Platelet function mediates both beneficial and harmful effects on human health, but few genes are known to contribute to variability in the process. We tested association of 2.5 million SNPs with platelet aggregation responses to 3 agonists (ADP, epinephrine and collagen) in two European-ancestry cohorts (N ≤ 2,753 in the Framingham Heart Study, N ≤ 1,238 in the Genetic Study of Atherosclerosis Risk), with replication (P < 0.05) in an African-American cohort (N ≤ 840 in the Genetic Study of Atherosclerosis Risk). We identified associations of seven loci with platelet aggregation, near/in GP6 (P = 4.6×10−13), PEAR1 (P = 3.4×10−12), ADRA2A (P = 3.3×10−11), PIK3CG (P = 3.1×10−9), JMJD1C (P = 1.6×10−8), MRVI1 (P = 2.0×10−8), and SHH (P = 4.5×10−8). Evidence of replication was found for all loci. In total these findings provide new functional insights into platelet aggregation pathways and may suggest novel anti-platelet therapeutic targets.
Aggregation of blood platelets, a critical physiological response to vessel injury, leads to platelet thrombus formation and preserves vascular integrity. Responses are triggered by shear-stress or receptor agonists including ADP, collagen, epinephrine, and thrombin. Aggregation responses are mediated by glycoprotein receptors, and intracellular signaling pathways that trigger receptor activation and release of granules and thromboxane, further mediating feedback signaling. While normal platelet responsiveness maintains homeostasis and promotes wound healing, platelet hyperactivity may promote abnormal thrombosis, and the activation of platelets by plaques is a well-known contributor to acute coronary syndrome or stroke. Thus, several platelet aggregation mechanisms are targets for anti-platelet therapies for the treatment and prevention of cardiovascular disease1. Inter-individual differences in anti-platelet responses suggest genetic variability, but studies to date, mainly focusing on candidate genes, have uncovered few consistent, replicated associations2,3.
The heritabilities of aggregation responses were previously established in the Framingham Heart Study (FHS)4 and the Genetic Study of Atherosclerosis Risk (GS)5. Rare platelet-related disorders are known6; however, with the exception of a limited scan in FHS7, no previous genome-wide scans have been reported for aggregation phenotypes. To investigate common genetic influences, we combined results from two cohorts’ GWAS for platelet aggregation responses to three agonists: ADP, collagen, and epinephrine. Our meta-analysis includes European-ancestry (EA) study populations from FHS and GS, with participants free of symptoms of coronary artery disease and not taking anti-platelet medication. We conducted replication in an African-ancestry (AA) cohort that is also part of GS. The aim was to discover and replicate genome-wide significant loci associated with platelet aggregation and provide new insights into platelet aggregation mechanisms and its variability in humans.
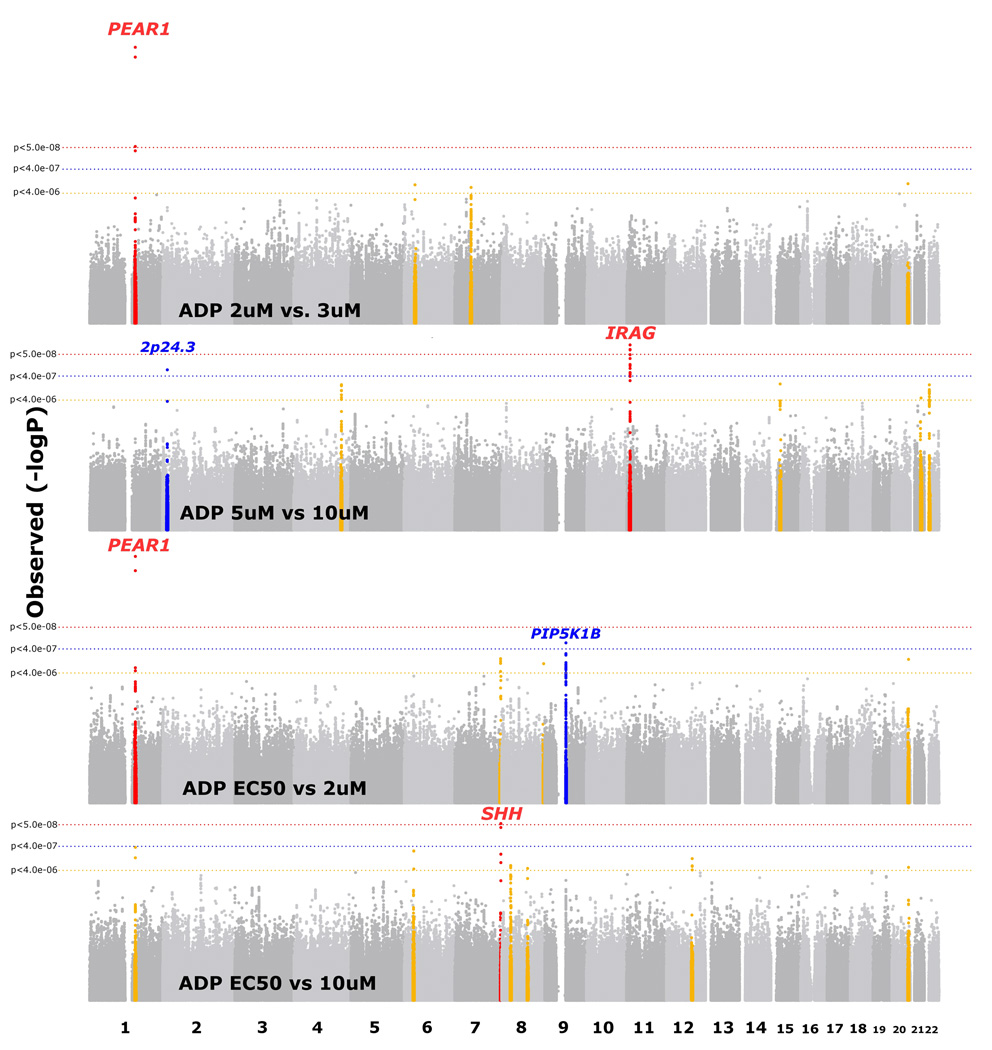
The two GS cohorts were younger, and the GS AA sample had higher BMI and a higher prevalence of smoking, diabetes and hypertension than the two EA samples (Supplementary Table 1). There was no evidence for inflation of test statistics for the meta-analyses conducted, with all λ ≤ 1.01. We observed 8 distinct platelet aggregation associations (Table 1) that met QC filters and surpassed a genome-wide significance threshold in meta-analysis (P < 5.0 × 10−8) with evidence for association in the same direction in both FHS and GS (P < 0.05 in both cohorts). Three regions were genome-wide significant for association with ADP-induced aggregation (Figure 1, Supplementary Figure 1a–c, Table 1): 1q23.1 (PEAR1 rs12566888 P = 3.4 × 10−12), 11p15.4 (MRVI1 rs7940646 P = 2.0 × 10−8), and 7q36.3 (SHH rs2363910 P = 4.5 × 10−8). The minor allele of the PEAR1 SNP was associated with a decrease in aggregation response, whereas the minor alleles of the MRVI1 and 7q36.3 variants were associated with increased responses (Table 2). All 3 regions showed evidence for replication (P < 0.05) in the African-ancestry sample based on genotyped SNPs that showed association with the same direction of effect (Table 3, PEAR1 rs12041331 P = 5.8 × 10−9, MRVI1 rs1874445 P = 9.9 × 10−3, SHH rs6943029 P = 2.4 × 10−3). Three additional loci were modestly associated with increased aggregation responses to ADP in EA and also showed evidence for association in AA at P < 0.05 (Supplementary Table 2).
Table 1.
Associations of top SNPs in loci with p<5.0×10−8 in European-ancestry only meta-analyses.
| SNPid | Chr | Nearest Genes | Coded allele |
FHS phenotype |
n | p-value | beta (sem) | MAF | GS phenotype |
n | p-value | beta (sem) | MAF | Combined EA meta-analysis p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loci associated with ADP aggregation | ||||||||||||||
| rs12566888 (G>T) | 1 | PEAR1 | T | ADP 3uM | 2753 | 6.7×10−8 | −0.06 (0.01) | 9.6% | ADP 2uM | 1110 | 5.6×10−6 | −8.62 (1.9) | 9.2% | 3.4×10−12 |
| rs7940646 (C>T) | 11 | MRVI1 | T | ADP 5uM | 1803 | 1.6×10−5 | 0.03 (0.007) | 31.2% | ADP 10uM | 1227 | 2.5×10−4 | 2.14 (0.58) | 31.8% | 2.0×10−8 |
| rs2363910 (G>T) | 7 | SHH | T | ADP EC50 | 2372 | 6.1×10−4 | −0.04 (0.01) † | 7.1% | ADP 2uM | 1110 | 2.6×10−6 | 4.52 (0.95) | 9.2% | 4.5×10−8 |
| Loci associated with epinephrine aggregation | ||||||||||||||
| rs4311994 (C>T) | 10 | ADRA2A | T | Epi EC50 | 2364 | 1.7×10−8 | 0.14 (0.02) † | 14.0% | Epi 2uM | 1238 | 2.8×10−4 | −7.24 (2.0) | 15.7% | 3.3×10−11 |
| rs12566888 (G>T) | 1 | PEAR1 | T | Epi EC50 | 2364 | 8.8×10−7 | 0.16 (0.03) † | 9.2% | Epi 2uM | 1238 | 1.3×10−4 | −9.77 (2.5) | 9.2% | 7.3×10−10 |
| rs342286 (A>G) | 7 | FLJ36031, PIK3CG | G | Epi EC50 | 2364 | 4.2×10−7 | 0.09 (0.02) † | 44.1% | Epi 2uM | 1238 | 1.3×10−3 | −4.47 (1.4) | 42.2% | 3.1×10−9 |
| rs10761741 (G>T) | 10 | JMJD1C | T | Epi EC50 | 2364 | 1.5×10−6 | −0.08 (0.02) † | 41.5% | Epi 2uM | 1238 | 2.0×10−3 | 4.05 (1.3) | 42.2% | 1.6×10−8 |
| Loci associated with collagen lag time | ||||||||||||||
| rs1671152 (G>T) | 19 | GP6 | T | Lag 190 ug/mL | 2310 | 9.1×10−14 | 0.03 (0.004) | 14.2% | Lag 2 ug/mL | 1162 | 0.037 | 0.017 (0.008) | 15.9% | 4.6×10−13 |
The betas presented in the Table relate directly to the phenotypes presented. Because threshold concentrations (EC50 traits) are inversely related with maximal aggregation response, the signs of the betas for EC50 traits were flipped before meta-analysis.
Figure 1.
Table 2.
Phenotype means by genotype estimates for each cohort.
| Phenotype means (s.d.) by genotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP id | Gene | GWAS P value | Genotypes | FHS | n† | FHS phenotype |
%var‡ | GS EA | n† | GS phenotype |
%var‡ |
| ADP SNPs with p<5.0×10−8 | |||||||||||
| rs12566888 | PEAR1 | 3.4×10−12 | GG | 68.6% (24) | 2238 | ADP 3uM | 1.14% | 21.3% (25) | 912 | ADP 2uM | 1.94% |
| GT | 61.8% (27) | 485 | 12.7% (23) | 188 | |||||||
| TT | 58.3% (23) | 30 | 0.44% (15) | 10 | |||||||
| rs7940646 | MRVI1 | 2.0×10−8 | CC | 75.5% (20) | 840 | ADP 5uM | 1.10% | 70.3% (15) | 572 | ADP 10uM | 1.06% |
| CT | 78.1% (18) | 799 | 72.2% (13) | 525 | |||||||
| TT | 82.6% (16) | 164 | 75.0% (11) | 130 | |||||||
| rs2363910 | SHH | 4.5×10−8 | GG | 3.32 uM (1.5) | 2047 | ADP EC50* | 0.40% | 18.8% (25) | 919 | ADP 2uM | 0.88% |
| GT | 3.22 uM (1.7) | 312 | 23.4% (26) | 184 | |||||||
| TT | 2.50 uM (1.3) | 13 | 34.2% (25) | 7 | |||||||
| Epinephrine SNPs with p<5.0×10−8 | |||||||||||
| rs4311994 | ADRA2A | 3.3×10−11 | CC | 1.74 uM (2.6) | 1749 | Epi EC50* | 1.44% | 35.5% (32) | 893 | Epi 2uM | 1.33% |
| CT | 2.46 uM (3.4) | 567 | 28.6% (34) | 305 | |||||||
| TT | 3.15 uM (4.1) | 48 | 17.5% (36) | 40 | |||||||
| rs12566888 | PEAR1 | 7.3×10−10 | GG | 1.85 uM (2.8) | 1939 | Epi EC50* | 0.99% | 34.9% (32) | 1023 | Epi 2uM | 1.45% |
| GT | 2.30 uM (3.1) | 402 | 25.8% (34) | 205 | |||||||
| TT | 3.07 uM (3.5) | 23 | 11.5% (38) | 10 | |||||||
| rs342286 | PIK3CG | 3.1×10−9 | AA | 1.65 uM (2.5) | 746 | Epi EC50* | 1.10% | 35.8% (33) | 398 | Epi 2uM | 0.91% |
| AG | 2.06 uM (3.0) | 1135 | 34.3% (33) | 597 | |||||||
| GG | 2.13 uM (3.0) | 483 | 26.5% (33) | 243 | |||||||
| rs10761741 | JMJD1C | 1.6×10−8 | GG | 2.13 uM (2.9) | 815 | Epi EC50* | 0.99% | 30.5% (33) | 387 | Epi 2uM | 0.71% |
| GT | 1.94 uM (2.9) | 1136 | 33.2% (33) | 637 | |||||||
| TT | 1.57 uM (2.6) | 413 | 38.3% (32) | 214 | |||||||
| Collagen lag SNPs with p<5.0×10−8 | |||||||||||
| rs1671152 | GP6 | 4.6×10−13 | GG | 80.45 s (19.2) | 1692 | Coll. lag | 2.31% | 104.76 s (32.5) | 821 | Coll. lag | 0.31% |
| GT | 85.95 s (21.8) | 580 | 108.12 s (33.8) | 317 | |||||||
| TT | 99.21 s (21.0) | 38 | 112.75 s (38.0) | 24 | |||||||
genotype numbers and categorization are based on rounded imputed dosages from MACH
the portion of phenotypic variation explained by the SNP was calculated by comparing models with and without the SNP
since threshold concentrations (EC50 traits) are inversely correlated with maximal aggregation responses, the trend of the phenotype means is expected to be opposite
Table 3.
Loci with replication evidence for platelet aggregation phenotypes based on genotyping in the GS African-ancestry (AA) cohort.
| SNPid | Gene | Top EA SNP1 |
p-value | r2 to top EA SNP |
FHS n† |
p-value | beta (s.e.m.) |
MAF | GS EA n† |
p-value | beta (s.e.m.) |
MAF | GS AA n† |
p-value | beta (s.e.m) |
MAF | EA meta- analysis p-value |
EA + AA meta- analysis p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loci associated with ADP aggregation | ||||||||||||||||||
| rs12041331 (G>A)* | PEAR1 | rs12566888 | 3.4×10−12 | 0.85 | 2372 | 3.6×10−6 | 0.06 (0.01) ‡ | 10.2% | 1110 | 6.1×10−6 | −8.52 (1.9) | 9.3% | 788 | 5.8×10−9 | −9.21 (1.6) | 35.8% | 2.2×10−10 | 3.8×10−16 |
| rs6943029 (G>A)* | SHH | rs2363910 | 4.5×10−8 | 0.70 | 2372 | 3.7×10−4 | −0.05 (0.01) ‡ | 6.7% | 1110 | 3.9×10−3 | 5.62 (1.9) | 9.8% | 788 | 2.4×10−3 | 5.35 (1.8) | 26.6% | 5.5×10−6 | 8.2×10−8 |
| rs1874445 (C>T)* | MRVI1 | rs7940646 | 2.0×10−8 | 0.58 | 1803 | 4.5×10−3 | 0.02 (0.007) | 38.1% | 1227 | 1.1×10−3 | 1.84 (0.56) | 40.2% | 836 | 9.9×10−3 | 2.29 (0.89) | 39.3% | 2.4×10−5 | 9.9×10−7 |
| Loci associated with ADP aggregation | ||||||||||||||||||
| rs12041331 (G>A)* | PEAR1 | rs12566888 | 7.3×10−10 | 0.85 | 2364 | 2.1×10−6 | 0.15 (0.03) ‡ | 10.3% | 1232 | 1.4×10−4 | −9.66 (2.5) | 9.3% | 837 | 8.3×10−17 | −17.9 (2.1) | 35.8% | 1.8×10−9 | 4.9×10−19 |
| rs869244 (G>A)* | ADRA2A | rs431194 | 3.3×10−11 | 0.28 | 2364 | 1.5×10−5 | 0.08 (0.02) ‡ | 35.3% | 1232 | 4.0×10−4 | −5.00 (1.4) | 35.3% | 837 | 2.2×10−6 | −9.03 (1.9) | 38.6% | 3.3×10−8 | 3.2×10−12 |
| rs2893923 (C>T)* | JMJD1C | rs10761741 | 1.6×10−8 | 0.69 | 2364 | 8.8×10−6 | −0.08 (0.02) ‡ | 33.6% | 1232 | 0.031 | 2.90 (1.3) | 31.2% | 837 | 8.8×10−3 | 6.36 (2.4) | 19.8% | 1.4×10−6 | 5.3×10−8 |
| Loci associated with collagen lag time | ||||||||||||||||||
| rs1671152 (G>T)* | GP6 | rs1671152 | 4.6×10−13 | n.a. | 2310 | 9.1×10−14 | 0.03 (0.004) | 14.2% | 1157 | 0.037 | 0.017 (0.008) | 15.9% | 763 | 0.048 | 4.82 (0.02) | 30.9% | 4.6×10−13 | 8.4×10−14 |
The top meta-analysis SNP and p-value in EA for the same trait, and LD with the selected replication SNP based on HapMap CEU using SNAP41.
The analyzed traits indicated by sample size are for FHS: ADP EC50 (n=2,372), ADP 5uM (n=1,803), Epi EC50 (n=2,364), collagen lag to 190 ug/mL (n=2,310), for GS: ADP 2uM (n=1,110 or 788), ADP 10uM (n=1,227 or 836), Epi 2uM (n=1,238 or 837), collagen lag to 2ug/mL (n=1,162 or 763)
The betas presented in the Table relate directly to the phenotypes presented. Because threshold concentrations (EC50 traits) are inversely related with maximal aggregation response, the signs of the betas for EC50 traits were flipped before meta-analysis.
The coded allele is the second allele listed. Multiple SNPs in the gene region indicate replication (p<0.05 with effect in the same direction) but only the most significant SNP in combined analysis is given
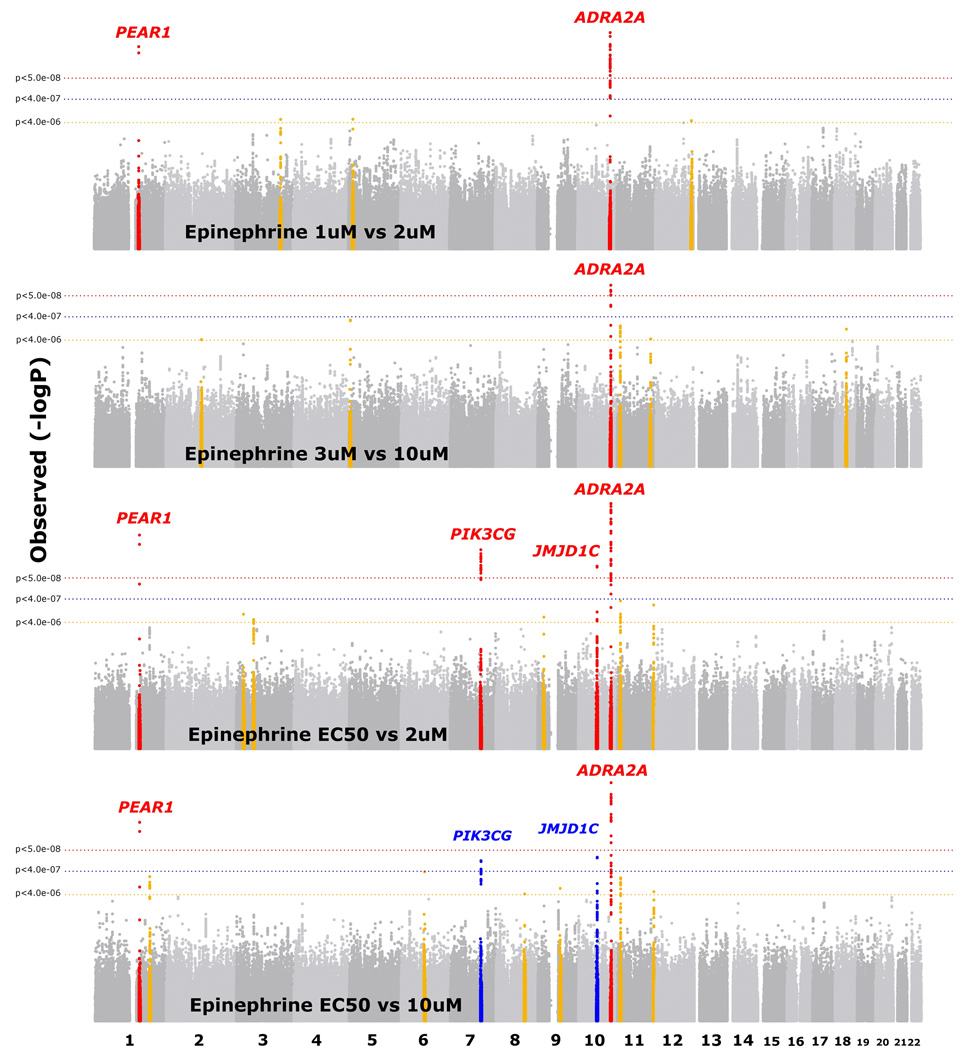
Four regions had genome-wide significant associations for epinephrine-induced platelet aggregation (Figure 2, Supplementary Figure 1d–g, Table 1): 10q25.2 (ADRA2A rs4311994 P = 3.3 × 10−11), 1q23.1 (PEAR1 rs12566888 P = 7.3 × 10−10), 7q22.3 (PIK3CG rs342286 P = 3.1 × 10−9), and 10q21.2 (JMJD1C rs10761741 P = 1.6 × 10−8). For the ADRA2A, PEAR1, and PIK3CG regions, the minor alleles were associated with reduced epinephrine-induced aggregation (Table 2), while the minor allele of the peak SNP nearest JMJD1C was associated with increased aggregation (Table 2). Three of the regions showed consistent results in the African-ancestry sample (Table 3, PEAR1 rs12041331 P = 8.3 × 10−17, ADRA2A rs869244 P = 2.2 × 10−6, JMJD1C rs2893923 P = 8.8 × 10−3). Four SNPs at 7q22.3 did not meet replication criteria (best SNP rs342296, P = 0.13), though none was the peak SNP in EA meta-analyses. Eight additional regions showed moderate associations with epinephrine-induced aggregation in EA along with association in AA (Supplementary Table 2).
Figure 2.
A single region, 19q13.42 (GP6 rs1671152, EA P =4.6 × 10−13, AA P = 0.048), was associated with log10 collagen lag time response at a genome-wide significant level in FHS with P = 9.1 × 10−14 (Supplementary Figure 1h, Supplementary Figure 2, Table 1). The peak associated SNP causes a Thr>Lys change at amino acid 323. The strong association of the Lys allele with decreased collagen response (increased lag time, Table 2) observed in FHS was weakly replicated in GS (EA, P = 0.037; AA, P = 0.048). Our first meta-analysis compared collagen doses of 190 ug/mL (FHS, calf-skin-derived collagen) with 2 ug/mL (GS, equine-tendon-derived collagen) since these provided the most similar lag time distributions (Supplementary Figure 3), consistent with several orders of magnitude higher efficacy of calf- vs. equine-derived collagen (pers. comm., BioData, Inc.). We additionally analyzed associations of the single FHS dose compared with results from three other doses in GS (1, 5, 10 ug/mL), but did not find any additional genome-wide significant loci or gain stronger replication evidence for the GP6 locus. Three additional loci with evidence of moderate association in the main meta-analysis for collagen lag time in the EA sample showed similar association in the AA sample (Supplementary Table 2).
Given that the three platelet function agonists analyzed here target partially overlapping mechanisms of platelet aggregation, we inspected whether significantly associated loci overlapped across agonists. Four regions showed association with aggregation phenotypes in both the EA and AA samples and showed evidence for platelet responses to ≥2 different agonists (Supplementary Table 3).
While an understanding of rare disorders of platelet aggregation has emerged6, the discovery of common genetic variations contributing to platelet aggregation has been marginally successful even though aggregation traits are heritable4,5. Prior studies were performed in modest sample sizes, utilized candidate gene approaches focusing on glycoprotein receptors, and often employed variable conditions in diseased populations. By adopting a GWAS approach in large cohorts of relatively healthy individuals and using similar platelet–rich plasma (PRP)-derived aggregation phenotypes, we discovered or replicated strong associations (P = 5.0 × 10−8) for 7 distinct loci with platelet aggregation, and found suggestive evidence for many additional loci (summarized in Table 4 and Supplementary Table 4). The findings for the PEAR18,9, ADRA2A10,11 and GP612,13 regions provide strong evidence in a much larger sample than past studies, while the associations in the regions of MRVI1, SHH, JMJD1C, and PIK3CG are novel.
Table 4.
Summary of loci associated with platelet aggregation and function.
| Locus | Representative gene | Effects of minor alleles on platelet aggregation to agonists |
Expression in platelets |
Expression in megakaryocytes† |
Other genes ± 60kb of peak SNP |
|---|---|---|---|---|---|
| Loci with p<5.0×10−8 | |||||
| 1q23.1 | PEAR1 (platelet endothelial aggregation receptor 1) | ↓ ADP, ↓ epinephrine | Yes9,14 | not measured | NTRK1, C1orf92, ARHGEF11, INSRR |
| 11p15.4 | MRVI1 (murine retrovirus integration site 1 homolog) | ↑ ADP, ↑ epinephrine | Yes22 | ↑ 1.1 fold42 | |
| 7q36.3 | SHH (sonic hedgehog homolog) | ↑ ADP | Unknown | ↓ 0.7 fold | |
| 10q25.2 | ADRA2A (adrenergic, alpha-2A-, receptor) | ↓ epinephrine | Yes15,16 | ↑ 2.3 fold | |
| 7q22.3 | PIK3CG (phosphoinositide-3-kinase, catalytic, gamma | ↓ epinephrine | Yes24 | ↑ 3.7 fold | FLJ36031 |
| 10q21.2 | JMJD1C (jumonji domain containing 1C) | ↑ epinephrine | Unknown | ↑ 4.0 fold | |
| 19q13.42 | GP6 (glycoprotein VI (platelet)) | ↑ collagen lag | Yes13,17,18 | ↑ 7.0 fold | NLRP2, RDH13 |
indicates the average fold intensity of a representative gene transcript relative to the mean background intensity across stem-cell derived megakaryocytes measured with the Illumina Human WG-6 v2 (n=4 individuals), further described in Watkins et al.43
Platelet endothelial aggregation receptor-1 (PEAR1) undergoes tyrosine phosphorylation after platelet-platelet contact14. A PEAR1 promoter region variant (rs2768759) was associated with increased aggregation in PRP, most strongly in response to epinephrine, and in both pre- and post-aspirin treatment conditions8. Recently a candidate gene study found association of PEAR1 SNPs with ADP and collagen responses in 500 whole blood-derived samples, and an increase in surface PEAR1 expression upon activation9. These candidate gene studies8,9 had limited coverage of the PEAR1 region. In our study, the prior SNPs8,9 were not among the strongest associations; instead, the peak associations with ADP and epinephrine response lie within a relatively conserved region of intron 1 of PEAR1.
Variation in ADRA2A receptor numbers and polymorphisms in ADRA2A that influence epinephrine-induced aggregation in diverse populations were reported nearly 15 years ago10,11. The association of ADRA2A expression with epinephrine response is logical, given that ADRA2A serves as the primary receptor for epinephrine on platelets. Additional reports in small samples have reproduced ADRA2A associations15, including recognition of complex population patterns in the region and effects on RNA levels in vitro16. Notably, unlike prior studies focused on the immediate gene region, the peak SNP associations we observed are somewhat distant and 3’ from the gene (EA, rs4311994, 63kb, P = 3.3 × 10−11; AA, rs869244, 70kb, P = 2.2 × 10−6) suggesting partial LD with causal variants close to the gene or possible long range regulatory elements.
The association of GP6 variants with collagen lag time is biologically plausible, as GP6 is the primary glycoprotein receptor that mediates collagen responses in platelets. The peak GP6 SNP in FHS, a nonsynonymous variant (Thr323Lys), was strongly associated with collagen lag time (rs1671152, P = 9.1 × 10−14). Notably rs1671152 is in LD with rs1613662 (Ser219Pro, HapMap CEU r2=1.0). Both variants have been associated with diminished collagen expression or downstream responses (e.g.,13,17). Due to multiple GP6 protein isoforms formed by splicing and a frameshift, Thr323Lys is alternatively His322Aln in a shorter isoform. Five nSNPs are in LD, including Ser219Pro and Thr323Lys/His322Aln, making it difficult to determine which are functional13,17, although a recent study supports an effect on receptor binding of Thr323Lys/His322Aln within this haplotype17. GP6 plays a role in thrombus formation18. Interestingly, two studies recently replicated association of the 219Pro allele with reduced risk for deep vein thrombosis, indicating potential clinical relevance for genetic findings in GP619,20. In our study, both Thr323Lys and Ser219Pro were similarly associated with collagen lag time (EA, P = 4.6 × 10−13 vs. P = 4.7 × 10−12, AA, P = 0.048 vs. P = 0.08).
MRVI1 (also known as IRAG), which showed both ADP- and epinephrine-induced associations (Table 3, Supplementary Table 3), has prior evidence of functions in platelet aggregation. MRVI1 is a member of a signaling complex which influences smooth muscle cell relaxation through negative regulation of INP3-induced calcium signaling21. In mice MRVI1 plays a direct role in the inhibition of platelet aggregation and in vivo thrombosis22. There is also prior evidence for platelet-related functions for some genes at other novel loci we report. In a human heterologous system SHH+ microvesicles induce differentiation along a megakaryocyte lineage suggesting a link to platelet biology23. Polymorphisms near PIK3CG (rs342293) were recently associated with decreased mean platelet volumes24. The SNP rs342286, associated here with epinephrine-induced aggregation (P < 3.1 × 10−9), is in a strong LD (HapMap CEU, r2=0.87) with rs342293. Our finding did not directly replicate in African-Americans (P = 0.13) although the direction of effect was similar. A direct function for PIK3CG in platelet signaling has been demonstrated25, making it a putative mediator of the effects observed in ours and the prior study24.
We note associations near two genes, MRVI1 and PIK3CG, with prior known platelet functions which are related to intracellular signaling pathways in platelets21,22,24,25. When we attempted to replicate regions with modest evidence for association (P < 1.0 × 10−4 in EA) we found further evidence for regions that encode proteins with known involvement in platelet signaling pathways, including RGS1826–29, RAP1B30–33, and RAPGEF234,35, as well as others with putative platelet functions including ST3GAL436,37 and PRNP38,39 (Supplementary Table 2, Supplementary Table 4). Evaluation of prior candidate SNPs and gene regions from the literature indicated evidence consistent with prior studies in the regions of P2RY12, a receptor that mediates ADP responses, TAOK1, previously associated with mean platelet volume40, and FCER1G, previously associated with collagen response9 and here associated with collagen lag time (Supplementary Table 5).
Combining our novel findings with prior studies, 7 loci are strongly implicated for genetic roles in platelet aggregation, with several more loci having consistent evidence in ≥2 populations (Supplemental Table 4). These additional loci contain genes with compelling biological links to platelet function and warrant further investigation. Known functions of genes at the novel loci suggest that in addition to glycoprotein receptors, proteins involved in intracellular signal transduction pathways and platelet homeostasis are also critical to mediating aggregation responses. Some variants from our study (e.g., GP6) have already been associated with clinically apparent cardiovascular disease. It will be important to conduct further functional and clinical studies to examine the clinically relevant function of genetic variants in these loci and the potential of corresponding proteins as targets for drug treatment, given the central role of platelet function in multiple disease etiologies including thrombosis, myocardial infarction, stroke, wound healing and response to infection.
Platelet aggregation phenotype collection
FHS is a community-based, prospective, longitudinal study following 3 generations of participants. The Offspring cohort studied here represents the second generation, including spouses44. GS is a family-based, prospective study. Apparently healthy subjects free of current aspirin or anti-platelet use were included in phenotype collection for FHS4,45 and GS8,46. FHS participants were excluded from analysis if they self-reported use of aspirin or anti-platelet medication, or in the absence of response to 5 mg/mL arachidonic acid which was presumed indicative of aspirin therapy4,45. GS families were identified from probands with documented premature (age <60) coronary artery disease (CAD) in one of ten Baltimore area hospitals; unaffected, apparently healthy siblings, offspring of the siblings and probands, and parents of the offspring were recruited from 2003–2006 for a study of platelet reactivity8,46. Eligible participants were free of coronary artery disease, had no history of any bleeding disorder or hemorrhagic event, and no serious comorbidities. Participants with a history of aspirin intolerance, abnormal platelet count, hematocrit, or white blood cell count, or current use of anticoagulants or antiplatelet agents were excluded. Use of aspirin and/or nonsteroidal anti-inflammatory drugs was prohibited for ten days before the study visit.
Both studies isolated platelet-rich plasma (PRP) from blood samples taken from participants after fasting, and measured platelet aggregation after addition of agonists using a four-channel aggregometer (BioData Corp., Horsham, PA). FHS samples were collected at exam cycle 5 from participants’ antecubital vein while in the supine position between 8AM and 9AM (previously described)4,45. Blood was placed in a 3.8% sodium citrate solution and centrifuged at 160 × g for 5 min at room temperature to separate out platelet-rich plasma (PRP). In GS participants, after an 8–12 h fast, blood was drawn and collected in vacutainer tubes containing sodium citrate (3.2%), after discarding the first 4 ml. PRP was prepared by centrifugation of whole blood at 180 × g for 15 min, and platelet poor plasma (PPP) was prepared by centrifugation at 2000 × g for 10 min. PRP was diluted with PPP to adjust platelet counts to 200,000/ul. All GS platelet function studies were completed within 2 h after the blood draw.
FHS tested aggregation for periods 4 min post-ADP (0.05, 0.1, 0.5, 1.0, 3.0, 5.0, 10.0, 15.0 uM), 5 min post-epinephrine (0.01, 0.03, 0.05, 0.1, 0.5, 1.0, 3.0, 5.0, 10.0 uM), and lag time (s) to aggregation with 190 ug/mL calf-skin-derived Type-I collagen (BioData Corp., Horsham, PA). Threshold concentrations (EC50) were determined as the minimal concentration of agonist required to produce a >50% aggregation. Testing was not conducted at higher concentrations if >50% aggregation was observed. The maximal aggregation response (% aggregation) was also determined for each participant at each concentration tested. GS recorded maximal aggregation (% aggregation) for periods 5 min post-ADP (2.0, 10.0 uM) and post-epinephrine (2.0, 10.0 uM), and lag time (s) to aggregation with equine-tendon-derived Type-I collagen (1, 2, 5 and 10 ug/mL, Chronolog Corp., Havertown, PA).
Genotyping and imputation
DNA was extracted and genotyped for consenting FHS participants with the Affymetrix 500K array and an additional gene-focused 50K array as part of the SNP Health Association Resource (SHARe) project. DNA was extracted and genotyped for the GS samples with the Illumina 1M (duo) array at deCODE Genetics (Reykjavik, Iceland). FHS and GS both used MACH to impute ~2.54 million SNPs based on the HapMap CEU phased haplotypes (release 22). SNPs were excluded from imputation in FHS that had MAF < 1%, HWE P < 1.0 × 10−6, SNP call rate < 97.0%, MISHAP test P < 1.0 × 10−9, Mendelian errors > 100, or were missing from the HapMap CEU population release 22. Two hundred unrelated individuals were selected from FHS who had low SNP missingness, low numbers of Mendelian errors and who did not show up as outliers in EIGENSTRAT 2.047 (default parameters). The 200 individuals were used to infer MACH model parameters first (MACH flags used: --rounds 100 –greedy), and subsequently applied on all 8,481 individuals (MACH flags used: --greedy --mle –crossovermap –errormap). FHS samples were excluded from GWAS analysis if they had genome-wide call rates < 97.0%, high Mendelian error rates or exhibited genome-wide heterozygosity > 5 s.d. away from the mean. In GS, participants with sex discrepancies or Mendelian errors > 2% were excluded from imputation. SNPs excluded from imputation had MAF < 1%, HWE P < 1.0 × 10−6, or call rate < 95.0%. GS selected 200 EA individuals (pre-screened to be unrelated) by prioritizing those individuals with low missingness, balanced in number of males and females; none of them identified as outliers by EIGENSTRAT 2.0. Similar to FHS, the 200 pre-selected individuals were used to infer model parameters first (MACH flags used: --rounds 100 --greedy), and subsequently that model was applied to all 1,991 EA individuals with genotypes (MACH flags used: --greedy --mle –crossovermap --errormap).
Genetic analyses in each cohort
Both cohorts evaluated age- and sex- adjusted models for aggregation phenotypes. FHS and GS included the principal components (PC) from EIGENSTRAT 2.047 (n=8 and n=2, respectively) as covariates to account for potential population admixture. Collagen lag times and EC50 concentrations were log10 transformed before analysis. Epinephrine maximal aggregations were BLOM-transformed due to non-Gaussian distributions. Linear mixed effects (LME) models were used in FHS and GS individually to test the association under an additive model between a SNP and specific phenotype adjusted for age, sex and PCs. The linear mixed effects (LME) model follows in a matrix form: Y = XB + ZU + ε, where Y is an m × 1 vector of responses; X is an m × p design matrix of the fixed effects; B is the parameter p × 1 vector of fixed effects; Z is an m × q incidence matrix of random effects, and U is a q × 1 vector of random effects with E(U) = 0, and covariance matrix G; 0 is an m × 1 vector of random effects with E(0) = 0 and covariance matrix R. In the fixed effects we included SNP genotypes using an additive model (0 for one major allele, 1 for the heterozygote, and 2 for the minor allele homozygote genotype) for the original genotypes and dosage (probabilistic estimations) for the imputed genotypes. We tested whether the SNP additive effects differed from zero. FHS used the R kinship and GWAF packages48, accounting for familial relatedness, while GS used PROC MIXED in SAS (v. 9.1.3 for Linux OS) with the option for EMPIRICAL variance49 and including the family identification number in the random effects to account for relatedness.
The agonist conditions and number of subjects analyzed in GWAS were as follows: for ADP: in FHS, EC50 (n=2,372), 3uM (n=2,753), 5uM (n=1,803) and in GS, 2uM (n=1,110), 10uM (n=1,227); for epinephrine: in FHS, EC50 (n=2,364), 1uM (n=2,166), 3uM (n=1,220), and for GS, 2uM (n=1,238), 10uM (n=1,232); for collagen lag time: in FHS (n=2,310) and in GS, 1, 2, 5 and 10 ug/mL (n=931, 1,162, 1,222, 1,223, respectively). In GS, participants from 230 families with African-ancestry were used in replication analyses: collagen lag 2 ug/mL (n=763), ADP 2uM (n=788) and 10uM (n=836), epinephrine 2uM (n=837) and 10uM (n=840).
Meta-analysis
SNPs considered in the meta-analyses did not have missing information for either cohort, and had MAF >= 1.0% and an imputation observed to expected ratio >= 0.30 in both cohorts. After this QC filtering ~2.33 million SNPs were included in the meta-analysis for each trait. Sample-size weighted meta-analysis was conducted with the software METAL combining the GS and FHS. The phenotypes used in meta-analyses were for the same agonists at the concentrations with the best available overlap (see Supplementary Table 1). Additionally, when meta-analyzing FHS threshold response (EC50) associations for ADP and epinephrine and GS maximal aggregation, the sign of the beta in FHS was flipped, since threshold response and maximal aggregation are inversely related. Results presented are based on individual cohort age-, sex- and PC-adjusted analyses, and meta-analyses corrected for individual study genomic control inflation rates. Regional association plots (Supplementary Figure 1a–h) were generated with SNAP41.
Replication analysis
We conducted testing for replication in an independent, African-ancestry sample within GS. Since LD patterns in general for African-ancestry individuals at the genome level are more complex and diverse than in populations that are primarily of European-ancestry, relying on single sentinel SNPs from European-ancestry individuals or on imputed or proxy SNPs in African-ancestry individuals for replication comparisons could lead to spurious associations. Thus, we chose to focus replication efforts on all SNPs in regions with evidence for association in the EA meta-analyses (P<1.0×10−4) that were directly genotyped with the Illumina 1M (duo) array and had MAF ≥ 1.0% in the AA replication sample. We searched for evidence of age- and sex-adjusted association in the AA samples only for the same platelet aggregation phenotypes corresponding to those in the main scan. Replication evidence was defined by SNPs with effects in the same direction in AA samples as in EA samples at a P<0.05 threshold.
Supplementary Material
Acknowledgements
This work was supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. This work was supported by the National Heart, Lung and Blood Institute through the PROGENI (U01 HL72518) and STAMPEED (R01 HL087698-01) consortia, and through R01-HL-48157. This research was conducted in part using the resources of the Johns Hopkins General Clinical Research Center, funded through the National Center for Research Resources, M01-RR000052, and the Washington University DSG cluster. We thank Georg Ehret and Santhi Ganesh for providing R code that was modified to generate plots displayed in Figures 1–3.
Footnotes
Database Accessions
HaemGen megakaryocyte results: ArrayExpress accession number E-TABM-633.
Author contributions
A.D. Johnson, L.R.Y., C.J.O and L.C.B. led the study. A.D. Johnson took primary responsibility for drafting the manuscript with contributions and editing from L.R.Y., M-H.C., N.F., M.G.L., G.T., A.T.K., Q.Y., D.M.B., C.J.O and L.C.B. G. Tofler, M.G.L. and C.J.O. were involved in the original guidance, collection and analysis of Framingham platelet phenotype data. L.R. Yanek, N.F., D.M.B. and L.C.B. were involved in the guidance, collection and analysis for the Genetic Study of Atherosclerosis Risk (GeneStar) phenotype data. S.J. Lin, A.T.K. and M.A.P. designed the database and analysis system used in GeneStar analyses. A.D. Johnson, L.R.Y., and M-H.C conducted genome-wide association analyses. A.D. Johnson, L.R.Y., M-H. C. and A.T.K. conducted additional analyses on SNP replication and the percent of variance explained. M-H. Chen conducted the meta-analyses. All authors read and approved the final version of the manuscript.
References
- 1.Wolff T, Miller T, Ko S. Aspirin for the primary prevention of cardiovascular events: an update of the evidence for the U.S. Preventive Services Task Force. Ann. Intern. Med. 2009;150:405–410. doi: 10.7326/0003-4819-150-6-200903170-00009. [DOI] [PubMed] [Google Scholar]
- 2.Quinn MJ, Topol EJ. Common variations in platelet glycoproteins: pharmacogenomic implications. Pharmacogenomics. 2001;2:341–352. doi: 10.1517/14622416.2.4.341. [DOI] [PubMed] [Google Scholar]
- 3.Faraday N, Becker DM, Becker LC. Pharmacogenomics of platelet responsiveness to aspirin. Pharmacogenomics. 2007;8:1413–1425. doi: 10.2217/14622416.8.10.1413. [DOI] [PubMed] [Google Scholar]
- 4.O’Donnell CJ, et al. Genetic and environmental contributions to platelet aggregation: the Framingham Heart Study. Circ. 2001;103:3051–3056. doi: 10.1161/01.cir.103.25.3051. [DOI] [PubMed] [Google Scholar]
- 5.Bray PF, et al. Heritability of platelet function in families with premature coronary artery disease. J. Thromb. Haemo. 2007;5:1617–1623. doi: 10.1111/j.1538-7836.2007.02618.x. [DOI] [PubMed] [Google Scholar]
- 6.Salles II, et al. Inherited traits affecting platelet function. Blood Rev. 2008;22:155–172. doi: 10.1016/j.blre.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Yang Q, Kathiresan S, Lin JP, Tofler GH, O’Donnell CJ. Genome-wide association and linkage analyses of hemostatic factors and hematological phenotypes in the Framingham Heart Study. BMC Med. Gen. 2007;8:S12. doi: 10.1186/1471-2350-8-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrera-Galeano JE, et al. A novel variant in the platelet endothelial aggregation receptor-1 gene is associated with increased platelet aggregability. Arterioscler. Thromb. Vasc. Biol. 2008;28:1484–1490. doi: 10.1161/ATVBAHA.108.168971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones CI, et al. on behalf of the Bloodomics consortium. A functional genomics approach reveals novel quantitative trait loci associated with platelet signaling pathways. Blood. 2009 May 8; doi: 10.1182/blood-2009-02-202614. published online. [DOI] [PubMed] [Google Scholar]
- 10.Freeman K, et al. Genetic polymorphism of the α2-adrenergic receptor is associated with increased platelet aggregation, baroreceptor sensitivity, and salt excretion in normotensive humans. Amer. J. Hyper. 1995;8:863–869. doi: 10.1016/0895-7061(95)00155-I. [DOI] [PubMed] [Google Scholar]
- 11.Kambayashi J, et al. Prevalence of impaired responsiveness to epinephrine in platelets among Japanese. Thromb. Res. 1996;81:85–90. doi: 10.1016/0049-3848(95)00216-2. [DOI] [PubMed] [Google Scholar]
- 12.Croft SA, et al. Novel platelet membrane glycoprotein VI dimorphism is a risk factor for myocardial infarction. Circ. 2001;104:1459–1463. doi: 10.1161/hc3801.096397. [DOI] [PubMed] [Google Scholar]
- 13.Joutsi-Korhonen L, et al. The low-frequency allele of the platelet collagen signaling receptor glycoprotein VI is associated with reduced functional responses and expression. Blood. 2003;101:4372–4379. doi: 10.1182/blood-2002-08-2591. [DOI] [PubMed] [Google Scholar]
- 14.Nanda N, et al. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. JBC. 2005;280:24680–24689. doi: 10.1074/jbc.M413411200. [DOI] [PubMed] [Google Scholar]
- 15.Yabe M, et al. Identification of ADRA2A polymorphisms related to shear-mediated platelet function. Biochem. Biophys. Res. Comm. 2006;347:1001–1005. doi: 10.1016/j.bbrc.2006.06.180. [DOI] [PubMed] [Google Scholar]
- 16.Small KM, Brown KM, Seman CA, Theiss CT, Liggett SB. Complex haplotypes derived from noncoding polymorphisms of the intronless α2A-adrenergic gene diversify receptor expression. PNAS. 2006;103:5472–5477. doi: 10.1073/pnas.0601345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trifiro E, et al. The low-frequency isoform of platelet glycoprotein VIb attenuates ligand-mediated signal transduction but not receptor expression or ligand binding. Blood. 114:1893–1899. doi: 10.1182/blood-2009-03-209510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massberg S, et al. A crucial role of glycoprotein VI for platelet recruitment to the injured arterial wall in vivo. J. Exp. Med. 2003;197:41–49. doi: 10.1084/jem.20020945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bezemer ID, et al. Gene variants associated with deep vein thrombosis. JAMA. 2008;299:1306–1314. doi: 10.1001/jama.299.11.1306. [DOI] [PubMed] [Google Scholar]
- 20.Tregouet DA, et al. Common susceptibility allele are unlikely to contribute as strongly as the FV and ABO loci to VTE risk: results from a GWAS approach. Blood. 2009 Mar 10; doi: 10.1182/blood-2008-11-190389. published online. [DOI] [PubMed] [Google Scholar]
- 21.Schlossmann J, et al. Regulation of intracellular calcium by a signaling complex of IRAG, IP3 receptor and cGMP kinase Ibeta. Nature. 2000;404:197–201. doi: 10.1038/35004606. [DOI] [PubMed] [Google Scholar]
- 22.Antl M, et al. IRAG mediates NO/cGMP-dependent inhibition of platelet aggregation and thrombus formation. Blood. 2007;109:552–559. doi: 10.1182/blood-2005-10-026294. [DOI] [PubMed] [Google Scholar]
- 23.Martinez MC, et al. Transfer of differentiation signal by membrane microvesicles harboring hedgehog morphogens. Blood. 2006;108:3012–3020. doi: 10.1182/blood-2006-04-019109. [DOI] [PubMed] [Google Scholar]
- 24.Soranzo N, et al. A novel variant on chromosome 7q22.3 associated with mean platelet volume, counts, and function. Blood. 2009;113:3831–3837. doi: 10.1182/blood-2008-10-184234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoenwaelder SM, et al. Identification of a unique co-operative phosphoinositide 3-kinase signaling mechanism regulating integrin αIIbβ3 adhesive function in platelets. J. Biol. Chem. 2007;282:28648–28658. doi: 10.1074/jbc.M704358200. [DOI] [PubMed] [Google Scholar]
- 26.Yowe D, et al. RGS18 is a myeloerythroid lineage-specific regulator of G-protein-signaling molecule highly expressed in megakaryocytes. Biochem. J. 2001;359:109–118. doi: 10.1042/0264-6021:3590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gagnon AW, Murray DL, Leadley RJ. Cloning and characterization of a novel regulator of G protein signaling in human platelets. Cell Signal. 2002;14:595–606. doi: 10.1016/s0898-6568(02)00012-8. [DOI] [PubMed] [Google Scholar]
- 28.Kim SD, et al. The expression patterns of RGS transcripts in platelets. Platelets. 2006;17:493–497. doi: 10.1080/09537100600758123. [DOI] [PubMed] [Google Scholar]
- 29.Garcia A, et al. Differential proteome analysis of TRAP-activated platelets: involvement of DOK-2 and phosphorylation of RGS proteins. Blood. 2004;103:2088–2095. doi: 10.1182/blood-2003-07-2392. [DOI] [PubMed] [Google Scholar]
- 30.Guidetti GF, et al. The Gi-coupled P2Y12 receptor regulates diacylglycerol-mediated signaling in human platelets. J. Biol. Chem. 2008;283:28795–28805. doi: 10.1074/jbc.M801588200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cannobbio I, et al. Genetic evidence for a predominant role of PI3Kβ catalytic activity in ITAM- and integrin-mediated signaling in platelets. Blood. 2009 Jun 10; doi: 10.1182/blood-2009-03-208074. published online. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, et al. Rap1b is critical for glycoprotein VI-mediated but not ADP receptor-mediated α2β1 activation. J. Thromb. Haem. 2009;7:693–700. doi: 10.1111/j.1538-7836.2009.03289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chrzanowska-Wodnicka M, et al. Rap1b is required for normal platelet function and hemostasis in mice. J. Clin. Invest. 2005;115:680–687. doi: 10.1172/JCI22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letschka T, et al. PKC-θ selectivity controls the adhesion-stimulating molecule Rap1. Blood. 2008;112:4617–4627. doi: 10.1182/blood-2007-11-121111. [DOI] [PubMed] [Google Scholar]
- 35.Severson EA, Lee WY, Capaldo CT, Nusrat A, Parkos CA. Junctional adhesion molecule A interacts with Afadin and PDZ-GEF2 to activate Rap1a, regulate β1 integrin levels, and enhance cell migration. Mol. Biol. Cell. 2009;20:1916–1925. doi: 10.1091/mbc.E08-10-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellies LG, et al. Sialyltransferase ST3Gal-IV operates as a dominant modifier of hemostasis by concealing asialoglycoprotein receptor ligands. PNAS. 2002;99:10042–10047. doi: 10.1073/pnas.142005099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grewal PK, et al. The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat. Med. 2008;14:648–655. doi: 10.1038/nm1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holada K, Glierova H, Simak J, Vostal JG. Expression of cellular prion protein on platelets from patients with gray platelet or Hermansky-Pudlak syndrome and the protein’s association with α-granules. Haematologica. 2006;91:1126–1129. [PubMed] [Google Scholar]
- 39.Jones M, et al. Human platelets as a substrate source for the in vitro amplification of the abnormal prion protein (PrPSc) associated with variant Creutzfeldt-Jakob disease. Transfusion. 2009;49:376–384. doi: 10.1111/j.1537-2995.2008.01954.x. [DOI] [PubMed] [Google Scholar]
- 40.Meisinger C, et al. A genome-wide association study identifies three loci associated with mean platelet volume. Am. J. Hum. Genet. 2009;84:66–71. doi: 10.1016/j.ajhg.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaughnessy JD, et al. Mrvil, a common MRV integration site in BXH2 myeloid leukemias, encodes a protein with homology to a lymphoid-restricted membrane protein Jaw1. Oncogene. 18:2069–2084. doi: 10.1038/sj.onc.1202419. [DOI] [PubMed] [Google Scholar]
- 43.Watkins NA, et al. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood. 2009;113:1–9. doi: 10.1182/blood-2008-06-162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families: the Framingham Offspring Study. Am. J. Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 45.Mukamal KJ, et al. Alcohol consumption and platelet activation and aggregation among women and men: The Framingham Offspring Study. Alc. Clin. Exp. Res. 2005;29:1906–1912. doi: 10.1097/01.alc.0000183011.86768.61. [DOI] [PubMed] [Google Scholar]
- 46.Becker DM, et al. Sex differences in platelet reactivity and response to low-dose aspirin therapy. JAMA. 2006;295:1420–1427. doi: 10.1001/jama.295.12.1420. [DOI] [PubMed] [Google Scholar]
- 47.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Gen. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 48.Chen MH, Yang Q. GWAF: an R package for genome-wide association analysis with family data. Bioinformatics. 2009 Dec 29; doi: 10.1093/bioinformatics/btp710. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Littell RC, Milliken GA, Stroup WW, Wolfinger RD. SAS System for Mixed Models. Cary, North Carolina: SAS Institute; 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.