Abstract
Rhizobium sp. strain NGR234 has an exceptionally broad host range and is able to nodulate more than 112 genera of legumes. Since the overall organization of the NGR234 genome is strikingly similar to that of the narrow-host-range symbiont Rhizobium meliloti strain 1021 (also known as Sinorhizobium meliloti), the obvious question is why are the spectra of hosts so different? Study of the early symbiotic genes of both bacteria (carried by the SymA plasmids) did not provide obvious answers. Yet, both rhizobia also possess second megaplasmids that bear, among many other genes, those that are involved in the synthesis of extracellular polysaccharides (EPSs). EPSs are involved in fine-tuning symbiotic interactions and thus may help answer the broad- versus narrow-host-range question. Accordingly, we sequenced two fragments (total, 594 kb) that encode 575 open reading frames (ORFs). Comparisons revealed 19 conserved gene clusters with high similarity to R. meliloti, suggesting that a minimum of 28% (158 ORFs) of the genetic information may have been acquired from a common ancestor. The largest conserved cluster carried the exo and exs genes and contained 31 ORFs. In addition, nine highly conserved regions with high similarity to Agrobacterium tumefaciens C58, Bradyrhizobium japonicum USDA110, and Mesorhizobium loti strain MAFF303099, as well as two conserved clusters that are highly homologous to similar regions in the plant pathogen Erwinia carotovora, were identified. Altogether, these findings suggest that ≥40% of the pNGR234b genes are not strain specific and were probably acquired from a wide variety of other microbes. The presence of 26 ORFs coding for transposases and site-specific integrases supports this contention. Surprisingly, several genes involved in the degradation of aromatic carbon sources and genes coding for a type IV pilus were also found.
The Proteobacteria contain many bacterial species that associate with plants either as symbionts or pathogens. Complete sequencing of the genomes of several of these microbes, including Agrobacterium tumefaciens (20, 49), Bradyrhizobium japonicum (28), Mesorhizobium loti (27), and Rhizobium (Sinorhizobium) meliloti (18), has made comparative genome analyses possible. Rhizobium sp. strain NGR234 belongs to the α-proteobacteria and is able to establish nitrogen-fixing symbiosis with many different legumes. Despite extensive study, the molecular mechanisms behind this broad host range are not fully apparent (6). Although R. meliloti has a very limited host range (5), it is phylogenetically close to NGR234 and the organization of both genomes is similar (14a, 31a). In both cases, the genome comprises three replicons (14a). Most symbiotic genes are carried on SymA plasmids of 0.54 Mb in strain NGR234 (17) and 1.35 Mb in R. meliloti (1). Both bacteria also possess a second group of plasmids, the so-called exo- or megaplasmids (pSymB). pNGR234b is estimated to be 2.2 Mb (31a), and the size of R. meliloti is 1.68 Mb (14). Chromosomes in both bacteria are about the same size (3.5 Mb in NGR234, 3.34 Mb in R. meliloti) (7, 31a). Also snapshot sequencing suggested that housekeeping and many metabolic genes are similar (48). Sequencing data also suggested that NGR234 differs in its gene content from R. meliloti, however.
We sequenced two large contigs of pNGR234b, one of which contains loci involved in extracellular polysaccharide (EPS) synthesis and, thus, in fine-tuning symbiosis. Altogether 575 open reading frames (ORFs) were identified, of which 222 appear to be organized into clusters of more than four genes. Comparative analyses indicated that NGR234 may have acquired large parts of the genetic content of pNGR234b from other soil- and plant-associated microbes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli was grown at 37°C on Luria-Bertani medium (42) supplemented with appropriate antibiotics, and Rhizobium sp. strain NGR234 (46) was grown on TY medium (0.5% tryptone, 0.25% yeast extract, and 10 mM CaCl2 [pH 7.0]).
Manipulation of DNA and construction of an ordered cosmid library.
On the basis of hybridization and sequence data, a minimal set of cosmids of the canonical ordered library of the NGR234 genome (37a) was selected for further analysis. Selected cosmids were partially digested with Sau3A. Fragments of 0.5 to 3.5 kb were isolated after electrophoretic separation on agarose gels, cloned into pTZ19R (Amersham, Essex, United Kingdom), and sequenced with standard primers. Sequencing was performed by using dye terminator technology on a model 377 sequencer (Applied Biosystems, Foster City, Calif.) or on capillary sequencers from Amersham. The GC-Phrap software package (http://www.jgi.doe.gov/Docs/JGI_Seq_Quality.html#_SeqQ.I) was used to assemble the sequences. Editing and finishing was facilitated by the Staden software package (http://www.mrc-lmb.cam.ac.uk/pubseq/staden_home.html). Sequencing of PCR-generated fragments was used to close single- and double-stranded gaps. ORFs were initially identified by the programs Glimmer (http://www.tigr.org/software/glimmer/) and GeneMarkS (4). The cutoff limit for ORFs without database homologues was 150 bp. Predicted ORFs and intergenic regions were used to interrogate nonredundant protein databases with Blast programs via the website http://www.ncbi.nlm.nih.gov/blast. ORFs were entered into the ERGO Integrated Genomics, Inc. (Chicago, Ill.) bioinformatics suite for genome annotation and metabolic reconstruction. The predicted ORFs were subjected to two initial rounds of annotation (one automatic and one manual). Proteins were categorized with a modified Riley classification (40). Analysis of the sequenced region resulted in the identification of 575 ORFs, which were arbitrary assigned identification numbers with the specific prefix ngr. The names of ORFs of pNGR234a that are also found on pNGR234b are followed by a superscript b; thus, the cysteine synthase cysM (y4xP) has a homologue y4xPb on pNGR234b and so on.
Nucleotide sequence accession numbers.
The nucleotide sequences were deposited in GenBank under accession numbers AY316746 and AY316747.
RESULTS AND DISCUSSION
General.
In total, 357,655 bp of contig1 and 234,455 bp of contig2 were sequenced, and 575 ORFs were identified (354 on contig1 and 221 on contig2). At 61.8 mol%, the average G+C content for both contigs is similar to that found for the entire genome (5a) but is also significantly higher than the value of 58.5 mol% calculated for pNGR234a (17). The gene density on both contigs was almost identical at one gene per 1.03 kb. Possible functions were assigned to 403 ORFs, which were grouped into eleven categories (Fig. 1 and Table 1).
FIG. 1.
Physical organization of the ORFs of pNGR234b. Coordinates are given in kilobases. Putative genes and ORFs are colored (grey-boxed area) according to putative functions. ORFs were named genes when BLAST-P searches of the National Center for Biotechnology Information database indicated an identity of <E−80. Several insertion sequence elements are indicated as regions boxed with dotted lines. Angled arrows indicate the locations of possible sigma-54-dependent promoters as well as the location of two possible ROSE elements. Conserved clusters of genes are shown as double-headed and horizontal arrows. Conserved clusters (≥3 ORFs) present in identical or highly similar orders in other bacteria are indicated. Colors show the highest similarity to the bacterium indicated in the boxed area. Only the microbe that showed the highest similarity and most conserved gene order over the entire range of the cluster is indicated.
TABLE 1.
Genes and ORFs identified on the sequenced contigs of pNGR234ba
| Functional class | No. of ORFs identified |
|---|---|
| Regulators | 58 |
| Cellular processes | 106 |
| Transporters | 79 |
| Others | 27 |
| Small-molecule metabolism | 129 |
| Macromolecule metabolism | 8 |
| DNA modification | 6 |
| Heat shock | 7 |
| Transposases and integrases | 26 |
| Protection responses | 14 |
| Exopolysaccharide synthesis and modification | 38 |
| Structural genes | 11 |
| Hypothetical | 172 |
| No function | 128 |
| No similarity and no function | 44 |
| Total | 575 |
Functions were assigned by using the ERGO software and the corresponding genome database. Data were obtained by DNA sequence analysis of 594 kb of pNGR234b (estimated total size, ≈2.2 Mb).
Transporters and genes involved in protection responses.
Altogether, 79 ORFs were classified as encoding transporters or proteins related to transport processes, a majority of which are ABC-type transporters. Surprisingly, a relatively high number of ORFs (11) were linked to the transport of spermidine or putrescine, possible osmoprotectants. Other predicted solute transporters include those for iron, amino acids, peptides (and oligopeptides), rhizopines, taurine, and other small molecules. This density of transporters is similar to that found on the R. meliloti pSymB (14, 18), but pNGR234b carries at least one component of a phosphotransferase system (ngr563) (18).
Other interesting features included 14 ORFs that play roles in protection responses. Among these are three genes encoding proteins involved in resistance to acriflavine (ngr226, ngr288, and ngr289), genes involved in detoxification of other small molecules (e.g., ngr065, ngr334, two copies of the multidrug resistance protein B), CopC (copper resistance protein C), and Ngr174, a macrolide glycosyltransferase. ngr153 encodes a homoserine lactone efflux protein, which belongs to the family of the RhtB proteins, that can confer resistance to elevated levels of exogenous l-threonine, l-homoserine, and analogues (53). Homologues of this protein are also found in A. tumefaciens and B. japonicum as well as several other gram-negative bacteria (e.g., Salmonella enterica serovar Typhi, E. coli O157:H7 Sakai, and Brucella melitensis) but not in R. meliloti.
Catabolic functions.
A significant number of genes encoding proteins that could be involved in oxidative metabolism were identified (Table 1). Among them are dehydrogenases, oxidoreductases, and dehydratases. Several predicted sugar kinase genes were also found. A number of ORFs were identified that encode proteins involved in the degradation of complex or aromatic carbon sources, including a protocatechuate 3,4,-dioxygenase (ngr051), an opine oxidase (ngr333 and ngr334, α- and β-subunit), and a hydroxyquinol 1,2-dioxygenase (ngr391). Other proteins possibly involved in the degradation of complex carbon compounds available to NGR234 include four myoinositol 2-dehydrogenases (ngr233, ngr250, ngr251, and ngr252) and two other proteins linked to the degradation of myoinositol (IolD and IolE). Also, one ORF encodes an octopine dehydrogenase subunit B (ngr446), two ORFs encode agmatinases (ngr257 and ngr540), and another encodes a metapyrocatechase (ngr571). The latter is involved in the degradation of naphthalene in Bacillus, Pseudomonas, and Rhodoccocus via the metacleavage pathway (29, 33). A homologue of the trihydroxytoluene oxygenase, which is involved in the catabolism of 2,4-dinitrotoluene, is encoded by ORF ngr570, and a protein involved in nitrilotriacetate catabolism (ngr138) (50) was found. Both ngr570 and ngr138 are part of a conserved cluster in B. japonicum, several Brucella species, Burkholderia pseudomallei, and Sphingomonas aromaticivorans. Obviously, pNGR234b is important in the catabolism of a remarkably wide spectrum of carbon and energy sources, including loci that are involved in the degradation of aromatic compounds (i.e., ngr570 and ngr571) but are not found in the R. meliloti genome.
Regulatory elements.
Another 58 ORFs encode possible regulatory proteins: two for possible polymerase sigma factors (sigJ and sigB), with the rest mostly belonging to the LysR, GntR, and TetR families. Two ORFs (ngr159 and ngr160) encode possible homologues of the two-component regulators NodV-NodW or NwsA-NwsB (Fig. 1. The nodVW genes of B. japonicum are involved in activation of nod gene expression in response to plant-produced isoflavones (20a, 30).
Chaperones and cofactor biosynthesis.
Two copies of the heat shock proteins GroES and GroEL, as well as several other ORFs encoding small heat shock proteins, two of which belong to the Hsp20 family (ngr309 and ngr311), were found (Fig. 1 and Table 1). All are required for rapid adaptation to heat stress (34). Although their transcription is commonly activated from sigma-70 promoters, it is also negatively regulated by cis-acting elements (ROSE [repression of heat-shock gene expression]) (32, 35). Two possible ROSE elements upstream of the groES genes are indicated in Fig. 1.
Other genes encode proteins involved in the biosynthesis of cofactors and vitamins. Examples include proteins involved in the biosynthesis of pyrroloquinone (PqqA to -E), pyridoxal phosphate (PdxA), a riboflavin-specific deaminase (ngr151), and the thiamine biosynthesis protein ThiD. Despite their well-known roles as classical cofactors, several of these compounds might be involved in promoting colonization of roots (44, 51). Two proteins involved in the biosynthesis of amino acids (AroC, the shikimate 5-dehydrogenase, and the possible cysteine synthase y4xPb [CysM] homologue) were identified; both, however, may have chromosomal homologues. Nevertheless, the observation that pNGR234b encodes pathways involved in cofactor and amino acid biosynthesis, indicates an essential role in cellular processes. The suggestion that pNGR234b might be essential was further supported by the discovery of a gene involved in the biosynthesis of the 30S ribosomal protein S21 A (RbsU). Interestingly, in both NGR234 and R. meliloti, the rbsU gene is close to the cold shock gene cspA. In R. meliloti, however, the corresponding homologues of rbsU and cspA are found on the bacterial chromosome and are transcribed as one operon (7, 36). To further verify that the pNGR234b is essential for NGR234 cell processes and metabolism, further tests in which the pNGR234b is cured from the host strain are necessary, as well as confirming that other genes essential for cell division are present on this replicon (41).
Macromolecular metabolism.
When an imbalance between carbon and nitrogen, phosphorus, or biotin occurs, many rhizobia sequester the excess carbon as polyhydroxybutyrate (13, 25, 45). Proteins involved in the degradation of polyhydroxybutyrate, BdhA (poly-3-hydroxybutyrate-dehydrogenase) and PhaZ (poly-3-hydroxybutyrate-depolymerase), were found. Degradation is initiated by the action of a polyhydroxybutyrate depolymerase that releases the monomer 3-hydroxybutyrate. A possible endoglucanase (ngr054) and two putative cellulose synthases (ngr055 and ngr066) are encoded on pNGR234b. Several putative proteins involved in cell wall biosynthesis as well as the degradation of polygalacturonate (ngr072) were discovered. The latter belongs to the family of 28 glycosyl hydrolases (23) that cleave 1,4-α-d-galactosiduronic linkages in pectate and other galacturonans. Pectinases and polygalacturonases are part of the armory of plant pathogens, including Erwinia chrysanthemi, where the expression of pectinases is directly linked to pathogenicity (24, 26). No polygalacturonases have been reported in the genome of R. meliloti, whereas A. tumefaciens (ORF ATU4560) and B. japonicum USDA110 (2a) possess putative polygalacturonases. The identification of polygalacturonases in NGR234 is thus intriguing and may suggest a role during the infection process.
Transposases and integrases.
Twenty-six ORFs encoding integrases and transposases were found. Their presence is probably linked to the high frequency of DNA rearrangements found in soil bacteria (21, 22). Short repeats (the largest is 140 bp) (positions are indicated in Fig. 1) are interspersed between the integrases and transposases. Interestingly, the G+C content of the DNA fragment framed by both repeats has a significantly lower G+C content (60.4%) than the remaining part of the contig1 (61.7%). These data suggest that lateral transfer of genetic material has occurred.
ORFs without assigned functions.
Finally, 128 ORFs were identified for which similarities were observed but functions could not be assigned, and 44 ORFs had no known homologues in the databases.
Comparative analyses of loci found in other plant-associated species.
Possible horizontal transfer of all the identified ORFs was examined by comparing pNGR234b with the genomes of other plant-associated microbes. All potential operons and gene clusters (≥3 ORFs), were compared with the genes of pNGR234a (17) and the complete genomes of the following members of the Rhizobiaceae: A. tumefaciens (20, 49), B. japonicum USDA110 (28), M. loti MAFF303099 (27), and R. meliloti 1021 (18). Available information on the plant pathogen Erwinia carotovora (http://www.sanger.ac.uk/Projects/E_carotovora/) was also included in the analysis.
Initial analyses indicate that at least 176 ORFs are part of paralogous clusters and 291 ORFs are part of 62 chromosomal clusters. Use of the ERGO suite to examine a number of these loci showed that 29 conserved gene clusters (comprising 222 ORFs) have similar or identical gene orders to those found in one or several plant-associated microbes (Fig. 1). Of these, 158 ORFs were identified in clusters or operons that are highly similar to clusters or operons of pSymB of R. meliloti.
Analysis of the exo-exs cluster.
Obvious structural similarities were seen among the exo and exs genes of pNGR234b and pRmSymB. This cluster contains 31 ORFs stretching from the thiD gene to the exsI gene (Fig. 1 and 2). DNA identities of about 80% extend across the cluster, and the orientation of the genes is the same. exo and exs genes are involved in the synthesis of low-molecular-weight EPSs, which are essential for nodule invasion (2, 37). Thus, Exo mutants of NGR234 are ineffective on the host plant Leucaena leucocephala (8). Profiling with restriction enzymes and comparison with previously sequenced exoX and exoY genes (GenBank accession number X16704) showed that the core of this cluster has been previously mapped (9). In addition, the pNGR234b exo cluster is similar to comparable loci in A. tumefaciens C58 and M. loti MAFF303099. The exoPNOMAL fragment is present in all four species (Fig. 2). The most striking differences between NGR234 and R. meliloti were found on both sides of the conserved exoI region. ORFs corresponding to exoH and the genes exoTWV were not found in the sequenced regions of pNGR234b, suggesting that two deletions occurred (Fig. 2). This possibility is supported by the identification of a 37-bp exoH fragment of NGR234 (upstream of exoK), which forms the left border of the deleted exoH region.
FIG. 2.
Physical organization of the exo-exs cluster of Rhizobium sp. strain NGR234 compared to that of A. tumefaciens strain C58, M. loti strain MAFF303099, and R. meliloti strain 1021. Asterisks mark deletions of exoH and exoTWV (see text).
In R. meliloti, the ExoH protein is a succinyl transferase that is required to produce succinoglycan. It seems likely that a common rhizobial ancestor produced succinoglycans, as R. meliloti does, whereas NGR234 evolved symbiotically active EPSs that lack succinyl groups. The repeating subunit of the NGR234 polysaccharide is a nonasaccharide that consists of a main chain [Gal-(Glu)5] and a side chain [(GlcA)2-Gal] decorated by an acetyl and a pyruvyl group on the terminal galactose (11). Based on the known function of the exo genes of R. meliloti (39), the exo cluster of pNGR234b is probably involved in the synthesis of the main chain [Gal-(Glu)5]. ExoF and ExoY are required for the first step of synthesis of the oligosaccharide subunit (transfer of galactose to a lipid carrier), and ExoA, ExoL, ExoM, ExoO, and ExoU are glycosyl transferases which subsequently add glucose units, forming the hexasaccharide [Gal-(Glu)5]. In R. meliloti, elongation of the oligosaccharide chain is continued by the glucosyl transferase ExoW. Based on the results obtained in R. meliloti, one can assume that in the absence of exoW, NGR234 is unable to link an additional glucose unit and completes the synthesis of its subunit by using two glucuronyl transferases and a galactosyl transferase, enzymes that are outside the sequenced regions.
ExoV modifies the terminal glucose of the R. meliloti succinoglycan subunit with a pyruvyl group. Although the NGR234 exo gene cluster lacks exoV, the nonreducing galactose of the subunit is also pyruvylated (11), suggesting that a nonidentified pyruvyl transferase must exist in the NGR234 genome. Interestingly, NGR234 also harbors the acetyltransferase exoZ. ExoZ of R. meliloti acts on the trisaccharide Gal-(Glu)2 (39), whereas the subunit of NGR234 is acetylated at the nonreducing galactose of the side chain [(GlcA)2-Gal] and at sites that have not been determined (10, 11). Thus, it is possible that the NGR234 ExoZ gained the ability to acetylate the third sugar of the side chain, thereby conserving the specificity for trisaccharides.
Once the succinoglycan subunits of R. meliloti are synthesized, they are polymerized and exported by ExoP, ExoQ, and ExoT (19). The pNGR234b exo cluster lacks exoT. This finding suggests that exoT is not required for acidic EPS synthesis in NG234 or that a functional exoT exists at another position in the genome. The symbiotically active succinoglycan of R. meliloti consists of low-molecular-weight succinoglycan, which is released from the polymer by the extracellular glycanases ExoK and ExsH (52). We identified a sequence encoding a putative ExoK glycanase, but exsH was not found. A promoter is located 211 bp upstream of the exoK start codon of R. meliloti (3). The corresponding sequence in NGR234 is located within the mutated region upstream of exoK. It is possible, therefore, that the promoter of exoK is not functional in NGR234. Low extracellular glycanase activity would explain why NGR234 produces relatively low amounts of low-molecular-weight EPS (10).
In summary, the organization of the exo cluster suggests that acquisition and deletion of genetic information has extensively shaped pNGR234b. It is tempting to speculate that the exoKHTWV is the original sequence of succinoglycan-producing members of the Rhizobiaceae. In A. tumefaciens, this arrangement has been maintained. Then a common ancestor of R. meliloti and NGR234 acquired a 2-kbp fragment containing ngr20301 (also known as smb20953), ngr011 (also known as smb20952), exoI, and ngr2014 (also known as smb21673). These imported DNA sequences seem not to be involved in the synthesis of EPS. Finally, two deletions (perhaps first exoTWV and later the now useless exoH) resulted in the organization of pNG234b shown in Fig. 2.
Genes encoding type IV pili.
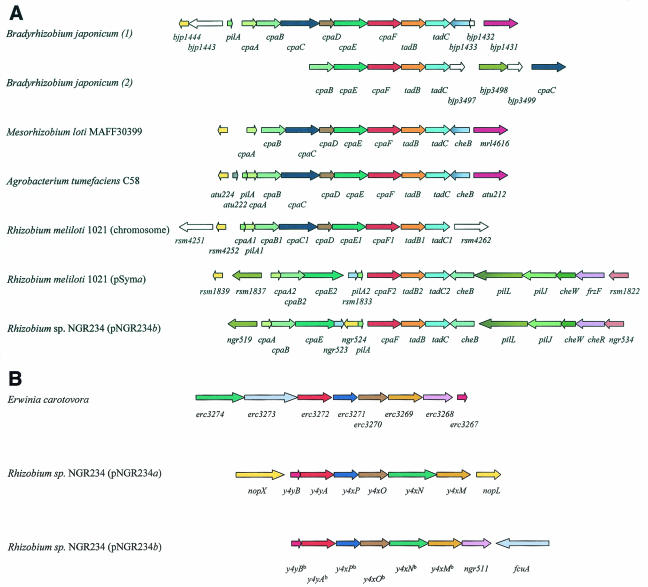
Among other interesting loci identified was a cluster encoding a type IV pilus. Type IV pili are unique structures on the bacterial surface that are found in many gram-negative bacteria (Fig. 3A), where they play an important role in bacterial adhesion to host cells, biofilm formation, conjugative DNA transfer, motility, and infection by bacteriophages (12). Pili are secreted through the inner and outer membranes. In Caulobacter crescentus, at least seven genes are required for pilus assembly, including pilA, cpaA, cpaB, cpaC, cpaD, cpaE, and cpaF.
FIG. 3.
(A) Physical organization and comparative analysis of the type IV pilus cluster of Rhizobium sp. strain NGR234. Same colors indicate similar predicted functions of the depicted ORF. No coloring indicates that no link to the pili biosynthesis cluster or to the larger gene cluster was observed. ORFs rsm4251 and rsm4262 encode transposases. (B) Physical organization and comparative analysis of the y4yB-y4xM clusters of Rhizobium sp. strain NGR234. Same colors indicate similar predicted functions of the depicted ORF.
Use of the ERGO suite highlighted clusters of similarly organized genes encoding type IV pili in plant-associated bacteria including A. tumefaciens, B. japonicum, M. loti, and R. meliloti (Fig. 3A). Surprisingly, B. japonicum and R. meliloti both carry two copies of most of the genes necessary for pili biosynthesis. One copy in R. meliloti is located on pSymA while the second is on the chromosome. Comprising at least 16 ORFs, the pSymA copy is strikingly similar to the copy identified on pNGR234b (Fig. 3A). Interestingly, cpaB and cpaC are apparently separated in both organisms on the copies found on the megaplasmids. A putative cpaC homologue was identified directly upstream of the pilA gene in NGR234 (ORF ngr524) and upstream of the cpaA gene on pRmSymA (ORF rsm1839). Nevertheless, both putative cpaC homologues encoded much shorter proteins than the cpaC copy on the R. meliloti chromosome, but a similar ORF was observed in several of the other rhizobial clusters (Fig. 3A). CpaC belongs to the PulD/pIV family of proteins, which are commonly referred to as secretins. CpaC might be involved in outer membrane channel formation and pilus secretion (43). A function has yet to be assigned to the cpaD gene product.
Two clusters with high similarity to loci of E. carotovora.
Two other clusters are highly homologous to loci of the plant pathogen E. carotovora. A contig2 cluster stretching from ORF ngr068 to ngr074 encodes genes probably involved in modification and/or degradation of plant cell walls. A second cluster on contig1 includes the ORFs y4yBb (ngr505) to fcuA, which encode genes important in iron transport and subsequent metabolism. Seven of the ORFs, y4yBb to ngr511, are found on the same strand with overlapping stop and start codons, suggesting they could be transcribed as an operon. fcuA is located approximately 300 bp downstream of ngr511 and on the opposite strand (Fig. 3B). y4yAb may encode a decarboxylase, whereas y4xPb (CysM) has homology to cysteine synthases. Both enzyme families require pyridoxal phosphate as a cofactor, and one of the genes (pdxA) required for the synthesis of this cofactor are located on the same contig. y4xOb shows weak homology to octopine dehydrogenases, y4xNb belongs to the IucA-IucC family of siderophore biosynthetic enzymes, y4xMb encodes a possible permease and possesses 11 predicted transmembrane domains, ngr511 has homology to iron(III) dicitrate binding proteins, the fcuA gene codes for a possible ferric siderophore receptor, and y4yBb is homologous to many hypothetical bacterial proteins of unknown function.
Interestingly, several of the ORFs within this cluster (y4yBb to y4xMb) are duplicated (85% identity and in the same order) on pNGR234a (Fig. 3B). Upstream of y4yB on pNGR234a is a 40-bp repeat that is highly similar to y4yBb on pNGR234b. Downstream of the pNGR234a y4xM ORF there is no sign of ngr511 nor any indication of sequences originating from pNGR234b. Interestingly, the duplicated ORFs on pNGR234a are found within a region containing the genes encoding a functional type III protein secretion system. Furthermore, they lie between nopX and nopL, two genes that encode proteins secreted by this system (47). It is possible that y4yB to y4xM could be coregulated with nopX, as there are no obvious transcriptional termination signals in the 185-bp (nopX to y4yB) intergenic region. Transcriptional analysis of pNGR234a showed that y4yB to y4xM are all strongly induced 24 h after flavonoid addition, indicating possible symbiotic functions (38) (it should be cautioned that some of the transcripts could have come from the other duplicated genes, however). Since y4yB to y4xM are within the type III secretion system cluster, a polar mutation in y4yB was generated, but it did not have any obvious symbiotic phenotype or effect on protein secretion (R. Dieckmann, C. Marie, X. Perret, W. J. Broughton, and W. J. Deakin, unpublished data). Obviously, the discovery of second copies of y4yB to y4xM suggests that a double mutant should now be created to answer this question.
Both B. japonicum USDA110 and M. loti MAFF303099 possess type III secretion systems, yet neither has homologues of y4yB to y4xM, perhaps suggesting that this locus is not essential for protein secretion. Several of the ORFs have homology to proteins involved in iron transport, including a siderophore receptor, implying that they may monitor the iron status of the environment. It is noteworthy that iron often limits bacterial virulence and, particularly, type III protein secretion. The role of iron in virulence of Erwinia has been well documented (15, 16), although homologues of the y4yB cluster have not been implicated. Type III protein secretion by the plant pathogen Ralstonia solanacearum is controlled by an outer membrane receptor (PrhA) with homology siderophore receptors (31). R. solanacearum also contains homologues of many of the genes in the y4yB cluster. Several genes of the y4yB-y4xM cluster are also found in other plant pathogens as well as the human pathogen Staphylococcus epidermidis ATTC 14990.
In summary, these observations suggest that 40% of the genes and operons identified on pNGR234b are not strain specific and may have been acquired from other related bacteria. This would explain why NGR234 is so successful in nodulating different legumes; symbiotic competence arises from a flexible genome that is able to efficiently integrate foreign DNA from other bacteria.
Acknowledgments
We thank G. Gottschalk and G. C. Walker for many helpful comments on the manuscript and D. Gerber for general support.
Research in LBMPS is financed by the Fonds National Suisse de la Recherche Scientifique (project 31-63893.00) and the Université de Genève. Research in Göttingen was funded by the Genomik Network of the BMBF and the FCI.
REFERENCES
- 1.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K. C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battisti, L., J. Lara, and J. Leigh. 1992. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc. Natl. Acad. Sci. USA 89:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Baumberger I. C., N. Fraefel, M. Göffert, and H. Hennecke. 2003. New NodW- or NifA-regulated Bradyrhizobium japonicum genes. Mol. Plant-Microbe Interact. 16:342-351. [DOI] [PubMed]
- 3.Becker, A., A. Kleickmann, M. Keller, W. Arnold, and A. Pühler. 1993. Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol. Gen. Genet. 241:367-379. [DOI] [PubMed] [Google Scholar]
- 4.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broughton, W. J. 2003. Roses by other names: taxonomy of the Rhizobiaceae. J. Bacteriol. 185:2975-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Broughton, W. J., M. J. Dilworth, and I. K. Passmore. 1972. Base ratio determination using unpurified DNA. Anal. Biochem. 46:164-172. [DOI] [PubMed]
- 6.Broughton, W. J., S. Jabbouri, and X. Perret. 2000. Keys to symbiotic harmony. J. Bacteriol. 182:5641-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Pühler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H., M. Batley, J. W. Redmond, and B. G. Rolfe. 1985. Alteration of the effective nodulation properties of a fast-growing broad host-range Rhizobium due to changes in exopolysaccharide synthesis. J. Plant Physiol. 120:331-349. [Google Scholar]
- 9.Chen, H., J. X. Gray, M. Nayudu, M. A. Djordjevic, M. Batley, J. W. Redmond, and B. G. Rolfe. 1988. Five genetic-loci involved in the synthesis of acidic exopolysaccharides are closely linked in the genome of Rhizobium sp. strain NGR234. Mol. Gen. Genet. 212:310-316.
- 10.Djordjevic, S. P., H. Chen, M. Batley, J. W. Redmond, and B. G. Rolfe. 1987. Nitrogen fixation ability of exopolysaccharide synthesis mutants of Rhizobium sp. strain NGR234 and Rhizobium trifolii is restored by the addition of homologous exopolysaccharides. J. Bacteriol. 169:53-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djordjevic, S. P., B. Rolfe, M. Batley, and J. W. Redmond. 1986. The structure of the exopolysaccharide from Rhizobium sp. strain ANU280 (NGR234). Carbohydr. Res. 148:87-99. [Google Scholar]
- 12.Dorr, J., T. Hurek, and B. Reinhold-Hurek. 1998. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol. Microbiol. 30:7-17. [DOI] [PubMed] [Google Scholar]
- 13.Encarnacion, S., M. Dunn, K. Willms, and J. Mora. 1995. Fermentative and aerobic metabolism in Rhizobium etli. J. Bacteriol. 177:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Flores, M., P. Mavingui, L. Girard, X. Perret, W. J. Broughton, E. Martinez-Romero, G. Davila, and R. Palacios. 1998. Three replicons of Rhizobium sp. strain NGR234 harbor symbiotic gene sequences. J. Bacteriol. 180:6052-6053. [DOI] [PMC free article] [PubMed]
- 15.Franza, T., I. Michaud-Soret, P. Piquerel, and D. Expert. 2002. Coupling of iron assimilation and pectinolysis in Erwinia chrysanthemi 3937. Mol. Plant-Microbe Interact. 15:1181-1191. [DOI] [PubMed] [Google Scholar]
- 16.Franza, T., C. Sauvage, and D. Expert. 1999. Iron regulation and pathogenicity in Erwinia chrysanthemi 3937: role of the Fur repressor protein. Mol. Plant-Microbe Interact. 12:119-128. [DOI] [PubMed] [Google Scholar]
- 17.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 18.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez, J. E., C. E. Semino, L. X. Wang, L. E. Castellano-Torres, and G. C. Walker. 1998. Biosynthetic control of molecular weight in the polymerization of the octasaccharide subunits of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 95:13477-13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 20a.Göttfert, M., P. Grob, and H. Hennecke. 1990. Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host speciifcity in Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA 87:2680-2684. [DOI] [PMC free article] [PubMed]
- 21.Hallet, B., and D. J. Sherratt. 1997. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol. Rev. 21:157-178. [DOI] [PubMed] [Google Scholar]
- 22.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 23.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herron, S. R., J. A. E. Benen, R. D. Scavetta, J. Visser, and F. Jurnak. 2000. Structure and function of pectic enzymes: virulence factors of plant pathogens. Proc. Natl. Acad. Sci. USA 97:8762-8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann, K., E. B. Heinz, T. C. Charles, M. Hoppert, W. Liebl, and W. R. Streit. 2000. Sinorhizobium meliloti strain 1021 bioS and bdhA gene transcriptions are both affected by biotin available in defined medium. FEMS Microbiol. Lett. 182:41-44. [DOI] [PubMed] [Google Scholar]
- 26.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213-257. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti (supplement). DNA Res. 7:381-406. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 29.Kulakov, L., V. Delcroix, M. Larkin, V. Ksenzenko, and A. Kulakova. 1998. Cloning of new Rhodococcus extradiol dioxygenase genes and study of their distribution in different Rhodococcus strains. Microbiology 144:955-963. [DOI] [PubMed] [Google Scholar]
- 30.Loh, J., M. Garcia, and G. Stacey. 1997. NodV and NodW, a second flavonoid recognition system regulating nod gene expression in Bradyrhizobium japonicum. J. Bacteriol. 179:3013-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marenda, M., B. Brito, D. Callard, S. Genin, P. Barberis, C. Boucher, and M. Arlat. 1998. PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol. 27:437-453. [DOI] [PubMed] [Google Scholar]
- 31a.Mavingui, P., M. Flores, X. Guo, G. Dávila, X. Perret, W. J. Broughton, and R. Palacios. 2002. Dynamics of genome architecture in Rhizobium sp. strain NGR234. J. Bacteriol. 184:171-176. [DOI] [PMC free article] [PubMed]
- 32.Munchbach, M., A. Nocker, and F. Narberhaus. 1999. Multiple small heat shock proteins in rhizobia. J. Bacteriol. 181:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakai, C., H. Kagamiyama, M. Nozaki, T. Nakazawa, S. Inouye, Y. Ebina, and A. Nakazawa. 1983. Complete nucleotide sequence of the metapyrocatechase gene on the TOI plasmid of Pseudomonas putida mt-2. J. Biol. Chem. 258:2923-2928. [PubMed] [Google Scholar]
- 34.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 35.Nocker, A., N. P. Krstulovic, X. Perret, and F. Narberhaus. 2001. ROSE elements occur in disparate rhizobia and are functionally interchangeable between species. Arch. Microbiol. 176:44-51. [DOI] [PubMed] [Google Scholar]
- 36.O'Connell, K. P., and M. F. Thomashow. 2000. Transcriptional organization and regulation of a polycistronic cold shock operon in Sinorhizobium meliloti RM1021 encoding homologs of the Escherichia coli major cold shock gene cspA and ribosomal protein gene rpsU. Appl. Environ. Microbiol. 66:392-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellock, B. J., H. P. Cheng, and G. C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Perret, X., W. J. Broughton, and S. Brenner. 1991. Canonical ordered cosmid library of the symbiotic plasmid of Rhizobium species NGR234. Proc. Natl. Acad. Sci. USA 88:1923-1927. [DOI] [PMC free article] [PubMed]
- 38.Perret, X., C. Freiberg, A. Rosenthal, W. J. Broughton, and R. Fellay. 1999. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 32:415-425. [DOI] [PubMed] [Google Scholar]
- 39.Reuber, T. L., and G. C. Walker. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269-280. [DOI] [PubMed] [Google Scholar]
- 40.Riley, M. 1993. Functions of the gene products of Escherichia coli. Microbiol. Rev. 57:862-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothfield, L., S. Justice, and J. Garcia-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Streit, W. R., C. M. Joseph, and D. A. Phillips. 1996. Biotin and other water-soluble vitamins are key growth factors for alfalfa root colonization by Rhizobium meliloti 1021. Mol. Plant-Microbe Interact. 9:330-338. [DOI] [PubMed] [Google Scholar]
- 45.Tombolini, R., S. Povolo, A. Buson, A. Squartini, and M. P. Nuti. 1995. Poly-beta-hydroxybutyrate (PHB) biosynthetic genes in Rhizobium meliloti 41. Microbiology 141:2553-2559. [DOI] [PubMed] [Google Scholar]
- 46.Trinick, M. J. 1980. Relationships amongst the fast-growing Rhizobium of Lablab purpureus, Leucena leucocephala, Mimosa sp., Acacia farnesiana, and Sesbania grandiflora and their affinities with other Rhizobium groups. J. Appl. Bacteriol. 49:39-53. [Google Scholar]
- 47.Viprey, V., A. Del Greco, W. Golinowski, W. J. Broughton, and X. Perret. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28:1381-1389. [DOI] [PubMed] [Google Scholar]
- 48.Viprey, V., A. Rosenthal, W. J. Broughton, and X. Perret. 2000. Genetic snapshots of the Rhizobium species NGR234 genome. Genome Biol. 1:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M.-J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z.-Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J.-F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 50.Xu, Y., M. W. Mortimer, T. S. Fisher, M. L. Kahn, F. J. Brockman, and L. Xun. 1997. Cloning, sequencing, and analysis of a gene cluster from Chelatobacter heintzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH:flavin mononucleotide oxidoreductase. J. Bacteriol. 179:1112-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, G., T. V. Bhuvaneswari, C. M. Joseph, M. D. King, and D. A. Phillips. 2002. Roles for riboflavin in the Sinorhizobium-alfalfa association. Mol. Plant-Microbe Interact. 15:456-462. [DOI] [PubMed] [Google Scholar]
- 52.York, G. M., and G. C. Walker. 1997. The Rhizobium meliloti exoK gene and prsD/prsE/exsH genes are components of independent degradative pathways which contribute to production of low-molecular-weight succinoglycan. Mol. Microbiol. 25:117-134. [DOI] [PubMed] [Google Scholar]
- 53.Zakataeva, N. P., V. V. Aleshin, I. L. Tokmakova, P. V. Troshin, and V. A. Livshits. 1999. The novel transmembrane Escherichia coli proteins involved in the amino acid efflux. FEBS Lett. 452:228-232. [DOI] [PubMed] [Google Scholar]