Abstract
A relatively rare aldehyde dehydrogenase 1 (ALDH1) positive “stem cell-like” subpopulation of tumor cells has the unique ability to initiate and perpetuate tumor growth; moreover it is highly resistant to chemotherapy and significantly associated with poor clinical outcomes. The development of more effective therapies for cancer requires targeting of this cell population. Using cDNA microarray analysis, we identified that the expression of the C. elegans lin-28 homolog (LIN28) was positively correlated with the percentage of ALDH1+ tumor cells; this was further validated in an independent set of tissue arrays (n=197). Both lose-of-function and gain-of-function studies demonstrated that LIN28 plays a critical role in the maintenance of ALDH1+ tumor cells. In addition, we found that there is a double negative feedback loop between LIN28 and let-7 in tumor cells, and that let-7 negatively regulates ALDH1+ tumor cells. Finally, we report that a LIN28/let-7 loop modulates self renewal and differentiation of mammary gland epithelial progenitor cells. Our data provide evidence that cancer stem cells may arise through a “reprogramming-like” mechanism. A rebalancing of the LIN28/let-7 regulatory loop could be a novel therapeutic strategy to target ALDH1+ cancer stem cells.
Keywords: LIN28, microRNA, let-7, cancer stem cells
Introduction
Aldehyde dehydrogenase 1 (ALDH1) catalyzes the irreversible oxidation of a range of aliphatic and aromatic aldehydes to their corresponding carboxylic acids. High endogenous ALDH1 activity has been detected in normal stem and progenitor cells of various lineages including hematopoietic, mesenchymal, neural, mammary, and prostate (1–3). Recently, ALDH1 has been successfully used to identify a unique “stem cell-like” subpopulation of cells from tumors, which shares novel characteristics with normal embryonic and somatic stem/progenitor cells: self-renewal and multi-potent differentiation (1). Preclinical studies have further demonstrated that this subpopulation of cells is highly tumorigenic and resistant to chemotherapy (4–7). Moreover, these cells promote and mediate tumor metastasis in vivo (8–11). Importantly, a high percentage of ALDH1+ cells in most types of epithelial tumors, such as breast (1, 10, 12, 13), lung (14), pancreatic (15), bladder (16), ovarian (7), and prostate (17), is associated with a poorer clinical outcome for these patients. This provides robust clinical evidence that the ALDH1+ tumor cells play a critical role in cancer initiation and progression. Therefore, more effective cancer therapies may be developed by targeting this cell population. However, our knowledge of the cellular and molecular regulation of ALDH1+ in cancer stem cells of human tumors is limited. In this study, an important ALDH1+ tumor cell regulatory loop between reprogramming factor LIN28 and miRNA let-7 was identified by high throughput profiling and was then further characterized in human breast and ovarian tumors.
Materials and Methods
Patients and specimens
The ovarian cancer specimens for microarray analysis (n=26, Table S1) were collected at the Universityof Turin, Italy. Detailed information is provided in the Supporting Methods.
Cell lines and cell culture
T47D and MCF7 cells were purchased from ATCC, A2780 and 2008 cells were obtained from the Ovarian Cancer Tissue & Cell Bank, and Hela tet-on cells were purchased from Clontech.
RNA isolation and cDNA microarray analysis
Total RNA was isolated with TRIzol reagent (Invitrogen). cDNA microarrays were performed on the human U133+ 2.0 GeneChip (Affymetrix). Detailed information is provided in the Supporting Methods.
Tissue microarray
The tissue microarray was constructed at the University of Helsinki. Detailed information is provided in the Supporting Methods.
Immunohistochemistry and image analysis
Immunohistochemistry was performed using the VECTASTAIN ABC Kit as described by the manufacturer (Vector). The following primary antibodies were used in this study: mouse anti-human ALDH1 (clone: 44/ALDH, 1:250, BD Pharmingen) and rabbit anti-human LIN28 (1:4,000, Abcam). Antibodies were incubated overnight at 4° C, and the immunoreaction was visualized using 3,3′-diaminobenzidine. The image was analyzed using Image-Pro Plus 4.1 software (Media Cybernetics).
Lentiviral transduction and generation of stable cell lines
Two individual lentiviral shRNA clones targeting LIN28 were purchased from OpenBiosystms. eGFP shRNA and non-target shRNA were used as controls. The pSin-EF2-LIN28 lentiviral expression vector was purchased from Addgene. Lentiviral vectorand packing vectors were transfected intothe packaging cell line 293T (ATCC) using FuGene6 Transfection Reagent (Roche). The media was changed 8 hrs post-transfection and the lentivirus containing media was collected 48 hrs later.
Protein Isolation and Western blots
Cells were lysed in 200μl mammalian protein extraction reagent (Pierce). Then, 15ug of total protein was separated by 10% SDS-PAGE under denaturing conditions, and transferred to a PVDF membrane (Millipore). Membranes were blocked in 5% non-fat milk (Biorad) and then incubated with an anti-LIN28 primary antibody (1:10,000, Abcam), followed by incubation in anti-rabbit secondary antibodyconjugated with horseradish peroxidase (HRP, 1:10,000; Amersham Biosciences) together with an HRP-conjugated primary antibody to beta-actin (1: 10,000; Sigma). Immunoreactive proteins were visualized using the LumiGLO chemiluminescent substrate (Cell Signaling).
ALDEFLUOR Assay and FACS analysis
ALDH1 activity was detected using the ALDEFLUOR assay kit (StemCell Technologies) as described by Ginestier et al (1).
Mammosphere culture
Mammosphere cultures were performed as described by Dontu et al (18). Detailed information is provided in the Supporting Methods.
RNA-immunoprecipitation
Detailed information is provided in the Supporting Methods.
Quantitative Real-time RT-PCR and TaqMan miRNA assay
Detailed information is provided in the Supporting Methods.
Let-7-responsive sensor construction and transfection
The let-7-sensor was constructed by introducing two copies of let-7b perfect complement sequences into the 3′ UTR region of the firefly luciferase gene in the psiCheck 2 vector (Promega).
miRNA in situ hybridization (ISH) and image analysis
In situ detection of miRNA expression was performed on tissue microarray sections by locked nucleic acid (LNA) probes (Exiqon). Detailed information is provided in the Supporting Methods.
Retroviral transduction and stable cell line generation
The retroviral human miRNA expression vector (Figure S1A) was purchased from GeneService. Detailed information is provided in the Supporting Methods.
Tet-on inducible cell lines
Stable cell lines inducibly expressing let-7b were generated using the retrovirus-based RevTet-On system (Clontech). The human genomic sequence of let-7b and an upstream reporter gene (DsRed) were cloned into the pRevTRE response vector downstream of the tetracycline-responsive element (TRE) (Figure S1B), then pRevTet-On and pRevTRE-DsRed-let-7b were separately introduced into HeLa cells by retroviral gene transfer. The reporter gene expression was monitored by fluorescent microscopy (Figure S1C) and FACS analysis (Figure S1D), and let-7b expression was measured using real-time RT-PCR (Applied Biosystems).
Transfection of let-7 mimic and inhibitor oligonucleotides
Pre-miR miRNA precursor and control oligos were purchased from Ambion, and miRCURY LNA miRNA inhibitors and control oligos were purchased from Exiqon. Transfections were performed using the Lipofectamine RNAiMAX transfection reagent (Invitrogen), and then cells were incubated in the media containing the transfection mixture for 72 hrs.
3’UTR reporter construct and assay
The full length sequence of human LIN28 3’ UTR was cloned from human genomic DNA. The PCR products were ligated to the PCR2.1 TOPO cloning vector (Invitrogen) and subcloned into psiCHECK2 reporter vector (Promega). Mutagenesis of miRNA binding sites on reporter vectors was performed by the approach of overlap extension by PCR. Detailed information is provided in the Supporting Methods.
Mammary gland epithelial cell isolation, infection, and colony forming assay
Detailed information is provided in the Supporting Methods.
Statistics
Statistical analysis was performed using the SPSS statistics software package (SPSS, Chicago, IL). All results were expressed as mean ± SD, with significance at p<0.05.
Results
LIN28 expression is positively correlated with a higher percentage of ALDH1+ tumor cells
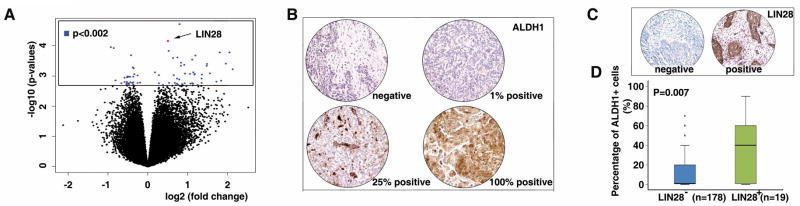
To explore the molecular mechanisms regulating ALDH1+ tumor cells, we chose 26 human ovarian tumor specimens (Table S1), in which the ALDH1 expression was characterized by immunohistochemistry, and the percentage of ALDH1+ tumor cells was scored by two independent investigators. Using the median value of ALDH1+ cells observed in these tumors (7.5%) as cut-off point, the tumors were divided into two groups: ALDH1 low (n=13, ALDH1+ = 2.6 ± 2.3 %) and ALDH1 high (n=13, ALDH1+ = 48.8 ± 37.5 %). An Affymetrix microarray was then used to characterize the transcriptional signature of these tumors. In summary, we found that the expression of 59 genes was significantly different between the ALDH1 high and ALDH1 low groups (p<0.002, Table S2, Figure 1A). Interestingly, we found that expression of the homolog of C. elegans lin-28 (LIN28) was remarkably higher in the ALDH1 high group (p=0.00007). LIN28 is an evolutionarily conserved RNA binding protein, which is highly expressed in embryonic stem (ES) cells, progenitor cells and developing tissues but not in most adult organs (19-24). Together with OCT4, SOX2 and NANOG, LIN28 can act as a reprogramming factor, reprogramming somatic cells to induced pluripotent stem cells (iPS cells) (25). LIN28/LIN28B has also been identified as an oncogene which is upregulated/reactivated in tumors, promoting transformation (26–30). These results suggest that LIN28 may be one of the genes involved in the regulation of ALDH1+ tumor cells. To validate this result, we analyzed LIN28 expression and the percentage of ALDH1+ tumor cells in an independent ovarian cancer tissue array set (n=197) using immunohistochemistry (Figure 1B and C). Consistent with the cDNA microarray result, the percentage of ALDH1+ tumor cells was significantly higher in LIN28+ tumors (n=19) compared to LIN28− tumors (n=178, p=0.007, Figure 1D). Similar correlation was also observed in a breast cancer tissue array set (n=69, p=0.045). This positive correlation of LIN28 expression with ALDH1+ tumor cells indicates that LIN28 may play a functional role in maintenance of ALDH1+ cells in human tumors.
Figure 1. LIN28 expression is positively correlated with a higher percentage of ALDH1+ tumor cells.
A. An Affymetrix cDNA microarray was used to identify the differential gene expression between ALDH1 high and ALDH1 low ovarian tumors (n=26). The results indicated 59 genes (74 probes) that were significantly different between these two groups (blue spots, p<0.002, Table S2). LIN28 was positively correlated with a higher percentage of ALDH1+ tumor cells (red spot, p=0.00007). B to C. An independent ovarian cancer tissue array was used to validate the cDNA microarray result. ALDH1 and LIN28 were detected by immunohistochemistry. D. The percentage of ALDH1+ tumor cells was significantly higher in the LIN28+ tumors compared to LIN28− tumors in the validation tissue array set (p=0.007, n=197).
LIN28 plays a functional role in the maintenance of ALDH1+ tumor cells
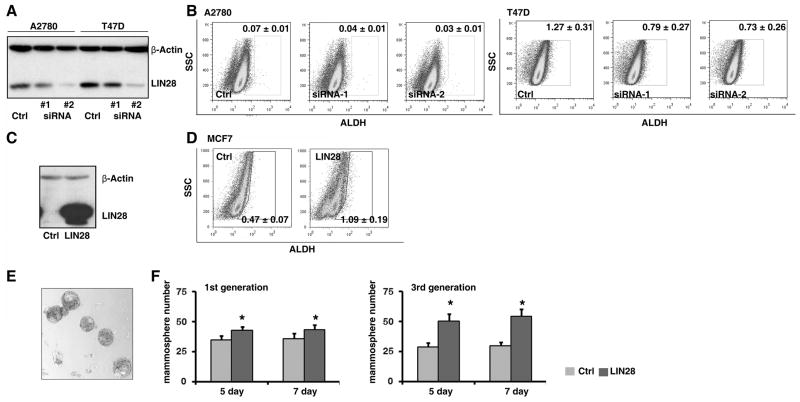
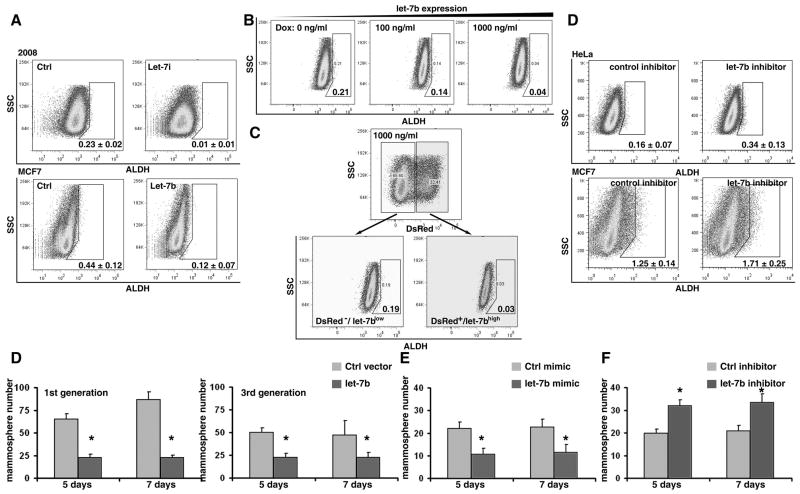
To futher examine whether LIN28 plays a role in the regulation of the ALDH1+ cell population, we chose two cancer cell lines (A2780 and T47D) which highly express LIN28. Using lentiviral shRNA vectors, LIN28 was specifically knocked down in these cell lines (Figure 2A). The precentage of ALDH1+ cells in these two cell lines was measured by the ALDEFLUOR, as assay which was originally developed to detect ALDH1 activity in hematopoietic tissues, and that has since been successfully applied to detect progenitor and cancer stem cells in non-hematopoietic tissues such as the mammary gland and breast cancer (1). We found that the knockdown of LIN28 expression in these cells significantly decreased the brightly fluorescent ALDH1 (ALDH1br) cell population (Figure 2B), and that this occurred in a dose-dependent manner; the two independent shRNA clones knocked down LIN28 expression to different levels, leading to different levels of ALDH1+ cells in each population. Similar observation was also found in vivo (Figure S3). To confirm this observation, we also ectopically expressed LIN28 in a LIN28 negative cell line (MCF7, Figure 2C). Consistent with the shRNA study, enforced expression of LIN28 remarkably increased the ALDH1br population (Figure 2D). Then, we examined the self-renewal capability of MCF7 cells using a mammosphere assay. Ectopically expressing LIN28 produced a significant increase in the number of spheres, especially in the third generation (Figure 2E and F). Finally, other cancer stem cell markers such as CD133 and CD24/CD44 were also examined (Figure S4). Taken together, these results demonstrate that LIN28 plays a functional role in the maintenance of the ALDH1+ cancer stem cell population in tumors.
Figure 2. LIN28 plays a functional role in the maintenance of ALDH1+ tumor cells.
A. LIN28 was knocked down by two independent lentiviral shRNA clones in two LIN28+ tumor cell lines (A2780 and T47D). Reduced protein expression of LIN28 was confirmed by Western blots. Full-length blots are presented in Supplemental Figure S1. B. The percentage of ALDH1br tumor cells in the above cell lines was measured using the ALDEFLUOR assay. Blocking endogenous LIN28 expression by shRNAs significantly reduced the percentage of ALDH1br tumor cells. C. LIN28 was ectopically expressed in MCF7 cells by lentiviral infection. Enforced protein expression of LIN28 was confirmed by Western blots. D. The percentage of ALDH1br tumor cells in the above cell lines was measured using the ALDEFLUOR assay. Enforced LIN28 expression significantly increased the percentage of ALDH1br tumor cells. E. Typical morphology of mammospheres in MCF7 cells. F. Enforced LIN28 expression significantly increased mammosphere numbers compared to the control cells. Left panel presents the result in the first generation of mammospheres; right panel presents the result in the third generation of mammospheres. All p-values <0.05.
LIN28 modulates the biogenesis of miRNA let-7 in tumor cells
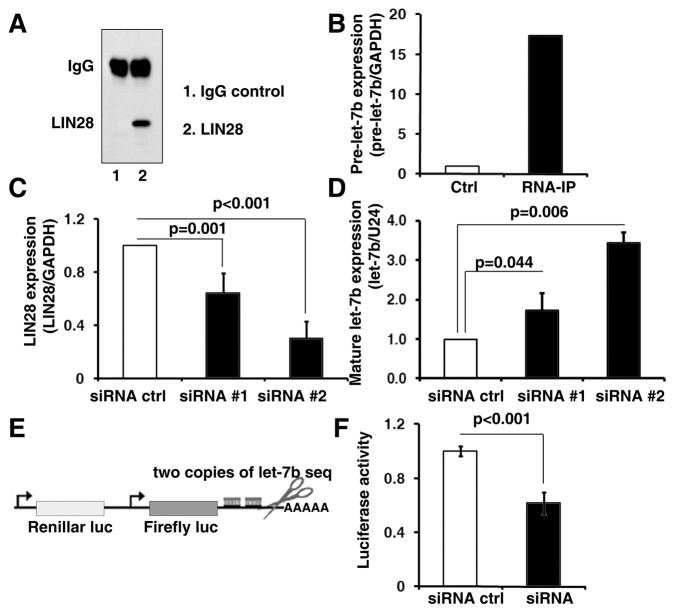
Next, we examined the mechanism by which LIN28 regulates the ALDH1+ cell population in tumors. Results from previous studies suggest that LIN28 maintains embryonic and somatic stem/progenitor cell pluripotency by blocking miRNA let-7 maturation (22, 31–36). However, it is still unknown whether upregulated/reactivated LIN28 also regulates let-7 maturation in cancer, since tumor cells maybe absent the molecular context for LIN28/let-7 regulation. To address this question, we used RNA immunoprecipitation and real-time RT-PCR to demonstrate that LIN28 was able to specifically bind to the let-7 precursor in A2780 cells (Figure 3A and B). In addition, we found that knocking down LIN28 expression by shRNAs significantly increased expression of mature let-7 in a dose-dependent manner in A2780 cells (Figure 3C and D). Finally, using a let-7 sensor assay, which contained a constitutively expressed reporter bearing sequences complementary to let-7 in the downstream 3′ untranslated region (3′ UTR), we demonstrated that blocking LIN28 in A2780 cells remarkably increased let-7 activity (Figure 3E and F). Similar observations were also found in T47D cells (Figure S5). Taken together, we conclude that as in embryonic and somatic stem/progenitor cells (22, 31–36), upregulated/reactivated LIN28 modulates miRNA let-7 maturation and activity in tumor cells.
Figure 3. LIN28 modulates the biogenesis of let-7 in tumor cells.
A. LIN28 binding RNAs in A2780 cells were precipitated by RNA immunoprecipitation using a LIN28 antibody (lane2). Rabbit IgG was used for the control immunoprecipitation (lane 1). B. The amount of let-7b precursor (pre-let-7b) in the precipitated RNAs was detected by real-time RT-PCR. Pre-let-7b was remarkably enriched in the LIN28-RNA-IP compared to the control IgG-RNA-IP (17.3-fold). C. LIN28 was knocked down by two independent lentiviral shRNA clones in A2780 cells. Reduced mRNA expression of LIN28 was confirmed by real-time RT-PCR. D. Mature let-7b expression in the shRNA and control cells was measured by stem-loop RT-PCR. Blocking endogenous LIN28 expression significantly increased mature let-7b expression in A2780 cells. E. A luciferase let-7b sensor assay, which contained a constitutively expressed firefly luciferase reporter bearing sequences complementary to let-7b in the downstream 3’UTR, was used to monitor the let-7b activity in A2780 cells. F. Blocking endogenous LIN28 expression significantly reduced luciferase activity of the let-7b sensor, indicating that let-7b function was remarkably increased in the LIN28 knock down cells.
Let-7 modulates the ALDH1+ tumor cell population in cancer
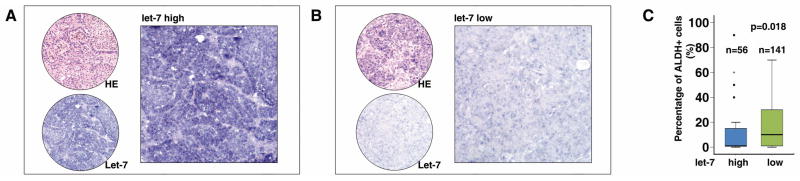
Given that let-7 has been shown to regulate the differentiation of somatic stem/progenitor cells, as well as cancer stem cells, into differentiated cells (37), we hypothesized that LIN28 may maintain ALDH1+ cell populations by modulating miRNA let-7 maturation. To test this hypothesis, we first asked whether there was a negative correlation between let-7 expression and ALDH1+ cell numbers in tumors. Using the same tissue array set as above (Figure 1B to C), let-7 expression was detected by in situ hybridization (Figure 4A and B), and the hybridization signals were classified into two groups: no staining/weak cytoplasmic signals (let-7 low); or moderate/strong cytoplasmic signals (let-7 high). We found that, in contrast to LIN28, the percentage of ALDH1+ tumor cells in the let-7 low group (n=141) was significantly higher than the let-7 high group (n=56; p=0.018, Figure 4C). Similar correlation was also observed in a breast cancer tissue array set (n=69, p=0.016). These observations indicate that let-7 may be involved in the differentiation of ALDH1+ tumor cells in cancer. To further functionally test our hypothesis, we stably over-expressed let-7 in tumor cell lines using retroviral vectors (Figure S3A). As expected, increased let-7 expression dramatically reduced the number of ALDH1br cells (Figure 5A). In order to confirm that this observation, we generated an inducible cell line in which let-7 expression was controlled by the administration of doxycycline (Figure S1B to E). We found that the ALDH1br cell population was negatively associated with increased let-7 expression in a dose-dependent manner (Figure 5B). Importantly, in our polyclonal inducible cells, only ~30% of the cells responded to doxycycline treatment, as detected by the reporter gene DsRed (Figure S1D and 5C). This allowed us to separate the doxycycline treated cells into two subpopulations: DsRed−/let-7low and DsRed+/let-7high (Figure 5C). We found that the percentage of ALDH1br cells was more than 6-fold lower in the let-7 induced cells (DsRed+/let-7high) compared to the non-induced let-7 cells (DsRed−/let-7low, Figure 5C). Additionally, using a let-7 inhibitor, we blocked endogenous let-7 expression in the above cell lines, and found that downregulation of let-7 significantly increased the number of ALDH1br cells (Figure 5D). Finally, we examined the effect of let-7 on the self-renewal ability of MCF7 cells using the mammosphere assay. In contrast to LIN28, over-expression of let-7 led to a decrease in the number of mammospheres, while a let-7 inhibitor led to an increase in the number of mammospheres in MCF7 cells (all p<0.05, Figure 5D to F). Taken together, these results demonstrate that the expression of mature let-7 modulates the ALDH1+ tumor cell population in cancer.
Figure 4. let-7 expression is negatively correlated with a higher percentage of ALDH1+ tumor cells.
A and B. Mature let-7 was detected by in situ hybridization using a LNA probe in the ovarian cancer tissue array. C. Summary of the negative correlation of let-7 expression and percentage of ALDH1+ tumor cells in the tissue array (n=197, p=0.018).
Figure 5. let-7 modulates the ALDH1+ tumor cell population in cancer.
A. Stably enforced expression of let-7 significantly decreased the ALDH1br cell population in 2008 and MCF7 cells. Overexpression of let-7 by retroviral miRNA expression vectors (Figure S3A). B. Induced let-7 expression remarkably decreased the ALDH1br cell population in HeLa cells in a dose-dependent manner. The doxycycline controlled let-7 inducible HeLa cells were generated by a retrovirus-based RevTet-On system (Clontech, Figure S3B). The expression of let-7 and reporter gene (DsRed) were induced by doxycycline treatment (Figure S3C to E). C. After treatment with 1,000ng/ml doxycycline, the reporter gene DsRed (DsRed/let-7) was detectable in about 30% of polyclonal HeLa cells (Figure S3 C and D). Taking this advantagethe DsRed−/letlow and DsRed+/let-7high cell populations were gated, and the percentage of ALDH1br cells was compared between these two groups. The percentage of ALDH1br cells was significantly lower in the DsRed+/let-7high population. D. LNA let-7 inhibitor was used to block let-7 expression in HeLa and MCF7 cells. Blocking endogenous let-7 significantly increased the ALDH1br cell population. D. Enforced let-7 expression significantly decreased the number of mammosphere compared to the control cells. The left side presents the result in the first generation of mammospheres; the right side presents the result in the third generation of mammospheres. E. To avoid artifacts induced by retroviral infection, let-7 was transiently overexpressed via the transfection of a by let-7 mimic. Enforced let-7 expression significantly decreased the number of mammospheres compared to the control cells. F. Endogenous let-7 was transiently blocked via the transfection of a let-7 inhibitor. Blocking let-7 expression significantly increased the number of mammospheres compared to the control cells. All p-values <0.05.
let-7 targets LIN28 expression in tumor cells
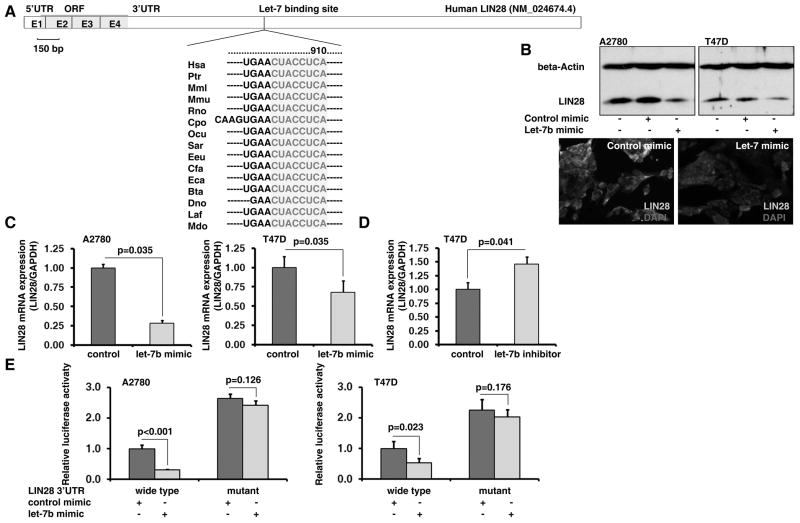
Interestingly, when we treated LIN28+ tumor cells (A2780 and T47D) with a let-7 mimic, we found that the expression of both LIN28 protein and mRNA was significantly decreased (Figure 6B and C), suggesting that let-7 was able to regulate LIN28 via a feedback loop in tumor cells. We predicted that there was a conserved let-7 binding site in the LIN28 3’ UTR by TargetScan (Figure 6A); this was then confirmed experimentally. Using a LIN28 3’ UTR reporter assay, we demonstrated that transfection of a let-7 mimic was able to significantly reduce luciferase activity in the wild type but not the let-7 binding site mutant LIN28 3’ UTR reporters (Figure 6E). The above results demonstrate the existence of a feedback regulatory loop between let-7 and LIN28 in tumor cells.
Figure 6. let-7 targets LIN28 expression in tumor cells.
A. A conserved let-7 binding site in the human LIN28 3’UTR was predicated by TargetScan. B. Transfection of a let-7 mimic remarkably deceased LIN28 protein expression in A2780 and T47D cells. C. Transfection with a let-7 mimic significantly deceased LIN28 mRNA expression in A2780 and T47D cells. D. Transfection with a let-7 LNA inhibitor significantly increased LIN28 mRNA expression in T47D cells. E. The LIN28 3’UTR reporter assay demonstrated that transfection of a let-7 mimic significantly reduced the luciferase activity in the wild type but not the let-7 binding site mutant LIN28 3’UTR reporters.
The LIN28/let-7 loop controlles mammary gland progenitor cell differentiation
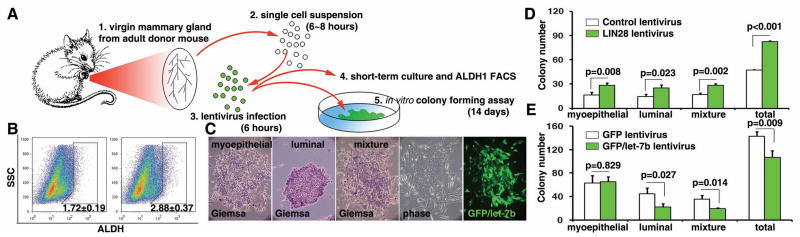
Our studies demonstrate that the LIN28/let-7 loop regulates the ALDH1+ cancer stem cell population in tumors, suggesting that this regulatory loop may also play an important role in maintenance of stem/progenitor cells under normal physiological conditions. To test this hypothesis, we used a mouse mammary gland epithelial cell model. A standard protocol was used to isolate the heterogeneous mouse mammary gland epithelial cells containing epithelial progenitor cells. Then, LIN28 was introduced into the mammary gland epithelial cells by lentiviral infection (Figure 7A). Protein expression was confirmed by Western blot analysis (data not shown). After short-term (48–72 hours) cultures the percentage of the ALDHbr population was analyzed by FACS. Enforced LIN28 expression significantly increased the percentage of ALDHbr cells in the population (Figure 7B). Another mammary gland progenitor cell marker CD24/CD49f was also examined (Figure S6). To further confirm that LIN28 indeed led to an increase in the number of mammary gland progenitor cells, we quantified the number of progenitors (colony-forming cells) using a mammary colony forming assay in which the number of clones reflects the self-renewal capacity, whereas the type of the clones indicates the bipotent differentiation ability (Figure 7C). In summary, LIN28 significantly increased the numbers of all three types of colonies (Figure 7D), suggesting that LIN28 was able to increase the self-renewal capacity of the mammary gland epithelial progenitor cells. In addition, we found that the enforced expression of let-7 led to the differentiation of progenitor cells and significantly decreased total colony numbers (Figure 7E). Interestingly, let-7 selectively decreased progenitor cells which had the ability to form the luminal or mixture type of colonies. Taken together, these results demonstrate that a LIN28/let-7 regulatory loop plays a functional role in the maintenance of mammary gland epithelial progenitor cells.
Figure 7. A LIN28/let-7 loop controls mammary gland progenitor cell differentiation.
A. Protocol of mammary gland epithelial cell isolation and lentiviral infection. B. Enforced expression of LIN28 significantly increased the ALDH1br cell population in mammary gland epithelial cells. C. Three types of colonies were identified by Giemsa staining. D. Enforced LIN28 expression led to a significant increase in the number of colonies. E. Enforced expression of let-7 led to a significant decrease in the luminal and mixture types of colonies.
Discussion
Recent research on iPS cells supports the hypothesis that cancer stem cells may arise through a “reprogramming-like” mechanism (38). By enforcing the expression of a set of genes, the so-called ‘reprogramming factors’, differentiated somatic cells can be converted to iPS cells, which have the same capabilities as ES cells to give rise to all tissue types of the body. Interestingly, most these reprogramming factors are overexpressed or upregulated in certain types of human tumors, and at least some of them (e.g. c-MYC, KLF4, SOX2 and LIN28) are established or putative oncogenes (38). Moreover, five independent studies have shown that disabling p53, an essential tumor-suppressor gene, remarkably improves the efficiency of iPS cell production (38). Therefore, there may be overlapping mechanisms that control the functions and maintenance of iPS cells and cancer stem cells (38). LIN28, one of reprogramming factors discussed above, is very restricted in its expression; it is found only in ES cells, developing tissues, and tumors (19–24, 26–30). In human tumors, LIN28/LIN28B is upregulated and functions as an oncogene promoting malignant transformation and tumor progression (26–30). We have shown that LIN28 is positively correlated with the percentage of ALDH1+ tumor cells in cancer, suggesting that LIN28 may play a role in regulation of ALDH1+ tumor cells. In further functional studies we have shown that LIN28 also functionally maintains this cell population. Our data provided evidence that cancer stem cells may arise through a “reprogramming-like” mechanism; reactivated reprogramming factors such as LIN28 may promote the conversion of epithelial cells to a more undifferentiated stage, and furthermore maintain a small subpopulation of tumor cells in this stem-like stage. In agreement with our findings, Peng et al. recently reported that LIN28 together with OCT4 identified a subpopulation of stem cell-like cells in ovarian carcinoma (30). However, several critical questions need to be addressed further, including the mechanisms by which these reprogramming factors are reactivated in tumors (since most of them are completely silenced after development), the interactions between these reprogramming factors during the ‘reprogramming’ process, and cellular safeguard mechanisms such as p53 respond to these reprogramming events.
The present study also suggests that the maintenance of the ALDH1+ tumor cell population by LIN28 is mediated by the regulation let-7, and that a LIN28/let-7 regulatory loop controls ALDH1+ cancer stem cells. The tumor suppressor role of let-7 in cancer was first demonstrated by the Slack laboratory (39). They found that the let-7 family negatively regulates let-60/RAS in C. elegans by binding to multiple let-7 complementary sites in its 3’UTR (39). Moreover, having found that let-7 expression is lower in lung tumors than in normal lung tissue, whereas RAS protein is significantly higher in lung tumors, let-7 was proposed as a tumor suppressor gene (39–41). Increasing evidence indicates that let-7 plays a functional role in normal and cancer stem cell differentiation. First, in C. elegans, let-7 times seam cell, the stem cells that divide asymmetrically during each larval stage, terminal differentiation, possibly by acting as a regulator of multiple genes required for cell cycle and proliferation (42–45). Second, in ES cells, mature let-7 is poorly expressed, although its precursor transcripts are readily detected (46). This may oppose the actions of a family of cell-cycle-regulating miRNAs that maintain self-renewal in ES cells (47). Third, in mammalian embryonic and somatic stem cells, let-7 interacts with two iPS genes, MYC and LIN28, and these auto-regulatory loops [MCY/let-7 (27) and LIN28/let-7 (22, 31–36)] may control stem cell self-renewal and differentiation (37). Forth, Nishino et al. have shown that during aging, elevated levels of let-7b blocks HMGA2 and contributes to declining neural stem cell function; in contrast, HMGA2 maintains neural stem cell function in young mice through repression of the Ink4a/Arf locus (48). Fifth, Ibarra et al. have found that let-7 is depleted in a population of self-renewing mammary epithelial progenitor cells that can reconstitute the mammary gland. Enforced expression of let-7 leads to a loss of these self-renewing cells from mixed cultures, suggesting that let-7 plays a role in the regulation of progenitor cell maintenance (3). Finally, by comparing miRNA expression in self-renewing and differentiated breast tumor cells, Yu et al. found that let-7 was markedly reduced in breast cancer stem cells but increased upon differentiation (49). These findings demonstrate that let-7 plays a functional role in normal and cancer stem cells. In support of these notions, we have shown that let-7 plays an opposing function to LIN28 in regulating the ALDH1+ tumor cell population. Importantly, we reported a double-negative feedback regulating loop of LIN28 and let-7 in tumor cells. It has been widely reported that the let-7 family of genes is globally downregulated in cancer (37, 39, 46), which may be one of mechanisms of LIN28 reactivation in tumors. Finally, LIN28 may regulate the ALDH1+ cell population through a let-7 independent pathway (20). For example, LIN28 may directly regulate other reprogramming factors in cancer stem cells, such as OCT4 (30, 50).
In summary we have identified a novel mechanism regulating ALDH1+ cancer stem cells, which could lead to new therapeutic strategies for targeting the ALDH1+ tumor cell population. For example, recent studies indicate that LIN28 may cooperate with a terminal uridylyl transferase (TUTase) to regulate let-7 maturation (23, 36). Given that polymerases are facile targets for pharmacological inhibition by small chemical compounds, TUT4 may prove to be a novel target for manipulating the LIN28/let-7 regulation loop (23). Additionally, nanoparticle-delivered LIN28 siRNA or let-7 mimics may also be an attractive therapeutic strategy to target ALDH1+ cancer stem cell population in tumors.
Supplementary Material
Acknowledgments
This work was supported by grants from the Breast Cancer Alliance, the Ovarian Cancer Research Found, the Mary Kay Ash Charitable Foundation, the National Cancer Institute (R01CA142776 and Ovarian Cancer SPORE P50CA83638-7951), and the US Department of Defense. Xiaojuan Lin was supported by the China Scholarship Council.
References
- 1.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu S, Ginestier C, Charafe-Jauffret E, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci U S A. 2008;105:1680–5. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–43. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dylla SJ, Beviglia L, Park IK, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS ONE. 2008;3:e2428. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanei T, Morimoto K, Shimazu K, et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res. 2009;15:4234–41. doi: 10.1158/1078-0432.CCR-08-1479. [DOI] [PubMed] [Google Scholar]
- 6.Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–9. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng S, Yang X, Lassus H, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croker AK, Goodale D, Chu J, et al. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2008 doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–13. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Hoogen C, van der Horst G, Cheung H, et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70:5163–73. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- 12.Morimoto K, Kim SJ, Tanei T, et al. Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci. 2009;100:1062–8. doi: 10.1111/j.1349-7006.2009.01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neumeister V, Agarwal S, Bordeaux J, Camp RL, Rimm DL. In situ identification of putative cancer stem cells by multiplexing ALDH1, CD44, and cytokeratin identifies breast cancer patients with poor prognosis. Am J Pathol. 2010;176:2131–8. doi: 10.2353/ajpath.2010.090712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang F, Qiu Q, Khanna A, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330–8. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasheed ZA, Yang J, Wang Q, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102:340–51. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su Y, Qiu Q, Zhang X, et al. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:327–37. doi: 10.1158/1055-9965.EPI-09-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Su Y, Mei Y, et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients' outcome. Lab Invest. 2010;90:234–44. doi: 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4. RNA Cell. 1997;88:637–46. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 20.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–38. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West JA, Viswanathan SR, Yabuuchi A, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–13. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- 23.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–9. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Zhu H, Shah S, Shyh-Chang N, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42:626–30. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 26.Viswanathan SR, Powers JT, Einhorn W, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–8. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang TC, Zeitels LR, Hwang HW, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–9. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let–7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dangi-Garimella S, Yun J, Eves EM, et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–58. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene. 2010;29:2153–9. doi: 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- 31.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Rybak A, Fuchs H, Smirnova L, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 35.Piskounova E, Viswanathan SR, Janas M, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–4. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 36.Heo I, Joo C, Kim YK, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–6. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trang P, Medina PP, Wiggins JF, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–7. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 43.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–69. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 44.Lin SY, Johnson SM, Abraham M, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–50. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 45.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans Dev Cell. 2005;8:321–30. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 46.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–6. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–39. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 50.Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–8. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.