Abstract
In this report we describe the cloning, organization, and promoter analysis of the Staphylococcus aureus thioredoxin (trxA) and thioredoxin reductase (trxB) genes and their transcription in response to changes in oxygen concentration and to oxidative stress compounds. Northern analysis showed that the S. aureus trxA and trxB genes were transcribed equally well in aerobic and anaerobic conditions. Several oxidative stress compounds were found to rapidly induce transcription of the trxA and trxB genes. The most pronounced effects were seen with diamide, a thiol-specific oxidant that promotes disulfide bond formation; menadione, a redox cycling agent; and τ-butyl hydroperoxide, an organic peroxide. In each case the induction was independent of the general stress sigma factor σB. These studies show that the S. aureus trxA and trxB genes are upregulated following exposure to these oxidative stress agents, resulting in increased disulfide bond formation. In contrast, no effect of hydrogen peroxide on induction of the trxA and trxB genes was seen. We also show that the S. aureus thioredoxin reductase appears to be essential for growth. This observation, coupled with structural differences between the bacterial and mammalian thioredoxin reductases, suggests that it may serve as a target for the development of new antimicrobials.
Staphylococcus aureus is a major human pathogen causing a wide variety of diseases, ranging from mild skin infections to life-threatening septicemia, meningitis, and toxic shock syndrome (28). Its abilities to colonize and exploit host functions and to cope with an often hostile environment make it a highly successful and opportunistic pathogen that is difficult to eradicate. Several factors contribute to its pathogenesis. In the first place, S. aureus determines a broad array of cell wall-associated proteins that enable it to evade host immune defense systems as well as numerous extracellular proteins, including hemolysins, toxins, and proteases, many of which are virulence factors (31). Second, S. aureus is able to counteract and eliminate a variety of reactive oxygen intermediates, such as superoxide anions and hydrogen peroxide, which pose a threat to its survival. These species are generated during physiological conditions from the incomplete reduction of oxygen or from exposure to light, radiation, redox active agents, or host phagocytes and result in damage to proteins, lipids, and DNA (47). Third, outside its host, S. aureus can withstand numerous environmental stresses, chief among which are desiccation, osmotic stress, starvation, and heat shock, which account for its persistence and survival in nature (6, 7).
In common with other facultative aerobes, S. aureus can grow in the absence of oxygen either by fermentation or by using an alternative terminal electron acceptor such as nitrate. Oxygen availability or the lack of it may be a key factor in S. aureus pathogenicity, potentially triggering events such as host cell adherence and invasiveness that are necessary for colonization on cell surfaces, tissues, and other environmental niches (9, 26). Indeed, the ability of S. aureus to adapt to extreme changes in external oxygen concentration implies the existence of redox-dependent processes that regulate the expression of genes in the transition from aerobic to anaerobic growth and vice versa.
Thiol-specific redox systems play a major role in the protection of cells against toxic oxygen species as well as in maintaining the intracellular thiol-disulfide balance and in providing reducing power to key reductive enzymes such as ribonucleotide reductases. In many prokaryotes this is achieved by a number of small proteins, thioredoxin or glutaredoxin, each of which contains a pair of redox active cysteines. Thioredoxin is maintained in its reduced form by thioredoxin reductase, while glutaredoxin is kept reduced by glutathione/glutathione reductase, in both cases at the expense of NADPH (3, 20). We have previously shown that S. aureus and many other gram-positive bacteria lack glutathione and its cognate enzymes (1, 32, 34) and must therefore employ thioredoxin and alternative thioredoxin-based systems, such as the peroxiredoxins and alkyl hydroperoxide reductases, to carry out the above cellular processes (17, 42). In some of these bacteria, in particular the actinomycetes, a novel low-molecular-weight glycothiol, mycothiol, is made in high concentrations with properties that suggest it has a role analogous to that of glutathione in intermediary metabolism (33).
In this paper we analyze the expression of the S. aureus thioredoxin (trxA) and thioredoxin reductase (trxB) genes in growth and in response to changes in oxygen concentration and to compounds that cause oxidative stress. We further show that the thioredoxin system is essential for growth.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. S. aureus strains were grown at 30°C or 37°C in tryptone soy broth (TSB; Difco) supplemented with erythromycin (12 μg ml−1) where appropriate. Transformants were selected on TSB agar plates containing antibiotics. Escherichia coli was grown in Luria-Bertani medium with the addition of ampicillin (100 μg ml−1) or kanamycin (50 μg ml−1) as needed.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli XL-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqlacZΔM15 Tn10(Tcr)] | Laboratory stock |
| S. aureus | ||
| Oxford (NCTC6571) | Wild-type laboratory strain | Laboratory stock |
| RN4220 | Restriction-deficient mutant of 8325-4 (UV cured of prophages) used as primary recipient for plasmids propagated in E. coli; spontaneous mutant with 11-bp deletion in rsbU | 12, 24 |
| SH1000 | Derivative of 8325-4 carrying intact rsbU gene | Gift of S. Foster |
| Plasmids | ||
| pAUL-A | Temperature-sensitive E. coli-S. aureus shuttle vector, lacZ Emr | 45 |
| pO17 | pAUL-A containing ∼0.7-kbp internal S. aureus Oxford trxB DNA fragment | This study |
| pO18 | pAUL-A containing ∼0.7-kbp internal S. aureus Oxford glnA DNA fragment | This study |
| pO115 | pAUL-A containing internal S. aureus trxB and glnA DNA fragments in tandem | This study |
Abbreviations: Emr, and Tcr, resistance to erythromycin/lincomycin and tetracycline, respectively.
S. aureus liquid cultures were grown aerobically at 37°C in an air orbital shaker at 250 rpm. For limiting oxygen conditions, the standard anaerobic growth conditions used for growth of cultures were agitation at 100 rpm in an orbital shaker at 37°C in TSB medium supplemented with cysteine (5.7 mM) to scavenge traces of oxygen and 0.001% resazurin as a redox indicator (29). Wheaton serum bottles (100-ml capacity) containing 60 ml of the above-described medium were purged with nitrogen gas for 4 min at a pressure of 0.75 atm prior to being autoclaved. Aerobic cultures were subcultured (0.5 ml) in 60 ml of the above-described medium and grown for 16 to 20 h to stationary phase (optical density at 600 nm [OD600], ≈2), and 2 ml was used to inoculate 60 ml of the same medium.
DNA manipulations.
For E. coli, preparation of plasmids, DNA manipulations, and transformation of competent cells were performed as previously described (44). For S. aureus, genomic DNA was prepared as described previously (35). Standard procedures were employed for restriction enzyme digestion, ligation, Southern blotting, and radiolabeling of oligodeoxynucleotides (44) unless otherwise stated. The nucleotide sequences of the DNA regions containing the S. aureus Oxford trxA and trxB genes were determined from both strands by the dideoxy procedure with the ABI Prism 377 automatic sequencer (Perkin-Elmer Biosystems) and the Prism dye terminator cycle sequencing kit (Applied Biosystems).
Construction of trxB insertion mutants.
To create a disruption of the S. aureus trxB gene, an internal fragment (≈0.7 kbp) of the Oxford trxB gene (nucleotides 1400 to 2076 of GenBank AJ223781) was amplified by PCR with the forward primer (with an added EcoRI restriction site) 5′ CCCCGAATTCACAGAAGAAGTAGAG-3′ and the reverse primer (with an added BamHI restriction site) 5′-CCCCGGATCCCTGGTACTGATGTTGTC-3′ and ligated into the corresponding sites of the E. coli-S. aureus temperature-sensitive shuttle vector pAUL-A (42) to give pOI7. S. aureus RN4220 was electroporated with pOI7, and transformants were selected for growth at 30°C (the permissive temperature) and at 40°C (the nonpermissive temperature) in the presence of erythromycin. Integration of pOI7 into the chromosome by a single reciprocal recombination event is expected to result in disruption of the chromosomal trxB gene.
Control plasmid pOI8 contains a ≈0.7-kbp internal DNA fragment of the glnA gene (nucleotides 1893 to 2592 of GenBank X76490) inserted in the HindIII polylinker site of pAUL-A; the DNA fragment was obtained from PCR amplification of S. aureus Oxford genomic DNA with 5′-GGGGAAGCTTTAGAGGATATGGGCTTCG-3′ as the forward primer (with an added HindIII restriction site) and 5′-GGGGAAGCTTTTTTAATAACTTCATTTTCACGC-3′ as the reverse primer (with an added HindIII restriction site). Control plasmid pOI15 contains internal DNA fragments of the trxB and glnA genes. It was obtained by inserting the HindIII glnA fragment into the corresponding site in the polylinker of pOI7. The structures of the pAUL-A disruption vectors were confirmed by restriction analysis and nucleotide sequencing.
RNA extraction.
Total RNA was isolated from cultures of exponentially grown S. aureus cells in TSB medium at 37°C as described (13) with modifications (29). Cells (50 mg wet weight) were lysed in 0.3 ml of TES buffer containing 150 mM NaCl, 78 mM EDTA,100 mM Tris-HCl, pH 7.5, and 100 μg of lysostaphin (Sigma) ml−1, and RNA was extracted with 1.5 ml of RNazol B (Tel-test). RNA concentrations were determined by A260 measurements, and RNA integrity was analyzed by agarose-formaldehyde gel electrophoresis (44).
Northern hybridization.
Northern blot analysis was used to quantify the amounts of trxA and trxB mRNA and to determine the sizes of transcripts in total RNA from S. aureus Oxford cultures grown under aerobic and anaerobic conditions as previously described (29). Internal fragments of the genes trxA (nucleotides 2333 to 2649 of GenBank AJ223480) and trxB (nucleotides 1424 to 2069 of GenBank AJ223781) were amplified by PCR and labeled with the DIG PCR probe synthesis kit (Roche Molecular Biochemicals). The effect of oxidative stress compounds on trxA and trxB transcription was determined for S. aureus 8325-4 and SH1000. Cultures of cells grown to the mid-exponential phase (OD600 ≈1.0) were incubated for 5 to 15 min with diamide, menadione, hydrogen peroxide, and τ-butyl hydroperoxide at the concentrations indicated in Results, and total RNA was isolated and analyzed as described above. The concentrations correspond to those described elsewhere (27, 36).
Primer extension.
Primer extension was carried out with avian myeloblastosis virus reverse transcriptase (Promega) essentially as described (29). The reverse primers used in reactions were trxA, 5-TTACATGGACCACACCATGTTGCCCAAAAA-3′, complementary to nucleotides 2402 to 2431 of GenBank AJ223480, and trxB, 5′-GTAATCATTTCGAAACC-3′, complementary to nucleotides 1424 to 1440 of GenBank AJ223781.
Sequence analysis, database search, and deduced protein analysis.
Sequence entry, primary analysis, and open reading frame (ORF) searches were performed with the National Center for Biotechnology Information server ORF Finder (http://www.ncbi.nim.nih.gov/gorf/.html) and the Clone Manager 7 program (Scientific & Educational Software, Durham, N.C.). Primary sequences of S. aureus trxA and trxB were identified in databases of the University of Oklahoma Advanced Center for Genome Technology (http://www.genome.ou.edu/staph.html/) (strain NCTC 8323), the Institute for Genomic Research (TIGR [http://www.tigr.org/]) (strain COL), and the Staphylococcus aureus Sequencing Group at the Sanger Centre [http://www.sanger.ac.uk/Projects/S_aureus/) (EMRSA-16 strain 252 and methicillin-susceptible strain 476) with BLAST algorithms BLASTn and tBLASTn (2). Pairwise and multiple sequence alignments were performed with the ClustalW program, version 1.84 (15), with the EMBL ClustalW server (http://www2.ebi.ac.uk/clustalw/?), and the Network Protein Sequence Analysis server (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html).
Other methods.
Signals from Northern blots were scanned and intensities were measured with the ImageMaster software system (Pharmacia).
Nucleotide sequence accession numbers.
The nucleotide sequences of the DNA regions containing the S. aureus Oxford trxA and trxB genes have been deposited in the GenBank database with accession nos. AJ223480 and AJ223781, respectively.
RESULTS
Organization of the S. aureus trxA and trxB genes and sequences of the promoter regions.
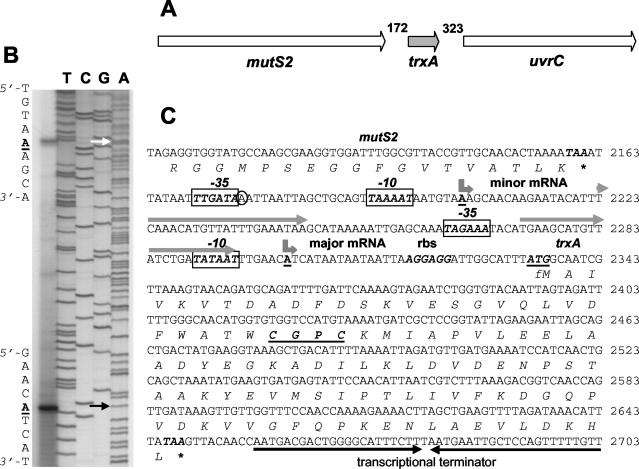
The starting point for this study was the isolation and sequencing of the S. aureus Oxford trxA and trxB genes and identification of their promoter regions. The S. aureus trxA and trxB genes (GenBank accession no. AJ223480 and AJ223781, respectively) were originally isolated with information derived from the known sequences of related genes. Figures 1A and 2A depict the DNA regions containing the trxA and trxB genes. Primer extension analysis identified two trxA transcription start points, each beginning with an adenine nucleotide (Fig. 1B). The nucleotide sequence of the 5′ upstream regulatory region of the S. aureus trxA gene is shown in Fig. 1C. The 5′ end of the major transcript maps 31 nucleotides upstream of the ATG translation start codon, and the 5′ end of the minor transcript occurs at 131 nucleotides from the ATG codon. The two start sites are separated by 99 nucleotides. The −10 sequences but not the −35 sequences upstream of the major and minor transcription start points closely matched the corresponding consensus motif in the promoters of gram-positive bacteria recognized by RNA polymerase containing the vegetative sigma factor σA (14). Two 22-bp imperfect direct repeats were located in the region containing the trxA promoters, one just downstream of the minor promoter and the other spanning and overlapping the major promoter.
FIG. 1.
Chromosomal organization, primer extension analysis, and sequence of the S. aureus Oxford trxA gene. (A) The gene organization is identical to that found in all of the S. aureus strains referred to in this work; the numbering of nucleotides in intergenic regions is for S. aureus Mu50, accession no.. NC_002758. The ≈5.3-kbp DNA region containing trxA and flanking genes is shown. Gene designations: mutS2 encodes a mismatch repair ATPase; trxA encodes thioredoxin; uvrC encodes a subunit of the endonuclease nucleotide excision repair system. (B) Total RNA was isolated from aerobic cultures of S. aureus Oxford. Primer extension was carried out as described in Materials and Methods, and the products were separated by electrophoresis under denaturing conditions alongside sequencing reactions with the same primers. Arrows point to the A nucleotides of the two trxA transcription start sites. (C) Nucleotide sequence of the trxA promoter (numbering according to GenBank AJ223480). Transcription start points are shown by bent arrows above the underlined boldface A nucleotides. The trxA ATG translational start codon (underlined) and its ribosome-binding site are shown in boldface italic letters. Putative −10 and −35 hexamer sequences are shown as boxed boldface italic letters. Two direct repeats are shown by grey arrows above the nucleotide sequences. The TAA stop codons of the mutS2 and trxA genes are marked by an asterisk. A putative rho-independent transcriptional terminator is indicated by two solid inverted arrows below the nucleotide sequence.
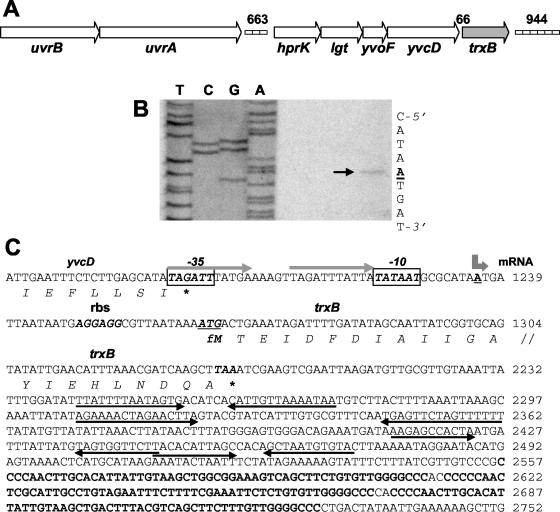
FIG. 2.
Chromosomal organization, primer extension analysis, and sequence of the S. aureus Oxford trxB gene. (A) The organization is identical to that found in all of the S. aureus strains referred to in this work; the numbering of nucleotides in intergenic regions is for S. aureus Mu50, accession no. NC_002758. The ≈11.4-kbp DNA region containing trxB and flanking genes is shown. Gene designations: uvrA and uvrB encode subunits of the endonuclease nucleotide excision repair system; hprK encodes a bifunctional kinase-phosphatase; lgt encodes a prelipoprotein diacylglyceroltransferase; yvoF encodes a putative acetyltransferase; yvcD encodes a hypothetical protein (designations according to reference 8); trxB encodes thioredoxin reductase. The directions of ORFs are indicated by arrows. Boxes indicate two intergenic regions that contain STAR tandem direct repeats. (B) Total RNA was isolated from aerobic cultures of S. aureus Oxford. Primer extension was carried as described in Materials and Methods, and the products were separated by electrophoresis under denaturing conditions alongside sequencing reactions with the same primers. An arrow points to the A nucleotide of the trxB transcription start point. (C) Nucleotide sequence of the trxB promoter (numbering according to GenBank AJ223781). The transcription start point is shown by a bent arrow above the underlined boldface A nucleotide. The trxB ATG translational start codon (underlined) and its ribosome-binding site are shown in boldface italic letters. Putative −10 and −35 hexamer sequences are shown as boxed boldface italic letters. The TAG and TAA translational stop codons of the yvcD and trxB genes, respectively, are marked by asterisks. Four pairs of inverted repeats located downstream of trxB are shown by arrows below the nucleotide sequence. Three long imperfect STAR direct repeats containing an ApaI endonuclease restriction site (GGGCCC) are indicated in bold letters.
Primer extension analysis identified a single trxB transcription start point (Fig. 2B). The 5′ end of the transcript begins with an adenine nucleotide and maps 29 nucleotides upstream of the ATG start codon. Figure 2C shows the nucleotide sequence of the 5′ upstream regulatory region of the trxB gene. The −10 sequence but not the −35 sequence upstream of the trxB transcription start site closely matched the corresponding consensus motif of σA-dependent promoters. Two 11-bp direct repeats containing one mismatch overlapped the −10 and −35 sequences of the trxB promoter.
Comparison of the trxA promoter regions of the Oxford strain from this work with that of the genomes of seven other S. aureus strains, NCTC8325, COL, EMRSA-16 strain 252, methicillin-susceptible strain 476, N315, Mu50, and MW2 (4, 25), revealed a very high degree of conservation. We noted just one difference in the Oxford trxA minor promoter region, a single nucleotide change located between the −10 and −35 RNA polymerase recognition motifs. Comparison of seven S. aureus trxB promoters showed them to be identical.
Inspection of the DNA region downstream of the S. aureus Oxford trxB gene revealed the presence of several STAR direct repeats that were noted previously (8), each containing an ApaI restriction site, and closely related to those present in each of the S. aureus strains referred to above. Similar repeats were also noted in the region upstream of trxB between the uvrA and hprK genes. Examination of the trxA upstream and downstream regions revealed the presence of additional STAR direct repeats, but these mapped ≈70 and ≈13 kb away. While the trxA and trxB genes mapped to quite different sites in the chromosome, they are both linked to genes involved in excision repair of DNA damage after UV irradiation. Thus, trxA is located in a cluster of conserved genes, mutS2-trxA-uvrC, and trxB is located in a gene cluster just downstream of the uvrAB genes. A similar organization of trx genes occurs in Staphylococcus epidermidis and Staphylococcus xylosus (21).
Transcription analysis of the trxA and trxB genes in aerobic and anaerobic growth conditions.
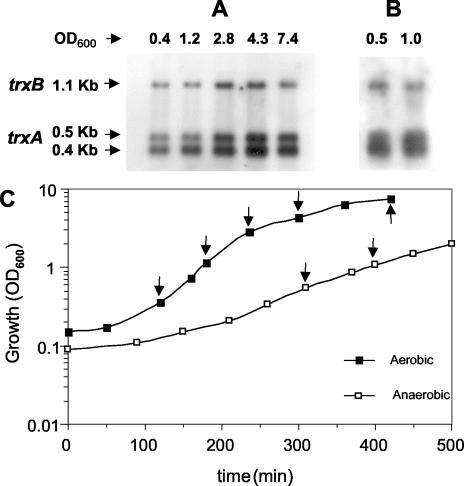
Northern analysis was used to quantify transcription of the S. aureus trxA and trxB genes in aerobic and anaerobic growth. Figure 3 shows the growth profiles and amount of trxA and trxB transcripts present in total RNA from aerobic and anaerobic cultures of S. aureus Oxford at different stages of growth. In aerobic conditions (Fig. 3A) a single trxB transcript was detected with the expected size of ≈1.1 kb. The amount of the trxB transcript was approximately constant in the early and mid-exponential phases of growth, increased two- to threefold at the late stage of exponential growth, and dropped gradually to the initial level in the stationary phase of growth. Two trxA transcripts were detected. Their sizes, ≈0.4 kb and ≈0.5 kb, corresponded to those predicted from the positions of the two trxA transcription start points. The amounts of the two transcripts were about the same, and their levels followed a profile similar to that of the trxB transcript. In anaerobic conditions (Fig. 3B), the sizes of the transcripts were identical to those found in aerobic growth, demonstrating that the same promoters are used for expression of the trxA and trxB genes under aerobic and anaerobic conditions. The relative amounts of trxA and trxB transcripts made under aerobic and anaerobic conditions were about the same, establishing that expression of the thioredoxin and thioredoxin reductase genes is not modulated by oxygen.
FIG. 3.
Northern hybridization analysis of S. aureus Oxford trxA and trxB transcripts in aerobic and anaerobic cultures. Total RNA was obtained from aerobic (A) and anaerobic (B) cultures at the early and mid-exponential phases and stationary phase, electrophoresed, blotted, and hybridized to trxA and trxB probes (see Materials and Methods). Sizes of transcripts are in kilobases; arrows indicate the positions of transcripts. (C) Growth profiles of S. aureus Oxford in aerobic and anaerobic conditions. The OD600 was used to follow the growth of cultures in TSB medium at 37°C. The arrows in panel C indicate the samples in panels A and B.
Transcriptional regulation of S. aureus trxA and trxB in response to disulfide and oxidative stress agents.
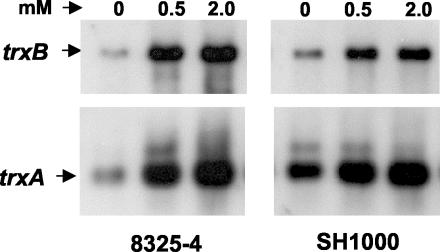
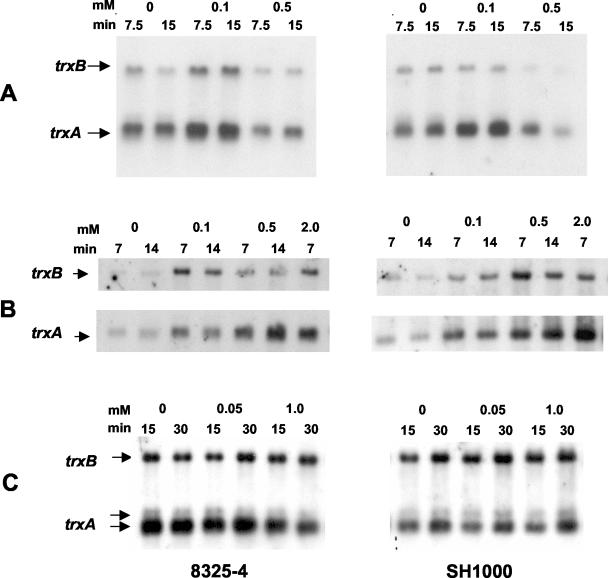
The effects of different disulfide and oxidative stress compounds on transcription of the S. aureus trxA and trxB genes are shown in Fig. 4 and 5. These experiments were carried out with isogenic strains 8325-4 and SH1000 in order to determine whether the general σB stress sigma factor plays a role in the transcriptional response. SH1000 possesses a functional σB stress factor, and 8325-4 lacks a functional σB stress factor. Figure 4 shows representative Northern analyses of the trxA and trxB genes following treatment of mid-exponential-phase cultures with diamide, a thiol-specific oxidizing agent. Figure 5 shows results for treatment with menadione, a redox cycling agent; τ-butyl hydroperoxide, an organic peroxide; and hydrogen peroxide.
FIG. 4.
Northern hybridization analysis of S. aureus 8325-4 and SH1000 trxA and trxB transcripts: effect of diamide. Cultures of 8325-4 and SH1000 were grown aerobically to the mid-exponential phase of growth and treated with 0.5 and 2.0 mM diamide for 15 min. Total RNA obtained from cultures at an OD600 of ≈1.0 was electrophoresed, blotted, and hybridized to trxA and trxB probes (see Materials and Methods). Arrows indicate the positions of transcripts.
FIG. 5.
Northern hybridization analysis of S. aureus 8325-4 and SH1000 trxA and trxB transcripts: effect of oxidative stress agents. Cultures of 8325-4 and SH1000 were grown aerobically to the mid-exponential phase of growth and treated with (A) menadione, (B) τ-butyl hydroperoxide, and (C) hydrogen peroxide at the concentrations and for the times shown. Total RNA obtained from cultures at an OD600 of ≈1.0 was electrophoresed, blotted, and hybridized to trxA and trxB probes (see Materials and Methods). Arrows indicate the positions of transcripts.
Diamide at 0.5 and 2.0 mM increased trxB transcription in both SH1000 and 8325-4 by three- to fivefold at 15 min after treatment and had a similar effect on trxA transcription. Only the smaller of the trxA transcripts was induced. Menadione at 0.1 mM increased trxB transcription in 8325-4 by three- to fourfold at 7.5 and 15 min but had little effect on trxB transcription in SH1000. The effects on trxA transcription in 8325-4 and SH1000 were approximately three- to fourfold and two- to threefold, respectively. As with diamide, the effect on trxA induction was confined to the smaller transcript. At higher concentrations, above 0.5 mM, menadione had an inhibitory effect on growth and on induction of trxA and trxB transcription. Paraquat, another redox cycling agent, had no significant effect on transcription of either trxA or trxB at up to 2 mM (results not shown).
Treatment with hydrogen peroxide and τ-butyl hydroperoxide gave quite different responses. Hydrogen peroxide at low (0.05 mM) and high (1 mM) concentrations had little or no effect after 15 to 30 min on transcription of trxA or trxB. In contrast, τ-butyl hydroperoxide at 0.1 to 2 mM increased trxB transcription in 8325-4 and SH1000 by about three- to fivefold at 7 to 14 min after treatment. The above concentrations of τ-butyl hydroperoxide had a similar effect on induction of trxA transcription in both strains. As before, only the smaller of the two trxA transcripts was induced. The results show that each of the oxidative stress compounds diamide, menadione, and τ-butyl hydroperoxide induced transcription of the S. aureus trxA and trxB genes and that the response was not dependent on the σB alternative sigma factor.
S. aureus trxB gene is essential for growth.
To establish whether the trxB gene is essential for growth, plasmid pOI7, which contains an internal ≈0.7-kbp fragment of the trxB gene lacking essential N- and C-terminal regions, was introduced by electroporation into S. aureus RN4220, and transformants were selected on plates containing erythromycin. Plasmid pOI7 is a derivative of the shuttle vector pAUL-A, which is temperature sensitive for replication at 40°C and carries an erythromycin resistance gene. A single homologous recombination event between the plasmid and the host chromosome is expected to result in disruption of the trxB gene.
One transformant out of several hundred that grew at the permissive temperature, 30°C, was able to grow at the nonpermissive temperature, 40°C. PCR analysis showed that it contained the wild-type trxB locus. Repeated attempts to isolate a trxB disruptant in this way were unsuccessful. Control experiments carried out with plasmid pOI8, which contains an internal ≈0.7-kbp fragment of the glutamine synthetase glnA gene, showed that all of the transformants that were isolated at 30°C grew at 40°C and that the latter contained an insertion of the plasmid at the expected glnA chromosomal site.
A second control plasmid, pOI15, was constructed to contain both the ≈0.7-kbp trxB and 0.7-kbp glnA internal DNA fragments inserted next to one another in the pAUL-A vector. Following electroporation, 24 of the transformants that grew at 30°C were tested for growth at 40°C. All of the transformants grew at 40°C. PCR analysis of several of them showed that they contained a wild-type trxB locus and an integrated copy of the plasmid at the glnA locus.
DISCUSSION
Originally, thioredoxin was identified in E. coli as the hydrogen donor system for ribonucleotide reductase (41). Soon after, E. coli was found to possess an alternative thiol redox system that utilizes glutathione and glutaredoxin and can effectively substitute for thioredoxin (19). E. coli mutants that fail to make a functional thioredoxin or thioredoxin reductase are able to grow because they use the glutathione-glutaredoxin system. In contrast, in the studies described here, we were unable to isolate an S. aureus trxB mutant, implying that the S. aureus thioredoxin system is essential for growth. We presume, therefore, that S. aureus lacks an alternative thiol redox system to replace the thioredoxin redox system. Similar conclusions were reached in studies with Bacillus subtilis (46) and Streptomyces coelicolor (unpublished results), where attempts to isolate insertional trxA and trxB mutants, respectively, were unsuccessful. These findings are compatible with our previous observations which demonstrated that certain gram-positive bacteria, including S. aureus, B. subtilis and S. coelicolor do not synthesize glutathione (1, 34).
Two other studies have shown that a functional thioredoxin trxA gene is essential for the phototrophic and heterotrophic growth of the cyanobacterium Synechocystis and for aerobic and anaerobic respiration of the facultative photosynthetic bacterium Rhodobacter sphaeroides (39). However, both of these gram-negative bacteria synthesize glutathione (34), and it is possible that some essential thioredoxin-mediated process in these organisms cannot be replaced by the glutathione-glutaredoxin system.
Transcriptional analysis showed that the S. aureus Oxford trxA and trxB genes were equally well expressed in aerobic and anaerobic conditions, demonstrating that the trx genes are not significantly modulated by the level of oxygen (Fig. 3). Since the thioredoxin system appears to be essential for aerobic growth, this result suggests that it is also likely to be needed for anaerobic growth. In aerobic conditions, an essential role for the S. aureus thioredoxin system is to transfer reducing equivalents from NADPH to the class Ib ribonucleotide reductase for the synthesis of deoxyribonucleotides from ribonucleotides. However, this does not appear to be the case for anaerobic growth because the nrdDG genes that encode the class III ribonucleotide reductase are expressed exclusively in the absence of oxygen (29), and the anaerobic ribonucleotide reductase derives its reducing power from formate (30). Clearly, the thioredoxin system must play a critical role in some other essential aspect of anaerobic cellular metabolism. It is therefore of interest that in Rhodobacter sphaeroides, expression of the trxA gene was found to be governed by oxygen tension (38).
Several oxidative stress compounds were found to cause rapid induction of transcription of the S. aureus trxA and trxB genes. The most pronounced effects were seen with diamide, menadione, and τ-butyl hydroperoxide. Diamide is a thiol-specific oxidant that reacts with free thiols in a stoichiometric manner to promote disulfide bond formation (23). In E. coli a functional thioredoxin or glutaredoxin system was found to be necessary to cope with diamide stress (43); similarly, in S. coelicolor inactivation of the σR factor that is responsible for modulating thioredoxin expression resulted in sensitivity to diamide (36, 37). The increased expression of the S. aureus trxA and trxB genes following diamide treatment therefore appears to be a response to combat increased disulfide bond formation. These results are in accordance with a recent study in B. subtilis which identified trxA and trxB among the genes that were strongly induced by exposure to diamide (27).
Menadione had an effect similar to that of diamide on induction of thioredoxin transcription. Menadione and other redox cycling agents are reduced in the cell by NADPH and oxidoreductases to produce superoxide anions. In E. coli thioredoxin can reduce menadione (18). If the S. aureus thioredoxin is able to reduce menadione, then its effect will be to withdraw electrons from the thioredoxin pathway. Consequently, the level of disulfides will increase, and increased trxA and trxB expression will, as with diamide, trigger a cellular response to overcome thiol oxidation.
The induction of trxA and trxB genes in response to τ-butyl hydroperoxide can be explained in the same way. Organic peroxides are often highly toxic compounds that oxidize thiols to disulfides and generate reactive free radicals that damage DNA and lipids. Peroxiredoxins that detoxify organic peroxides are well characterized in numerous bacteria. Alkylhydroperoxide reductases comprise two subunits; the larger protein, AhpF, is highly homologous to thioredoxin reductase and can use NADPH as the direct electron donor. Thus, exposure of S. aureus to alkylhydroperoxides will, it is expected, effectively drain away electrons from the thioredoxin pathway and lead to increased formation of disulfides and result in induction of trxAB expression. Unexpectedly, we were unable to detect any marked effects of hydrogen peroxide on induction of the trxA and trxB genes. Two recent reports of studies in B. subtilis and Oenococcus oeni showed that hydrogen peroxide caused a severalfold induction of the trxA gene (22, 46). We presume that the presence of a potent catalase in S. aureus effectively eliminates hydrogen peroxide (6).
A variety of studies have shown that the S. aureus σB factor positively controls the transcription of a family of genes in response to environmental stresses, including recovery from heat shock and biofilm expression, and also that it influences expression of some virulence-associated genes but is not required for resistance to ethanol or osmotic shock or for starvation survival (5, 11, 40). Most of the 27 genes belonging to the S. aureus σB regulon were shown to be homologous to B. subtilis genes, several of which are known to be regulated in response to stress by σB (11). In B. subtilis, σB was found to mediate increased transcription of trxA in response to heat and salt stress and ethanol treatment (46). Transcription of the trxA gene occurs at two promoters, one utilizing the general stress factor σB and the other the vegetative σA factor. Both promoters were induced in response to ethanol and heat, whereas the σA-dependent promoter was induced with hydrogen peroxide.
In this study we were unable to find any marked effect of the alternative σB factor on the transcription of the S. aureus trxA and trxB genes in response to exposure to disulfide and oxidative stress compounds. The S. aureus trxA gene is transcribed from two promoters, albeit at different levels; in the Oxford strain, the two trxA transcripts were made in similar amounts, and in 8325-4 and SH1000 the smaller transcript was the dominant one. However, in contrast to the situation in B. subtilis, both were σA dependent, and in both their transcription in response to oxidative stress was independent of a functional σB factor. Possibly, the existence of multiple promoters in these bacteria serves to ensure sufficient levels of thioredoxin to protect cells in response to both specific oxidative stresses and general nonspecific stresses.
Thiol redox processes play a central role in a broad range of cellular processes. In addition to its role in maintaining the cellular redox homeostasis, for protection against reactive oxygen species, and in provision of electrons to reductive enzymes, it has been shown, for example, to be implicated in mammalian cells in controlling cell proliferation and development and in apoptosis (10). One aspect of particular interest focuses on the possibility that the thioredoxin system may provide a new target for the development of compounds directed against gram-positive bacteria. This idea is premised on the fact that the bacterial and mammalian thioredoxin systems differ in some important ways. Thus, while the S. aureus thioredoxin and thioredoxin reductase proteins are very similar to those of other gram-positive bacteria and have the same active-site motifs, CGPC and CAV/TC, respectively, the redox active site of the human thioredoxin reductase is a hexapeptide, CVNVGC, which is identical to that present in the active site of the human glutathione reductase (16). Also, the NADPH and flavin adenine dinucleotide nucleotide binding domains of the human thioredoxin reductase are more similar to those of the human glutathione reductase than to their prokaryotic counterpart. Furthermore, the human thioredoxin reductase, like the glutathione reductase, is a much larger molecule than the bacterial enzyme due to the presence of a dimer interface domain.
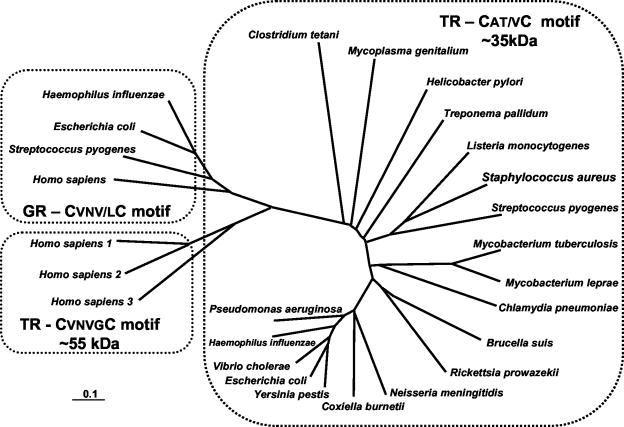
Figure 6 shows the molecular phylogeny of bacterial and human glutathione and thioredoxin reductases and clearly distinguishes between the bacterial and human thioredoxin reductases The bacterial species shown are those of selected pathogenic organisms. Because S. aureus contains a thioredoxin system that appears to be essential for growth, lacks an alternative glutathione based system, and differs from the human enzyme in key structural and mechanistic respects, it may prove to be an attractive target for the development of a new class of antistaphylococcal compounds.
FIG. 6.
Phylogeny of bacterial and human thioredoxin and glutathione reductases (TR and GR, respectively). The deduced amino acid sequences of thioredoxin reductase were determined for the proteins from Brucella suis (GenBank AE014445), Chlamydia pneumoniae (SwissProt Q9Z8M4), Clostridium tetani (SwissProt Q890T3), Coxiella burnetii (GenBank X75627), Escherichia coli (SwissProt P09625), Haemophilus influenzae (SwissProt P43788), Helicobacter pylori (SwissProt P56431), Homo sapiens 1 (SwissProt Q16881), H. sapiens 2 (GenBank AF171055), H. sapiens 3 (GenBank AF171054), Listeria monocytogenes (SwissProt O32823), Mycobacterium leprae (SwissProt P46843), Mycobacterium tuberculosis (SwissProt P52214), Mycoplasma genitalium (SwissProt P47348), Neisseria meningitides (GenBank AL162756), Pseudomonas aeruginosa (SwissProt Q9I0M2), Rickettsia prowazekii (SwissProt Q9ZD33), Staphylococcus aureus (SwissProt O54079), Streptococcus pyogenes (SwissProt Q878I8), Treponema pallidum (SwissProt O83790), Vibrio cholerae (SwissProt Q9KSS4), and Yersinia pestis (Swiss Prot: Q8ZGC9). Deduced amino acid sequences of glutathione reductases used were from Escherichia coli (SwissProt P06715), Haemophilus influenzae (SwissProt P43783), Homo sapiens (SwissProt P00390), and Streptococcus pyogenes (GenBank AE014147).
Acknowledgments
This work was supported in part by the Israel Science Foundation (787/01). We are grateful to the University of Oklahaoma Advanced Center for Genome Technology, TIGR, and the Sanger Center for the use of preliminary S. aureus genomic sequence data.
We thank Simon Foster and Steven Projan for providing strains and plasmids.
REFERENCES
- 1.Aharonowitz, Y., Y. Av-Gay, R. Schreiber, and G. Cohen. 1993. Characterization of a broad-range disulfide reductase from Streptomyces clavuligerus and its possible role in beta-lactam antibiotic biosynthesis. J. Bacteriol. 175:623-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arner, E. S., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267:6102-6109. [DOI] [PubMed] [Google Scholar]
- 4.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 5.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor sigma B controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clements, M. O., and S. J. Foster. 1999. Stress resistance in Staphylococcus aureus. Trends Microbiol. 7:458-462. [DOI] [PubMed] [Google Scholar]
- 7.Clements, M. O., S. P. Watson, R. K. Poole, and S. J. Foster. 1999. CtaA of Staphylococcus aureus is required for starvation survival, recovery, and cytochrome biosynthesis. J. Bacteriol. 181:501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramton, S. E., N. F. Schnell, F. Gotz, and R. Bruckner. 2000. Identification of a new repetitive element in Staphylococcus aureus. Infect. Immun. 68:2344-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickinson, D. A., and H. J. Forman. 2002. Glutathione in defense and signaling: lessons from a small thiol. Ann. N. Y. Acad. Sci. 973:488-504. [DOI] [PubMed] [Google Scholar]
- 11.Gertz, S., S. Engelmann, R. Schmid, A. K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the sigma(B) regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giachino, P., S. Engelmann, and M. Bischoff. 2001. Sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart, M. E., M. S. Smeltzer, and J. J. Iandolo. 1993. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J. Bacteriol. 175:7875-7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. With CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 16.Hirt, R. P., S. Muller, T. M. Embley, and G. H. Coombs. 2002. The diversity and evolution of thioredoxin reductase: new perspectives. Trends Parasitol. 18:302-308. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann, B., H. J. Hecht, and L. Flohe. 2002. Peroxiredoxins. Biol. Chem. 383:347-364. [DOI] [PubMed] [Google Scholar]
- 18.Holmgren, A. 1979. Reduction of disulfides by thioredoxin. Exceptional reactivity of insulin and suggested functions of thioredoxin in mechanism of hormone action. J. Biol. Chem. 254:9113-9119. [PubMed] [Google Scholar]
- 19.Holmgren, A. 1985. Thioredoxin. Annu. Rev. Biochem. 54:237-271. [DOI] [PubMed] [Google Scholar]
- 20.Holmgren, A. 2000. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid. Redox. Signal. 2:811-820. [DOI] [PubMed] [Google Scholar]
- 21.Huynh, P. L., I. Jankovic, N. F. Schnell, and R. Bruckner. 2000. Characterization of an HPr kinase mutant of Staphylococcus xylosus. J. Bacteriol. 182:1895-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jobin, M. P., D. Garmyn, C. Divies, and J. Guzzo. 1999. Expression of the Oenococcus oeni trxA gene is induced by hydrogen peroxide and heat shock. Microbiology 145:1245-1251. [DOI] [PubMed] [Google Scholar]
- 23.Kosower, N. S., and E. M. Kosower. 1987. Formation of disulfides with diamide. Methods Enzymol. 143:264-270. [DOI] [PubMed] [Google Scholar]
- 24.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 26.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. 87:4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leichert, L. I., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 29.Masalha, M., I. Borovok, R. Schreiber, Y. Aharonowitz, and G. Cohen. 2001. Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. J. Bacteriol. 183:7260-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulliez, E., S. Ollagnier, M. Fontecave, R. Eliasson, and P. Reichard. 1995. Formate is the hydrogen donor for the anaerobic ribonucleotide reductase from Escherichia coli. Proc. Natl. Acad. Sci. 92:8759-8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newton, G. L., R. C. Fahey, G. Cohen, and Y. Aharonowitz. 1993. Low-molecular-weight thiols in streptomycetes and their potential role as antioxidants. J. Bacteriol. 175:2734-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newton, G. L., C. A. Bewley, T. J. Dwyer, R. Horn, Y. Aharonowitz, G. Cohen, J. Davies, D. J. Faulkner, and R. C. Fahey. 1995. The structure of U17 isolated from Streptomyces clavuligerus and its properties as an antioxidant thiol. Eur. J. Biochem. 230:821-825. [DOI] [PubMed] [Google Scholar]
- 34.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. Delcardayre, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 36.Paget, M. S., J. G. Kang, J. H. Roe, and M. J. Buttner. 1998. Sigma R, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). EMBO J. 17:5776-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paget, M. S., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigma R regulon. Mol. Microbiol. 42:1007-1020. [DOI] [PubMed] [Google Scholar]
- 38.Pasternak, C., K. Assemat, A. M. Breton, J. D. Clement-Metral, and G. Klug. 1996. Expression of the thioredoxin gene (trxA) in Rhodobacter sphaeroides Y is regulated by oxygen. Mol. Gen. Genet. 250:189-196. [DOI] [PubMed] [Google Scholar]
- 39.Pasternak, C., K. Assemat, J. D. Clement-Metral, and G. Klug. 1997. Thioredoxin is essential for Rhodobacter sphaeroides growth by aerobic and anaerobic respiration. Microbiology 143:83-91. [DOI] [PubMed] [Google Scholar]
- 40.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor sigma B is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reichard, P. 1993. From RNA to DNA, why so many ribonucleotide reductases? Science 260:1773-1777. [DOI] [PubMed] [Google Scholar]
- 42.Rhee, S. G., S. W. Kang, T. S. Chang, W. Jeong, and K. Kim. 2001. Peroxiredoxin, a novel family of peroxidases. IUBMB Life 52:35-41. [DOI] [PubMed] [Google Scholar]
- 43.Ritz, D., H. Patel, B. Doan, M. Zheng, F. Aslund, G. Storz, and J. Beckwith. 2000. Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J. Biol. Chem. 275:2505-2512. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Schaferkordt, S., and T. Chakraborty. 1995. Vector plasmid for insertional mutagenesis and directional cloning in Listeria spp. BioTechniques 19:720-725. [PubMed] [Google Scholar]
- 46.Scharf, C., S. Riethdorf, H. Ernst, S. Engelmann, U. Volker, and M. Hecker. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 180:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zahrt, T. C., and V. Deretic. 2002. Reactive nitrogen and oxygen intermediates and bacterial defenses: unusual adaptations in Mycobacterium tuberculosis. Antioxid. Redox. Signal. 4:141-159. [DOI] [PubMed] [Google Scholar]