Abstract
Two-component signal transduction pathways play a major role in the response of bacteria to external cues. These pathways are initiated by large collection of histidine kinases (HKs) containing a sensor domain that perceives the environmental signal followed by an HK domain that triggers a histidine-aspartate phosphorelay. Previous phylogenetic analyses identified 11 major families of two-component HKs by comparing signature motifs within the HK domain. Here we describe a new family with homology to Agrobacterium tumefaciens BphP2, an HK first discovered by the presence of a phytochrome sensor domain involved in light perception. Members of this sensor HK family differ from most others by the absence of a recognizable F box and the presence of several uniquely conserved residues, including a histidine in the N box and a tryptophan-X-glutamic acid sequence in the G1 box, which we have used to define the family (HWE). At least 81 members were identified in a variety of α- and γ-proteobacteria, with a significant enrichment in the Rhizobiaceae family. Several representatives were shown to have HK activity in vitro, supporting their proposed participation in phosphorelays. One or more domains related to signal transduction were evident N-terminal to the HK domain, including chemotactic methyltransferase domains, suggesting that this family has multiple roles in environmental signaling. The discovery of the HWE family further extends the diversity within the HK superfamily and expands the importance of two-component signaling in bacteria.
Protein kinase cascades are widely used by both prokaryotes and eukaryotes to help them sense and respond to external and internal signals. One cascade type commonly employed by bacteria to adapt to environmental changes involves histidine-aspartate phosphorelays. These relays, which are also referred to as two-component signal transduction pathways, invariably involve two signaling proteins/modules, a sensor histidine kinase (HK) and a paired response regulator (RR) (17, 27). Upon receipt of a specific stimulus by a sensor domain within the HK, an associated kinase domain is activated, resulting in autophosphorylation of a conserved histidine within the HK domain. This high-energy phosphate is then donated to a conserved aspartate within a cognate RR. For some RRs, this phosphorylation directly activates an associated output domain that then initiates the response. The most common responses involve transcriptional up or down regulation of target genes mediated by activation of a DNA-binding domain directly appended to the RR. For other RRs, further transfer of the aspartyl phosphate to a histidine within a histidine phosphotransferase (HPT) followed by donation to an aspartate within a second RR is employed, thus creating a four-step His-Asp-His-Asp relay (2).
Arguably the most widespread signal transduction pathways in bacteria, these HK systems influence numerous cellular processes, including chemotaxis, osmoregulation, anaerobic respiration, photosynthesis, nitrogen and phosphate acquisition, sporulation, host recognition by pathogens, antibiotic production, the cell cycle, and viability (17, 27). The modular organization of the two-component kinase cascades allows individual components to house various permutations of sensor, HK, RR, HPT, and output modules. For example, a number of hybrid HKs exists which contain the sensor, HK, and RR domains together in a single polypeptide. A linear phosphorelay is generated by some signals. For others, multiple relays are activated by several HKs to induce an anastomosing web of responses (17, 22). Some HKs even exhibit phosphatase activity towards their cognate RR as an additional way to regulate phosphotransfer through RRs (16).
Sensor HKs are typically organized as homodimers with the sensor domain at the N terminus and the HK domain, which also contains the sites for intermolecular contact, at the C terminus. Formation of the phosphohistidine intermediate actually occurs in trans, by using one member of the HK dimer to bind ATP and its partner to provide the histidine phosphoacceptor. Whereas the sensor domain is highly variable among members of the superfamily, the HK domain is more conserved, especially within a set of six recognizable motifs or boxes designated H, N, F, G1, G2, and G3 after the invariant amino acid residue(s) in each (10, 14). For example, the H box contains a conserved histidine that serves as the initial phosphoacceptor from ATP, whereas, the N and G1 to G3 boxes contain an asparagine and several glycine residues, respectively, that help define the nucleotide-binding cleft (10).
Structural comparisons indicate that much of the HK domain shares features with other ATPase modules, including those from GyrB, Hsp90, and MutL, and thus have been included in a larger collection of kinase-ATPase proteins, termed the GHKL superfamily (10). The unifying feature of this superfamily is the presence of a distinctive ATP-binding pocket, called the Bergerat fold (5). It is an α/β sandwich consisting of a four-stranded mixed β sheet and three α helices. These α and β elements constitute the structural framework of the ATP-binding site, whereas the amino acids that contact the bound ATP cluster in highly conserved surface loops connecting these elements (10).
The accelerating release of finished genomic sequences has greatly expanded the number of known two-component HKs in the bacterial kingdom and even demonstrated a limited presence in archaea, plants, fungi, and protozoans (17, 27). Phylogenetic analyses have tentatively divided the HK superfamily into at least 11 families (12) that can be arranged in three superclusters (17). Sequence comparisons among individual families have shown that the organization of the HK domain is not absolute and that substantial divergence from the canonical HK domain can be tolerated. Whereas all appear to have the recognizable H and N boxes, some families are either missing or have distinct F and G1 to G3 boxes (12). Whether this heterogeneity translates into distinct nucleotide preferences, activities, and/or functions is not yet known.
During our analysis of AtBphP2, a unique bacteriophytochrome (BphP) from Agrobacterium tumefaciens, we discovered that it has HK activity in vitro even though the region presumed to be responsible is substantially different from that found in typical HKs (18). Using the amino acid sequence from this region as a query, we subsequently identified a group of 81 related proteins in a wide array of other α- and γ-proteobacteria, many of which were not annotated by protein motif prediction programs to be HKs. The HK domain from this group can be distinguished from those of other sensor HK families by the absence of a recognizable F box and the presence of signature H and W-E residues within the presumed N and G1 boxes, respectively. Based on this conservation, we have classified these HKs as the HWE family. A variety of predicted sensing domains are upstream of the HK domain, including the pocket used by BphPs to bind light-sensing bilins and the methyltransferase domain used by components of chemotaxis signaling pathways to modulate the response to attractants and repellants. The discovery of the HWE family further expands the superfamily of sensor HKs used in bacterial signaling.
MATERIALS AND METHODS
Alignments and phylogenetic analysis.
Database searches were performed with National Center for Biotechnology Information BLASTP (1) with the HK domain from AtBphP2 as the query. Protein sequence alignments and unrooted phylogenetic trees were generated with CLUSTALX V1.8 (29) by using the predicted HK domain beginning ∼10 residues before the presumed histidine phosphorylation site in the H box and ending ∼11 residues after the most C-terminal glycine in the G-rich box (His523 and Gly693, respectively, in AtBphP2). An alignment of the full domain is available upon request. Other protein motifs were predicted by SMART (24). Phylogenetic comparisons to other HK families used classifications and representatives as defined by reference 12.
Cloning and expression of HWE-HK proteins.
Coding regions for AtBphP2, AtExsG, and SmSMa2063 were amplified from A. tumefaciens strain C58 and Sinorhizobium meliloti strain 2011 genomic DNA by PCR with TaKaRa Ex-Taq polymerase (PanVera, Madison, Wis.). The PCR products were inserted directly into pGEMT and then introduced into pET21b (Novagen) as NdeI and XhoI fragments. The resulting open reading frames begin with the original start codon and end with six additional His codons followed by a stop codon. Recombinant proteins were expressed in E. coli strain BL21-Codon Plus(DE3)-RIL (Stratagene, La Jolla, Calif.) and purified by nickel chelate affinity chromatography as described previously (6). Site-directed mutations for AtBphP2 (H523K, H611K, W651Y, and E653D) were introduced by QuikChange (Stratagene). Each coding region was sequenced in its entirety by the dideoxy method to confirm introduction of the appropriate mutation.
In vitro phosphorylation assay.
Protein kinase assays were performed with [γ-32P]ATP as described previously (18), with the exception that the incubation time was for 2 h to ensure saturation of the phosphorylated intermediates. Protein levels were verified by Ponceau staining of the membranes prior to autoradiography.
RESULTS
Identification of the HWE HK Family.
The founding member of the HWE HK family, BphP2 from A. tumefaciens, was discovered during our search for proteins related to Deinococcus radiodurans BphP in various finished bacterial genomic sequences (6). BphPs are a family of photoreceptors that contain an N-terminal chromophore pocket, comprised of both a GAF (cyclic GMP, adenylyl cyclase, FhlA domain) and a phytochrome (phy) motif that autocatalytically attaches bilins such as biliverdin (6, 20, 30). Through an as yet to be defined set of chromophore-protein interactions, BphPs attain a set of distinctive red-far red light photochromic spectral properties that allow these pigments to become photoreversible switches in light perception. Most BphPs identified in the search also contain the recognizable array of H, N, F, and G1 to G3 boxes in their C-terminal halves, strongly suggesting that they function as light-regulated sensor HKs (6, 9). This activity was confirmed by in vitro kinase assays of several representatives that demonstrated both autophosphorylation of the presumed histidine and subsequent transfer to a cognate RR, which is often encoded within the same operon (6, 15, 18, 33).
One unique member of the BphP family is AtBphP2. It contains the GAF-phy sequence that binds biliverdin, followed by a domain with little similarity to the canonical HK domain, and terminates with an RR, suggesting that it is a unique type of a hybrid HK (18). The recombinant protein was demonstrated to have kinase activity, with deletion analysis identifying the region between the GAF-phy and RR motifs as essential (reference 18 and data not shown). To help locate the residues important for the phosphotransferase activity, we compared this region to the HK domains from previously described sensor HKs. The closest match was a group of HKs from the thermophilic archaeon Methanobacterium thermoautotrophicum (26). Their consensus H boxes aligned with a histidine-containing sequence in AtBphP2, but the distal ∼200 residues of AtBphP2 aligned poorly, suggesting that the N, F, and G1 to G3 motifs were either not sufficiently related or absent.
Using the internal sequence of AtBphP2 as a query, we searched by BLAST (1) for related proteins. Surprisingly, 81 proteins from a variety of bacteria were identified (as of 1 April 2003) that also contain this sequence near their C-termini. As can be seen from the alignment in Fig. 1, the group contained a number of conserved regions. The most proximal region resembled the H box from the Methanobacterium group of HKs, including the presence of the presumed active site histidine that is always followed by an arginine, but had low similarity to H boxes from all other HK types (12). Closer analysis of the conserved regions downstream of the H box located a likely N box defined by the presence of a conserved asparagine (underlined) that is flanked by an unusual consensus HELATNAXKYGALS motif. No obvious F box was detected. Likewise, the C-terminal region showed several distinctive features. Three potential G boxes were identifiable in an ∼50-amino-acid glycine-rich stretch, but their sequences differed from the canonical G1 to G3 boxes. The consensus GXGXG motif employed by the G2 domain to bind the phosphates of ATP (10) was replaced by a near unanimous GFGXXL/V motif, whereas the consensus DXGXG sequence used by the G1 domain to bind the adenine moiety (10) was apparently replaced by a nearly invariant WXEXGGP sequence (Fig. 1). A number of other conserved residues were also evident, including a second conserved tryptophan in the intervening sequence between the N and G1 boxes (data not shown). Given that the histidine in the N box and the tryptophan-X-glutamic acid residues within the G1 box appeared to help define this group of potential HKs, we designated them the HWE family for simplicity.
FIG. 1.
Amino acid sequence alignment of portions of the kinase region of the HWE family of HKs. The 81 members were aligned by ClustalX and displayed with MACBOXSHADE by using a threshold of 60%. Only the sequences encompassing the H-, N-, and G-rich boxes are shown. Reverse type and gray boxes denote identical and similar amino acids, respectively. Dots denote gaps. The residue numbers are for AtBphP2 (18). The closed arrowhead identifies the histidine predicted to form the phosphohistidine intermediate. Open arrowheads identify the positionally conserved histidine, tryptophan, and glutamic acid residues that help distinguish this group. The diamond identifies the positionally conserved asparagine in the N box. Circles identify glycines within the G-rich box. Each sequence is listed as an abbreviated species name followed by the accession number. Sequences are from A. tumefaciens (At), M. loti (Ml), Caulobacter crescentus (Cc), S. meliloti (Sm), R. palustris (Rp), R. sphaeroides (Rs), B. japonicum (Bj), Novosphingobium aromaticivorans (Na), Magnetospirillum magnetotacticum (Mm), X. axonopodis (Xa), X. campestris (Xc), B. melitensis (Bm), Brucella suis (Bs), Rhodospirillum rubrum (Rr), R. leguminosarum (Rl), P. putida (Pp), and P. syringae (Ps). GenBank accession numbers are as follows: AtBphP2, NP_355125; RlBphP, CAC95194; PpBphP2, AAL50633; PsBphP2, ZP_00126919.
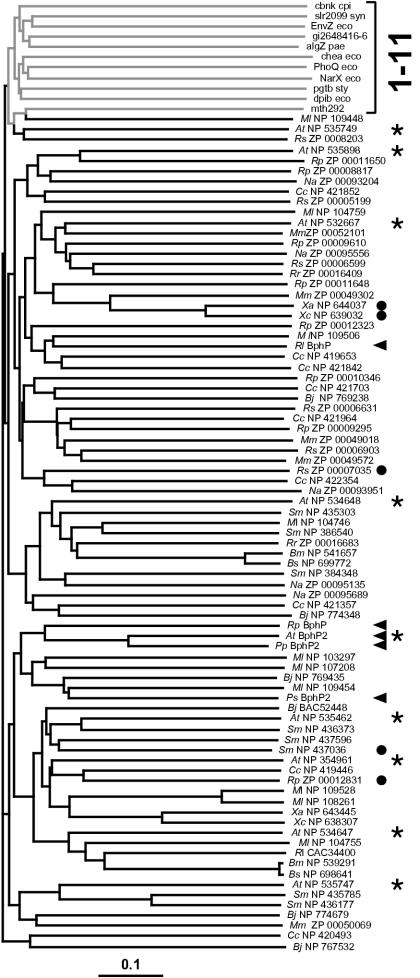
To provide further support for the idea that the HWE family represents a new type of HKs, we generated an unrooted phylogenetic tree by the neighbor-joining algorithm (13) by using all 81 members of the family along with single representatives of the 11 previously described HK families (12). To improve the alignment, only the sequences between the presumed H and G3 boxes were used. Based on a bootstrap value of 1,000, the HWE members all clustered together separate from most other HK families (Fig. 2). The only exception was the representative of the group 11 HKs (MNP 109448) (12), which emerged from one branch of the HWE cluster. However, when more members of group 11 were included in random bootstrap replicates, group 11 clearly behaved as a distinct out-group separate from the HWE family (data not shown). Although containing a similar organization of their G1 to G3 boxes, the HKs of group 11 have the DXGXG and not WXEXGGP motif in the G1 box and are missing the conserved histidine in the N box. The only exception was A. tumefaciens ExsG. Although first assigned to group 11 HKs (12), our comparisons indicate that ExsG more closely aligns with the HWE group, having both the WXEXGGP motif and the N box histidine.
FIG. 2.
Phylogenetic comparison of the HWE-HK family with representatives of the previously described HK families. The tree was generated by using the predicted HK domain by the ClustalX neighbor-joining method. The bar represents a branch length equivalent to 0.1 amino acid change per residue. The bracket on the right identifies representatives of the 11 previously defined HK families (gray lines). Black lines indicate the 81 representatives of the HWE HK family. Arrowheads and circles identify members with GAF-phy and potential chemotaxis methyltransferase sensor domains, respectively, upstream of the HK domain. The asterisks identify members of the HWE-HK family from A. tumefaciens. Abbreviations are as defined in the legend to Fig. 1.
Members of the HWE HK family have kinase activity.
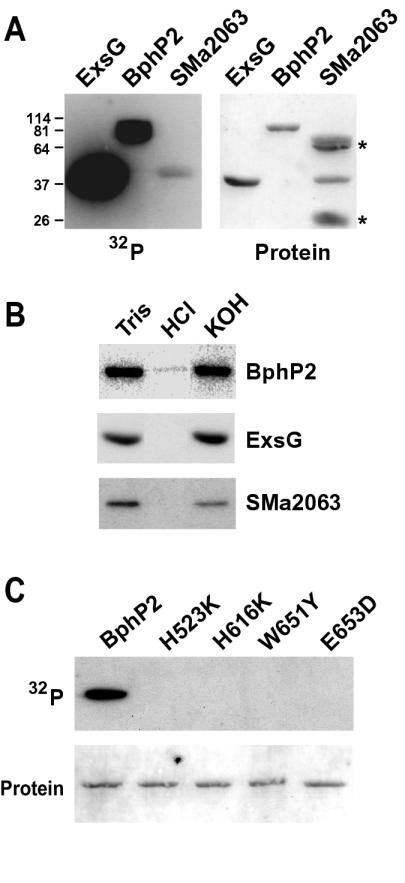
To confirm that the HWE proteins are bona fide HKs, we tested several representatives for kinase activity in vitro. Recombinant proteins were expressed with His6 tags in Escherichia coli and purified by nickel chelate affinity chromatography. The proteins were then incubated with [γ-32P]ATP and assayed for the formation of the phosphohistidine intermediate by autoradiography following sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the proteins. Full-length BphP2 and ExsG from A. tumefaciens (with their appended RRs) and open reading frame SMa2063 from S. meliloti (previously not noted as a potential HK) all showed detectable HK activity (Fig. 3A). Furthermore, the phosphorylated forms were base stable but acid labile, consistent with the formation of a phosphohistidine intermediate (Fig. 3B). Formation of the intermediate was clearly less robust for SMa2063. This could reflect a lower intrinsic activity of SMa2063 or that less of the recombinant SMa2063 polypeptide folded into its active conformation. Alternatively, because AtBphP2 and AtExsG contain an appended RR domain, it was also possible that both these HKs were labeled with two phosphates, one attached to the HK histidine and the second attached to the RR aspartate.
FIG. 3.
Kinase activity of representatives from the HWE HK family. (A) Autophosphorylation of AtExsG, AtBphP2, and SmSMa2063. Recombinant proteins were incubated with [γ-32P]ATP and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the gels were either subjected to autoradiography (left) or stained for protein (right). Asterisks identify contaminants in the SMa2063 protein preparation. Numbers on the left are apparent molecular masses (in kilodaltons) of proteins used to calibrate the gel. (B) The stability of the autophosphorylated form of each after incubation for 2 h at 22°C in 50 mM Tris (pH 7.0), 1 M HCl, or 3 M KOH. (C) Importance of the phosphoacceptor site (His523) and the signature HWE residues (His616, Trp651, and Glu653) to the kinase activity of AtBphP2. Recombinant proteins containing the indicated mutation at each position were tested for kinase activity as described in the legend for panel A. (Top) Autoradiogram; (bottom) protein staining. The positions of the amino acids are indicated in Fig. 1.
To help support a role for the signature HWE residues in the HK activity of the group, we tested their importance in the phosphotransferase reactions by using appropriate site-directed mutations of AtBphP2. To minimize perturbations of hydrophobicity and/or charge of the resulting polypeptides, His616, Trp651, and Glu653 (Fig. 1) were substituted for somewhat conservative lysine, tyrosine, and aspartic acid residues, respectively. Importantly, all three point mutations retained their ability to bind biliverdin and became red-far red light photochromic, indicating that the substitutions did not drastically perturb the structure of the photoreceptor. Like replacement of phosphoacceptor His523 in the H box for a lysine, replacement of the other three residues eliminated autophosphorylation (Fig. 3C). No HK activity was detected despite extended incubation times with ATP and prolonged exposures of the autoradiograms, indicating that these three residues, in addition to the active site histidine, are critical for phosphotransfer.
Distribution of the HWE HKs.
As can be seen from the list of species that contain HWE HKs, these kinases can be found in a range of α- and γ-proteobacteria (Table 1). However, a clear lineage-specific gene expansion was evident. For example, many were discovered in members of the Rhizobiaceae family, which includes S. meliloti, Mesorhizobium loti, Rhizobium leguminosarum, Rhodopseudomonas palustris, and Brucella melitensis in addition to A. tumefaciens. Both M. loti and A. tumefaciens contain a large collection of HWE HKs, with 10 and 9 detected in their respective genomic sequences (Table 1). For A. tumefaciens, at least 51 predicted HKs were found, with the other 42 belonging to the more typical HKs of groups 1 to 4 (12). However, the HWE HK domain is not universally present in bacteria, as we were unable to find related sequences in the complete genomic sequence of a number of cyanobacteria, archaea, and other bacteria (e.g., E. coli and Bacillus subtilis). The motif was also undetectable in all available fungal, plant, and animal genomes, suggesting that these proteins have not radiated by horizontal transfer into eukaryotes.
TABLE 1.
Complete list of HWE-HK proteins
| Organisma | Family | Descriptionb,c | Accession no. |
|---|---|---|---|
| α-Proteobacteria | |||
| M. loti (10) | Rhizobiaceae | PAS | NP_103297, NP_109506, NP_435303 |
| TM | NP_109454, NP_104759, NP_107208 | ||
| GAF | NP_104746 | ||
| NP | NP_109528, NP_108261, NP_355125 | ||
| A. tumefaciens (9) | Rhizobiaceae | PAS | NP_535898, NP_535747, NP_535749 |
| phy | NP_355125 | ||
| HAMP | NP_534648 | ||
| RR | NP_532667, NP_354961 | ||
| NP | NP_534647, NP_535462 | ||
| R. palustris (9) | Rhizobiaceae | PAS | ZP_00009295, ZP_00012323, ZP_00010346, ZP_00009610 |
| phy | ZP_00010576 | ||
| GAF | ZP_00011648, ZP_00008817 | ||
| Methyltransferase | ZP_00012831 | ||
| NP | ZP_00011650 | ||
| S. meliloti (8) | Rhizobiaceae | PAS | NP_435303 |
| GAF | NP_437596, NP_436177, NP_435785, NP_386540 | ||
| HAMP | NP_384348 | ||
| Methyltransferase | NP_437036 | ||
| NP | NP_436373 | ||
| B. japonicum (6) | Rhizobiaceae | PAS | NP_774348 |
| TM | NP_767532, NP_769435 | ||
| GAF | NP_774679 | ||
| RR | NP_769238 | ||
| NP | BAC52448 | ||
| B. melitensis (2) | Rhizobiaceae | PAS | NP_539291, NP_541657 |
| B. suis (2) | Rhizobiaceae | PAS | NP_698641, NP_699772 |
| R. leguminosarum (2) | Rhizobiaceae | phy | CAC95194 |
| NP | CAC34400 | ||
| C. crescentus (9) | Caulobacteriaceae | PAS | NP_419653, NP_421842, NP_421357, NP_421964 |
| GAF | NP_419446 | ||
| HAMP | NP_420493, NP_421703 | ||
| RR | NP_422354, NP_421852 | ||
| R. sphaeroides (6) | Rhodobacteriaceae | PAS | ZP_00005199, ZP_00006599, ZP_00006631 |
| TM | ZP_00008203 | ||
| Methyltransferase | ZP_00007035 | ||
| NP | ZP_00006903 | ||
| N. aromaticivorans (5) | Sphingomonadaceae | PAS | ZP_00093951, ZP_00095689 |
| TM | ZP_00095135, ZP_00095556, ZP_00093204 | ||
| M. magnetotacticum (5) | Rhodospirillaceae | PAS | ZP_00049018, ZP_00049572, ZP_00050069, ZP_00049302, ZP_00052101 |
| R. rubrum (2) | Rhodospirillaceae | PAS | ZP_00016409 |
| TM | ZP_00016683 | ||
| γ-Proteobacteria | |||
| X. campestris (2) | Xanthomonadaceae | PAS | NP_638307 |
| Methyltransferase | NP_639032 | ||
| X. axonopodis (2) | Xanthomonadaceae | PAS | NP_643445 |
| Methyltransferase | NP_644037 | ||
| P. putida (1) | Pseudomonadaceae | phy | AAL50633 |
| P. syringae (1) | Pseudomonadaceae | phy | ZP_00126919 |
The number of members of the HWE family in each species is indicated in parentheses.
NP, no predicted domains found by SMART.
Even though each protein was placed in a single class, many had multiple motifs predicted by SMART.
Sensor domains associated with members of the HWE HK family.
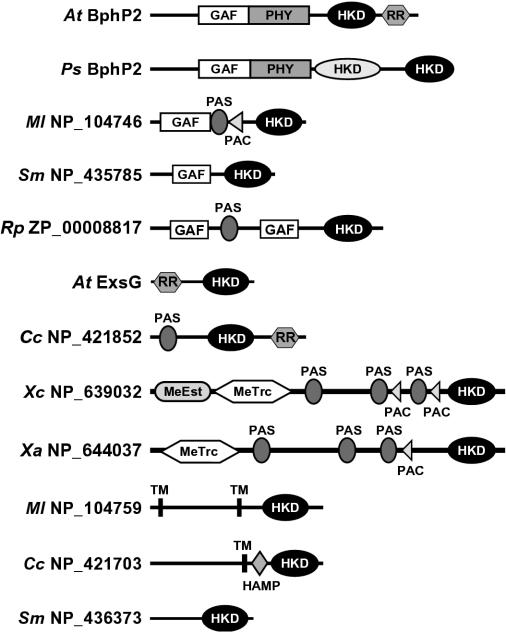
Examination of the sequences upstream of the HWE HK domain by SMART (24) identified a number of domains that presumably participate in environmental sensing (Fig. 4). In addition to AtBphP2, four other BphPs from R. leguminosarum, R. palustris, Pseudomonas putida, and Pseudomonas syringae contain a GAF-phy region used to bind bilin chromophores in addition to the HWE HK domain. For A. tumefaciens BphP2 and presumably the others, this connection helps the photoreceptors modulate an HK phosphorelay upon phototransformation of the holoproteins between the Pr and Pfr forms (30). The P. syringae BphP2 is particularly interesting because the polypeptide contains both the HWE and a more typical HK domain, suggesting that light perceived by the GAF-phy domain regulates two separate phosphorelays (Fig. 4).
FIG. 4.
Structural diversity within the HWE family of HKs. Shown are diagrams of representative members with the position of additional domains potentially important for sensing, location, and kinase activity as identified by the SMART database. Abbreviations: HKD, HK domain; MeEst, methylesterase; MeTrc, methyltransferase; TM, transmembrane. The HWE HKD is shown in black.
Other predicted domains include GAF without a phy motif and sensor modules like PAS (Per-Arnt-Sim domain), PAC (PAS-associated C-terminal domain), methyltransferase, and HAMP (HK, adenylyl cyclase, methyl binding protein-phosphatase domain) (Fig. 4). Such lone GAF domains without a companion phy domain are found in a number of signaling proteins, including cyclic nucleotide phosphodiesterases, adenylate cyclases, and the transcription factor FhlA (3). PAS domains are structurally related to GAF domains and are common among a variety of transcription regulators (34). The PAC sequence motif is typically located C-terminal to a subset of PAS motifs and is proposed to contribute to the PAS domain fold (23, 34). Like other sensor proteins, individual HWE HKs often contain multiple copies of the GAF, PAS, and PAC domains. The HAMP domain is found in other HKs, phosphatases, nucleotidyl cyclases, and chemoreceptors. While their functions remain unclear, HAMP domains can interact with HK domains and thus may regulate their activity in cis (4, 21). Six of the HKs are predicted to contain membrane-spanning helices within the N-terminal region that are also expected to function in signal perception. In addition to AtBphP2, several hybrid HKs were also identified that presumably use the appended RR to continue the phosphorelay from His to Asp (Fig. 4).
The most intriguing discovery was our detection of methyltransferase-methylesterase domains in five members of HWE HK family that appear related to that within E. coli CheR (Rhodobacter sphaeroides Rsph_p_2965, R. palustris Rpa1_p_4555, S. meliloti SMB20515, Xanthomonas axonopodis NP_644037, and Xanthomonas campestris NP_639032) (Fig. 4 and Table 1). The methylation activity of CheR is used to modulate the family of methylated chemotaxis proteins (MCPs) that serve as receptors for various attractant and repellant chemicals. For related members in the HWE HK family, the positionally conserved lysine and arginine residues necessary for CheR to interact with MCPs are present (25). Our finding of proteins containing both a CheR-type methyltransferase and HWE HK domains implies that dual function methyltransferase-phosphotransferase proteins may be used in some species to modulate chemical attraction and repulsion.
Reanalysis of the HWE HK phylogeny with respect to the nature of the predicted sensor domain(s) failed to detect any synonymous clustering with the various types of sensor domains spread throughout the tree. For example, HWE HK members that also contained the methyltransferase domain were dispersed in several distinct clades (Fig. 2). An exception is the BphP family where four of the five BphPs were located on the same branch, with R. leguminosarum BphP as the only outlier. Likewise, we failed to detect any clustering based on the species. The nine HWE HKs from A. tumefaciens were spread throughout the phylogram (Fig. 2). This lack of clustering suggests that the HWE-HK family is rapidly evolving.
DISCUSSION
Analyses of the complete sequences of more than 40 microbial genomes representing 8 of the 10 main bacterial phyla and both major branches of archaea have shown that two-component signal transduction systems driven by sensor HKs are major routes for environmental sensing in prokaryotes (17, 27). Currently, the HK superfamily contains more than 350 members, with some species containing over 60 separate proteins that presumably measure a variety of external signals (reference 12 and data not shown). Their modular architecture, which uses separate sensing, HK, RR, HPT, and output domains that can be arranged in numerous permutations, provides a facile way to create new or overlapping sensory networks. From our analysis of the BphPs, we discovered another set of kinase signaling systems employing the HWE HKs. While they appear mechanistically similar to other HKs, these HWE kinases use an HK domain bearing substantially different HK modules. Site-directed mutagenesis of one member, BphP2 from A. tumefaciens, demonstrated that the signature histidine, tryptophan, and glutamate residues can be critical for the autophosphorylation activity of this group. However, we acknowledge that other conserved amino acids are also likely to be important and thus provide additional distinguishing features to this family.
Our identification of 81 members from a variety of α- and γ-proteobacteria indicates that the HWE HK system is widespread. However, it is not universal, being undetectable in archaea and many bacteria. Their closest relatives are a group of HKs from the archaeon M. thermoautotrophicum (group 11 as designated in reference 12). The lack of similar HKs in other archaea would argue that group 11 is a more recent addition to the HK superfamily, possibly emerging from the HWE HKs following lateral gene transfer.
Bacterial species containing HWE HKs show great metabolic flexibility; they can be found in a very wide range of natural environments and can grow as aerobes or anaerobes or as phototrophs or heterotrophs. A number are noteworthy for their ability to interact either beneficially or detrimentally with eukaryotes. For example, several species of the Rhizobiaceae family (e.g., Bradyrhizobium japonicum, M. loti, R. leguminosarum, and S. meliloti) form symbiotic relationships with the roots of leguminous plants, with the bacterium providing a source of fixed nitrogen and the plant providing the carbon source (31). A. tumefaciens, P. syringae, and X. campestris are pathogens of many plant species. A. tumefaciens is particularly interesting because it uses a unique interkingdom conjugation system to introduce a set of bacterial genes into the plant that ultimately transforms both the development and biochemistry of the host for the pathogen's benefit (11). Given the prevalence of such bacterium-plant associations, it is possible that the HWE HKs provide signal transduction system(s) that encourage sensing, association, and/or cohabitation with the appropriate eukaryotic host(s).
Although HKs share characteristic amino acid motifs, they nevertheless show considerable variability even within their conserved H, N, F, and G1 to G3 boxes involved in phosphotransfer (12). The differences between the HWE-HK family and most of the other 11 characterized HK families include the absence of an obvious F box and significant changes within the N, G1, and G2 boxes, which collectively form the ATP-binding site. While all the other HK groups have signature DXGXG and GXGXG sequences in the G1 and G2 boxes, respectively (12), the HWE HKs contain conserved WXEXGGP and GFGXXL/V sequences in similar positions. How these changes impact phosphotransfer remains to be determined. Given their location within important parts of the Bergerat fold, these changes likely impact the folding, nucleotide binding, and/or ATPase activity of the family (10). For example, substitution of the aspartate for a glutamate in the G1 box could substantially affect ATP binding, given the importance of this aspartate in forming a hydrogen bond with the adenine moiety of ATP (19).
The linkage of the HWE HK domain with a variety of modules involved in sensing and signal transduction indicates that this family of kinases participates in a number of responses to external cues. The discovery of methyltransferase domains associated with several HWE HKs in particular suggests that these proteins participate in a hybrid type of chemical sensing relay. The best understood chemotactic response system is that from E. coli, which involves a family of MCP receptors, the CheA kinase, the CheB methylesterase, and the CheR methyltransferase (8). Detection of a repellent signal by the MCPs, for example, activates the CheA kinase. CheA then initiates a two-component signal transduction system through the CheY RR that ultimately stimulates a tumbling behavior to help the bacterium avoid the repellent. The system adapts to the repellent by CheA also phosphorylating CheB, which demethylates MCPs and thus reduces their ability to activate CheA, and a smooth swimming behavior is resumed. The system is reset by the constitutive methylation of MCPs by CheR (28, 32). For CheA, the kinase activity is not provided by its HK domain, which is instead used for homodimerization, but by an appended HPT domain (7).
Our detection of HWE HK domains associated with CheR-type methyltransferase domains in S. meliloti, R. sphaeroides, R. palustris, and Xanthomonas strains implies the participation of a dual kinase-methyltransferase component in chemical signaling for these species. Here, one can imagine that signaling by MCPs simultaneously activates both domains, leading to transduction of the signal through HWE HK autophosphorylation and methylation-dependent modulation of MCPs. However, we caution that we have not yet been able to demonstrate HK activity for any of the CheR-type HWE HKs. Preliminary attempts with CheR of M. loti were unsuccessful, most likely because the recombinant protein failed to assemble properly. As a result, we cannot rule out the possibility that the HWE HK domain in these hybrid proteins functions like that of CheA, having lost its phosphotransferase activity but retained its role in receptor dimerization (7).
The identification of the large family of HWE HKs further expands the role of two-component signal transduction systems in bacteria. Clearly, genetic analyses are now required to determine how these prokaryotic hosts use these sensor kinases to exploit specific ecological niches. Biochemical and structural comparisons of these kinases with the 11 previously identified groups should also help reveal differences important for phosphotransfer signaling among the HK superfamily.
Acknowledgments
This work was supported by a U.S. National Science Foundation grant (IBN 0091413) to R.D.V. and a United States-Israel Binational Research Development Fellowship (FI-316-2001) to B.K.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, and W. Miller. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleby, J. L., J. S. Parkinson, and R. B. Bouret. 1996. Signal transduction via the multi-step phosphorelay: not necessarily road less traveled. Cell 86:845-848. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., and C. P. Ponting. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22:458-459. [DOI] [PubMed] [Google Scholar]
- 4.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signaling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 5.Bergerat, A., B. De Massy, D. Gadelle, P. C. Varoutas, A. Nicolas, and P. Forterre. 1997. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386:414-417. [DOI] [PubMed] [Google Scholar]
- 6.Bhoo, S. H., S. J. Davis, J. Walker, B. Karniol, and R. D. Vierstra. 2002. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414:776-779. [DOI] [PubMed] [Google Scholar]
- 7.Bilwes, A. M., L. A. Alex, B. R. Crane, and M. I. Simon. 1999. Structure of CheA, a signal-transducing histidine kinase. Cell 96:131-141. [DOI] [PubMed] [Google Scholar]
- 8.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 9.Davis, S. J., A. V. Vener, and R. D. Vierstra. 1999. Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 286:2517-2520. [DOI] [PubMed] [Google Scholar]
- 10.Dutta, R., and M. Inouye. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25:24-28. [DOI] [PubMed] [Google Scholar]
- 11.Gelvin, S. B. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67:16-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grebe, W. T., and J. B. Stock. 1999. The histidine protein kinase family. Adv. Microb. Physiol. 41:141-227. [DOI] [PubMed] [Google Scholar]
- 13.Higgins, S. K., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 14.Hoch, J., and T. Silhavy. 1995. Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 15.Hubschmann, T., H. J. Jorissen, T. Borner, W. Gartner, and N. Tandeau de Marsac. 2001. Phosphorylation of proteins in the light-dependent signalling pathway of a filamentous cyanobacterium. Eur. J. Biochem. 268:3383-3389. [DOI] [PubMed] [Google Scholar]
- 16.Igo, M. M., A. J. Ninfa, J. B. Stock, and T. J. Silhavy. 1989. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 3:1725-1734. [DOI] [PubMed] [Google Scholar]
- 17.Inouye, M., and R. Dutta (ed.). 2002. Histidine kinases in signal transduction. Academic Press, New York, N.Y.
- 18.Karniol, B., and R. D. Vierstra. 2003. The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. USA 100:2807-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marina, A., C. Mott, A. Auyzenberg, W. A. Hendrickson, and C. D. Waldburger. 2001. Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. J. Biol. Chem. 276:41182-41190. [DOI] [PubMed] [Google Scholar]
- 20.Montgomery, B. L., and J. C. Lagarias. 2002. Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci. 7:357-366. [DOI] [PubMed] [Google Scholar]
- 21.Park, H., and M. Inouye. 1997. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J. Bacteriol. 179:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perego, M. 1998. Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Microbiol. 6:366-370. [DOI] [PubMed] [Google Scholar]
- 23.Ponting, C. P., and L. Aravind. 1997. PAS: a multifunctional domain family comes to light. Curr. Biol. 7:R674-R677. [DOI] [PubMed] [Google Scholar]
- 24.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiomi, D., I. B. Zhulin, M. Homma, and I. Kawagishi. 2002. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J. Biol. Chem. 277:42325-42333. [DOI] [PubMed] [Google Scholar]
- 26.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. Mcdougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J.-I. Mao, P. Rice, J. Nolling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 28.Swanson, R. V., R. B. Bourret, and M. I. Simon. 1993. Intermolecular complementation of the kinase activity of CheA. Mol. Microbiol. 8:435-441. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vierstra, R. D. 2002. Cyanophytochromes, bacteriophytochromes, and plant phytochromes: light-regulated kinases related to bacterial two-component regulators, p. 273-295. In M. Inouye and R. Dutta (ed.), Histidine kinases in signal transduction. Academic Press, New York, N.Y.
- 31.Weidner, S., A. Pühler, and H. Küste. 2003. Genomics insights into symbiotic nitrogen fixation. Curr. Opin. Biotechnol. 14:200-205. [DOI] [PubMed] [Google Scholar]
- 32.Wolfe, A. J., and R. C. Stewart. 1993. The short form of the CheA protein restores kinase activity and chemotactic ability to kinase-deficient mutants. Proc. Natl. Acad. Sci. USA 90:1518-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh, K.-C., S.-H. Wu, J. T. Murphy, and J. C. Lagarias. 1997. A cyanobacterial phytochrome two-component light sensory system. Science 277:1505-1508. [DOI] [PubMed] [Google Scholar]
- 34.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 9:331-333. [DOI] [PubMed] [Google Scholar]