Abstract
Overexpression of either heterologous or homologous proteins that are routed to the periplasm via the twin-arginine translocation (Tat) pathway results in a block of export and concomitant accumulation of the respective protein precursor in the cytoplasm. Screening of a plasmid-encoded genomic library for mutants that confer enhanced export of a TorA signal sequence (ssTorA)-GFP-SsrA fusion protein, and thus result in higher cell fluorescence, yielded the pspA gene encoding phage shock protein A. Coexpression of pspA relieved the secretion block observed with ssTorA-GFP-SsrA or upon overexpression of the native Tat proteins SufI and CueO. A similar effect was observed with the Synechocystis sp. strain PCC6803 PspA homologue, VIPP1, indicating that the role of PspA in Tat export may be phylogenetically conserved. Mutations in Tat components that completely abolish export result in a marked induction of PspA protein synthesis, consistent with its proposed role in enhancing protein translocation via Tat.
Secretion of proteins across lipid bilayer membranes is a process fundamental to life. The bacterial general secretory (Sec) pathway and its eukaryotic counterpart are responsible for the membrane translocation of the majority of secreted proteins. However, 6 years ago a fundamentally different pathway for protein translocation was discovered, first in plants and then in bacteria. In the latter organisms, the new pathway was termed the twin-arginine translocation (Tat) pathway because of the signature RR dipeptide found in most of the leader peptides of proteins that use this mode of export (5). A combination of biochemical and genetic studies has identified the key features that distinguish the Tat pathway from the Sec mechanism of protein export. (i) The Tat pathway is able to transport proteins that have attained a substantial degree of tertiary or even quaternary structure in the cytoplasm prior to export (16, 33, 34). (ii) The Tat translocase consists of the Tat(A/E)BC proteins, which are completely distinct and share, at most, little homology with the components of the Sec translocon (SecYEG) (7). (iii) Tat-specific leader peptides possess a number of significant differences relative to Sec sorting signals (6, 7, 13). (iv) Whereas the translocation of proteins by the Sec pathway requires ATP hydrolysis, the Tat pathway is solely dependent on the proton motive force, ΔμH+ (26, 28).
Recently, in vitro translocation assays with purified (inverted) inner membrane vesicles from Escherichia coli were developed by Yahr and Wickner (47) and independently by Alami et al. (2). It was shown that disruption of the H+ gradient abolishes export whereas ATP has little effect. Moreover, Alami et al. showed that functional membrane association of a Tat precursor with the Tat apparatus requires an intact Arg-Arg signal but is independent of the H+ gradient. However, these groups reported that translocation of SufI could not be observed in inside-out inner membrane vesicles prepared from wild-type (WT) E. coli cells. Translocation could only be detected in membrane vesicles that had been prepared from cells overexpressing TatABC from a strong T7 promoter, and even under these conditions, the efficiency was low. The same authors also reported that the precursor proteins became translocation incompetent as a function of time, a rather peculiar finding for a dedicated posttranslational pathway. For nearly all heterologous protein fusions to Tat leaders, e.g., green fluorescent protein (GFP) (15, 16, 41), chloramphenicol acetyltransferase (40), and even certain native Tat substrates (11, 24), the efficiency of export in vivo is also well below 100%. As a result, upon subcellular fractionation, a sizable fraction of the preprotein is retained in the cytoplasm. For example, with ssTorA-GFP fusions, about 50% of the preprotein is found in the spheroplast fraction of exponential-phase cells (4, 41).
Collectively, these observations suggest that while Tat(A/E)BC are the only essential components of the translocon, factors other than these proteins might help maintain export competence and/or enhance translocation efficiency in vivo. This would be analogous to the Sec pathway, where soluble (e.g., SecB) and membrane (SecD and SecF) proteins play distinct roles in the secretion process but do not represent essential components of the translocon (18). To date, the only auxiliary factors found to affect Tat transport are substrate-specific chaperones such as DmsD (29). DmsD binds to the leader peptides of precursor DmsA and TorA but not to mature DmsA and TorA. On the basis of this observation, a dual role for this chaperone was initially proposed in which DmsD assists in molybdopterin cofactor attachment and guides the preprotein to the translocation channel (29, 35). While DmsD is found associated with the inner membrane through an interaction with TatB and TatC (30), it is not required for efficient translocation of a DmsA signal peptide-GFP chimera or the authentic Tat substrate TorA (32). Thus, it appears that DmsD is a substrate-specific chaperone that does not play a generalized role in Tat pathway transport efficiency.
We have developed a genetic system for the isolation of multicopy E. coli genes that enhance the export of Tat substrate proteins in vivo. As was discussed above, a significant portion of ssTorA-GFP remains in the cytoplasm in a folded, fluorescent conformation. Fusion of an SsrA C-terminal extension to ssTorA-GFP results in the degradation of export-incompetent cytoplasmic protein. Therefore, in cells expressing ssTorA-GFP-SsrA, only protein that is exported from the cytoplasm via the Tat pathway is rescued from degradation and contributes to cell fluorescence (15). We sought to isolate chromosomal genes that confer enhanced cell fluorescence, and thus improved Tat export, when expressed from a multicopy plasmid. A gene fragment encoding phage shock protein A (pspA) was found to markedly increase the export efficiency of not only TorA-GFP-SsrA but also native Tat protein substrates (SufI, CueO) that accumulate in an export-incompetent form when expressed from multicopy plasmids. A similar effect was observed with the Synechocystis sp. pspA homologue, VIPP1, indicating that the role of PspA in Tat export may be phylogenetically conserved. Finally, we show that mutations in Tat components that completely abolish export result in a marked induction of PspA synthesis.
The pspA gene is the first gene in the pspABCDE operon that is induced upon infection by filamentous phage and numerous other stresses. pspA encodes a 26-kDa polypeptide that is approximately equally distributed between the cytoplasm and the inner membrane fraction (9). The PspA protein exhibits multiple functions. First, it serves as a negative regulator of transcriptional enhancer protein PspF, and thus, it negatively regulates its own expression (25). Second, PspA assists in maintenance of the proton motive force, which is thought to help the cell cope with membrane-related stresses (i.e., osmotic shock, lipid depletion, and blockage of the Sec pore) (21, 25). Most importantly, the transport of various Sec pathway precursors is less efficient in vivo and in vitro in the absence of PspA while expression of the pspA gene stimulates efficient Sec protein export (22). In accordance, its role in protein secretion and ΔμH+ maintenance may be a direct result of its association with the inner membrane.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. E. coli strain XL1-Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15Tn10 (Tetr)]) was used for screening of the genomic library by fluorescence-activated cell sorter (FACS). Plasmids pSufI-FLAG and pCueO-FLAG were constructed by PCR amplification of E. coli K-12 genomic DNA with primers SufI-For (5′-GCGATGGAGCTCTTAAAGAGGAGAAAGGTCATGTCACTC AGTCGGCGT-3′) and SufI-Rev (5′-GCTCTAGATTATCCCTTGTCGTCATCGTCC TTGTAGTCTGCTCCCGGTACCGGATTGACCAA-3′) and primers CueO-For (5′-GCGATGGAGCTCTTAAAGAGGAGAAAGGTCATGCAACGTCGTGATTTC-3′) and CueO-Rev (5′-GCTCTAGATTATCCCTTGTCGTCATCGTCCTTGT AGTCTGCTCCTACCGTAAACCCTAACAT-3′), where the sequence for the FLAG affinity tag was incorporated into the reverse primers. PCR products were digested with SacI and XbaI and ligated into the same sites of pBAD33. Plasmids pVipp1 and pV-236 were constructed by PCR amplification of Synechocystis sp. strain PCC6803 genomic DNA with primers Vfor (5′-GCGGCGTCATGATAGGATTATTTGACCGTTTAGGC-3′) and either Vrev (5′-GCGGCGCCCGGGTTATCCGTGATG GTGATGATGATGTGCTCCCAGATTATTTAACCGACG-3′) or V236rev (5′-GCGGCGCCCGGGTTAAGAGGTTCCCGGTAATGC-3′). PCR products were digested with BspHI and HindIII and ligated into the same sites of pTrc99. All plasmid constructs were confirmed by sequencing.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or properties | Source or reference |
|---|---|---|
| Strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lac1q ZΔM15 Tn10 (Tetr)] | Laboratory stock |
| L1 | HfrH relA1 spoT | 43 |
| L2 | L1 pspA::kan | 43 |
| CE1224 | F− thr leu Δ(proA-proB-phoE-gpt) his thi argE lacY galK xyl rpsL supE ompR | 21 |
| CE1343 | CE1224 pspA::kan | 21 |
| CE1419 | CE1224 ΔpspB-pspC::kan | 21 |
| K561 | HfrC λ+relA1 spoT T2r (ompF627 fadL701) lac1q | 20 |
| K1527 | K561 pspF::mTn10-tet (Tetr) | 20 |
| J134 | K561 pspABC::kan | 20 |
| MC4100 | F−araD139 Δ(argF-lac)U169 flbB5301 deoC1 ptsF25 relA1 rbsR22 rpsL150 thiA | Laboratory stock |
| BφD | MC4100 ΔtatB | 38 |
| BlLKφ | MC4100 ΔtatC | 8 |
| DADE | MC4100 ΔtatA-DΔE | 45 |
| JARV16 | MC4100 ΔtatA ΔtatE | 38 |
| ELV15 | MC4100 ΔtatA | 36 |
| J1M1 | MC4100 ΔtatE | 36 |
| Plasmids | ||
| pTGS | TorA-GFP-SsrA fusion in pBAD33 | 15 |
| pTG | TorA-GFP fusion in pBAD33 | 15 |
| pSufI-FLAG | sufI gene with C-terminal FLAG tag in pBAD33 | This study |
| pCueO-FLAG | cueO gene with C-terminal FLAG tag in pBAD33 | This study |
| pTrc99 | trc promoter; ColE1 ori Ampr | Amersham Biosciences |
| pPspA | E. coli pspA gene in pTrc99 | This study |
| pKK-SufI | E. coli sufl gene with RR → KK mutation in pTrc99 | This study |
| pTorA-AP | E. coli TorA ss fused to AP(Δ1-22) in pTrc99 | 16 |
| pVipp1 | Synechocystis sp. strain PCC6803 VIPP1 gene in pTrc99 | This study |
| pV-236 | VIPP1 gene truncated after amino acid 236 in pTrc99 | This study |
Cultures were routinely grown aerobically at 30°C in Luria-Bertani (LB) medium, and antibiotic selection was maintained at the following concentrations, as required: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml. Protein synthesis was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG; 0.1 mM) and/or arabinose (0.2%) when the cells reached an optical density at 600 nm of ∼0.5.
Generation of a genomic library and library screening.
A library of random 2- to 3-kbp genomic fragments was constructed by digestion of XL1-Blue genomic DNA with Sau3AI, gel purification of the 2- to 3-kbp products, and ligation into the BamHI site of plasmid pTrc99 (Amersham Biosciences). The ligation products were transformed into XL1-Blue cells carrying pTGS (15) and plated on LB agar containing 0.2% glucose and ampicillin and chloramphenicol at the concentrations indicated above. The resulting cell library (>105 clones) was harvested from plates and subcultured directly into liquid LB medium containing the appropriate antibiotics. Cells were grown at 30°C until mid-log phase (optical density at 600 nm, ∼0.5), and synthesis of ssTorA-GFP-SsrA and of polypeptides encoded within genomic DNA inserts was induced with arabinose and IPTG, respectively. Following 3 h of induction, the cells were washed once with phosphate-buffered saline and a 5-μl aliquot was diluted into 1 ml of phosphate-buffered saline and labeled with propidium iodide for flow cytometric detection of nonviable cells (14). FACS sorting was performed with a Becton-Dickinson FACSort, and the desired cell population was gated by setting appropriate SSC, FL1, and FL2 windows (side scatter is used to trigger the cell events, whereas FL1 is used to monitor GFP fluorescence and FL2 is used to monitor propidium iodine fluorescence). Typically, ca. 3 × 106 cells were examined in 30 min and 250 to 1,000 events were collected. The collected solution was sterilely filtered (0.45-μm pore size; Millipore), and the filters were placed on LB medium plates with ampicillin and chloramphenicol. After 12 h of incubation at 30°C, individual colonies were inoculated into LB medium with ampicillin and chloramphenicol in triplicate 96-well plates. Following 12 h of growth at 30°C, cells were similarly subcultured in triplicate into 96-well plates containing LB medium with ampicillin, chloramphenicol, 0.1 mM IPTG, and 0.2% arabinose and grown for 6 h at 30°C. Individual clones were screened via flow cytometry and on a fluorescent plate reader (Bio-Tek FL600; Bio-Tek Instruments, Winooski, Vt.) for verification of the fluorescent phenotype.
Cell fractionation.
Periplasmic and spheroplast fractions were prepared by subjecting equivalent amounts of cells to the lysozyme-EDTA-cold osmotic shock procedure (31). The resulting spheroplasts were resuspended in 10 ml of TE (10 mM Tris-Cl [pH 7.5], 2.5 mM Na-EDTA) and lysed by sonication, and intact cells and cellular debris were removed by centrifugation (5 min at 10,000 × g). Lysed spheroplasts, including soluble and insoluble fractions, were diluted in sodium dodecyl sulfate-polyacrylamide gel electrophoresis buffer and subjected to electrophoretic analysis, or alternatively, they were centrifuged and the supernatant was retained as the soluble cytoplasmic fraction. For isolation of membrane fractions, the lysate of E. coli cells was centrifuged at 27,000 × g for 20 min at 4°C. The membrane fraction was obtained by further centrifugation of the 27,000 × g supernatant at 100,000 × g for 1 h at 4°C. The supernatant was carefully removed, and the membranes were gently resuspended in morpholinepropanesulfonic acid (MOPS) buffer (50 mM, adjusted to pH 8.0 with KOH) containing 5 mM β-mercaptoethanol and 10 mM MgCl2 at a protein concentration of 20 mg/ml. Protein concentrations were determined with a Bio-Rad protein assay reagent kit with bovine serum albumin as the standard. β-Galactosidase activity was used as a cytoplasmic marker of fractionation efficiency (17). Only data from fractionation experiments in which ≥95% of the β-galactosidase activity was in the cytoplasmic fraction are reported. To analyze total cellular proteins, collected cells were resuspended in TE and homogenized in a French press cell (Carver) at 20,000 lb/in2.
Western blotting analysis.
Western blotting was performed as described previously (12). The following primary antibodies were used: polyclonal rabbit anti-PspA (gift from J. Tommassen), monoclonal mouse anti-GFP (Clontech) diluted 1:5,000, monoclonal mouse anti-FLAG (Sigma) diluted 1:3,000, polyclonal rabbit anti-SufI (10) diluted 1:3,000, monoclonal rabbit anti-DsbC (gift from John Joly, Genentech) diluted 1:10,000, and monoclonal rabbit anti-GroEL (Sigma) diluted 1:10,000. The secondary antibody was 1:10,000 goat anti-mouse-horseradish peroxidase or goat anti-rabbit-horseradish peroxidase. Membranes containing fractionated samples were first probed with anti-GFP, anti-FLAG, or anti-SufI antibodies and then, following development, stripped in Tris-buffered saline-2% sodium dodecyl sulfate-0.7 M β-mercaptoethanol. Stripped membranes were reblocked and probed with anti-DsbC and anti-GroEL antibodies simultaneously. Relative band intensities of Western blots were calculated with ImageJ v1.29, which was obtained from http://rsb.info.nih.gov/ij/.
RESULTS
FACS-based isolation of multicopy E. coli genes that enhance Tat secretion.
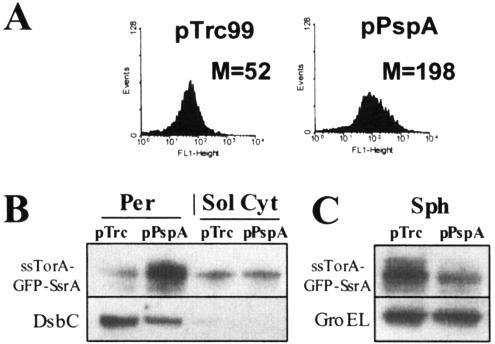
Earlier we had shown that the whole-cell fluorescence of XL1-Blue cells expressing ssTorA-GFP-SsrA is determined by the amount of protein exported to the periplasmic space by the Tat apparatus (15). Since a portion of ssTorA-GFP-SsrA is export incompetent and remains in the cytoplasm, where it is degraded, we reasoned that genes capable of enhancing export would increase the amount of protein that is sequestered in the periplasm. As a result, a larger fraction of GFP-SsrA would be rescued from degradation and therefore confer increased cell fluorescence. Cells containing a pBAD33-based plasmid expressing the ssTorA-GFP-SsrA gene (15) were cotransformed with an E. coli genomic DNA library consisting of 2- to 3-kbp fragments of insert DNA ligated into plasmid pTrc99. About 105 independent transformants were grown in liquid culture, and synthesis of ssTorA-GFP-SsrA and the polypeptides encoded within the genomic fragment inserts was induced by arabinose and IPTG, respectively. Clones conferring increased fluorescence (relative to cells cotransformed with the empty vector) were isolated by FACS. Highly fluorescent clones were isolated, and the gene inserts responsible for the increase in fluorescence were identified by DNA sequencing. Two independent clones that exhibited about fourfold higher whole-cell fluorescence contained genomic fragments that included the entire pspA gene. To further study the role of this gene, we amplified E. coli pspA by PCR and cloned it into pTrc99 downstream from the Trc promoter. Synthesis of ssTorA-GFP-SsrA and PspA was induced by arabinose and IPTG, respectively, and 3 h later, cell fluorescence was determined by flow cytometry. The mean fluorescence (M) of cells expressing PspA (M = 198) was ca. fourfold greater than that of control cells transformed with the empty vector (M = 52; Fig. 1A). The amount of ssTorA-GFP-SsrA in the osmotic shock fraction of cells coexpressing PspA was determined by Western blotting. In these experiments, efficiency of fractionation was evaluated by examining the distribution of periplasmic enzyme DsbC between the osmotic shock (periplasmic) and spheroplast fractions (Fig. 1B). Further, >95% of the β-galactosidase activity was localized within the spheroplast fraction, indicating a negligible amount of cell lysis during cell fractionation. Consistent with the FACS data, expression of PspA from pPspA resulted in an approximately fourfold increase in the intensity of the GFP-SsrA band in the osmotic shock fluid but not in the cytoplasmic fraction (Fig. 1B). Furthermore, only a very low level of intact ssTorA-GFP-SsrA was found within the soluble fraction of lysed spheroplasts, consistent with the expected degradation of the protein by the Clp machinery. Examination of intact spheroplasts, consisting of the soluble components and the inner membrane, revealed the presence of an appreciable amount of ssTorA-GFP-SsrA. Isolation of the membrane fraction further confirmed that the ssTorA-GFP-SsrA intact spheroplast fraction is indeed membrane associated and nonfluorescent (data not shown). Evidently, the membrane-associated protein is present in a form that is not fully folded and is protected from degradation by ClpXP. A similar accumulation of inactive ssTorA-GFP (expressed without an SsrA tag) in the membrane fraction was also reported by Barrett et al. (4). Coexpression of PspA resulted in a reduction in the amount of membrane-associated ssTorA-GFP-SsrA (Fig. 1C). Densitometric analysis of the protein band in intact spheroplasts with and without PspA expression revealed a fourfold greater amount in the latter samples.
FIG. 1.
(A) FACS histograms of cells coexpressing TorA-GFP-SsrA with the pspA gene, which confers an increase in Tat transport of GFP-SsrA to the periplasm. Mean cell fluorescence (M) is given for the genes isolated from a genomic library and for the negative control (pTrc99). All reported values of M are the average of three replicate experiments where each sample was assayed in triplicate (n = 9; standard error, <5%). (B) Western blot assay comparing GFP-SsrA transport in cells coexpressing pspA from pPspA to that in negative control cells (pTrc99), where osmotic shock (Per), soluble spheroplast (Sol Cyt), and (C) intact spheroplast (Sph) fractions are shown. Identical levels of total protein from the osmotic shock and spheroplast fractions were loaded per lane. DsbC and GroEL confirmed that equivalent amounts of periplasmic protein were loaded per lane. The quality of all fractionations was confirmed by β-galactosidase activity assays (see Materials and Methods).
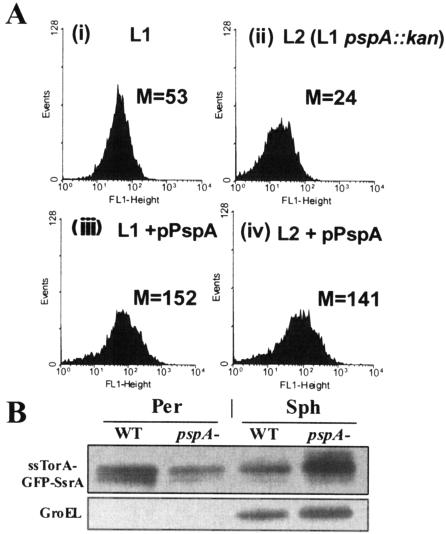
We next determined whether deletion of the pspA gene affected the translocation of ssTorA-GFP-SsrA via the Tat pathway. E. coli strain L2 (pspA::kan) exhibited 50% lower fluorescence relative to its isogenic parent strain, L1 (Fig. 2A). Transformation of E. coli L2 (pspA::kan) with pPspA, followed by induction of protein synthesis by IPTG, resulted in a fluorescence signal comparable to that obtained with the parental strain under the same conditions. Consistent with these results, Western blot analysis showed that in the pspA mutant background, the level of GFP-SsrA in the osmotic shock fraction was approximately threefold (based on densitometry) lower than that in the control (Fig. 2B) and that this was accompanied by a proportional increase in the amount of ssTorA-GFP-SsrA in intact spheroplasts. In contrast to these results, deletion of pspBC and pspF did not have any effect on the export efficiency of the GFP-SsrA fusion protein (data not shown).
FIG. 2.
(A) FACS histograms of cells expressing TorA-GFP-SsrA in WT cells (i), pspA mutant cells (pspA::kan) (ii), WT cells coexpressing pspA from pPspA (iii), and pspA::kan cells coexpressing pspA from pPspA (iv). M refers to mean cell fluorescence in all cases. All reported M values are the average of three replicate experiments in which each sample was assayed in triplicate (n = 9; standard error, <5%). (B) Western blot assay comparing GFP-SsrA transport in WT cells with that in pspA mutant cells. Identical levels of total protein from the osmotic shock (Per) and intact spheroplast (Sph) fractions were loaded per lane. GroEL Western blot assay confirmed the quality of cell fractionations and also that equivalent amounts of proteins from the total spheroplast fraction were loaded per lane.
Efficiency of protein export through the Tat pathway in vivo and in vitro is enhanced when inner membrane vesicles are prepared from cells in which the Tat translocon components TatABC have been expressed from a strong promoter (37, 47). Western blot analysis revealed no difference in the level of TatA, TatB, or TatC in cells containing pPspA and induced with IPTG relative to that in cells transformed with the empty vector (data not shown). Thus, PspA affects Tat export by a process that does not involve induction of Tat protein synthesis.
PspA enhances export of overexpressed endogenous Tat substrates.
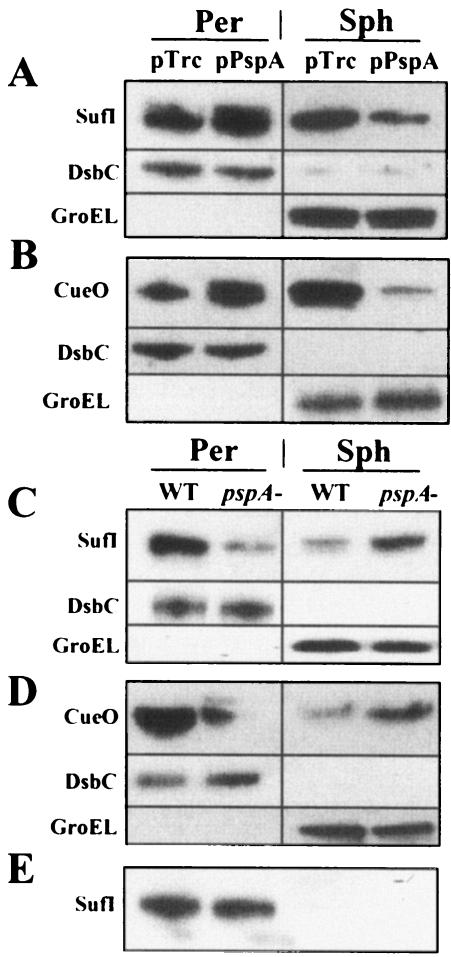
We examined the effect of pspA on the export of the well-characterized endogenous E. coli Tat substrate SufI (39). Recently, Chanal et al. reported that overproduction of SufI leads to the saturation of its own transport but has no effect on other Tat substrates (11). Consistent with this observation, we confirmed that in cells expressing a FLAG-tagged version of SufI from a multicopy plasmid, an appreciable amount of pre-SufI remains in the spheroplast fraction (Fig. 3A). A similar result was obtained in cells expressing native SufI without a FLAG tag, indicating that the presence of the C-terminal eight-amino-acid epitope does not have any effect on the efficiency of export (data not shown). We also found that the export of a second Tat substrate, the copper chaperone CueO, is also saturated when the protein is overproduced.
FIG. 3.
Western blot analysis of SufI (A) or CueO (B) transport in cells coexpressing pspA relative to control cells (pTrc99). Identical levels of total protein from osmotic shock (Per) and intact spheroplast (Sph) samples were loaded per lane. Western blot assays were used to compare SufI (C) and CueO (D) transport in WT and pspA mutant cells. (E) Chromosomal SufI levels in WT and pspA mutant cells. DsbC and GroEL served as quantitation markers and confirmed the quality of fractionations. The quality of all fractionations was also confirmed by β-galactosidase activity assays (see Materials and Methods).
Coexpression of PspA increased the accumulation of both SufI and CueO in the periplasmic fraction (50 and 100%, respectively) with a concomitant reduction in the amounts of pre-SufI and pre-CueO in intact spheroplasts (Fig. 3A and B). As was the case with ssTorA-GFP-SsrA, PspA specifically caused a reduction in the amount of insoluble, presumably membrane-bound, pre-SufI and pre-CueO (data not shown). Finally, consistent with the results shown in Fig. 2B, the efficiency of export of both SufI and CueO was severely reduced in the pspA mutant strain of E. coli (Fig. 3C and D).
SufI synthesized from its own promoter from the chromosomal gene was fully exported into the periplasm, and no precursor accumulation was observed in the intact spheroplast fraction. Similarly, no accumulation of pre-SufI was observed in a pspA background (Fig. 3E). This result indicates that PspA affects the localization of Tat substrate proteins only when export has become saturated, as was the case when ssTorA-GFP-SsrA, SufI, and CueO were expressed at high levels.
PspA is induced in Tat mutants.
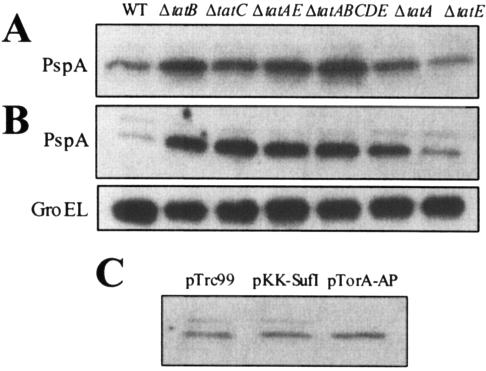
Induction of PspA synthesis has been shown to occur in response to a variety of membrane stresses, including osmotic shock, filamentous phage infection, jamming of the Sec apparatus (25), and most recently, upon depletion of inner membrane protein YidC, which is involved in the membrane integration of membrane-spanning α-helices (42). Given the effect of PspA on the export of Tat substrates described above, it was of interest to examine whether partial or complete blockage of Tat export also results in induction of PspA. Cells carrying mutations that block Tat export completely (tatB, tatC, tatAE, and tatABCDE) exhibited markedly higher levels of PspA protein (Fig. 4A). Interestingly, the ΔtatA mutant had somewhat lower induction of PspA synthesis, which is consistent with the fact that Tat export is not completely abolished in that strain (36). The tatA homologue tatE is expressed at a low level in E. coli and so far has not been found to significantly affect the export of any of the Tat proteins tested (36). Consistent with this observation, induction of PspA did not occur in the ΔtatE mutant strain. Overexpression of SufI exacerbated the effect of mutations in the tat genes (Fig. 4B). Notably, in ΔtatA mutant cells, expression of SufI resulted in a more dramatic increase in the level of PspA, reaching about 60% of that observed in the tatABCE mutant carrying a complete deletion of the Tat apparatus. Once again, no induction of PspA was observed in ΔtatE cells. Expression of SufI alone in WT cells did not result in a higher level of PspA (compare Fig. 4A and B), indicating that the limited blockage of Tat export that occurs under these conditions (11) is not sufficient for induction of PspA synthesis. Similarly, neither (i) expression of SufI carrying an RR→KK mutation in the leader peptide that completely abolishes export (39) nor (ii) fusion of a Tat signal sequence (ssTorA) with alkaline phosphatase (ssTorA-AP), which was previously shown to be misfolded and export incompetent (16), resulted in induction of PspA (Fig. 4C). The latter result is particularly noteworthy because expression of reduced, nonexported ssTorA-AP partially blocks the export of both SufI and CueO (M. P. DeLisa and G. Georgiou, unpublished data), yet this effect was not sufficiently strong to cause PspA induction. Thus, an increase in PspA synthesis is only caused by mutations that abolish export nearly completely.
FIG. 4.
Western blot analysis of PspA protein levels probed in the WT and six different tat mutant strains (A). (B) Same as panel A, but all cells carried pSufI-FLAG and were induced to express full-length SufI. An equivalent number of cells was loaded per lane, and GroEL served as a quantitation marker. (C) PspA protein levels in WT cells expressing the empty-vector control (pTrc99; lane 1), SufI carrying an RR→KK mutation in the leader peptide in pTrc99A (lane 2), and TorA-AP in pTrc99 (lane 3).
The VIPP1 plant homologue enhances Tat export and complements the export defect of pspA null mutants.
The VIPP1 protein found in photosynthetic bacteria and plant chloroplasts has homology with bacterial PspA. Interestingly, the occurrence of PspA and/or VIPP1 is closely linked to the occurrence of the Tat system, with both present in plant chloroplasts, archaea, and bacteria but notably lacking in mycoplasma and mammals (46, 48). In Arabidopsis thaliana, disruption of the VIPP1 gene has detrimental effects on the plant's ability to form properly structured thylakoids and to carry out photosynthesis (23). To examine whether the function of VIPP1 is phylogenetically conserved, we tested for the ability of VIPP1 to substitute for the E. coli pspA-encoded mutant protein. The VIPP1 gene was amplified from Synechocystis sp. strain PCC6803 genomic DNA and cloned into pTrc99 with a C-terminal histidine tag for detection. Cells transformed with pTrc99 VIPP1 and induced with IPTG produced a protein of the expected molecular weight that, similar to PspA, was localized both in the cytoplasm and in intact spheroplasts (data not shown). The presence of VIPP1 resulted in increased periplasmic accumulation of ssTorA-GFP-SsrA to a degree comparable to that observed when PspA was expressed in a similar manner (compare Fig. 5A with 1B). It has been postulated that the C-terminal extension that discriminates VIPP1 proteins from PspA is important for its function in thylakoid formation (44). Therefore, the C-terminal domain would be unlikely to have a role in the stimulation of Tat export in E. coli. Consistent with this hypothesis, a truncated version of VIPP1 (V-236) was coexpressed with ssTorA-GFP-SsrA and found to enhance periplasmic accumulation of GFP-SsrA at a level only slightly lower than that observed for full-length VIPP1 (Fig. 5A). Thus, only amino acids 1 to 236 of VIPP1 are required in order to relieve the saturation of Tat export caused by overexpression of translocation-competent substrate proteins. In addition, both full-length VIPP1 and V-236 were able to restore the export of ssTorA-GFP-SsrA in E. coli pspA mutant cells to the level observed in WT cells, conferring WT levels of cell fluorescence (data not shown).
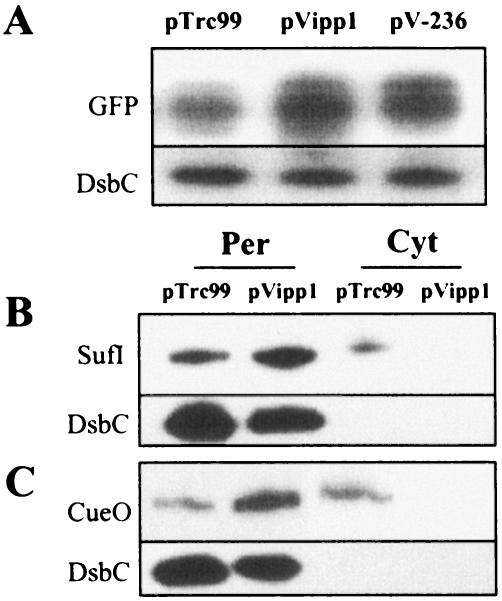
FIG. 5.
Effects of VIPP1 and V-236 on export of GFP-SsrA. (A) Western blot analysis of the periplasmic fraction from WT cells coexpressing TorA-GFP-SsrA and either the empty pTrc99 vector (lane 1), full-length VIPP1 (lane 2), or a truncated version of VIPP1 (V-236; lane 3). Western blot analysis of SufI (B) or CueO (C) transport in WT cells coexpressing VIPP1 from pTrc99 relative to that in control cells carrying empty pTrc99. DsbC and GroEL served as quantitation markers and confirmed the quality of fractionations. The quality of all fractionations was also confirmed by β-galactosidase activity assays (see Materials and Methods).
VIPP1 was also able to relieve the export saturation observed upon overexpression of the native substrates SufI and CueO. As shown in Fig. 5B and C, VIPP1 restored the export efficiency of both proteins to nearly 100%. Rather remarkably, in separate studies, we found that purified E. coli PspA was able to stimulate the in vitro chloroplast Tat-mediated export of proteins across the thylakoid membrane of Pisum sativum in a protein concentration-dependent manner (J. V. Perea and S. M. Theg, unpublished data). This result indicates that bacterial PspA exerts an effect on the export of proteins via the plant equivalent of the Tat system, possibly by supplementing the function of native VIPP1.
DISCUSSION
Extensive in vitro and in vivo studies have clearly established that Tat(A/E)BC are the only essential components of the Tat translocon. However, several lines of evidence indicate that, in addition to the Tat proteins, other cellular components can affect the export process. In vitro, the efficiency of protein translocation into membrane vesicles is typically at least an order of magnitude lower than that observed in the cell (47). Export efficiency can be increased by preparing membrane vesicles from cells that overexpress the Tat proteins, but even under these conditions, translocation proceeded at an efficiency of only about 20%. In vivo, the export of a number of heterologous proteins and endogenous Tat substrates becomes saturated when their expression is elevated (4, 11, 16). In part, this saturation of export might be related to the energetic requirement of Tat translocation. Alder and Theg recently reported that the cost of in vitro cpTat protein transport in the plant thylakoid is the energetic equivalent of approximately 104 ATP molecules per molecule transported, a value that represents almost 3% of the total energy output of the chloroplast (3). If indeed the energetic cost for protein translocation via the bacterial Tat pathway is equally high, then it is easy to imagine how a high level of expression of precursor protein will lead to saturation of export. Factors additional to Tat(A/E)BC might be able to enhance the efficiency of coupling between the flux of protons across the membrane and protein translocation.
We have carried out a genetic analysis for multicopy genes that relieve the saturation of export of ssTorA-GFP-SsrA, in turn resulting in higher cell fluorescence that enabled the selection of the respective clones by flow cytometry. In addition to pspA, which is the subject of the present investigation, the genetic screen also led to isolation of the gene encoding the Tat translocon component TatC and several additional genes whose precise role in Tat export is the subject of ongoing studies. The diverse functions exhibited by these genes suggest that several distinct processes might be contributing to the saturation of protein export.
We found that expression of PspA relieves the export saturation that occurs upon high-level expression of heterologous (ssTorA-GFP-SsrA) and native (SufI, CueO) Tat substrates. Saturation of export is manifest with the accumulation of the preprotein in the insoluble portion of the spheroplast fraction. As a result of the action of PspA, the amount of insoluble precursor is reduced concomitant with an increase in the protein in the periplasmic fraction. We also found that, for all of the proteins tested, the export saturation was exacerbated in strains deficient in pspA. It is important to note that pspA affects efficiency of protein export only under conditions in which the transport machinery is saturated. When SufI is expressed at a low level from the chromosomal copy, no accumulation of precursor protein is observed in the spheroplast fraction either in WT cells or in a pspA mutant.
The precise mechanism by which PspA is able to mediate transfer of the protein from the insoluble spheroplast fraction to the periplasm is not clear. PspA may be affecting any of a number of processes, including, for example, the folding of the protein into a form competent for export, improved coupling of the proton flux with secretion, proteolysis of membrane-associated translocation-incompetent precursors, or even release of the newly translocated protein from the periplasmic side of the membrane.
Whatever the precise mechanism of the action of PspA might be, it is clearly not related to the regulatory role of the protein as an inhibitor of the enhancement of transcription by PspF (25). Deletion of the other genes in the psp operon, including the transcriptional activator pspF or the two positive regulators pspBC, did not exacerbate the saturation of ssTorA-GFP-SsrA export. Thus, although PspA has been shown to bind PspB, PspC, and PspF in vivo (1, 19), these interactions are not involved in the observed PspA-dependent increase in Tat export efficiency.
The level of PspA was markedly increased when Tat export was nearly or completely abolished as a result of the inactivation of Tat genes (i.e., in ΔtatA, ΔtatB, ΔtatC, or ΔtatABCDE mutant strains). Consistent with these findings, Kleerebezum et al. found that depletion of proteins involved in Sec translocation occurring after the protein reaches the membrane (i.e., in SecD, SecF, and SecA) led to induction of PspA. These authors postulated that the PspA-inducing signal was dissipation of the ΔμH+, such as occurs upon entrance of a precursor protein into and subsequent blockage of the export apparatus in the inner membrane. Similarly, Van der Laan et al. demonstrated that depletion of YidC also resulted in elevated levels of PspA (42). Induction of PspA expression upon depletion of YidC proved to be a reliable indicator of a reduced proton motive force, which is in line with previous suggestions that PspA senses membrane damage and/or a reduction of the proton motive force. Since the translocation defects observed in a psp mutant strain are caused by a drop in proton motive force (ΔμH+) and at least one function of PspA appears to be maintenance of ΔμH+ under these stress conditions (21), it is tempting to speculate that stimulation of Tat transport by PspA might be through maintenance of ΔμH+ during translocation of Tat precursors.
In conclusion, we note that the identification of factors that can increase protein flux and enhance the yield of secreted proteins via the Tat pathway is significant from a biotechnology standpoint. The Sec pathway has served as the primary conduit for the secretion of many industrially important proteins. However, the observation that many Sec substrates can become stuck in the translocation pore is problematic when seeking high recombinant yields. To date, there have been no reported cases of precursors becoming jammed in the Tat machinery. In fact, efforts to develop genetic constructs that form membrane-spanning translocation intermediates have proved largely unsuccessful (27). The primary limitation of the Tat pathway relative to the Sec pathway is the relatively poor export efficiency of proteins targeted to the Tat pathway. Therefore, strategies whereby cellular factors such as those identified herein are coexpressed with recombinant proteins of interest should help alleviate the inefficiency of Tat transport.
Acknowledgments
We are especially grateful to P. Model for the generous gift of psp mutants and to J. Tommassen for anti-PspA antisera. We thank G. Buchanan for assistance with pulse-chase assays.
This work was supported by a grant from the Foundation for Research to G.G. P.L. is supported by a BBSRC-funded Ph.D. studentship, and T.P. is a Royal Society Research Fellow.
REFERENCES
- 1.Adams, H., W. Teertstra, J. Demmers, R. Boesten, and J. Tommassen. 2003. Interactions between phage-shock proteins in Escherichia coli. J. Bacteriol. 185:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alami, M., D. Trescher, L. F. Wu, and M. Muller. 2002. Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J. Biol. Chem. 277:20499-20503. [DOI] [PubMed] [Google Scholar]
- 3.Alder, N. N., and S. M. Theg. 2003. Energetics of protein transport across biological membranes. a study of the thylakoid ΔpH-dependent/cpTat pathway. Cell 112:231-242. [DOI] [PubMed] [Google Scholar]
- 4.Barrett, C. M., N. Ray, J. D. Thomas, C. Robinson, and A. Bolhuis. 2003. Quantitative export of a reporter protein, GFP, by the twin-arginine translocation pathway in Escherichia coli. Biochem. Biophys. Res. Commun. 304:279-284. [DOI] [PubMed] [Google Scholar]
- 5.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 6.Berks, B. C., T. Palmer, and F. Sargent. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47:187-254. [DOI] [PubMed]
- 7.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 8.Bogsch, E. G., F. Sargent, N. R. Stanley, B. C. Berks, C. Robinson, and T. Palmer. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273:18003-18006. [DOI] [PubMed] [Google Scholar]
- 9.Brissette, J. L., M. Russel, L. Weiner, and P. Model. 1990. Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchanan, G., E. Leeuw, N. R. Stanley, M. Wexler, B. C. Berks, F. Sargent, and T. Palmer. 2002. Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol. Microbiol. 43:1457-1470. [DOI] [PubMed] [Google Scholar]
- 11.Chanal, A., C. L. Santini, and L. F. Wu. 2003. Specific inhibition of the translocation of a subset of Escherichia coli TAT substrates by the TorA signal peptide. J. Mol. Biol. 327:563-570. [DOI] [PubMed] [Google Scholar]
- 12.Chen, G., A. Hayhurst, J. G. Thomas, B. R. Harvey, B. L. Iverson, and G. Georgiou. 2001. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS). Nat. Biotechnol. 19:537-542. [DOI] [PubMed] [Google Scholar]
- 13.Cristobal, S., J. W. de Gier, H. Nielsen, and G. von Heijne. 1999. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 18:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daugherty, P. S., G. Chen, B. L. Iverson, and G. Georgiou. 2000. Quantitative analysis of the effect of the mutation frequency on the affinity maturation of single chain Fv antibodies. Proc. Natl. Acad. Sci. USA 97:2029-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLisa, M. P., P. Samuelson, T. Palmer, and G. Georgiou. 2002. Genetic analysis of the twin arginine translocator secretion pathway in bacteria. J. Biol. Chem. 277:29825-29831. [DOI] [PubMed] [Google Scholar]
- 16.DeLisa, M. P., D. Tullman, and G. Georgiou. 2003. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc. Natl. Acad. Sci. USA 100:6115-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derman, A. I., J. W. Puziss, P. J. Bassford, Jr., and J. Beckwith. 1993. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J. 12:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Driessen, A. J., P. Fekkes, and J. P. van der Wolk. 1998. The Sec system. Curr. Opin. Microbiol. 1:216-222. [DOI] [PubMed] [Google Scholar]
- 19.Elderkin, S., S. Jones, J. Schumacher, D. Studholme, and M. Buck. 2002. Mechanism of action of the Escherichia coli phage shock protein PspA in repression of the AAA family transcription factor PspF. J. Mol. Biol. 320:23-37. [DOI] [PubMed] [Google Scholar]
- 20.Jovanovic, G., L. Weiner, and P. Model. 1996. Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress-induced psp operon. J. Bacteriol. 178:1936-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleerebezem, M., W. Crielaard, and J. Tommassen. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 15:162-171. [PMC free article] [PubMed] [Google Scholar]
- 22.Kleerebezem, M., and J. Tommassen. 1993. Expression of the pspA gene stimulates efficient protein export in Escherichia coli. Mol. Microbiol. 7:947-956. [DOI] [PubMed] [Google Scholar]
- 23.Kroll, D., K. Meierhoff, N. Bechtold, M. Kinoshita, S. Westphal, U. C. Vothknecht, J. Soll, and P. Westhoff. 2001. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. USA 98:4238-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikhaleva, N. I., C. L. Santini, G. Giordano, M. A. Nesmeyanova, and L. F. Wu. 1999. Requirement for phospholipids of the translocation of the trimethylamine N-oxide reductase through the Tat pathway in Escherichia coli. FEBS Lett. 463:331-335. [DOI] [PubMed] [Google Scholar]
- 25.Model, P., G. Jovanovic, and J. Dworkin. 1997. The. Escherichia coli phage-shock-protein (psp) operon. Mol. Microbiol. 24:255-261. [DOI] [PubMed] [Google Scholar]
- 26.Mori, H., and K. Cline. 2002. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J. Cell Biol. 157:205-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musser, S. M., and S. M. Theg. 2000. Characterization of the early steps of OE17 precursor transport by the thylakoid ΔpH/Tat machinery. Eur. J. Biochem. 267:2588-2598. [DOI] [PubMed] [Google Scholar]
- 28.Musser, S. M., and S. M. Theg. 2000. Proton transfer limits protein translocation rate by the thylakoid ΔpH/Tat machinery. Biochemistry 39:8228-8233. [DOI] [PubMed] [Google Scholar]
- 29.Oresnik, I. J., C. L. Ladner, and R. J. Turner. 2001. Identification of a twin-arginine leader-binding protein. Mol. Microbiol. 40:323-331. [DOI] [PubMed] [Google Scholar]
- 30.Papish, A. L., C. L. Ladner, and R. J. Turner. 2003. The twin-arginine leader-binding protein, DmsD, interacts with the TatB and TatC subunits of the Escherichia coli twin-arginine translocase. J. Biol. Chem. 278:32501-32506. [DOI] [PubMed] [Google Scholar]
- 31.Randall, L. L., and S. J. Hardy. 1986. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell 46:921-928. [DOI] [PubMed]
- 32.Ray, N., J. Oates, R. J. Turner, and C. Robinson. 2003. DmsD is required for the biogenesis of DMSO reductase in Escherichia coli but not for the interaction of the DmsA signal peptide with the Tat apparatus. FEBS Lett. 534:156-160. [DOI] [PubMed] [Google Scholar]
- 33.Rodrigue, A., A. Chanal, K. Beck, M. Muller, and L. F. Wu. 1999. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J. Biol. Chem. 274:13223-13228. [DOI] [PubMed] [Google Scholar]
- 34.Sanders, C., N. Wethkamp, and H. Lill. 2001. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol. Microbiol. 41:241-246. [DOI] [PubMed] [Google Scholar]
- 35.Sargent, F., B. C. Berks, and T. Palmer. 2002. Assembly of membrane-bound respiratory complexes by the Tat protein-transport system. Arch. Microbiol. 178:77-84. [DOI] [PubMed] [Google Scholar]
- 36.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sargent, F., U. Gohlke, E. De Leeuw, N. R. Stanley, T. Palmer, H. R. Saibil, and B. C. Berks. 2001. Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur. J. Biochem. 268:3361-3367. [DOI] [PubMed] [Google Scholar]
- 38.Sargent, F., N. R. Stanley, B. C. Berks, and T. Palmer. 1999. Sec-independent protein translocation in Escherichia coli. A distinct and pivotal role for the TatB protein. J. Biol. Chem. 274:36073-36082. [DOI] [PubMed] [Google Scholar]
- 39.Stanley, N. R., T. Palmer, and B. C. Berks. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 275:11591-11596. [DOI] [PubMed] [Google Scholar]
- 40.Stanley, N. R., F. Sargent, G. Buchanan, J. Shi, V. Stewart, T. Palmer, and B. C. Berks. 2002. Behaviour of topological marker proteins targeted to the Tat protein transport pathway. Mol. Microbiol. 43:1005-1021. [DOI] [PubMed] [Google Scholar]
- 41.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 42.Van Der Laan, M., M. L. Urbanus, C. M. Ten Hagen-Jongman, N. Nouwen, B. Oudega, N. Harms, A. J. Driessen, and J. Luirink. 2003. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. USA 100:5801-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiner, L., J. L. Brissette, and P. Model. 1991. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on sigma 54 and modulated by positive and negative feedback mechanisms. Genes Dev. 5:1912-1923. [DOI] [PubMed] [Google Scholar]
- 44.Westphal, S., L. Heins, J. Soll, and U. C. Vothknecht. 2001. Vipp1 deletion mutant of Synechocystis: a connection between bacterial phage shock and thylakoid biogenesis? Proc. Natl. Acad. Sci. USA 98:4243-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wexler, M., F. Sargent, R. L. Jack, N. R. Stanley, E. G. Bogsch, C. Robinson, B. C. Berks, and T. Palmer. 2000. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J. Biol. Chem. 275:16717-16722. [DOI] [PubMed] [Google Scholar]
- 46.Wu, L. F., B. Ize, A. Chanal, Y. Quentin, and G. Fichant. 2000. Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J. Mol. Microbiol. Biotechnol. 2:179-189. [PubMed] [Google Scholar]
- 47.Yahr, T. L., and W. T. Wickner. 2001. Functional reconstitution of bacterial Tat translocation in vitro. EMBO J. 20:2472-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yen, M. R., Y. H. Tseng, E. H. Nguyen, L. F. Wu, and M. H. Saier, Jr. 2002. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 177:441-450. [DOI] [PubMed] [Google Scholar]