Abstract
Pseudomonas syringae pv. tomato strain DC3000 is a pathogen of tomato and Arabidopsis. The hrp-hrc-encoded type III secretion system (TTSS), which injects bacterial effector proteins (primarily called Hop or Avr proteins) into plant cells, is required for pathogenicity. In addition to being regulated by the HrpL alternative sigma factor, most avr or hop genes encode proteins with N termini that have several characteristic features, including (i) a high percentage of Ser residues, (ii) an aliphatic amino acid (Ile, Leu, or Val) or Pro at the third or fourth position, and (iii) a lack of negatively charged amino acids within the first 12 residues. Here, the well-studied effector AvrPto was used to optimize a calmodulin-dependent adenylate cyclase (Cya) reporter system for Hrp-mediated translocation of P. syringae TTSS effectors into plant cells. This system includes a cloned P. syringae hrp gene cluster and the model plant Nicotiana benthamiana. Analyses of truncated AvrPto proteins fused to Cya revealed that the N-terminal 16 amino acids and/or codons of AvrPto are sufficient to direct weak translocation into plant cells and that longer N-terminal fragments direct progressively stronger translocation. AvrB, tested because it is poorly secreted in cultures by the P. syringae Hrp system, was translocated into plant cells as effectively as AvrPto. The translocation of several DC3000 candidate Hop proteins was also examined by using Cya as a reporter, which led to identification of three new intact Hop proteins, designated HopPtoQ, HopPtoT1, and HopPtoV, as well as two truncated Hop proteins encoded by the naturally disrupted genes hopPtoS4::tnpA and hopPtoAG::tnpA. We also confirmed that HopPtoK, HopPtoC, and AvrPphEPto are translocated into plant cells. These results increased the number of Hrp system-secreted proteins in DC3000 to 40. Although most of the newly identified Hop proteins possess N termini that have the same features as the N termini of previously described Hop proteins, HopPtoV has none of these characteristics. Our results indicate that Cya should be a useful reporter for exploring multiple aspects of the Hrp system in P. syringae.
Pseudomonas syringae is a host-specific bacterial pathogen of plants whose various pathovars cause necrotic lesions on leaves or fruits of susceptible plants and elicit the hypersensitive response (HR), a defense-associated localized programmed cell death, in resistant plants. P. syringae interactions with plants depend on the hrp (for “hypersensitive response and pathogenicity”) gene cluster, which encodes a type III secretion system (TTSS) or Hrp system that injects bacterial proteins into plant cells (1). Similar TTSSs exist in a variety of other gram-negative plant and animal pathogens (13, 28). The P. syringae proteins injected into host cells by the Hrp system are primarily known as effectors or Avr (avirulence) or Hop (Hrp-dependent outer protein) proteins. Hop proteins may contribute to P. syringae virulence by suppressing plant immunity. For example, AvrB, AvrRpm1, and AvrRpt2 all interfere with the function of RIN4, a plant protein implicated in the regulation of plant defense responses (3, 36, 37). However, certain plants are able to resist P. syringae infection through the actions of R proteins, which detect effectors or their activities (45). For example, the Arabidopsis R protein RPM1 triggers the HR when AvrB or AvrRpm1 modifies RIN4 (37).
Like the effectors found in other plant and animal pathogens, P. syringae Hop proteins are targeted to the Hrp secretion apparatus by N-terminal sequences (2, 39, 40, 47). The N termini of most Hop proteins do not exhibit significant amino acid homology, and the mechanism by which Hops are targeted to the Hrp system has not been elucidated. Although recent studies of the Yersinia effector protein YopE suggest that amphipathic N-terminal amino acid sequences are important for directing this protein to the Yersinia TTSS, there is evidence that mRNA sequences additionally contribute to effector targeting (34, 35, 42). Type III chaperones, which are small, acidic proteins that bind to and stabilize specific effectors in the bacterial cytoplasm, also play a role in targeting some effector proteins to the type III secretion apparatus (18).
P. syringae Hop proteins have been identified in a variety of different pathovars and strains, and the complete inventory of these proteins in any single strain is not yet known. However, the recent completion of the genome sequence of P. syringae pv. tomato strain DC3000 has facilitated bioinformatic and genetic studies that have led to identification of many Hops in this organism (9, 12, 22). In several recent reports, new Hops were identified on the basis that they have several common characteristics. First, most hop genes appear to be regulated by HrpL, an alternative sigma factor that activates the Hrp regulon (4, 30, 46, 50, 51). Because HrpL interacts with hrp box (5′-GGAACC-N16-CCACNNA-3′) sequences or variants of these sequences, searches were conducted to identify all of the potential hrp promoters in the DC3000 genome (19, 52). Several open reading frames (ORFs) with homology to known hop genes were identified downstream of such promoter sequences, and many of the encoded proteins were confirmed to be secreted by the DC3000 Hrp system (19, 24, 41, 52). Second, the N-terminal regions of most Hops appear to have common characteristics. For example, the first 50 amino acids of known effector proteins have a high serine content compared to the serine contents of a set of random housekeeping proteins in P. syringae (24). In addition to the high serine content, the N termini of most Hops are amphipathic, have an aliphatic amino acid (Ile, Val, or Leu) or Pro at the third or fourth position, and have no acidic amino acids (Asp or Glu) in the first 12 amino acids (41). Genomic searches for ORFs with these characteristics have yielded many potential hop genes (24, 41). Although some of these ORFs were confirmed to encode Hops in secretion or translocation assays, many of the hop candidates have yet to be tested.
Several methods have been used to show that Hop candidate proteins are secreted or translocated by the P. syringae Hrp system. Immunoblot analyses have revealed that certain Hop proteins are present in the culture supernatants of P. syringae grown in hrp-inducing media (40, 49). The drawback of this assay is that it does not distinguish translocated effectors from other proteins that may function as extracellular components of the Hrp secretion apparatus. In addition, certain known Hops, such as AvrB, are secreted poorly by P. syringae in culture (11, 49).
P. syringae Hop proteins have also been identified by using the reporter protein ′AvrRpt2. ′AvrRpt2 is the C-terminal functional domain of AvrRpt2, an effector protein that elicits the HR in plants containing the RPS2 R gene. Although ′AvrRpt2 itself cannot be translocated into plants, the N-terminal secretion signals of other effectors can substitute for the native AvrRpt2 secretion signal (23, 39). Thus, new Hops have been discovered by searching for proteins that, when fused to ′AvrRpt2, allow translocation of the hybrids into plant cells in an hrp-dependent manner (24, 52). Two potential problems with this method are that it is not quantitative and that it is formally possible that residual sequences in the ′AvrRpt2 reporter could contribute to translocation and yield false-positive results.
Finally, the calmodulin-dependent adenylate cyclase (Cya) domain of the cyclolysin toxin from Bordetella pertussis has also been exploited as a reporter for translocation of effector proteins (32). The Cya reporter was initially used to demonstrate the translocation of YopE into animal cells by Yersinia enterocolitica and subsequently was utilized to show that AvrBs2 is delivered into pepper plants by Xanthomonas campestris pv. vesicatoria (10, 48). Cya possesses two features that allow it to be a reporter for type III translocation: (i) it is not active in bacterial cytoplasm because bacteria do not possess calmodulin, and (ii) it is not secreted or translocated by the TTSS. However, when the N-terminal portion of an effector is fused to Cya, bacteria can deliver the resulting hybrid protein into the cytosol of host cells, where it can bind to calmodulin and produce cyclic AMP (cAMP) from ATP.
We are interested in identifying the complete set of Hop proteins translocated into plants by P. syringae pv. tomato strain DC3000, as well as in gaining a better understanding of Hop targeting signals and the translocation process. To that end, we optimized Cya as a reporter for P. syringae Hrp-mediated translocation using the known Hop proteins AvrPto and AvrB and truncated derivatives of these proteins. We then used Cya to test whether several HrpL-regulated hop candidate genes encode proteins that are translocated by the Hrp system into plant cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli TOP10 and DH5α were used as hosts for all cloned plasmids except pCPP3234, which was maintained in E. coli DB3.1. E. coli was grown in Luria-Bertani or terrific broth at 37°C unless otherwise specified, and P. syringae and Pseudomonas fluorescens were grown in King's B medium (KB) or hrp-derepressing fructose minimal medium (hrpMM) at 30°C (29, 31, 43). For E. coli and P. syringae, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; rifampin, 50 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 10 μg/ml. Because P. fluorescens 55 is resistant to spectinomycin and low levels of tetracycline, plasmids were maintained in this strain by using 100 μg of streptomycin per ml and 40 μg of tetracycline per ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant phenotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80lacZΔM15 ΔlacZYA-argF)U169 deoR recA1 endA1 hsdR17 phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| TOP10 | F−mcrA Δmrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7679 galU galK rpsL endA1 nupG | Invitrogen |
| DB3.1 | F−gyrA462 endA1 Δ(sr1-recA) mcrB mrr hsdS20 supE44 ara-14 galK2 lacY1 proA2 rpsL20 xyl-5 λ−leu mtl-1 | Invitrogen |
| P. syringae pv. tomato strains | ||
| DC3000 | Wild type, Rifr | 15 |
| CUCPB5114 | DC3000 ΔhrpK-hrpR::ΩCm, Rifr Cmr | 20 |
| P. fluorescens 55 | Wild type, Nxr | 27 |
| Plasmids | ||
| pLN18 | pLAFR3 derivative containing 25-kb P. syringae pv. syringae 61 hrc-hrp cluster with shcA and hopPsyA replaced by an nptII cassette, Tcr Kmr | 30a |
| pCPP3297 | pLN18 containing an unmarked deletion in hrcC | This study |
| pUFRO34 | Broad-host-range cloning vector expressing NptII, Kmr | 16 |
| pVLT35 | Broad-host-range cloning vector containing tac promoter and laclq, Spr/Strr | 17 |
| pMJH20 | pWSK29 containing codons 2 to 406 of cya, Ampr | 38 |
| pCPP3214 | pVLT35 containing codons 2 to 406 of cya, Spr/Strr | This study |
| pCPP3234 | pVLT35 containing Gateway reading frame B cassette and codons 2 to 406 of cya, Spr/Strr Cmr | This study |
| pCPP3221 | pVLT35 expressing AvrPto(1-164)-Cya | This study |
| pCPP3222 | pVLT35 expressing AvrPto(1-135)-Cya | This study |
| pCPP3223 | pVLT35 expressing AvrPto(1-100)-Cya | This study |
| pCPP3224 | pVLT35 expressing AvrPto(1-50)-Cya | This study |
| pCPP3225 | pVLT35 expressing AvrPto(1-16)-Cya | This study |
| pCPP3236 | pVLT35 expressing HopPtoC-Cya | This study |
| pCPP3271 | pVLT35 expressing ShcV-Cya | This study |
| pCPP3288 | pVLT35 expressing AvrB(1-30)-Cya | This study |
| pCPP3290 | pVLT35 expressing AvrB(1-321)-Cya | This study |
| pCPP3298 | pVLT35 expressing PSPTO4589 and PSPTO4588-Cya | This study |
| pCPP3299 | pVLT35 expressing PSPTOB003-Cya | This study |
| pCPP3300 | pVLT35 expressing PSPTO0524-Cya | This study |
| pCPP3302 | pVLT35 expressing ShcV and HopPtoV-Cya | This study |
| pCPP3303 | pVLT35 expressing HopPtoQ-Cya | This study |
| pCPP3304 | pVLT35 expressing PSPTO0834-Cya | This study |
| pCPP3305 | pVLT35 expressing PSPTO0836-Cya | This study |
| pCPP3306 | pVLT35 expressing PSPTO0837-Cya | This study |
| pCPP3307 | pVLT35 expressing PSPTO2695-Cya | This study |
| pCPP3308 | pVLT35 expressing AvrPphEPto-Cya | This study |
| pCPP3309 | pVLT35 expressing HopPtoK-Cya | This study |
| pCPP3310 | pVLT35 expressing HopPtoT1-Cya | This study |
| pCPP3311 | pVLT35 expressing PSPTO4781-Cya | This study |
| pCPP3312 | pVLT35 expressing PSPTO0835-Cya | This study |
| pCPP3313 | pVLT35 expressing HopPtoW′-Cya | This study |
| pLN401 | pVLT35 expressing ShcS4 and HopPtoS4′-Cya | This study |
DNA manipulations.
Plasmid DNA was isolated and manipulated by using standard methods (43). Restriction enzymes were obtained from New England Biolabs. PCR was performed with either ExTaq (Takara) or Vent (New England Biolabs) polymerase used according to the manufacturer's instructions. DNA sequencing was performed at the Cornell Biotechnology Center with an automated 3700 DNA analyzer (Applied Biosystems).
Construction of plasmids.
Plasmids expressing AvrPto-Cya fusion proteins were created in two steps. First, the template pCPP2329 DNA (25), which contained avrPto from P. syringae pv. tomato JL1065, and the avrPto primer pairs listed in Table 2 were used to generate PCR products that were digested with XbaI and SspI and cloned into the XbaI and SmaI sites of pMJH20. The reulting plasmids were then digested with XbaI and HindIII, and the avrPto-cya fragments were cloned into pVLT35.
TABLE 2.
Primers used to construct the cya gene fusions in this study
| cya fusion gene | 5′ (upstream) primer | 3′ (downstream) primer |
|---|---|---|
| avrPto(1-164) | 5′-GTTCTAGAGTGAGCGGATAACAATTTCAC-3′ | 5′-GTCCATAATATTGACTTGCCAGTTACGGTACG-3′ |
| avrPto(1-135) | 5′-GTTCTAGAGTGAGCGGATAACAATTTCAC-3′ | 5′-TGGTTTAATATTCGGTGGGATGTCGGCATG-3′ |
| avrPto(1-100) | 5′-GTTCTAGAGTGAGCGGATAACAATTTCAC-3′ | 5′-CTCACAAATATTCATTCCCGGATTGATTCCTG-3′ |
| avrPto(1-50) | 5′-GTTCTAGAGTGAGCGGATAACAATTTCAC-3′ | 5′-AGTTCAATATTTGGTAGACCAGCAGACTC-3′ |
| avrPto(1-16) | 5′-GTTCTAGAGTGAGCGGATAACAATTTCAC-3′ | 5′-GTCTGCAATATTCACCTGATGGGCCATC-3′ |
| avrB(1-321) | 5′-CACCATGGGCTGCGTCTCGTCAAAAAG-3′ | 5′-AAAGCAATCAGAATCTAGCAAGCT-3′ |
| avrB(1-30) | 5′-CACCATGGGCTGCGTCTCGTCAAAAAG-3′ | 5′-GGGGAGTGCTCTGAAAGACGTAC-3′ |
| hopPtoT1 | 5′-CACCATGAAAACAGTCAGCAA-3′ | 5′-TGACTTTTGAGCCGCCT-3′ |
| hopPtoQ | 5′-CACCATGCATCGTCCTATCACCG-3′ | 5′-ATCTGGGGCTACCGTCGA-3′ |
| hopPtoS4′ | 5′-CACCTTGCTAGGTATTAATGTCATG-3′ | 5′-GACCTTCCCAAGCTCTGG-3′ |
| hopPtoAG′ | 5′-CACCGCGCAGGAGTGCTCAACAATG-3′ | 5′-CTTCAGACCTTCCTTAACTACCCG-3′ |
| hopPtoV | 5′-CACCATGAGCTTATCGCCGACG-3′ | 5′-CTTTCTATCCGGAATAACTTTTTTCC-3′ |
| shcV | 5′-CACCATGAGCTTATCGCCGACG-3′ | 5′-GTTAAAACGAAAATGTGTCGCTG-3′ |
| PSPTO4588 | 5′-CACCATGATCGCGTTCGCAACCGG-3′ | 5′-GCCAGATGAGCTCGCCACAG-3′ |
| PSPTO2695 | 5′-CACCATGACTCAGACCGCCCTG-3′ | 5′-GGGAATCAGCCGATCACGAC-3′ |
| PSPTO4781 | 5′-CACCGTGCTCGCTTTTGCA-3′ | 5′-AGGCCGTCCTTCCATCCTC-3′ |
| PSPTOB003 | 5′-CACCATGAACAGGCTTCACAAGACC-3′ | 5′-GTCCTCGTTTAGCCGCAGC-3′ |
| PSPTO0834 | 5′-CACCATGAAAGCACTGGGCTTAATG-3′ | 5′-AAGATAAGCGAGATTTGGTGTGA-3′ |
| PSPTO0835 | 5′-CACCAGGAATAAAGGAGTAACAATGAAAG-3′ | 5′-GTACGGGCTCCGATTGTTCGA-3′ |
| PSPTO0836 | 5′-CACCATGCAGGCGATAGGGGC-3′ | 5′-TTTTTCTGCAAACCATTGCTTG-3′ |
| PSPTO0837 | 5′-CACCATGAAAAAACTCAATTTTTCCGAGC-3′ | 5′-GAAACGGGAGGAGGTATGC-3′ |
| PSPTO0524 | 5′-CACCATGAAAAAATGTATTGCTCTGCTCC-3′ | 5′-CTGGTTACCATATAGCGGCTCAG-3′ |
Other cya fusions were created by using the Gateway cloning technology (Invitrogen). In this system, an effector gene contained in an entry clone can be transferred by recombination into a cya destination vector, creating an in-frame cya gene fusion. Entry clones were constructed by cloning the PCR products obtained with the primer pairs listed in Table 2 (excluding the avrPto primers) into the pENTR/SD/D-TOPO vector (Invitrogen) according to the manufacturer's directions. pCPP2327 (21) was used as the template for the avrB PCRs, and DC3000 genomic DNA was used as the template for all other reactions. The cya destination vector, pCPP3234, was constructed in two steps. First, pMJH20 was digested with SacI and HindIII, and the cya-containing fragment was cloned into pVLT35 to create pCPP3214. The Gateway reading frame B cassette was then cloned into the SmaI site of pCPP3214 to create pCPP3234. Plasmids expressing cya gene fusions were created by site-specific recombination reactions (or LR reactions) between entry clones and pCPP3234. DNA sequencing confirmed that the effector or effector candidate sequences in all of the cya fusion plasmids did not contain any mutations in the 5′ region (∼first 100 codons) important for targeting proteins to the hrp secretion system.
The hrcC deletion in pCPP3297 was constructed by crossover PCR as described by Link et al. (33) by using pLN18 as the template DNA, the upstream primers 5′-CACTCGAGGAAGCCCTGGCATTGATTG-3′ and 5′-GGCAACCGGGAGTTTGAAGAGAGCCGGCCATCACTG-3′, and the downstream primers 5′-CAGTGATGGCCGGCTCTCTTCAAACTCCCGGTTGCC-3′ and 5′-CACTCGAGACATCGCCAACAGTTTGCTG-3′. The ∼2.4-kb ΔhrcC crossover PCR fragment was digested with XhoI and cloned into pLD55. The resulting plasmid was then digested with PstI, and the ΔhrcC fragment was cloned into the temperature-sensitive plasmid pMAK705 (26) to create pCPP3296. pCPP3296 was transformed into E. coli MC4100 containing pLN18, and replacement of the wild-type hrcC gene with the ΔhrcC allele was carried out by sequential temperature shifts as described by Hamilton et al. (26). pLN18 mutants that acquired the ΔhrcC mutation were confirmed by PCR and restriction enzyme analysis.
Preparation of protein samples.
DC3000 strains carrying plasmids expressing AvrPto-Cya fusions and neomycin phosphotransferase II (NptII) were scraped off KB plates, washed twice in hrpMM, and resuspended in 43 ml of hrpMM containing 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) and appropriate antibiotics. Wild-type and Δhrp mutant bacteria were resuspended at optical densities at 600 nm (OD600) of 0.15 and 0.12, respectively. These cultures were grown at 22°C with shaking until the OD600 were ∼0.5. Cellular protein fractions were collected by pelleting 1 ml of a culture and resuspending the bacteria in 100 μl of protein sample buffer (43). To collect extracellular protein fractions, 40-ml portions of cultures were centrifuged at 21,000 × g for 1 h, and the upper 30 ml of each supernatant was removed and filtered through a 0.45-μm-pore-size polysulfone syringe filter (Pall Life Sciences). Three milliliters of trichloroacetic acid was added to each of the supernatant fractions, and proteins were precipitated at 4°C overnight. Samples were then centrifuged at 21,000 × g for 1 h, and the pellets were washed twice with 1 ml of ice-cold acetone. Each pellet was resuspended in 105 μl of protein sample buffer. After boiling for 5 min, 15-μl portions of cellular protein samples and 45-μl portions of supernatant protein samples were loaded onto a sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gel electrophoresis (PAGE) gel.
P. fluorescens 55 isolates expressing Cya fusion proteins were grown on KB plates for 2 days. Bacteria were scraped off the plates, resuspended at an OD600 of 0.1 in liquid KB supplemented with streptomycin and 200 μM IPTG, and grown at 28°C for 7 h. After the OD600 of each culture was recorded, 1.5 ml was centrifuged with a microcentrifuge, and the pellet was suspended in protein sample buffer. Equal amounts of cells, based on OD600, were loaded onto SDS-PAGE gels.
Immunoblot analysis.
Protein samples were separated by electrophoresis on SDS-PAGE gels and transferred to Immobilon-P membranes (Millipore) by using a Semiphor semidry transfer system (Hoefer). Western blot analysis was carried out by using a Western-Light Plus kit (Tropix) according to the manufacturer's instructions. Cya fusion proteins were detected by using primary anti-Cya (3D1) mouse monoclonal immunoglobulin G (IgG) antibodies (Santa Cruz Biotechnology) at a dilution of 1:5,000 and secondary anti-mouse IgG-alkaline phosphatase conjugate antibodies (Sigma) at a dilution of 1:30,000. NptII was detected by using primary anti-NptII rabbit polyclonal IgG antibodies (United States Biological, Swampscott, Mass.) at a dilution of 1:5,000 and secondary anti-rabbit IgG-alkaline phosphatase conjugate antibodies (Sigma) at a dilution of 1:30,000.
Plant growth and bacterial inoculation.
Tomato (Lycopersicon esculentum cv. Money Maker) and wild tobacco (Nicotiana benthamiana) plants were grown under greenhouse conditions and transferred to the laboratory 1 day prior to inoculation. During experiments, plants were maintained in the laboratory at room temperature (24°C) with illumination. Bacteria grown on KB plates for ∼24 h were prepared for inoculation by suspension in 5 mM morpholinoethanesulfonic acid (MES) (pH 5.5) at an OD600 of 0.3 (1 × 108 CFU/ml) for adenylate cyclase assays, at an OD600 of 0.4 for AvrB-Cya HR assays, or at an OD600 of 0.8 for all other HR assays. Unless otherwise noted, each bacterial inoculum also contained 100 μM IPTG to induce expression of cya fusions. Bacteria were infiltrated into the fully expanded upper leaves of plants as previously described (27). The area of infiltration was marked to ensure that the leaf tissue subsequently collected for cAMP assays contained bacterial inoculum.
Adenylate cyclase assays.
To assay adenylate cyclase activity in plant tissue, leaf disks were collected with a 1-cm-diameter cork borer, frozen in liquid nitrogen, ground to a powder, and suspended in 300 μl of 0.1 M HCl. For the time course experiments, samples were collected in an identical manner except that a 0.8-cm-diameter cork borer was used and the ground tissue was suspended in 250 μl of 0.1 M HCl.
To assay the adenylate cyclase activity of Cya fusion proteins expressed in E. coli, strains were grown in 5 ml of Luria-Bertani medium containing 100 μM IPTG to an OD600 of 0.6 to 0.8. The cultures were centrifuged, and the pellets were washed and resuspended in sonication buffer (20 mM Tris-HCl [pH 8.0], 10 mM MgCl2). The bacteria were disrupted by sonication with a microtip for 2 min, and the cellular debris was pelleted by centrifugation with a microcentrifuge. Adenylate cyclase activity was determined in the presence or absence of calmodulin by using 5 μl of each lysate, as previously described (48).
cAMP was quantified in bacteria or leaf samples by using a Correlate-EIA cAMP immunoassay kit (Assay Designs) according to the manufacturer's directions. The amount of each sample used for quantification was adjusted so that it was in the detection range of the assay. The protein content of each sample was determined by the Bio-Rad protein assay (Bio-Rad).
RESULTS
Characterization of AvrPto-Cya and AvrB-Cya hybrid proteins.
To determine whether Cya can be used as a reporter to study P. syringae type III-mediated protein translocation into plants, we constructed C-terminal translational Cya fusions to the well-characterized P. syringae effector proteins AvrPto (164 amino acids) and AvrB (322 amino acids). The avrPto(1-X)-cya gene fusions, which contained the first 164, 135, 100, 50, or 16 codons of avrPto fused to codons 2 to 406 of B. pertussis cya, were cloned downstream from the tac promoter in the broad-host-range plasmid pVLT35 (Table 1). The resulting plasmids were conjugated into P. syringae pv. tomato DC3000 and CUCPB5114 (DC3000 ΔhrpK-hrpR::ΩCm), and Hrp-mediated secretion of the AvrPto(1-X)-Cya hybrid proteins into the culture supernatant was examined after bacteria were grown in hrpMM (Fig. 1). As a control for cellular lysis, each strain also contained plasmid pUFRO34, which constitutively expresses the cytoplasmic NptII protein. Cellular and supernatant fractions were separated by SDS-PAGE, and the AvrPto(1-X)-Cya and NptII proteins were detected by Western analysis by using antibodies to Cya and NptII. AvrPto(1-164)-Cya, AvrPto(1-135)-Cya, AvrPto(1-100)-Cya, and AvrPto(1-50)-Cya were all secreted into the culture supernatant in an Hrp-dependent manner, indicating that Cya does not inhibit secretion through the P. syringae Hrp system. AvrPto(1-16)-Cya secretion was not detected even though this protein was expressed at levels comparable to those of all of the other hybrid proteins. In some cases, the Cya antibodies detected multiple protein species on the immunoblot (Fig. 1). The additional AvrPto-Cya species could have been degradation products or proteins with alternative translation start sites. The additional AvrPto-Cya species did not appear to affect secretion of the full-length AvrPto(1-X)-Cya proteins.
FIG. 1.
Secretion of AvrPto-Cya hybrid proteins from P. syringae pv. tomato DC3000. DC3000 and CUCPB5114 (DC3000 ΔhrpK-hrpR::ΩCm) strains containing plasmids that express AvrPto(1-X)-Cya fusion proteins (where X is 164, 135, 100, 50, or 16) were grown in culture under conditions that induce hrp-mediated protein secretion, as described in Materials and Methods. These strains also contained pUFRO34, a plasmid that expresses the cytoplasmic NptII protein. Cultures were separated into cellular and supernatant fractions by centrifugation, and an immunoblot analysis was performed with protein samples electrophoresed on an SDS-12.5% PAGE gel. The supernatant samples loaded on the gel were 86-fold more concentrated than the cellular samples. The AvrPto(1-X)-Cya and NptII (29.1 kDa) proteins were detected by using antibodies to Cya and NptII, respectively. Lanes 1, 6, 11, and 16, AvrPto(1-164)-Cya (62.1 kDa); lanes 2, 7, 12, and 17, AvrPto(1-135)-Cya (58.6 kDa); lanes 3, 8, 13, and 18, AvrPto(1-100)-Cya (54.8 kDa); lanes 4, 9, 14, and 19, AvrPto(1-50)-Cya (49.0 kDa); lanes 5, 10, 15, and 20, AvrPto(1-16)-Cya (45.3 kDa). The positions of prestained protein standards on the gel are indicated on the right.
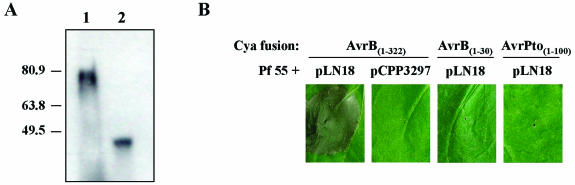
Two avrB(1-X)-cya gene fusions, avrB(1-321)-cya and avrB(1-30)-cya, were also constructed in pVLT35. The plasmid expressing AvrB(1-321)-Cya was toxic to DC3000 and caused cellular lysis in liquid culture (data not shown). Therefore, we expressed AvrB(1-321)-Cya and AvrB(1-30)-Cya in P. fluorescens containing the cosmid pLN18, which encodes the complete Hrp system of P. syringae pv. syringae 61 (Fig. 2A). pLN18 is a ΔshcAhopPsyA::ΩKm derivative of pHIR11, which allows P. fluorescens to deliver Avr-Hop proteins into plants (27, 30a), P. fluorescens(pLN18) expressing AvrB(1-321)-Cya induced an HR in N. benthamiana, showing that Cya does not inhibit the translocation or activity of the full-length AvrB protein (Fig. 2B). AvrB(1-321)-Cya delivery was Hrp dependent, as this protein did not cause the HR when it was expressed in P. fluorescens containing pCPP3297, a ΔhrcC derivative of pLN18. The HR caused by AvrB(1-321)-Cya was not due to its Cya domain because AvrPto(1-100)-Cya, which was translocated into N. benthamiana (as shown below), did not cause the HR in N. benthamiana. AvrB(1-30)-Cya also did not induce the HR in N. benthamiana.
FIG. 2.
HRs elicited by AvrB-Cya fusion proteins in N. benthamiana after delivery by P. fluorescens 55 expressing the P. syringae pv. syringae 61 Hrp system. (A) Expression of AvrB-Cya hybrid proteins in P. fluorescens. P. fluorescens 55 strains containing plasmids that express the Hrp system from P. syringae pv. syringae 61 (pLN18) and AvrB(1-321)-Cya (pCPP3290) or AvrB(1-30)-Cya (pCPP3288) were grown in culture as described in Materials and Methods. Protein samples were separated on an SDS-12.5% PAGE gel, and an immunoblot analysis was performed by using antibodies to Cya. Lane 1, AvrB(1-321)-Cya (79.5 kDa); lane 2, AvrB(1-30)-Cya (46.8 kDa). The positions of prestained protein standards on the gel are indicated on the left. (B) AvrB(1-321)-Cya induces an HR in N. benthamiana. N. benthamiana leaves were infiltrated with suspensions (OD600, 0.4) of P. fluorescens 55 (Pf 55) strains expressing AvrB(1-321)-Cya, AvrB(1-30)-Cya, or AvrPto(1-100)-Cya and a wild-type Hrp system from pLN18 or a ΔhrcC mutant Hrp system from pCPP3297. Photographs were taken 48 h after inoculation.
Optimization of AvrPto(1-164)-Cya delivery into plants.
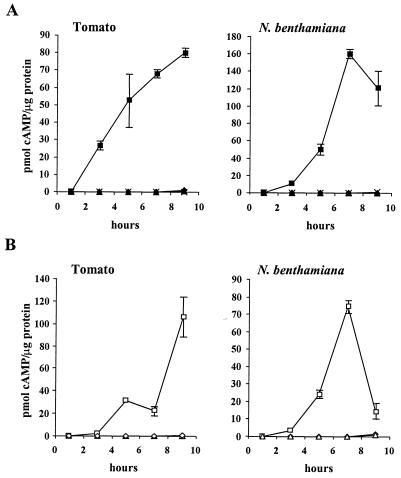
To determine the optimal conditions for studying P. syringae translocation of Cya hybrid proteins into plant cells, we analyzed AvrPto(1-164)-Cya translocation with regard to the kinetics of cAMP accumulation, the delivery system, and the test plant. The kinetics of cAMP accumulation in plant leaves were examined following inoculation with DC3000 or CUCPB5114 (DC3000 ΔhrpK-hrpR::ΩCm) strains expressing AvrPto(1-164)-Cya (Fig. 3A). As an additional control, plants were inoculated with DC3000 containing pCPP3214, a pVLT35-based plasmid that includes codons 2 to 406 of cya and does not express Cya due lack of a translation start site. cAMP was extracted from leaf samples taken 1, 3, 5, 7, and 9 h postinoculation and quantified. In tomato (L. esculentum cv. Money Maker), a susceptible host for DC3000, cAMP levels increased over time only in leaf samples inoculated with DC3000 expressing a wild-type Hrp system and AvrPto(1-164)-Cya (Fig. 3A). Similar results were observed in N. benthamiana, even though DC3000 induces the HR in this plant (Fig. 3A). However, we did not observe any increases in cAMP levels that were indicative of AvrPto(1-164)-Cya translocation into Nicotiana tabacum cv. Xanthi leaves (H.-S. Oh and A. Collmer, unpublished data). To rule out the possibility that AvrPto(1-164)-Cya present in the plant apoplast could have caused cAMP accumulation, plants were inoculated with a lysate prepared from an E. coli culture expressing AvrPto(1-164)-Cya. This lysate did not cause cAMP accumulation in tomato or N. benthamiana leaves (Fig. 3A), supporting the conclusion that AvrPto(1-164)-Cya is translocated into plant cells via the DC3000 Hrp secretion system. Plants inoculated with bacteria expressing AvrPto containing a C-terminal FLAG tag instead of Cya also did not induce cAMP accumulation, indicating that the Cya domain of AvrPto(1-164)-Cya was responsible for the production of cAMP (data not shown).
FIG. 3.
Time course of cAMP accumulation in plants after inoculation with Pseudomonas or E. coli cells expressing AvrPto(1-164)-Cya. (A) Tomato (L. esculentum cv. Money Maker) and N. benthamiana plants were inoculated with DC3000 bacteria (OD600, 0.3) containing pCPP3214 [Cya(2-406)] (♦) or pCPP3221 [AvrPto(1-164)-Cya] (▪) or with CUCPB5114 (DC3000 ΔhrpK-hrpR::ΩCm) containing pCPP3221 (▴). Plants were also infiltrated with a cleared lysate prepared from a culture of E. coli containing pCPP3221 (×). (B) Tomato (L. esculentum cv. Money Maker) and N. benthamiana plants were inoculated with P. fluorescens 55 cells (OD600, 0.3) containing pLN18 and pCPP3214 (⋄), pLN18 and pCPP3221 (□), or pCPP3297 and pCPP3221 (▵). pLN18 and pCPP3297 contain the wild-type and ΔhrcC hrp system genes from P. syringae pv. syringae 61, respectively. Leaf samples were collected with a 0.8-cm-diameter cork borer 1, 3, 5, 7, and 9 h postinoculation. cAMP was quantified in triplicate for each sample, and the standard deviations are indicated by error bars. The graphs are based on data from one representative experiment. Repeated experiments on different days yielded similar results, although the cAMP levels varied by up to 50% for each strain.
Because plasmids expressing certain Cya fusion proteins were toxic to DC3000, we examined whether P. fluorescens containing the cosmid pLN18 could be used as an alternative delivery system for translocation of Cya fusion proteins into plant cells. P. fluorescens expressing AvrPto(1-164)-Cya increased cAMP levels in tomato leaves when bacteria also contained pLN18 but not when bacteria also contained the ΔhrcC mutant pCPP3297 (Fig. 3B). In addition, P. fluorescens(pLN18) containing pCPP3214 (pVLT35::cya) did not cause cAMP accumulation in tomato leaves. These results indicate that like DC3000, P. fluorescens(pLN18) translocates AvrPto(1-164)-Cya into tomato in an Hrp-dependent manner. Similar results were observed in N. benthamiana (Fig. 3B). N. benthamiana was used as the host plant in subsequent experiments due to the ease of infiltrating its large leaves. Because P. fluorescens(pLN18) does not cause the HR in N. benthamiana and because plasmids expressing Cya fusion proteins did not appear to be as toxic to this strain, this delivery system was used in the experiments described below unless otherwise indicated.
Characterization of the translocation domains of AvrPto-Cya and AvrB-Cya hybrid proteins.
To determine the minimal N-terminal region needed for AvrPto-Cya translocation into plant cells, cAMP accumulation was analyzed in N. benthamiana after infiltration with DC3000 or P. fluorescens(pLN18) strains expressing AvrPto(1-X)-Cya hybrid proteins. AvrPto(1-16)-Cya caused a weak increase in cAMP production in N. benthamiana, and AvrPto(1-50)-Cya and AvrPto(1-100)-Cya caused progressively stronger cAMP production (Table 3). However, AvrPto(1-135)-Cya and AvrPto(1-164)-Cya did not appear to be translocated better than AvrPto(1-100)-Cya, which suggests that all of the targeting information is present in the first 100 amino acids of AvrPto, with the minimal signal being in the first 16 amino acids and/or codons. AvrB(1-30)-Cya and AvrB(1-321)-Cya resulted in a similar trend in cAMP production in N. benthamiana. In E. coli lysates supplemented with calmodulin, AvrPto(1-16)-Cya and AvrB(1-30)-Cya were able to produce cAMP levels comparable to those produced by Cya fusions with larger N-terminal fragments of these effectors (Table 3). Thus, the nominal signal produced by these fusions in N. benthamiana was presumably the result of weak translocation. These observations indicate that N-terminal signals are both sufficient and required for the Hrp-mediated translocation of AvrPto-Cya and AvrB-Cya into plants. Furthermore, although the minimal information for AvrPto translocation is in the first 16 amino acids, information in the first 100 residues contributes cumulatively to translocation efficacy.
TABLE 3.
Calmodulin-dependent adenylate cyclase activity of AvrPto-Cya and AvrB-Cya hybrid proteins in vitro and in planta
| AvrPto or AvrB-Cya fusion protein | In vitro activity in E. coli lysates (nmol of cAMP/μg of protein)a,d
|
In planta activity in N. benthamianad
|
||||
|---|---|---|---|---|---|---|
| Translocation by DC3000 (pmol of cAMP/μg of protein)b
|
Translocation by P. fluorescens (pmol of cAMP/μg of protein)c
|
|||||
| With calmodulin | Without calmodulin | Wild-type | Δhrp | pLN18 | pCPP3297 | |
| AvrPto(1-164) | 140.6 ± 22.0 | 0.6 ± 0.1 | 157.1 ± 7.7 | 0.0 ± 0.0 | 103.6 ± 12.9 | 0.0 ± 0.0 |
| AvrPto(1-135) | 179.5 ± 14.9 | 1.0 ± 0.3 | 101.6 ± 14.9 | 0.0 ± 0.0 | 114.3 ± 33.2 | 0.1 ± 0.0 |
| AvrPto(1-100) | 186.1 ± 26.2 | 1.9 ± 0.0 | 122.3 ± 0.0 | 0.0 ± 0.0 | 56.6 ± 2.8 | 0.0 ± 0.0 |
| AvrPto(1-50) | 145.0 ± 16.5 | 1.1 ± 0.2 | 66.7 ± 10.9 | 0.0 ± 0.0 | 12.3 ± 2.9 | 0.0 ± 0.0 |
| AvrPto(1-16) | 134.2 ± 1.8 | 0.6 ± 0.2 | 2.0 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| AvrB(1-321) | 114.3 ± 16.6 | 0.3 ± 0.0 | NDe | ND | 73.1 ± 12.7 | 0.0 ± 0.0 |
| AvrB(1-30) | 275.7 ± 8.1 | 0.7 ± 0.1 | ND | ND | 3.4 ± 0.3 | 0.0 ± 0.0 |
cAMP was quantified in sonicated cellular lysates of E. coli in the presence or absence of exogenous calmodulin as described in Materials and Methods.
cAMP was quantified in N. benthamiana leaf samples 7 h after infiltration with wild-type or Δhrp (CUCPB5114) DC3000 at an OD600 of 0.3.
cAMP was quantified in N. benthamiana leaf samples 7 h after infiltration with P. fluorescens (OD600, 0.3) containing a cosmid expressing either a wild-type (pLN18) or ΔhrcC mutant (pCPP3297) Hrp system from P. syringae pv. syringae 61.
cAMP was quantified in triplicate for each sample, and the values are means ± standard deviations. Repeated experiments yielded similar results.
ND, not determined.
Analysis of the production and stability of candidate Hop-Cya fusions.
Because Cya was successfully used as a reporter for AvrPto and AvrB translocation into plant cells, we sought to use it as a tool to identify novel Hop proteins in DC3000. We compiled a list of potential hop genes by analyzing the coding regions downstream of HrpL-regulated promoters identified in previous reports (19, 41). Eighteen genes were chosen for further study, and in-frame cya fusions to each entire ORF were constructed in the broad-host-range plasmid pVLT35 (Table 4). Three of the hop candidate genes (PSPTO4597, PSPTO4588, and PSPTO4720) were located downstream of ORFs encoding small, acidic proteins that may be dedicated type III chaperones (PSPTO4599, PSPTO4589, and PSPTO4721, respectively). Because these potential chaperones might be required for secretion of their putative target Hops, they were expressed along with the appropriate Cya fusion proteins. Every Cya hybrid protein exhibited calmodulin-dependent adenylate cyclase activity in E. coli lysates (data not shown), indicating that all of them had the potential to induce cAMP accumulation in planta. Western blot analysis also confirmed that each Cya hybrid protein was expressed in P. fluorescens (Fig. 4). Although a hybrid protein of the expected size was detected in each lane of the immunoblot, the protein levels varied, and multiple bands were present in some cases. These results may have been due to protein degradation or translation from alternative start sites.
TABLE 4.
Hop candidates tested in Cya translocation assay
| Effector or candidatea | Alternative designation
|
Homolog(s) and/or likely function (BLASTP E value; GenBank accession no.) | |
|---|---|---|---|
| Petnicki-Ocwieja et al.b | Guttman et al.c | ||
| HopPtoK (PSPTO0044) | HopPtoK | HolPtoAB | AvrRps4 of P. syringae pv. pisi (2e-44; AAB51082) |
| HopPtoC (PSPTO0590) | HopPtoC | AvrPpiC2Pto | AvrPpiC2 of P. syringae pv. pisi (1e-148; CAC16701) |
| AvrPphEPto (PSPTOA0012) | AvrPphEPto | AvrPphEPto | AvrPphE of P. syringae pv. phaseolicola (1e-117; AAA67930) |
| PSPTO0877 (HopPtoQ) | Orf19 | HolPtoQ | Hypothetical protein of X. campestris pv. campestris (1e-134; NP_636447), hypothetical protein of X. axonopodis pv. citri (1e-128; NP_644627), hypothetical protein of R. solanacearum (3e-92; CAD13773) |
| PSPTOA0019 (HopPtoT1) | Orf16 | HolPtoU | HolPtoU2 of P. syringae pv. tomato (1e-156; AAL84264) |
| PSPTO0901 (HopPtoAG′) | Orf28 | NAd | HolPsyAG of P. syringae pv. syringae B728A (1e-58; DAA00387) |
| PSPTO4599 + PSPTO4597 (ShcS4 + HopPtoS4′) | Orf25 + Orf26 | ChpPtoZ + HolPtoZ | PSPTO4588 (6e-6) |
| PSPTO 4589 + PSPTO4588 | Orf14 + Orf15 | NA | HopPtoS4′ (6e-6) |
| PSPTO2695 | NA | NA | Hypothetical protein of X. campestris pv. campestris (1e-99; NP_637485), hypothetical protein of X. axonopodis pv. citri (2e-99; NP_642404), Arabidopsis thaliana wound-induced protein (9e-60; AAM64873) |
| PSPTOB003 | Orf18 | NA | Probable exported protein YPO3534 of Y. pestis (2e-6; AG0429) |
| PSPTO4721 (ShcV) | Orf6 | NA | Chaperone for HopPtoV? |
| PSPTO4721 + PSPTO4720 (ShcV + HopPtoV) | Orf6 + Orf17 | NA | None |
| PSPTO0834 | Orf22 | NA | Mannitol dehydrogenase of Leuconostoc mesenteroides (3e-22; AAM09029) |
| PSPTO0835 | Orf23 | NA | Riboflavin-specific deaminase of Aquifex aeolicus (1e-37; G70339) |
| PSPTO0836 | Orf24 | NA | Predicted phosphatase of Clostridium acetobutylicum (2e-40; AAK78609) |
| PSPTO0837 | NA | NA | Conserved hypothetical protein of Vibrio cholerae (5e-41; A82470) |
| PSPTO4781 | Orf8 | NA | Esterase of Pyrobaculum calidifontis (1e-16; BAC06606) |
| PSPTO0524 | Orf27 | NA | Carboxypeptidase G2 of Bacillus halodurans (1e-28; BAB04071) |
Coding sequences are indicated by the name and/or P. syringae pv. tomato ORF (PSPTO) number in the complete genome sequence of DC3000 (GenBank accession number AE016853).
See reference 41.
See reference 24.
NA, not applicable.
FIG. 4.
Expression of DC3000 effector-Cya or effector candidate-Cya fusion proteins in P. fluorescens 55. P. fluorescens 55 strains containing plasmids that express the Hrp system from P. syringae pv. syringae 61 (pLN18) and the different Cya fusion proteins were grown in culture as described in Materials and Methods. Protein samples were separated on an SDS-7% PAGE gel, and an immunoblot analysis was performed by using antibodies to Cya. The estimated molecular masses of the hybrid proteins and the positions of prestained protein standards on the gel are indicated.
Analysis of the translocation of class I Hop candidate Cya fusions.
The Hop candidate proteins were divided into three classes based on whether they had the following common characteristics of type III secreted proteins in P. syringae: (i) an aliphatic amino acid (Ile, Leu, or Val) or Pro at the third or fourth position, (ii) a relatively high serine content (more than 10%) in the first 50 amino acids, and (iii) no negatively charged amino acids (Asp or Glu) in the first 12 amino acids. Class I, II, and III Hop candidate proteins were defined as proteins that had all three, two, or less than two of these characteristics, respectively. To determine whether the Hop candidates were translocated into plants, cAMP was quantified in N. benthamiana leaf samples infiltrated with P. fluorescens 55 strains expressing one Cya hybrid protein and either a wild-type or nonfunctional Hrp system. We predicted that proteins in class I (HopPtoK-Cya, HopPtoC-Cya, AvrPphEPto-Cya, PSPTO0877-Cya, PSPTOA0019-Cya, PSPTO0901-Cya, and PSPTO4597-Cya) would be translocated into plants because they had all three of the characteristics of P. syringae Hops listed above (Table 5).
TABLE 5.
In planta adenylate cyclase activity in N. benthamiana of DC3000 Hop candidate-Cya fusion proteins
| Cya fusion proteina | First 12 residuesb | % of Serc | In planta activity (pmol of cAMP/μg of protein) after translocation byd:
|
|
|---|---|---|---|---|
| P. fluorescens (pLN18) | P. fluorescens (pCPP3297) | |||
| Class I | ||||
| HopPtoK (PSPTO0044) | MNRISTSSVNSS | 18 | 771.9 ± 27.5 | 0.7 ± 0.2 |
| HopPtoC (PSPTO0590) | MTIVSGHIGKHP | 16 | 21.5 ± 2.3 | 0.6 ± 0.6 |
| AvrPphEPto (PSPTOA0012) | MKIHNAGLTPPL | 14 | 1,109.5 ± 109.0 | 1.9 ± 0.3 |
| HopPtoQ (PSPTO0877) | MHRPITAGHTTS | 24 | 1,928.6 ± 94.0 | 0.3 ± 0.2 |
| HopPtoT1 (PSPTOA0019) | MKTVSNHSIPST | 14 | 84.0 ± 13.7 | 0.2 ± 0.1 |
| HopPtoAG′ (PSPTO0901) | MNPITHSFSHLG | 12 | 151.0 ± 10.6 | 1.9 ± 0.4 |
| ShcS4 + HopPtoS4 (PSPTO4599 + PSPTO4597) | MKISGSTSPAHT | 24 | 944.7 ± 77.5 | 0.9 ± 0.1 |
| Class II | ||||
| PSPTO4589 + PSPTO4588 | MKKSGAGTQAYA | 14 | 0.2 ± 0.1 | 0.2 ± 0.1 |
| PSPTO2695 | MTQTALVVGASG | 10 | 0.0 ± 0.0 | 0.6 ± 0.1 |
| PSPTOB003 | MNRLHKTSLLAA | 8 | 0.0 ± 0.0 | 0.1 ± 0.1 |
| ShcV (PSPTO4721) | MSLSPTLQKLTN | 8 | 0.0 ± 0.0 | 0.1 ± 0.0 |
| Class III | ||||
| ShcV + HopPtoV (PSPTO4721 + PSPTO4720) | MLDKVGNSARVDe | 6 | 378.2 ± 27.1 | 0.6 ± 0.3 |
| PSPTO0834 | MKALGLMDNQRLe | 4 | 0.6 ± 0.4 | 0.4 ± 0.3 |
| PSPTO0835 | MKVTVFSQISID | 8 | 0.0 ± 0.0 | 0.4 ± 0.1 |
| PSPTO0836 | MQAIGAVIFDMDe | 8 | 0.3 ± 0.1 | 0.2 ± 0.1 |
| PSPTO0837 | MKKLNFSELNWI | 6 | 0.0 ± 0.0 | 0.2 ± 0.1 |
| PSPTO4781 | MLAFAYVSLIRE | 6 | 0.0 ± 0.0 | 0.7 ± 0.4 |
| PSPTO0524 | MKKCIALLLTLV | 4 | 1.7 ± 0.7 | 1.5 ± 0.9 |
Coding sequences fused to cya are indicated by the name and/or P. syringae pv. tomato ORF (PSPTO) number in the complete genome sequence of DC3000 (GenBank accession number AE016853). In cases where two ORFs are listed, cya was fused to the second ORF. Plasmids expressing these hybrid proteins also contained the preceding ORFs, which encoded small (14- to 17-kDa), acidic proteins that may have acted as type III chaperones.
Acidic residues are underlined. The Ile, Leu, Val, or Pro residues located at the third or fourth position of the proteins are in bold type.
Percentage of Ser residues in the first 50 amino acids of the predicted protein.
cAMP measurement experiments were performed in triplicate with N. benthamiana leaf samples collected 7 h after infiltration with P. fluorescens strains (OD600, 0.3) containing pLN18 or pCPP3297. The values are means ± standard deviations, and repeated experiments yielded similar results.
The sequence had more than one potential translation start site, and the most N-terminal methionine is shown.
All seven of the strains expressing class I hybrid proteins generated cAMP in N. benthamiana in an hrp-dependent manner, suggesting that they were translocated into plant cells by the P. syringae Hrp system (Table 5). The lower levels of cAMP induced by HopPtoC-Cya may have been due to the low levels of this protein present in P. fluorescens (Fig. 4). As indicated in previous reports, two of the proteins in class I, HopPtoK and HopPtoC, travel the Hrp pathway, as does an AvrPphEPto homolog in another P. syringae strain. Specifically, a HopPtoK-′AvrRpt2 fusion is translocated by DC3000, HopPtoC is secreted in culture by DC3000, and an AvrPphEPma-′AvrRpt2 fusion is translocated by P. syringae pv. maculicola (24, 41). All of the remaining Hrp system-translocated proteins in class I were not tested previously for secretion in culture or translocation into plants and therefore were given Hop designations. The proteins encoded by PSPTO0877 and PSPTOA0019 were designated HopPtoQ and HopPtoT1, respectively.
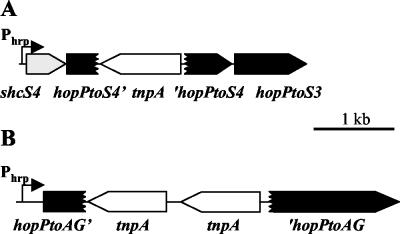
When the sequences surrounding PSPTO4597 and PSPTO0901 on the DC3000 chromosome were analyzed, transposon insertions were found following both of these ORFs (Fig. 5). In the PSPTO4597 region, the ORF following the transposon insertion encodes a predicted protein with similarity to the C-terminal portion of HopPtoS3. Therefore, we propose that PSPTO4597 should be designated hopPtoS4::tnpA and that its product should be designated HopPtoS4′ to indicate that it most likely is the N-terminal portion of a truncated effector protein. We also designated the protein encoded by PSPTO4599 ShcS4 to reflect its potential role as a chaperone for HopPtoS4. Two transposase genes follow PSPTO0901. PSPTO0901 and the ORF following the transposase gene insertions encode predicted proteins with high levels of homology to the N and C termini, respectively, of the same protein encoded by P. syringae pv. syringae B728A. Greenberg and Vinatzer recently designated this B728A ORF holPsyAG (hop-like), and our results further suggest that this gene encodes a Hop (22). PSPTO0901 was designated hopPtoAG::tnpA and its product was designated HopPtoAG′ to indicate that the gene is a truncated effector gene in DC3000 and the protein is a homolog of HolPsyAG.
FIG. 5.
The hopPtoS4 and hopPtoAG genes in P. syringae pv. tomato DC3000 are interrupted by transposon insertions. Schematic diagrams of the hopPtoS4 (A) and hopPtoAG (B) regions of the DC3000 chromosome show the positions of coding sequences for effectors (solid arrows and boxes), transposases (open arrows), and a predicted chaperone (gray arrow). The hopPtoS4::tnpA and hopPtoAG::tnpA genes encode truncated products containing 118 and 152 amino acids, respectively. The bent arrows indicate the positions of promoters containing hrp boxes (Phrp).
Analysis of the translocation of class II Hop candidateCya fusions.
The proteins in class II (PSPTO4588-Cya, PSPTO2695-Cya, PSPTOB003-Cya, and PSPTO4721-Cya) had two of the three characteristics of P. syringae Hop proteins described above. Although the protein encoded by PSPTO4588 had a low level of similarity to HopPtoS4′ and was encoded downstream from a potential chaperone (encoded by PSPTO4589), this hybrid protein did not induce cAMP accumulation in N. benthamiana. In addition, Cya fusions to the proteins encoded by PSPTO2695 and PSPTOB003, which are similar to a wound-induced protein in Arabidopsis and a protein annotated as a probable exported protein in the genome of Y. pestis, respectively (Table 4), did not induce accumulation of cAMP in N. benthamiana (Table 5). Therefore, proteins having only two of three characteristics of Hops may not be targeted to the Hrp system for translocation into plants. PSPTO4721-Cya also did not induce cAMP in planta, suggesting that this protein remains in the bacterial cytoplasm. This result was not surprising, as the PSPTO4721 gene (ORF6) encodes a small (14.7-kDa), negatively charged protein that has the characteristics of type III chaperones.
Analysis of the translocation of class III Hop candidate Cya fusions.
We predicted that proteins in class III (PSPTO4720-Cya, PSPTO834-Cya, PSPTO835-Cya, PSPTO836-Cya, PSPTO837- Cya, PSPTO4781-Cya, and PSPTO0524-Cya) would not be translocated into plants because they have none or only one of the three characteristics of P. syringae Hops. Although six of these proteins were apparently not translocated into plant cells, PSPTO4720-Cya generated cAMP in N. benthamiana in an Hrp-dependent manner. Thus, PSPTO4720 was designated hopPtoV, and the ORF directly upstream (PSPTO4721), encoding a potential chaperone for HopPtoV, was designated shcV. These results demonstrate that most, but not all, Hops have the characteristics listed above.
Elicitation of the HR in N. benthamiana by HopPtoK-Cya and HopPtoQ-Cya.
None of translocated Cya hybrid proteins caused the macroscopic HR, which is manifested by visible tissue collapse, in N. benthamiana at the cell density (OD600, 0.3) used for the adenylate cyclase experiments (data not shown). However, when bacteria were inoculated at a higher cell density (OD600, 0.8), HopPtoK-Cya and HopPtoQ-Cya induced the hrp-dependent HR in N. benthamiana (Fig. 6). These results confirmed that high levels of cAMP can accumulate at 7 h postinoculation in N. benthamiana even when the Cya hybrid protein is capable of eliciting the HR, and they provided further evidence that Cya fusions do not disrupt the biological activity of these particular effectors. None of the other Cya fusion proteins consistently caused the HR in N. benthamiana after delivery by P. fluorescens containing pLN18 (Fig. 6 and data not shown).
FIG. 6.
HopPtoK-Cya and HopPtoQ-Cya induce HRs in N. benthamiana. N. benthamiana leaves were infiltrated with suspensions (OD600, 0.8) of P. fluorescens 55 strains expressing HopPtoK-Cya, HopPtoQ-Cya, or HopPtoT1-Cya and a wild-type P. syringae pv. syringae 61 Hrp system from pLN18 or a mutant P. syringae pv. syringae 61 Hrp system from pCPP3297. The photographs were taken 48 h after inoculation.
DISCUSSION
Cya has been used as a reporter to study TTSS-mediated translocation of effector proteins by Yersinia, Salmonella, enteropathogenic E. coli, and Xanthomonas (10, 14, 38, 48). In this study, the Cya reporter system was optimized in P. syringae by using the well-studied effector protein AvrPto. AvrPto-Cya induced cAMP accumulation in tomato or N. benthamiana when it was delivered by either DC3000 or P. fluorescens expressing a P. syringae Hrp system. This system permitted demonstration that the well-studied effector AvrB is translocated into plants despite its poor secretion in culture. Similar to translocation of other effector proteins, translocation of both AvrPto-Cya and AvrB-Cya was dependent upon N-terminal sequences. The Cya reporter also enabled further exploration of the targeting signals in DC3000 TTSS substrates and identification of several new effector proteins that are translocated into plant cells, two of which are truncated due to natural transposon insertions.
Cya as a reporter for P. syringae Hrp-mediated translocation into plants.
Cya was developed several years ago as a reporter for TTSS translocation of Yersinia effector proteins into animal cells, yet this enzyme was not used to investigate Hrp-mediated translocation of phytopathogen effector proteins until recently. X. campestris pv. vesicatoria expressing AvrBs2-Cya causes a substantial increase in cAMP levels in susceptible pepper plants as soon as 3 h postinoculation (10). Our results show that AvrPto(1-164)-Cya produces comparable results in susceptible tomato plants and N. benthamiana when it is expressed in P. syringae pv. tomato DC3000. P. fluorescens 55 containing pLN18 was also tested as a delivery system for effectors because some Cya fusion proteins were toxic to DC3000 and because the use of pLN18 permits translocation to be studied in the absence of other DC3000 proteins. P. fluorescens (pLN18), which expresses the Hrp system of P. syringae pv. syringae 61, translocates AvrPto(1-164)-Cya into tomato and N. benthamiana almost as well as DC3000. Although P. fluorescens(pLN18) is slightly less efficient than DC3000 at delivering truncated AvrPto-Cya fusions [AvrPto(1-100)-Cya, AvrPto(1-50)-Cya, and particularly AvrPto(1-16)-Cya] into N. benthamiana, it may be more suitable for testing candidate effectors because of its ability to tolerate overexpression of effectors and because translocation of individual effectors can be studied in the absence of potential competition from other effectors, which may complicate analysis of some TTSS effectors (8).
Casper-Lindley et al. reported that AvrBs2-Cya induction of cAMP accumulation is significantly reduced when bacteria are inoculated into a resistant pepper cultivar (10). We observed similar results in a resistant tomato cultivar infected with DC3000 expressing AvrPto(1-164)-Cya (data not shown). However, cAMP accumulation induced by AvrPto(1-164)-Cya was not inhibited in N. benthamiana even though DC3000 elicited the HR in this plant. In addition, AvrB-Cya, HopPtoK-Cya, and HopPtoQ-Cya each induced cAMP accumulation in N. benthamiana when it was delivered by P. fluorescens(pLN18) despite eliciting the HR. Thus, N. benthamiana, which is easily infiltrated and widely used in studies of plant-microbe interactions (5), appears to be useful as a universal host for Cya reporter studies of Hops that may cause the HR in plants. It is possible that induction of the HR occurs more slowly in N. benthamiana, allowing cAMP accumulation induced by Cya hybrid proteins to be quantified before resistance signaling pathways are activated. Alternatively, the resistance response mechanisms in N. benthamiana may be different from those present in tomato or pepper plants.
′AvrRpt2 has also been utilized as a reporter fusion to identify P. syringae proteins that are translocated into plant cells by the Hrp system (23, 52), but our results demonstrate that Cya has two advantages. First, as previously noted (10), adenylate cyclase assays appear to be more sensitive than HR assays. In order to observe the HRs caused by AvrB-Cya, HopPtoK-Cya, and HopPtoQ-Cya, it was necessary to inoculate plants with more bacteria than were used for Cya assays. Second, adenylate cyclase assays are at least semiquantitative, whereas HR assays are qualitative. Thus, unlike HR assays, Cya assays can be used to compare the amounts of effectors that are translocated into plants. However, the Cya reporter has limitations. In our studies, the amount of cAMP produced by a particular Cya fusion protein could vary as much as 50% in different experiments, although the relative cAMP levels produced by different fusion proteins in the same experiment were consistent. Therefore, quantitative comparisons can only be made with data from the same experiment.
Targeting signals of type III secreted proteins.
Many studies have shown that N-terminal sequences are required for effectors to be secreted by TTSSs (2, 7, 39, 44, 47). We have previously shown that the first 10 codons of AvrPto are required for Hrp-mediated secretion in culture (2). Here, we found that the first 16 codons of AvrPto are sufficient to direct weak Hrp-mediated translocation into plant cells. Similarly, the first 15 codons of AvrB were shown to be necessary for Hrp-mediated secretion in culture (as determined by using a cloned Erwinia chrysanthemi Hrp system that appears to secrete P. syringae effectors more permissively than the P. syringae Hrp system) (2), and here we found that the first 30 codons of AvrB (the smallest fragment tested) are sufficient to direct translocation of a Cya fusion protein into plant cells.
Thus, consistent with findings for effectors such as the Y. enterocolitica YopE and enteropathogenic E. coli Tir proteins, residues and/or codons 1 to 15 of at least one plant pathogen effector are sufficient to direct translocation of the Cya reporter into eukaryotic cells, although translocation directed by this minimal TTSS targeting signal can be very weak (8, 14). Previous work with translocation of AvrBs2-Cya fusions into pepper leaf cells by X. campestris pv. vesicatoria suggested that the first 41 residues of AvrBs2 were required for translocation, and no translocation was directed by smaller N-terminal fragments (10). However, it is possible that the weaker translocation of smaller fragments was below the threshold of detection in the apparently less sensitive pepper system.
The N termini of most known Hop proteins have a high serine content, an aliphatic amino acid (Ile, Val, or Leu) or Pro at the third or fourth position, and no acidic amino acids (Asp or Glu) in the first 12 amino acids (24, 41). To determine whether newly identified Hop proteins have these characteristics, we used the Cya reporter to test translocation of 18 Hop candidates that are encoded downstream of hrp promoter sequences. Seven Hop candidates had N termini that have all of the characteristics of Hop targeting signals, and all of these Cya hybrid proteins were translocated into N. benthamiana. In contrast, the Hop candidates that did not have all three of the characteristics of Hop N termini (with the exception of HopPtoV) were not translocated. Thus, the previously observed characteristics of known Hops also generally apply to new Hops. However, one newly identified Hop, HopPtoV, had none of the characteristics of Hop N termini (even when potential alternative start sites were considered), suggesting that some Hop proteins may have unique targeting signals that do not conform to any previously observed pattern. The ORF upstream of hopPtoV, which was designated shcV, appears to encode a putative type III chaperone. It is possible that ShcV binds and targets HopPtoV to the Hrp system, which may explain why HopPtoV has an unusual targeting signal, although other Hops with apparent chaperones have N termini that are typical of most Hops.
Interestingly, the PSPTO4588-Cya fusion protein (a class II candidate lacking an aliphatic amino acid at the third or fourth position) was not translocated into N. benthamiana even though the PSPTO4588-encoded protein has a low level of similarity to HopPtoS4′ and contains a high percentage of serine in its first 50 residues. Like hopPtoV, the PSPTO4588 gene is located downstream from a potential type III chaperone gene. Although it is possible that Cya may have prevented translocation of the PSPTO4588 protein or that the PSPTO4588 protein may have inhibited the adenylate cyclase activity of Cya, we favor and are currently testing the idea that the N-terminal region of the PSPTO4588 protein may have acquired mutations that eliminated the ability of this protein to be targeted to the type III secretion apparatus. In addition, we are exploring the possibility that sequences in other regions of the PSPTO4588 protein may block translocation despite the presence of otherwise functional targeting signals at the N terminus.
Expansion of the effector inventory in DC3000.
One of the goals of our laboratory is to determine the complete inventory of Hop proteins in DC3000. In a previous study, genetic and bioinformatic methods were used to identify hrp promoter sequences in the DC3000 genome, which led to the discovery of many candidate hop genes (19). A subsequent study revealed that the Hrp system of DC3000 secretes several of the Hops into the culture supernatant, but many Hop candidates were not tested (41). In this study, the Cya reporter system was used to test whether several of the candidate hop genes encode TTSS substrates. Our results confirm the previous conclusions or assumptions that HopPtoK, HopPtoC, and AvrPphEPto are translocated into plant cells. We also identified five new Hops in DC3000 that are translocated into plant cells, which increases the number of confirmed Hrp system-secreted proteins in this organism to 40.
HopPtoQ is of particular interest because it may be an important virulence factor in many phytopathogens; homologs are present in X. campestris pv. campestris, Xanthomonas axonopodis pv. citri, and Ralstonia solanacearum (Table 4). Furthermore, HopPtoQ-Cya produced far more cAMP in N. benthamiana than any other Cya fusion protein tested produced, despite the fact that HopPtoQ-Cya also elicited the HR in N. benthamiana. In contrast to HopPtoQ, HopPtoT1 and HopPtoV are unique to DC3000 according to the current databases, although at least two other copies of hopPtoT1-like genes are present in the DC3000 genome.
Two of the new hop genes identified in this study, hopPtoS4::tnpA and hopPtoAG::tnpA, encode proteins that are truncated due to natural transposon insertions. Seven percent of the genes in DC3000 are predicted to be mobile genetic elements, which may account for the large number of inactivated and duplicate hop genes present in this organism (9). Our results indicate that the naturally truncated proteins HopPtoS4′ and HopPtoAG′ are still targeted for translocation by the P. syringae Hrp system. Furthermore, the HopPtoS4′ gene is one of the DC3000 genes previously identified as being both induced during infection and activated by HrpL in culture (6). Hence, it seems likely that genetic rearrangements involving the 5′ regions of hop genes could lead to generation of new effector genes in DC3000. The genome of P. syringae pv. tomato DC3000 contains several regions that are enriched for genes encoding additional complete and disrupted effector candidates. The Cya reporter, used in conjunction with the cloned P. syringae Hrp system and the experimentally tractable N. benthamiana system that we describe here, should provide a useful resource for functional testing of these genes in DC3000 and other P. syringae genomes.
Acknowledgments
We thank Hye-Sook Oh for providing some of the Gateway entry vectors used in this study and Kent Loeffler for assistance with photography.
This work was supported by NSF Plant Genome Research Program Cooperative Agreement DBI-0077622, by NSF grant MCB-9982646 (to A.C.), by USDA NRI grant 2001-02751 (to L.M.S.), and by USDA NRI grant 01-35319-10019 (to J.R.A.).
REFERENCES
- 1.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. M., D. E. Fouts, A. Collmer, and O. Schneewind. 1999. Reciprocal secretion of proteins by the bacterial type III machines of plant and animal pathogens suggests universal recognition of mRNA targeting signals. Proc. Natl. Acad. Sci. 96:12839-12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axtell, M. J., and B. J. Staskawicz. 2003. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112:369-377. [DOI] [PubMed] [Google Scholar]
- 4.Badel, J. L., A. O. Charkowski, W.-L. Deng, and A. Collmer. 2002. A gene in the Pseudomonas syringae pv. tomato Hrp pathogenicity island conserved effector locus, hopPtoA1, contributes to efficient formation of bacterial colonies in planta and is duplicated elsewhere in the genome. Mol. Plant-Microbe Interact. 15:1014-1024. [DOI] [PubMed] [Google Scholar]
- 5.Baulcombe, D. C. 1999. Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2:109-113. [DOI] [PubMed] [Google Scholar]
- 6.Boch, J., V. Joardar, L. Gao, T. L. Robertson, M. Lim, and B. N. Kunkel. 2002. Identification of Pseudomonas syringae genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 44:73-88. [DOI] [PubMed] [Google Scholar]
- 7.Boland, A., M.-P. Sory, M. Iriarte, C. Kerbourch, P. Wattiau, and G. R. Cornelis. 1996. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolicotica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB,D,N delivery apparatus. EMBO J. 15:5191-5201. [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd, A. P., I. Lambermont, and G. R. Cornelis. 2000. Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J. Bacteriol. 182:4811-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Vanaken, T. V. Feldblyum, M. D'Ascenzo, W.-L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casper-Lindley, C., D. Dahlbeck, E. T. Clark, and B. J. Staskawicz. 2002. Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc. Natl. Acad. Sci. 99:8336-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collmer, A., A. O. Charkowski, W.-L. Deng, D. E. Fouts, J. H. Ham, A. H. Rehm, K. van Dijk, and J. R. Alfano. 2001. Bacterial Avr proteins: secreted agents of parasitism and elicitors of plant defense, p. 36-45. In N. T. Keen, S. Mayama, J. E. Leach, and S. Tsuyumu (ed.), Delivery and perception of pathogen signals in plants. APS Press, St. Paul, Minn.
- 12.Collmer, A., M. Lindeberg, T. Petnicki-Ocwieja, D. J. Schneider, and J. R. Alfano. 2002. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 10:462-469. [DOI] [PubMed] [Google Scholar]
- 13.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 14.Crawford, J. A., and J. B. Kaper. 2002. The N-terminus of enteropathogenic Escherichia coli (EPEC) Tir mediates transport across bacterial and eukaryotic cell membranes. Mol. Microbiol. 46:855-868. [DOI] [PubMed] [Google Scholar]
- 15.Cuppels, D. A. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Feyter, R., C. I. Kado, and D. W. Garbriel. 1990. Small, stable shuttle vectors for use in Xanthomonas. Gene 88:65-72. [DOI] [PubMed] [Google Scholar]
- 17.de Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 18.Feldman, M. F., and G. R. Cornelis. 2003. The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219:151-158. [DOI] [PubMed] [Google Scholar]
- 19.Fouts, D. E., R. B. Abramovitch, J. R. Alfano, A. M. Baldo, C. R. Buell, S. Cartinhour, A. K. Chatterjee, M. D'Ascenzo, M. L. Gwinn, S. G. Lazarowitz, N.-C. Lin, G. B. Martin, A. H. Rehm, D. J. Schneider, K. van Dijk, X. Tang, and A. Collmer. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. 99:2275-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouts, D. E., J. L. Badel, A. R. Ramos, R. A. Rapp, and A. Collmer. 2003. A Pseudomonas syringae pv. tomato DC3000 Hrp (type III secretion) deletion mutant expressing the Hrp system of bean pathogen P. syringae pv. syringae 61 retains normal host specificity for tomato. Mol. Plant-Microbe Interact. 16:43-52. [DOI] [PubMed] [Google Scholar]
- 21.Gopalan, S., D. W. Bauer, J. R. Alfano, A. O. Loniello, S. Y. He, and A. Collmer. 1996. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell 8:1095-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg, J. T., and B. A. Vinatzer. 2003. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr. Opin. Microbiol. 6:20-28. [DOI] [PubMed] [Google Scholar]
- 23.Guttman, D. S., and J. T. Greenberg. 2001. Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol. Plant-Microbe Interact. 14:145-155. [DOI] [PubMed] [Google Scholar]
- 24.Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler, and J. T. Greenberg. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722-1726. [DOI] [PubMed] [Google Scholar]
- 25.Ham, J. H., D. W. Bauer, D. E. Fouts, and A. Collmer. 1998. A cloned Erwinia chrysanthemi Hrp (type III protein secretion) system functions in Escherichia coli to deliver Pseudomonas syringae Avr signals to plant cells and to secrete Avr proteins in culture. Proc. Natl. Acad. Sci. 95:10206-10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, H.-C., R. Schuurink, T. P. Denny, M. M. Atkinson, C. J. Baker, I. Yucel, S. W. Hutcheson, and A. Collmer. 1988. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J. Bacteriol. 170:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 30.Innes, R. W., A. F. Bent, B. N. Kunkel, S. R. Bisgrove, and B. J. Staskawicz. 1993. Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J. Bacteriol. 175:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Jamir, Y., M. Guo, H.-S. Oh, T. Petnicki-Ocwieja, S. Chen, X. Tang, M. B. Dickman, A. Collmer, and J. R. Alfano. Identification of Pseudomonas syringae type III secreted effectors that suppress programmed cell death in eukaryotes. Plant J., in press. [DOI] [PubMed]
- 31.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 32.Ladant, D., and A. Ullmann. 1999. Bordatella pertussis adenylate cyclase: a toxin with multiple talents. Trends Microbiol. 7:172-176. [DOI] [PubMed] [Google Scholar]
- 33.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd, S., M. Sjostrom, S. Andersson, and H. Wolf-Watz. 2002. Molecular characterization of type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol. Microbiol. 43:51-59. [DOI] [PubMed] [Google Scholar]
- 35.Lloyd, S. A., M. Norman, R. Rosqvist, and H. Wolf-Watz. 2001. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol. Microbiol. 39:520-532. [DOI] [PubMed] [Google Scholar]
- 36.Mackey, D., Y. Belkhadir, J. M. Alonso, J. R. Ecker, and J. L. Dangl. 2003. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112:379-389. [DOI] [PubMed] [Google Scholar]
- 37.Mackey, D., B. F. Holt, A. Wiig, and J. F. Dangl. 2002. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108:743-754. [DOI] [PubMed] [Google Scholar]
- 38.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 39.Mudgett, M. B., O. Chesnokova, D. Dahlbeck, E. T. Clark, O. Rossier, U. Bonas, and B. J. Staskawicz. 2000. Molecular signals required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc. Natl. Acad. Sci. 97:13324-13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mudgett, M. B., and B. J. Staskawicz. 1999. Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol. 32:927-941. [DOI] [PubMed] [Google Scholar]
- 41.Petnicki-Ocwieja, T., D. J. Schneider, V. C. Tam, S. T. Chancey, L. Shan, Y. Jamir, L. M. Schechter, C. R. Buell, X. Tang, A. Collmer, and J. R. Alfano. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. 99:7652-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramamurthi, K. S., and O. Schneewind. 2002. Type III protein secretion in Yersinia species. Annu. Rev. Cell Dev. Biol. 18:107-133. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Schesser, K., E. Frithz-Lindsten, and H. Wolf-Watz. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178:7227-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider, D. S. 2002. Plant immunity and film noir: what gumshoe detectives can teach us about plant-pathogen interactions. Cell 109:537-540. [DOI] [PubMed] [Google Scholar]
- 46.Shen, H., and N. T. Keen. 1993. Characterization of the promoter of avirulence gene D from Pseudomonas syringae pv. tomato. J. Bacteriol. 175:5916-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sory, M.-P., A. Boland, I. Lambermont, and G. R. Cornelis. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. 92:11998-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sory, M.-P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 49.van Dijk, K., D. E. Fouts, A. H. Rehm, A. R. Hill, A. Collmer, and J. R. Alfano. 1999. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J. Bacteriol. 181:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao, Y., S. Heu, J. Yi, Y. Lu, and S. W. Hutcheson. 1994. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J. Bacteriol. 176:1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao, Y., and S. Hutcheson. 1994. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J. Bacteriol. 176:3089-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwiesler-Vollick, J., A. E. Plovanich-Jones, K. Nomura, S. Brandyopadhyay, V. Joardar, B. N. Kunkel, and S. Y. He. 2002. Identification of novel hrp-regulated genes through functional genomic analysis of the Pseudomonas syringae pv. tomato DC3000 genome. Mol. Microbiol. 45:1207-1218. [DOI] [PubMed] [Google Scholar]