Abstract
Prognosis for patients with non-small cell lung cancer (NSCLC) is poor. The potential value of modulating EGFR for treatment is reflected by the recent approval of specific drugs that inhibit its activity. Mutations in EGFR were reported in lung cancer and generated interest, once they enable the identification of lung cancers likely to respond to various targeted small molecules.
We tested 3 key genetic and epigenetic alterations (EGFR, RASSF1A, and BRAF) of this pathway on a series of primary NSCLC [Total 111; adenocarcinoma 49, squamous cell carcinoma (SCC) 48 and others 14]. The mutational status of KRAS (and p53) was known for these samples. The purpose of this study was to define the pattern of erbB pathway alterations in NSCLC and to test for associations with clinical parameters.
Five EGFR mutations were identified: 3 in adenocarcinoma (6 %), 1 in SCC (2%) and 1 in adenocarcinoma with bronchoalveolar component tumor (7%). EGFR mutations included 3 in-frame deletions in exon 19 and 2 point mutations in exon 21. Promoter methylation of RASSF1A was detected in 25 of 45 adenocarcinomas and 18 of 46 SCC. Mutations of EGFR, BRAF and KRAS in adenocarcinoma were mutually exclusive and inversely correlated with RASSF1A methylation (p = −0.394; p=0.007). Overall, genetic and/or epigenetic alterations of erbB pathway genes were detected in 80% (39/49) of adenocarcinomas.
Nearly half of primary adenocarcinoma harbor molecular alterations of the erbB pathway. Careful characterization of these alterations and response to anti-EGFR therapies is warranted to determine better and accurate determinants of clinical response.
Keywords: EGFR mutation, DNA methylation/epigenetics, RASSF1A
INTRODUCTION
Approximately 1 million new cases of lung cancer are diagnosed each year worldwide, resulting in more than 900,000 deaths1. Of these, approximately 220,000 new cases and 160,000 deaths occur annually in the United States2. The survival rates for lung cancers have changed little over the past two decades. A major factor in the high mortality of lung cancer patients is the presence of metastatic tumors in approximately two-thirds of patients at the time of diagnosis3 and no curative therapies exist for metastatic disease. Precise molecular information is required to the development of individualized therapeutic intervention for each tumor type.
The erbB pathway involves a family of tyrosine kinases (EGFR, HER2, etc.) and has contributed to resistance to radiation and chemotherapy in many tumor types4. After binding of several specific ligands, EGFR dimerizes and phosphorylates several tyrosine residues5. These phosphorylated tyrosines serve as the binding sites for several signal transducers that initiate multiple signaling pathways including the RAS-RAF-MEK-ERK pathway5. Oncogenic mutations in the RAS signaling pathway are found in the majority of human cancer. K-RAS leads to signaling of at least three different pathways6. In a recent genomic scale mutational analysis of genes encoding elements of RAS signal transduction pathways, activating mutations of BRAF were identified in several common human cancers, including melanoma, colon cancer, and others7. Interestingly, mutations of KRAS and BRAF were mutually exclusive in tumor types in which both occur, suggesting that KRAS and BRAF provide an equivalent or at least redundant oncogenic stimulus in cancer pathogenesis8. The RASSF1A gene is a candidate tumor suppressor gene at 3p21.3, and resides in one of the most common regions showing loss of heterozygocity in lung cancer9. RASSF1A is inactivated by the hypermethylation of CpG islands in many human cancers, including nasopharyngeal cancer, ovarian cancer, thyroid cancer, colorectal cancer, breast cancer and lung cancer6, 9–15. The presence of a RAS association domain suggests that RASSF1A may be involved in the RAS signaling pathway. Furthermore, it was reported that RASSF1A binds RAS, and its expression induces apoptosis16–18.
Promising characteristics of the EGFR as a molecular target for cancer therapy have prompted an extensive drug development effort to design pharmacologic inhibitors of EGFR signaling. Two first reports19, 20 described somatically acquired mutations in the tyrosine kinase domain of the EGFR that predict for clinical response to gefitinib. In addition, these mutations appeared more frequently in adenocarcinoma, women, non-smokers, and in Asians. In vitro studies demonstrated that the mutant EGFRs retained response to ligand and showed increased sensitivity to gefinitib.
In an effort to identify more comprehensively the genetic and epigenetic alterations of erbB signalling, we investigated 3 key members (EGFR, RASSF1A, and BRAF) of this pathway in a series of NSCLC for which the mutational status of p53 and KRAS had previously been determined21, 22. The objective was to define the pattern of the frequency of these alterations in NSCLC and establish whether any associations exist among these alterations and common pathologic subtypes of lung cancer.
MATERIALS AND METHODS
Patients
A total of 111 patients undergoing surgical resection of a primary NSCLC at The Johns Hopkins Hospital, the Johns Hopkins Bayview Medical Center, or the Medical College of Wisconsin/ Froedtert Memorial Hospital were included in this study. Among these specimens, 49 were adenocarcinoma (including those with bronchoalveolar components n=12), 48 were squamous cell carcinoma (SCC) and 14 were others including large cell carcinoma, adenosquamous and tumor classified as poorly differentiated non-small cell lung cancer.
The clinicopathological characteristics of these patients are summarized in Table 1. All samples were obtained as anonymized material under approval from the Johns Hopkins University Institutional Review Board.
Table 1.
Characteristics of 111 non small cell lung cancer patients.
| Variables | Subgroup | Number of Patients |
|---|---|---|
| Age | ||
| ≤ 66 years | 57 | |
| > 66 years | 54 | |
| Ethnicity | ||
| Caucasian | 82 | |
| African American | 27 | |
| Unknown | 2 | |
| Gender | ||
| Male | 64 | |
| Female | 47 | |
| Tumor Cell Type | ||
| Adenocarcinoma | 49 | |
| Squamous Cell | 48 | |
| Others* | 14 | |
| Pathologic Grade | ||
| Well differentiated | 11 | |
| Moderately differentiated | 48 | |
| Poorly differentiated | 41 | |
| Unknown | 11 | |
| Pathologic Stage | ||
| Stage I | 61 | |
| Stage II | 31 | |
| Stage III | 18 | |
| Stage IV | 1 | |
| Cigarette Smoking | ||
| Nonsmoker | 6 | |
| Smoker | 102 | |
| Unknown | 3 |
Others include large cell, adenosquamous and tumor classified as poorly differentiated non-small cell lung cancer and bronchoalveolar carcinoma
DNA extraction
H&E-stained sections were histologically examined at every 20 sections for the presence or absence of tumor cells, as well as for tumor density. Only sections that showed more than 70% of tumor cells were used for DNA extraction and microdisected tissues were digested with 1% SDS and 50μg/ml proteinase K (Boehringer Mannheim, Germany) at 48°C overnight, followed by phenol/chloroform extraction and ethanol precipitation of DNA as previously described23.
EGFR and BRAF mutation analysis
EGFR mutations were analyzed in exons 18, 19 and 21. These exons were chosen because most of the reported EGFR mutations occurred at these regions. PCR was performed with 5–10 ng of genomic DNA as a template by using the same PCR primers as reported previously20. PCR conditions are as follows: 95°C for 5 min, followed by 35–38 cycles of 95°C for 1 min, 55°C 1 min and 72°C for 1 min. The PCR products were gel purified using a Qiagen PCR product purification kit (Qiagen, Valencia, CA), and the purified PCR products were sequenced with Bigdye Terminator Cycle sequencing Ready Reaction kits (PE Applied Biosystems, Foster City, CA), all according to the manufacturer's instructions. Sequencing was performed in both directions using the forward and reverse PCR primers. The purified products were run on an ABI 310 PRISM Genetic Analyzer (PE Applied Biosystems). The data were collected and analyzed using the Applied Biosystems sequencing analysis software. BRAF mutations were analyzed in exon 15 using the Trimgen Kit as described previously24.
P53 and KRAS gene Mutations Analysis
Mutations at codons 12 and 13 of the KRAS gene were determined using a mismatch ligation assay22. A 270-base pair fragment of exon1 of the KRAS gene was amplified from the tumor DNA. This fragment was used as the template for four separate ligation assays to detect all possible mutations at KRAS codon positions 12a, 12b, 13a and 13b. The ligation products were separated on 12% denaturing polyacrylamide gels. The oligonucleotides used in the ligation assay have been reported previously22. Positive control (DNA with known KRAS mutation) and negative control (cloned polymerase chain reaction products from reactions devoid of DNA and non-neoplastic lung DNA) samples were included with each assay. Mutational status of p53 gene was determined by sequence analysis as described previously21, 25.
Analyses of EGFR Copy Number
The number of copies corresponding to the EGFR locus was determined by real-time quantitative PCR with a 7900 Sequence detector (Perkin-Elmer Applied Biosystems). Briefly, Fluorogenic PCRs were carried out in a reaction volume of 20 μl consisting of 600 nM concentrations of forward and reverse primers; 200 nM probe; 0.6 U of platinum Taq polymerase (Invitrogen, Frederick, MD); 200 μM concentrations each of dATP, dCTP, dGTP and dTTP; and 6.7 mM MgCl2. 20 ng of DNA were used in each real-time PCR reaction. The conditions used for amplification were: one cycle of 95°C for 3 min, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. Reactions were performed in triplicate and the average of the threshold cycle values was calculated. DNA content was normalized to that of Line-1—a repetitive element for which copy numbers per diploid genome are similar in normal or neoplastic human cells26, 27. Changes in copy number were calculated as: 2(Dt–Dline)–(Nt–Nline) as previously published27, where Dt is the average threshold cycle number for experimental primer in DNA extracted from tumor cells, Dline is the average threshold cycle number for Line-1 primer in DNA extracted from tumor cells, Nt is the threshold cycle number in reference DNA extracted from ARPE cells (a cell line derived from retinal pigment epithelial, used as a negative control), and Nline is the threshold cycle number for Line-1 primer in reference DNA extracted from ARPE26, 27. A cell line derived from human epidermoid carcinoma of the skin (A431) was used as a positive control for amplified EGFR. Primer sequences for each region analyzed in this study are included in Table 2.
Table 2.
Primer and probes sequences for Copy Number Analysis
| Gene | Chromossomal Location |
Accession Number (nucleotides) |
Forward 5′-3′ | Probe 6FAM 5′-3′ TAMRA |
Reverse 5′-3′ | Annealing Temperature |
|---|---|---|---|---|---|---|
| EGFR | 7p12 | NT 033968.6 (4848715–4848864) | GAATTCGGATGCA GAGCTTC | CGACGGTCCTCCA AGTAGTTCATG | GACATGCTGCG GTGTTTTC | 60 |
| LINE1 | 1q | U09116 (23927138–23927238) | AACTAGAACTCAGG ATTAAGAATCTCACTC | ATGTACCCAGTAG TCATTCAGGAGC | GCTTTTCTAGTTC TTTTAATTGTGATGTT | 64 |
Bisulfite treatment
DNA from primary tumor was subjected to bisulfite treatment, as described previously with little modification28, 29. Briefly, 2 μg of genomic DNA was denatured in 0.2 M NaOH for 20 min at 50°C. The denatured DNA was diluted in 500 μl of freshly prepared solution of 10 mM hydroquinone and 3 M sodium bisulfite, and incubated for 3 hours at 70° C. After incubation, the DNA sample was desalted through a column (Wizard DNA Clean-Up System, Promega), treated with 0.3 M NaOH for 10 min at room temperature, and precipitated with ethanol. The bisulfite-modified genomic DNA was resuspended in 120 μl of LoTE (2.5 mM EDTA, 10mM Tris-HCL) and stored at −80°C.
Methylation analysis of RASSF1A
Templates were amplified by a fluorescence based-real-time PCR (Taqman) as previously described30. In brief, primers and probes were designed to specifically amplify the bisulfite-converted promoter of the gene of interest: RASSF1A. The ratios between the values of RASSF1A and the internal reference gene, β -actin, obtained by Taqman analysis were used as a measure for representing the relative level of methylation in the particular sample (RASSF1A/β –actin × 1000). Fluorogenic PCRs were carried out in a reaction volume of 20 μl consisting of 600 nM of each primer, 200 nM of probe, 0.6 units of Taq Polymerase, 200 μM each of dATP, dCTP, dGTP; and 200 of μM dTTP; and 6.7 mM MgCl2. Three microliters of treated DNA solution were used in each real-time MSP reaction. Amplifications were carried out in 384-well plates in a 7900 Sequence detector (Perkin-Elmer Applied Biosystems). Each plate consisted of patient samples and multiple water blanks, as well as positive and negative controls. Leukocyte DNA from a healthy individual was methylated in vitro with excess SssI methyltransferase (New England Biolabs Inc., Beverly, MA) to generate completely methylated DNA, and serial dilutions (90–0.009 ng) of this DNA were used to construct a calibration curve for each plate. All samples were within the assay's range of sensitivity and reproducibility based on amplification of an internal reference standard (threshold cycle [CT] value for β –actin of ≤40). The relative level of methylated DNA for each gene in each sample was determined as a ratio of methylation specific PCR-amplified gene (RASSF1A) to β –actin (reference gene) and then multiplied by 1000 for easier tabulation (average value of triplicates of gene of interest divided by the average value of triplicates of β –actin × 1000). The samples were categorized as unmethylated or methylated based on the sensitivity of the assay. In addition to our cancer cases, we also analyzed DNA from 10 different non-neoplastic lung samples for methylation of RASSF1A. An empiric cut off value were established for the determination of methylation of RASSF1A yielding a specificity of 100%.
Statistical analysis
Associations among the alterations of different genes and the clinicopathological parameters were analyzed using the Wilcoxon's rank-sum test for continuous variables Fisher's exact test for categorical variables. Probability values below 0.05 were considered to be statistically significant. The effect of individual gene or combinations thereof on patients survival were estimated using the Kaplan-Meier method, and the difference between the survival curves of the different groups was analyzed by the log-rank test. The Cox proportional hazard model was used to estimate the HR of independent factors, influencing patient's survival, after controlling for potential confounding factors such as age, sex, and disease stage. Due to non-linear pattern of distribution in our variables, correlation coefficients and respective p values were obtained by Spearman rank correlation test. All p values were based on two-sided statistical analysis. The software package Stata version 10 was used for all of the above statistical tests.
RESULTS
The clinical characteristics of 111 patients undergoing pulmonary resection for NSCLC are shown in Table 1. 61 patients had stage I disease, 31 patients had stage II and 19 patients with had stage III or stage IV disease. The histologic type of 111 tumors included squamous cell cancer (n=48), adenocarcinoma (n=49), and others (n=14).
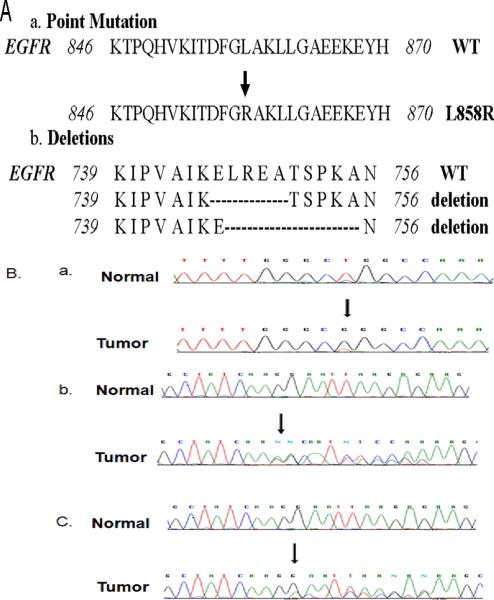
EGFR mutations were detected in 3 (6.1%) of 49 adenocarcinoma patients. One patient with squamous cell and one with poorly differentiated NSCLC showed EGFR mutation. Of the observed mutations in EGFR, 3 were found in exon 19 and rest 2 in exon 21. DNA was also sequenced for corresponding normal of all 5 tumors, which confirmed that all these mutations were somatic. Details of the resulting changes in EGFR protein as a consequence of these mutations are illustrated in Figure 1 and Table 3. We found BRAF mutation (V600E) in only one adenocarcinoma. Methylated RASSF1A allele was present in 50 (48%) of 104 (missing data n = 7 samples) tumors tested including 25 (55%) of 45 adenocarcinoma, 18 (39%) of 46 squamous cell carcinoma. A summary of the genetic and epigenetic alterations are shown in Table 4. No significant relationship was found between pathologic stage and the presence of EGFR mutations, BRAF mutations and RASSF1A methylation (data not shown). The EGFR copy number in tumor cells ranged from 0.14 to 121.2. Increased copy number was observed in 13% of the cases (cutoff of ≥3 copy per cell) or 22% (cutoff of ≥2.5 copy per cell). We also analysed EGFR copy number in lymphocytes of 54 normal subjects. It ranged from 0.04 to 3.31. Increased copy number was observed in 3.7% (cutoff of ≥3 copy per cell) or 5.6% (cutoff of ≥2.5 copy per cell). No correlations were observed between the EGFR copy number and RASSF1A methylation. The Spearman correlation coefficient between these two variables was 0.01619 (p=0.8).
Figure 1.
Analysis of EGFR mutations in the TK domain of the EGFR gene found in unselected cases with lung cancer. A) Sequence and position of EGFR mutations in the TK domain found in unselected cases with lung cancer B) (a) Sequencing chromatogram showing EGFR exon 21 wild type (on top) and mutant L858R (on bottom) from an adenocarcinoma. The arrow indicates the nucleotide substitution. (b) Sequencing chromatogram showing EGFR exon 19 wild type (on top) and mutant delE746-A750 (on bottom) from an adenocarcinoma. The arrow indicates the starting point of the deletion. (C) Sequencing chromatogram showing EGFR exon 19 wild type (on top) and mutant delE747-A755 (on bottom) from an adenocarcinoma. The arrow indicates the starting point of the deletion.
Table 3.
Genetic alterations of EGFR in NSCLC
| Sample ID | Alteration | Nucleotide | Cancer Type | ||
|---|---|---|---|---|---|
| Exon 19 | 1 | delE746-A750 | 2235–2249 | Adenocarcinoma | |
| 2 | delL747-A755 | 2239–2264 | Adenocarcinoma | ||
| 3 | delE746-A750 | 2236–2250 | Adenocarcinoma | ||
| Exon 21 | 4 | L858R | 2573 T>G | Adenocarcinoma | |
| 5 | L858R | 2573 T>G | Adenocarcinoma | ||
| 6 | Polymorphism | 836 | 2508C>T | Adenocarcinoma | |
| 7 | 836 | 2508C>T | SCC | ||
| 8 | 836 | 2508C>T | SCC |
Table 4.
Frequency of genetic and epigenetic alterations of markers examined
| Genes | Overall | Adeno | SCC | Others |
|---|---|---|---|---|
| EGFR | 5/111 (4.5%) | 3/49 (6.1%) | 1/48 (2.08%) | 1/14 (7.1%) |
| BRAF | 1/111 (0.9%) | 1/49 (2.04%) | 0/48 (0%) | 0/14 (0%) |
| K-RAS | 18/63 (28.6%) | 17/43 (39.5%) | 1/11 (9%) | - |
| p53 | 63/111 (56.7%) | 22/49 (44.9%) | 33/48 (68.7%) | 8/14 (57.1%) |
| RASSF1A Cutoff ⩾ 10 | 50/104 (48%) | 25/45 (55.5%) | 18/46 (39.1%) | 7/13 (53.8%) |
| Any Genetic alteration in ERB pathway | 24/111 (21.6%) | 21/49 (42.8%) | 2/48 (4.1%) | 1/14 (7.1%) |
| Any Genetic and/or epigenetic alteration (including p53 mutations) | 94/111 (84.7%) | 44/49 (89.8%) | 40/48 (83.3%) | 10/14 (71.4%) |
| Any Genetic and/or epigenetic alteration (excluding p53 mutations) | 66/111 (59.4%) | 39/49 (79.6%) | 20/48 (41.7%) | 7/14 (50%) |
Genetic and epigenetic alterations in adenocarcinoma
Tumor specific molecular alterations in erbB pathway genes were identified in 39 (79.6%) of 49 patients with lung adenocarcinoma. Of these 39 tumors, 3 (7.7%) harbored mutations in EGFR, 11 (28.2%) in K-RAS mutations, and BRAF mutation was found in one (2.5%). RASSF1A methylation was detected in 24/39 (61.5%) of these 39 adenocarcinoma patients. Overall, 21 (43%) out of 49 adenocarcinoma showed genetic alterations in any of the 3 genes (EGFR, BRAF and KRAS) in the erbB pathway. Mutations of EGFR, KRAS and BRAF are mutually exclusive. Mutation of KRAS and methylation of RASSF1A were inversely correlated (Spearman rank correlation: ρ = −0.386; p=0.014). Mutations of all erbB pathway genes (EGFR, KRAS, BRAF) were also inversely correlated with RASSF1A methylation (ρ = −0.394; p=0.007). However, in 5 cases we found both KRAS mutation and RASSF1A methylation. Among these 5 cases, all patients were smokers and 4 presented high stage tumors. The coexistence or mutually exclusive occurrence of these alterations can be visualized in Supplementary Figure 1.
ERB pathway alterations in squamous cell carcinoma
Consistent with the previous findings, mutations of erbB pathway genes were rare in squamous cell carcinoma. RASSF1A methylation occurred in 39.1% of primary tumors. Kaplan-Meier estimator of the survivorship function was used to examine the effect of individual gene and in combinations on patient's survival. The survival groups with altered erbB pathway in adenocarcinoma were compared by using the log-rank test. No significant relationship was found between erbB pathway genes alterations and patient survival. Representative Kapplan Meyer curves for individual alterations as well as combinations are shown in Supplementary Figure 2.
erbB pathway alterations and p53 status
We compared erbB pathway alterations and p53 status in these tumors. As expected for an independent pathway, we found no correlation between KRAS mutation (ρ = −0.20; p=0.19) or RASSF1A methylation (p = 0.13; p=0.39) and p53 status in these tumors. Moreover, we found no association between erbB pathway alterations (EGFR/KRAS/RASSF1A/BRAF) and p53 status in SCC or adenocarcinoma with EGFR copy number.
DISCUSSION
Elucidation of the molecular pathways involved in NSCLC is essential for our understanding of the pathogenesis of the disease, and, hence, for more precise diagnosis, accurate prognosis and better management of patients. In this study, we evaluated the molecular status of four key genes (EGFR, KRAS, RASSF1A and BRAF) in the erbB signaling pathway in primary NSCLC.
The frequency of oncogenic mutations of EGFR gene in our study is similar to previous findings19, 20. We found three in frame deletion on exon 19 (delE 746-A750; delL747-A755) and 2 point mutations on exon 21(L858R). The oncogenic potential and therapeutic response of novel deletion (delL747-A755 on exon 19) identified in this study need to be further evaluated. We found silent polymorphic changes in three additional cases on exon 21 (codon 836, Nucleotide 2508 C>T) (Table 3) and pathophysiologic role of these polymorphism not yet known in NSCLC. (THIS PARAGRAPH IS REWRITTEN)
The prevalence of RASSF1A methylation in adenocarcinoma in this study occurred at a frequency similar to that found by others9, 10, 30. On the basis of many observations suggesting that RASSF1A mediates RAS-dependent apoptosis (1, 6, 8, 11), and it was hypothesized that RASSF1A inactivation is closely related to RAS activation in human cancers and thus contributes to malignant transformation by inhibiting RAS-mediated apoptosis. Our findings of inverse correlation between RASSF1A methylation and KRAS mutations support this hypothesis.
Our data are not consistent with the two adenocarcinoma studies31, 32 pertaining to the association between mutation of KRAS and methylation of RASSF1A. In the latter states, tumors from Asian patients were tested by conventional MSP. In contrast, we tested samples from American patients, mostly Caucasian and used Quantitative MSP (QMSP). In our previous work in thyroid cancer, a 10% cutoff for RASSF1A methylation correlated inversely with mutations of the reference tyrosine kinase pathway33. Our findings do agree with van Engeland et al.12, which found that RASSF1A methylation occurs mainly in colorectal cancers without KRAS mutation, which implied that RASSF1A methylation is an alternative way of affecting RAS signaling. In 5 cases, we found an overlap between KRAS activation and RASSF1A methylation. Although our data did not reach statistical significance, RASSF1A methylation was more common in higher stage tumors. Therefore, it is possible that synergistic inactivation of RASSF1A and KRAS activation may drive poor prognosis. We found a stronger inverse correlation (p=0.007) when we compared the mutation status of any erbB pathway gene with RASSF1A methylation in adenocarcinoma, supporting the notion that all four genes are involved in RAS signaling and facilitate tumorigenesis.
We found a BRAF mutation in only one sample which is consistent with our previous observation34. However, we analyzed only the most common BRAF mutation in humans (T1796A). Interestingly, we did not found any other molecular alterations in this sample. In support of this notion, the BRAF mutation exhibits a trend towards mutual exclusion with KRAS and RASSF1A in human tumors7, 8, 33, 35. In the present study, we examined only codon V600 for BRAF mutation. Based on their findings, Marcia et al36 hypothesized that BRAF-related tumorigenesis in NSCLC is qualitatively different from that in melanomas with codon 600 mutations.
In our adenocarcinoma cohort we found that 29 out of 49 (80 %) cases showed alterations in erbB pathway genes. Virtually all adenocarcinomas may acquire alterations in erbB signaling at the receptor kinase or through alterations at key downstream molecules. Several markers have been identified that predict response to the EGFR-specific tyrosine kinase inhibitors (EGFR-TKI) in patients with NSCLC. Activating mutations in the EGFR tyrosine kinase domain (exons 18–21), increased EGFR copy number, and increased EGFR protein expression have been associated with favorable response to EGFR-TKIs19, 20, 37–45. In contrast, KRAS gene mutation, which occurs in 20% to 30% of NSCLCs, mainly in adenocarcinomas (~40%) and smokers46, has been reported to be associated with poor response to EGFR-TKIs47–49. It will be interesting to see if BRAF mutation and RASSF1A methylation also predict resistance to upstream targeted therapy. Characterization of all of these key genetic and epigenetic events may thus help to decide optimal therapy for NSCLC patients. Additionally identified (e.g. PI3K, 4% mutation in lung cancer)50 and unidentified genes may be responsible for signaling alterations in the remaining cases of adenocarcinoma. Consistent with the previous findings, our data suggests that genetic alterations of erbB pathway genes are rare in lung squamous cell carcinoma. However, RASSF1A methylation was detected in 39% of SCC cases and may identify EGFR signaling in these tumors or another role for RASSF1A in a different histologic subtype51, 52.
We have thus confirmed the importance of the erbB pathway in NSCLC development and identified one novel deletion mutation of EGFR in lung adenocarcinoma. Although EGFR and BRAF mutations are rare, in lung cancer, they may identify sensitive and resistant patients to targeted therapy, resulting in significantly improved outcomes for patients treated appropriately. The importance of RASSF1A methylation in determining anti-EGFR therapy needs to be further elucidated in clinical trials.
Supplementary Material
Acknowledgments
This work was partially supported by National Cancer Institute Grant U01-CA84986, Oncomethylome Sciences, SA. The funding agencies had no role in the design of the study, data collection, analysis, interpretation of the results, preparation of the manuscript, or the decision to submit the manuscript for publication. Under a licensing agreement between Oncomethylome Sciences, SA and the Johns Hopkins University, D.S. is entitled to a share of royalty received by the University upon sales of diagnostic products described in this article. D.S. owns Oncomethylome Sciences, SA stock, which is subject to certain restrictions under University policy. D.S. is a paid consultant to Oncomethylome Sciences, SA and is a paid member of the company's Scientific Advisory Board. The Johns Hopkins University in accordance with its conflict of interest policies is managing the terms of this agreement. Dr. Hoque is supported by Young Investigator Award from the International Association for the Study of Lung Cancer. Dr. Begum and Dr. Hoque are supported by a Young Clinical Scientist Award from the Flight Attendant Medical Research Institute.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Parkin DM, Pisani P, Ferlay J. Global cancer statistics. CA Cancer J Clin. 1999;49:33–64. 31. doi: 10.3322/canjclin.49.1.33. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Wingo PA, Ries LA, Giovino GA, et al. Annual report to the nation on the status of cancer, 1973–1996, with a special section on lung cancer and tobacco smoking. J Natl Cancer Inst. 1999;91:675–690. doi: 10.1093/jnci/91.8.675. [DOI] [PubMed] [Google Scholar]
- 4.Chakravarti A, Chakladar A, Delaney MA, et al. The epidermal growth factor receptor pathway mediates resistance to sequential administration of radiation and chemotherapy in primary human glioblastoma cells in a RAS-dependent manner. Cancer Res. 2002;62:4307–4315. [PubMed] [Google Scholar]
- 5.Arteaga CL. Overview of epidermal growth factor receptor biology and its role as a therapeutic target in human neoplasia. Semin Oncol. 2002;29:3–9. doi: 10.1053/sonc.2002.35642. [DOI] [PubMed] [Google Scholar]
- 6.Shields JM, Pruitt K, McFall A, et al. Understanding Ras: `it ain't over 'til it's over'. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 7.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 8.Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 9.Dammann R, Li C, Yoon JH, et al. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 10.Burbee DG, Forgacs E, Zochbauer-Muller S, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo KW, Kwong J, Hui AB, et al. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res. 2001;61:3877–3881. [PubMed] [Google Scholar]
- 12.van Engeland M, Roemen GM, Brink M, et al. K-ras mutations and RASSF1A promoter methylation in colorectal cancer. Oncogene. 2002;21:3792–3795. doi: 10.1038/sj.onc.1205466. [DOI] [PubMed] [Google Scholar]
- 13.Ibanez de Caceres I, Battagli C, Esteller M, et al. Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 2004;64:6476–6481. doi: 10.1158/0008-5472.CAN-04-1529. [DOI] [PubMed] [Google Scholar]
- 14.Hoque MO, Rosenbaum E, Westra WH, et al. Quantitative assessment of promoter methylation profiles in thyroid neoplasms. J Clin Endocrinol Metab. 2005;90:4011–4018. doi: 10.1210/jc.2005-0313. [DOI] [PubMed] [Google Scholar]
- 15.Hoque MO, Begum S, Sommer M, et al. PUMA in head and neck cancer. Cancer Lett. 2003;199:75–81. doi: 10.1016/s0304-3835(03)00344-6. [DOI] [PubMed] [Google Scholar]
- 16.Vos MD, Ellis CA, Bell A, et al. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem. 2000;275:35669–35672. doi: 10.1074/jbc.C000463200. [DOI] [PubMed] [Google Scholar]
- 17.Khokhlatchev A, Rabizadeh S, Xavier R, et al. Identification of a novel Ras-regulated proapoptotic pathway. Curr Biol. 2002;12:253–265. doi: 10.1016/s0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- 18.Ortiz-Vega S, Khokhlatchev A, Nedwidek M, et al. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene. 2002;21:1381–1390. doi: 10.1038/sj.onc.1205192. [DOI] [PubMed] [Google Scholar]
- 19.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 20.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 21.Ahrendt SA, Hu Y, Buta M, et al. p53 mutations and survival in stage I non-small-cell lung cancer: results of a prospective study. J Natl Cancer Inst. 2003;95:961–970. doi: 10.1093/jnci/95.13.961. [DOI] [PubMed] [Google Scholar]
- 22.Ahrendt SA, Decker PA, Alawi EA, et al. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001;92:1525–1530. doi: 10.1002/1097-0142(20010915)92:6<1525::aid-cncr1478>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 23.Hoque MO, Lee CC, Cairns P, et al. Genome-wide genetic characterization of bladder cancer: a comparison of high-density single-nucleotide polymorphism arrays and PCR-based microsatellite analysis. Cancer Res. 2003;63:2216–2222. [PubMed] [Google Scholar]
- 24.Benoit NE, Goldenberg D, Deng SX, et al. Colorimetric approach to high-throughput mutation analysis. Biotechniques. 2005;38:635–639. doi: 10.2144/05384PF01. [DOI] [PubMed] [Google Scholar]
- 25.Ahrendt SA, Halachmi S, Chow JT, et al. Rapid p53 sequence analysis in primary lung cancer using an oligonucleotide probe array. Proc Natl Acad Sci U S A. 1999;96:7382–7387. doi: 10.1073/pnas.96.13.7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang TL, Maierhofer C, Speicher MR, et al. Digital karyotyping. Proc Natl Acad Sci U S A. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 28.Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoque MO, Begum S, Topaloglu O, et al. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64:5511–5517. doi: 10.1158/0008-5472.CAN-04-0799. [DOI] [PubMed] [Google Scholar]
- 30.Topaloglu O, Hoque MO, Tokumaru Y, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–2288. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- 31.Kim DH, Kim JS, Park JH, et al. Relationship of Ras association domain family 1 methylation and K-ras mutation in primary non-small cell lung cancer. Cancer Res. 2003;63:6206–6211. [PubMed] [Google Scholar]
- 32.Endoh H, Yatabe Y, Shimizu S, et al. RASSF1A gene inactivation in non-small cell lung cancer and its clinical implication. Int J Cancer. 2003;106:45–51. doi: 10.1002/ijc.11184. [DOI] [PubMed] [Google Scholar]
- 33.Xing M, Cohen Y, Mambo E, et al. Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer Res. 2004;64:1664–1668. doi: 10.1158/0008-5472.can-03-3242. [DOI] [PubMed] [Google Scholar]
- 34.Cohen Y, Xing M, Mambo E, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 35.Yuen ST, Davies H, Chan TL, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res. 2002;62:6451–6455. [PubMed] [Google Scholar]
- 36.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 37.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 38.Cortes-Funes H, Gomez C, Rosell R, et al. Epidermal growth factor receptor activating mutations in Spanish gefitinib-treated non-small-cell lung cancer patients. Ann Oncol. 2005;16:1081–1086. doi: 10.1093/annonc/mdi221. [DOI] [PubMed] [Google Scholar]
- 39.Han SW, Kim TY, Hwang PG, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23:2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- 40.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 41.Takano T, Ohe Y, Sakamoto H, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 42.Taron M, Ichinose Y, Rosell R, et al. Activating mutations in the tyrosine kinase domain of the epidermal growth factor receptor are associated with improved survival in gefitinib-treated chemorefractory lung adenocarcinomas. Clin Cancer Res. 2005;11:5878–5885. doi: 10.1158/1078-0432.CCR-04-2618. [DOI] [PubMed] [Google Scholar]
- 43.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 44.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 45.Hirsch FR, Varella-Garcia M, Bunn PA, Jr., et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 46.Graziano SL, Gamble GP, Newman NB, et al. Prognostic significance of K-ras codon 12 mutations in patients with resected stage I and II non-small-cell lung cancer. J Clin Oncol. 1999;17:668–675. doi: 10.1200/JCO.1999.17.2.668. [DOI] [PubMed] [Google Scholar]
- 47.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 49.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 50.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 51.Oh HJ, Lee KK, Song SJ, et al. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006;66:2562–2569. doi: 10.1158/0008-5472.CAN-05-2951. [DOI] [PubMed] [Google Scholar]
- 52.Vos MD, Martinez A, Elam C, et al. A role for the RASSF1A tumor suppressor in the regulation of tubulin polymerization and genomic stability. Cancer Res. 2004;64:4244–4250. doi: 10.1158/0008-5472.CAN-04-0339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.