/MEK/ERK signaling pathway also antagonizes the PI-3k/AKT signaling pathway. The latter favors proliferation over differentiation by suppressing Rag transcription13,59 and by stabilizing cyclin D3 protein30. ERK may oppose the activities of Akt on Rag transcription as ERK can phosphorylate and activate the FoxO family members60 which Akt target for degradation13,59. Our data demonstrate that ERK opposes the activities of PI-3k on cyclin D3 by silencing Ccnd3 transcription. Additional undefined signaling pathways may influence cell cycle progression through the regulation of the inhibitors p21 and p27.
From our data, a picture emerges in which crucial molecular determinants of B lymphopoiesis are regulated by opposing signaling pathways that intersect at specific targets. By coordinately upregulating E2A and suppressing Ccnd3 transcription, the Ras/MEK/ERK pathway plays a necessary and central role in the transition from proliferation to Igκ recombination. This Ras-mediated differentiation program is modulated by STAT 5 which suppresses the activities of specific downstream ERK targets. It is through these, and other signaling networks30,59, that the balance between IL-7R and pre-BCR signaling determines if a pre-B cell divides or undergoes Igκ recombination.
METHODS
Mice
WT, Irf4−/−Irf8−/−, Aiolos−/− and Id3−/− (C57BL/6) and Rag2−/− (BALB/c) mice were housed in the animal facilities of the University of Chicago. Mice were used at 6–10 weeks of age and experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the University of Chicago.
Isolation and culture of Rag-2−/− pro-B and Irf4−/−Irf8−/− pre-B cells
CD19+ cells were isolated from bone marrow (6–10 week old mice) using a magnetic-activated cell sorter (MACS) separation column (Miltenyi Biotec). The resulting pro-B cells were cultured in complete Opti-MEM containing 7.5% fetal bovine serum and IL-7 (10ng/ml)30. Purified CD19+ cells from Irf4−/−Irf8−/− mice were overlaid on OP9 stromal cells and cultured in complete medium with 7.5ng/ml of IL-7 (+IL-7) or 0.1ng/ml of IL-7 (−IL-7)22. Cells were typically >99% CD19+, pre-BCR+. For the inhibitor studies we used 10µM PD98059 (MEK inhibitor) and 5µM ERK inhibitor (both Calbiochem).
Retroviral gene transduction
cDNAs encoding murine CA-Ras (Ras61L), DN-Ras (RasN17N), CA-STAT-5B (Dr. T. Gajewski, University of Chicago), DN-MEK (MEKK1-8E-K97M, Dr. Natalie G. Ahn, University of Colorado at Boulder), Aiolos, Igµ32or human ER-Id3 were sub-cloned into MIGR1. Myristylated Akt (CA-Akt) was used as described61. Control MIGR1 retrovirus or MIGR1 retroviruses containing a gene of interest were generated by calcium phosphate transfection of 293T cells and infection of the B cell progenitors was performed as described61. After 48 hours, GFP+ cells were isolated by FACS and cultured in complete medium.
Quantitative PCR analysis
Total cellular RNA was isolated with RNeasy kit (Qiagen) according to the manufacturer’s protocol. RNA was reverse-transcribed with the Superscript III reverse transcriptase (Invitrogen). qPCR was performed in a total volume of 25µl containing 1µl cDNA template, 0.5 µM of each primer and SYBR Green PCRMaster Mix (Applied Biosystems). Gene expression was analyzed with the ABI PRISM 7300 Sequence Detector and ABI Prism Sequence Detection Software version 1.9.1 (Applied Biosystems). Sample results were normalized by division of the value for the unknown gene by that obtained for beta 2 microglobulin (B2m). Quadruplicate reactions were done with all cDNA samples. The primers used are presented in Supplementary Table 1.
Immunoblot
Total NP-40 cell lysates were prepared61 and 30µg of each lysate were resolved by 4–20% SDS-PAGE and then transferred to Immuno-Blot PVDF membranes (BioRad)30. Protein blots were probed with anti-Cyclin D3 (sc-182), anti-Cyclin D2 (sc-593), anti-Cyclin E (sc-481) (all from Santa Cruz Biotechnology), anti-mouse Id3 (BD Pharmingen), anti-E47 (sc763, Santa Cruz Biotechnology), anti-total ERK (Zymed Laboratories), anti-phospho ERK (Phospho-p44/42 MAPK, Cell Signal), anti-STAT 5 (sc-835, Santa Cruz Biotechnology) and anti-Actin (Chemicon) as an internal loading control.
Flow cytometry
Single cell bone marrow suspensions were treated with Ack lysis buffer, resuspended in 3% FBS/PBS and stained with monoclonal antibodies to B220 (RA3–6B2), pre-BCR (SL156), CD43 (S7), CD25 (3C7), IgM (R6–60.2), CD11c (HL3), c-Kit(2B8), CD19 (1D3), NK1.1 (PK136), TCRβ (H57–597), Ter119 (TER-119), Mac-1 (M1/70), Gr-1 (RB6–8C5)(all from BD Biosciences Pharmingen). Anti-Igκ (H139-52.1) and its isotype Rat IgG1κ were from Southern Biotech. For intracellular staining with anti-phospho ERK antibodies (20A), cells were fixed with BD Cytofix buffer and permeabilized with BD Phosflow Perm Buffer III as recommended by the manufacturer (BD Biosciences Pharmingen). Monoclonal antibodies were directly coupled to fluorescein isothiocyanate, phycoerythrin, phycoerythrin-indotricarbocyanine, allophycocyanin or biotin. Surface marker expression on progenitor and mature B cells was acquired using a FACSCalibur (Becton Dickinson) and analyzed with FlowJo and CELLQuest software (Becton Dickinson). Cell sorting was done with a MoFlo cell sorter (Cytomation).
Cell Cycle Analysis
Cells were incubated in a solution containing propidium iodide and then analyzed on a FACScan (Becton Dickinson) as previously described30. Proportions of cells in G1, S, and G2/M phases of the cell cycle were analyzed using FlowJo and Cell Quest software (Becton Dickinson).
PCR analysis of Igκ rearrangements
Semiquantitative PCR using genomic DNA was done as described22. For PCR analysis of Igκ rearrangements indicated Irf4−/−Irf8−/− cell populations were cultured in IL-7 as described for 48 hours. PCR reactions employed degenerate Vκ and Igκ intron primers22 and 5-fold template dilutions. A region upstream of the Igκ intron was amplified to control for amount of genomic DNA22. DNA from WT splenic IgM+ B cells was used as positive control. PCR products were resolved by agarose gel electrophoresis, transferred to Hybond-N membranes (Amersham) and were quantitatively analyzed by Southern blotting using an Igκ intron probe. Products were quantified using a Phosphoimager22. Primer information is provided in Supplementary Table 1.
Electrophoretic Mobility Shift Assay
Nuclear extracts were prepared as described previously22. Prior to the addition of biotin-labeled double-strand DNA probe, 5µg nuclear extracts were incubated for 20 minutes on ice in 20 µl of reaction buffer containing 1X binding buffer, 1 µg of double-stranded poly(dI-dC), 2.5% glycerol, 0.05% NP-40 and 1 µg of bovine serum albumin. Samples were incubated with biotin-labeled probes (20 fmol/each) for 20 minutes at room temperature. In competition experiments, nuclear extracts were pre-incubated with 100-fold molar excess of unlabeled, double-stranded oligonucleotides for 20 minutes on ice. In super-shift experiments, the extracts were pre-incubated with antibodies anti-STAT 5 (sc-835 from Santa Cruz Biotechnology) or anti-E47 (sc763 from Santa Cruz Biotechnology) for 60 minutes on ice. Protein-DNA complexes were separated on 6% nondenaturing TBE gels (Invitrogen) and observed by the LightShift Chemiluminescent EMSA kit following the manufacturer's procedure (Pierce). Oligonucleotide probes used are provided in Supplementary Table 1.
Chromatin Immunoprecipitations (ChIP)
The assays were performed using the ChIP Assay Kit according to the manufacturer’s instructions (Upstate) with some modifications61. STAT 5 was immunoprecipitated with the antibody sc-835x and E47 with sc763x (both from Santa Cruz Biotechnology). Quantitative Real time PCRs were then performed on the purified DNA using using the primers provided in Supplementary Table 1.
Statistical analysis
Data were analyzed by unpaired t test and analysis of variance followed by the test of least significant difference for comparisons within and between groups. All categories within each analyzed experimental panel were compared with significant p values (<0.05) provided. All p values <0.001 were rounded to facilitate comparisons between results.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Thomas Gajewski, University of Chicago, IL, USA for kindly providing the cDNA encoding CA-Ras, CA-STAT 5B and DN-Ras and Dr. Natalie G. Ahn, University of Colorado, Boulder, CO, USA for kindly providing cDNA encoding DN-MEK; M. Veselits for technical assistance; R. Duggan and D. Leclerc for cell sorting services. We would also like to thank Dr. Fotini Gounari for many helpful discussions.
Footnotes
AUTHOR CONTRIBUTIONS
The majority of the experiments in this manuscript were designed, performed, interpreted and readied into figures by Malay Mandal. He also assisted in preparing the initial manuscript. Sarah Powers assisted in the design and analysis of many of the experiments. Kyoko Ochiai assisted with the chromatin immunoprecipitations. Katia Georgopoulos provided the Aiolos specific reagents, scientific advice and assisted in the preparation of the manuscript. Barbara Kee provided E2A specific reagents, contributed to the design of some experiments, provided scientific advice and edited the manuscript. Harinder Singh assisted in establishing many of the experimental systems necessary for this project. He also provided substantial scientific input and edited the final manuscript. Marcus Clark oversaw the entire project and prepared the final manuscript.
References
- 1.Shimizu T, Mundt C, Licence S, Melchers F, Martensson IL. VpreB1/VpreB2/lambda 5 triple-deficient mice show impaired B cell development but functional allelic exclusion of the IgH locus. J Immunol. 2002;168:6286–6293. doi: 10.4049/jimmunol.168.12.6286. [DOI] [PubMed] [Google Scholar]
- 2.Pelanda R, Braun U, Hobeika E, Nussenzweig MC, Reth M. B cell progenitors are arrested in maturation but have intact V(D)J recombination in the absence of Ig-alpha and Ig-beta. J Immunol. 2002;169:865–872. doi: 10.4049/jimmunol.169.2.865. [DOI] [PubMed] [Google Scholar]
- 3.Geier JK, Schlissel MS. Pre-BCR signals and the control of Ig gene rearrangements. Semin Immunol. 2006;18:31–39. doi: 10.1016/j.smim.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Clark MR, Cooper AB, Wang LD, Aifantis I. The pre-B cell receptor in B cell development: recent advances, persistent questions and conserved mechanisms. Curr Top Microbiol Immunol. 2005;290:87–103. doi: 10.1007/3-540-26363-2_5. [DOI] [PubMed] [Google Scholar]
- 5.Erlandsson L, et al. Both the pre-BCR and the IL-7Ralpha are essential for expansion at the pre-BII cell stage in vivo. Eur J Immunol. 2005;35:1969–1976. doi: 10.1002/eji.200425821. [DOI] [PubMed] [Google Scholar]
- 6.Bassing CH, Alt FW. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst) 2004;3:781–796. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Melchers F. B cell development and its deregulation to transformed states at the pre-B cell receptor-expressing pre-BII cell stage. Curr Top Microbiol Immunol. 2005;294:1–17. doi: 10.1007/3-540-29933-5_1. [DOI] [PubMed] [Google Scholar]
- 8.Melchers F. The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol. 2005;5:578–584. doi: 10.1038/nri1649. [DOI] [PubMed] [Google Scholar]
- 9.van Loo PF, Dingjan GM, Maas A, Hendriks RW. Surrogate-light-chain silencing is not critical for the limitation of pre-B cell expansion but is for the termination of constitutive signaling. Immunity. 2007;27:468–480. doi: 10.1016/j.immuni.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 10.Jumaa H, Mitterer M, Reth M, Nielsen PJ. The absence of SLP65 and Btk blocks B cell development at the preB cell receptor-positive stage. Eur J Immunol. 2001;31:2164–2169. doi: 10.1002/1521-4141(200107)31:7<2164::aid-immu2164>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 11.Xu S, Lee KG, Huo J, Kurosaki T, Lam KP. Combined deficiencies in Bruton tyrosine kinase and phospholipase Cgamma2 arrest B-cell development at a pre-BCR+ stage. Blood. 2007;109:3377–3384. doi: 10.1182/blood-2006-07-036418. [DOI] [PubMed] [Google Scholar]
- 12.Bai L, et al. Phospholipase Cgamma2 contributes to light-chain gene activation and receptor editing. Mol Cell Biol. 2007;27:5957–5967. doi: 10.1128/MCB.02273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herzog S, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat Immunol. 2008;9:623–631. doi: 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- 14.Young F, et al. Influence of immunoglobulin heavy- and light-chain expression on B-cell differentiation. Genes Dev. 1994;8:1043–1057. doi: 10.1101/gad.8.9.1043. [DOI] [PubMed] [Google Scholar]
- 15.Flemming A, Brummer T, Reth M, Jumaa H. The adaptor protein SLP-65 acts as a tumor suppressor that limits pre-B cell expansion. Nat Immunol. 2003;4:38–43. doi: 10.1038/ni862. [DOI] [PubMed] [Google Scholar]
- 16.Iritani BM, Forbush KA, Farrar MA, Perlmutter RM. Control of B cell development by Ras-mediated activation of Raf. EMBO J. 1997;16:7019–7031. doi: 10.1093/emboj/16.23.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw AC, Swat W, Ferrini R, Davidson L, Alt FW. Activated Ras signals developmental progression of recombinase-activating gene (RAG)-deficient pro-B lymphocytes. J Exp Med. 1999;189:123–129. doi: 10.1084/jem.189.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw AC, Swat W, Davidson L, Alt FW. Induction of Ig light chain gene rearrangement in heavy chain-deficient B cells by activated Ras. Proc Natl Acad Sci U S A. 1999;96:2239–2243. doi: 10.1073/pnas.96.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 2003;17:1703–1708. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma S, Pathak S, Trinh L, Lu R. Interferon regulatory factors 4 and 8 induce the expression of Ikaros and Aiolos to down-regulate pre-B-cell receptor and promote cell-cycle withdrawal in pre-B-cell development. Blood. 2008;111:1396–1403. doi: 10.1182/blood-2007-08-110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma S, Turetsky A, Trinh L, Lu R. IFN regulatory factor 4 and 8 promote Ig light chain kappa locus activation in pre-B cell development. J Immunol. 2006;177:7898–7904. doi: 10.4049/jimmunol.177.11.7898. [DOI] [PubMed] [Google Scholar]
- 22.Johnson K, et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Lazorchak AS, Schlissel MS, Zhuang Y. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin kappa locus in pre-B cells. Mol Cell Biol. 2006;26:810–821. doi: 10.1128/MCB.26.3.810-821.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokoyoda K, Egawa T, Sugiyama T, Choi BI, Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20:707–718. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Schlissel MS. Regulation of activation and recombination of the murine Igkappa locus. Immunol Rev. 2004;200:215–223. doi: 10.1111/j.0105-2896.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- 26.Romanow WJ, et al. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol Cell. 2000;5:343–353. doi: 10.1016/s1097-2765(00)80429-3. [DOI] [PubMed] [Google Scholar]
- 27.Inlay MA, Tian H, Lin T, Xu Y. Important roles for E protein binding sites within the immunoglobulin kappa chain intronic enhancer in activating Vkappa Jkappa rearrangement. J Exp Med. 2004;200:1205–1211. doi: 10.1084/jem.20041135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazorchak A, Jones ME, Zhuang Y. New insights into E-protein function in lymphocyte development. Trends Immunol. 2005;26:334–338. doi: 10.1016/j.it.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Kee BL, Quong MW, Murre C. E2A proteins: essential regulators at multiple stages of B-cell development. Immunol Rev. 2000;175:138–149. [PubMed] [Google Scholar]
- 30.Cooper AB, et al. A unique function for cyclin D3 in early B cell development. Nat Immunol. 2006;7:489–497. doi: 10.1038/ni1324. [DOI] [PubMed] [Google Scholar]
- 31.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 32.Zhang M, et al. Ubiquitinylation of Ig-beta dictates the endocytic fate of the B cell antigen receptor. J Immunol. 2007;179:4435–4443. doi: 10.4049/jimmunol.179.7.4435. [DOI] [PubMed] [Google Scholar]
- 33.Fleming HE, Paige CJ. Pre-B cell receptor signaling mediates selective response to IL-7 at the pro-B to pre-B cell transition via an ERK/MAP kinase-dependent pathway. Immunity. 2001;15:521–531. doi: 10.1016/s1074-7613(01)00216-3. [DOI] [PubMed] [Google Scholar]
- 34.Anderson LJ, Longnecker R. EBV LMP2A provides a surrogate pre-B cell receptor signal through constitutive activation of the ERK/MAPK pathway. J Gen Virol. 2008;89:1563–1568. doi: 10.1099/vir.0.2008/001461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halcomb KE, et al. Btk and phospholipase Cgamma2 can function independently during B cell development. Eur J Immunol. 2007;37:1033–1042. doi: 10.1002/eji.200636451. [DOI] [PubMed] [Google Scholar]
- 36.Morgan B, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson EC, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26:335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Schuh W, Meister S, Herrmann K, Bradl H, Jack HM. Transcriptome analysis in primary B lymphoid precursors following induction of the pre-B cell receptor. Mol Immunol. 2008;45:362–375. doi: 10.1016/j.molimm.2007.06.154. [DOI] [PubMed] [Google Scholar]
- 39.Wang JH, et al. Aiolos regulates B cell activation and maturation to effector state. Immunity. 1998;9:543–553. doi: 10.1016/s1074-7613(00)80637-8. [DOI] [PubMed] [Google Scholar]
- 40.Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol. 1997;15:155–176. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- 41.Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson RB, et al. Repression of immunoglobulin enhancers by the helix-loop-helix protein Id: implications for B-lymphoid-cell development. Mol Cell Biol. 1991;11:6185–6191. doi: 10.1128/mcb.11.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer KB, Skogberg M, Margenfeld C, Ireland J, Petterson S. Repression of the immunoglobulin heavy chain 3' enhancer by helix-loop-helix protein Id3 via a functionally important E47/E12 binding site: implications for developmental control of enhancer function. Eur J Immunol. 1995;25:1770–1777. doi: 10.1002/eji.1830250643. [DOI] [PubMed] [Google Scholar]
- 44.Sun L, et al. Expression of dominant-negative Ikaros isoforms in T-cell acute lymphoblastic leukemia. Clin Cancer Res. 1999;5:2112–2120. [PubMed] [Google Scholar]
- 45.Palmer MJ, et al. Interleukin-7 receptor signaling network: an integrated systems perspective. Cell Mol Immunol. 2008;5:79–89. doi: 10.1038/cmi.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onishi T, et al. Identification and characterization of a constitutively active STAT 5 mutant that promotes cell proliferation. Mol and Cell Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goetz CA, Harmon IR, O'Neil JJ, Burchill MA, Farrar MA. STAT 5 activation underlies IL7 receptor-dependent B cell development. J Immunol. 2004;172:4770–4778. doi: 10.4049/jimmunol.172.8.4770. [DOI] [PubMed] [Google Scholar]
- 48.Kelly E, Won A, Refaeli Y, Parijs LV. IL-2 and related cytokines can promote T cell survival by activating AKT. J Immunol. 2002;168:597–603. doi: 10.4049/jimmunol.168.2.597. [DOI] [PubMed] [Google Scholar]
- 49.Bertolino E, et al. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT 5. Nat Immunol. 2005;6:836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- 50.Crespo P, Leon J. Ras proteins in the control of the cell cycle and cell differentiation. Cell Mol Life Sci. 2000;57:1613–1636. doi: 10.1007/PL00000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bain G, et al. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 52.Hsu LY, et al. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 2003;19:105–117. doi: 10.1016/s1074-7613(03)00181-x. [DOI] [PubMed] [Google Scholar]
- 53.Jiang H, et al. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 54.Rainey MD, Zachos G, Gillespie DA. Analyzing the DNA damage and replication checkpoints in DT40 cells. Subcell Biochem. 2006;40:107–117. doi: 10.1007/978-1-4020-4896-8_8. [DOI] [PubMed] [Google Scholar]
- 55.Yasuda T, et al. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity. 2008;28:499–508. doi: 10.1016/j.immuni.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 56.Ebisuya M, Kondoh K, Nishida E. The duration, magnitude and compartmentalization of ERK MAP kinase activity: mechanisms for providing signaling specificity. J Cell Sci. 2005;118:2997–3002. doi: 10.1242/jcs.02505. [DOI] [PubMed] [Google Scholar]
- 57.Walker SB, Nelson EA, Frank DA. STAT 5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2006;26:224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 58.Henninghausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT 5A and STAT 5B. Genes Dev. 2008;22:711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asada S, et al. Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cell Signal. 2007;19:519–527. doi: 10.1016/j.cellsig.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 61.Mandal M, et al. Regulation of lymphocyte progenitor survival by the proapoptotic activities of Bim and Bid. Proc Nat Acad Sci, USA. 2008;105:20840–20845. doi: 10.1073/pnas.0807557106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.