Abstract
A gene cluster encoding the alternative sigma factor σB, three predicted regulators of σB (RsbV, RsbW, and RsbY), and one protein whose function is not known (Orf4) was identified in the genome sequence of the food pathogen Bacillus cereus ATCC 14579. Western blotting with polyclonal antibodies raised against σB revealed that there was 20.1-fold activation of σB after a heat shock from 30 to 42°C. Osmotic upshock and ethanol exposure also upregulated σB, albeit less than a heat shock. When the intracellular ATP concentration was decreased by exposure to carbonyl cyanide m-chlorophenylhydrazone (CCCP), only limited increases in σB levels were observed, revealing that stress due to ATP depletion is not an important factor in σB activation in B. cereus. Analysis of transcription of the sigB operon by Northern blotting and primer extension revealed the presence of a σB-dependent promoter upstream of the first open reading frame (rsbV) of the sigB operon, indicating that transcription of sigB is autoregulated. A second σB-dependent promoter was identified upstream of the last open reading frame (orf4) of the sigB operon. Production of virulence factors and the nonhemolytic enterotoxin Nhe in a sigB null mutant was the same as in the parent strain. However, σB was found to play a role in the protective heat shock response of B. cereus. The sigB null mutant was less protected against the lethal temperature of 50°C by a preadaptation to 42°C than the parent strain was, resulting in a more-than-100-fold-reduced survival of the mutant after 40 min at 50°C.
Bacillus cereus is a spore-forming gram-positive rod that is increasingly being recognized as a food-borne pathogen. It may cause illness through the production of a range of virulence factors. The most important virulence factors are a heat-stable emetic toxin, which causes vomiting, and several enterotoxins that cause diarrhea (14, 23). The symptoms of food-borne disease caused by B. cereus are generally mild and self-limiting, but in rare instances they can also be life-threatening, as was shown in 1998 when a food-poisoning outbreak in France, which was attributable to B. cereus, caused the deaths of three persons. The B. cereus strain that caused this outbreak produced a novel cytotoxin, CytK, which caused necrotic enteritis (30). B. cereus can also be the causative agent of other diseases, such as periodontitis, fulminant endophthalmitis, and meningitis in immunocompromised patients (2, 11, 12).
Because of the ubiquitous presence of B. cereus in the environment, it can easily contaminate food production or processing systems (23). B. cereus has the potential for multiple adaptive response pathways (20). These pathways may contribute to survival of the cells during food processing and storage and thus may contribute to the importance of B. cereus as a food-borne pathogen. Vegetative cells of B. cereus also play an important role in the pathogenesis of food-borne illness, because they produce the diarrheal enterotoxins in the host small intestine (31). In this situation, B. cereus has to deal with the stresses that it experiences in the gastrointestinal tract. Indeed, for some food-borne pathogens, the ability to mount a stress response is a prerequisite for virulence in the gastrointestinal tract (10).
Taxonomically, B. cereus is closely related to Bacillus thuringiensis and Bacillus anthracis. Together with Bacillus weihenstephanensis and Bacillus mycoides, these organisms form the B. cereus group. The members of this group are genotypically so similar that it has been proposed that the members of the B. cereus group should be considered members of the same species (17). However, the phenotypic differences among B. cereus, B. anthracis, and B. thuringiensis are substantial. While B. cereus causes generally mild cases of food-borne illness, B. anthracis is the etiological agent of the often lethal disease anthrax (22). B. thuringiensis, on the other hand, is generally considered a beneficial microorganism; it produces insecticidal toxins and is widely used as a biological control agent to counter insect pests in agriculture (41). Whole-genome sequencing of B. cereus ATCC 14579 (20) and B. anthracis Ames (39) and suppressive subtraction hybridization (38) have revealed some distinct genomic differences that distinguish B. cereus and B. thuringiensis from B. anthracis, but these differences do not seem to explain the phenotypic disparities in the B. cereus group mentioned above. The functional properties that differentiate these organisms are thought to be mostly caused by genes carried on plasmids or, possibly, by altered gene expression among strains (39).
Previously, a number of stress-induced proteins of B. cereus were identified by two-dimensional gel electrophoresis. These proteins included RsbV, the antagonist of the anti-sigma factor of σB, which was found to be upregulated during heat shock (33). This strongly suggested that a σB response is triggered during heat shock and potentially also under other stress conditions. σB has been studied extensively in several gram-positive bacteria. This protein is a secondary subunit of RNA polymerase that is known to play an important role in regulating gene expression when there are major changes in the environment. The model organism for study of σB is Bacillus subtilis (see reference 36 for a recent review). sigB null mutants of B. subtilis have decreased resistance to heat, acid, ethanol, salt, and oxidative stress (35). Similar effects have also been described for sigB null mutants of the human pathogens Listeria monocytogenes and Staphylococcus aureus (1, 5, 7, 8).
The regulatory network leading to expression of σB in B. subtilis has been extensively studied for a number of years. Two differentially regulated pathways lead to activation of σB in B. subtilis. The first pathway is induced under environmental stress conditions (like ethanol exposure and osmotic shock), and the second pathway is induced by a decrease in the level of intracellular ATP (36, 48). The regulatory network leading to σB activation and repression functions by a so-called partner switching mechanism. In this system, interactions between the anti-sigma factor of σB (RsbW) and the anti-sigma factor antagonist (RsbV) and more regulators further upstream in the regulatory cascade are controlled by serine phosphorylation and dephosphorylation. This leads to the formation or dissociation of protein-protein complexes, which can finally lead to the release of σB from an RsbW-σB complex (35). More than 200 general stress response genes are under the control of σB in B. subtilis, and these genes encode proteins with a wide variety of cellular functions (19, 34, 37). In B. anthracis the alternative sigma factor σB was shown to be a minor virulence factor and to be activated during the stationary growth phase and after a heat shock (9).
In this paper, we describe the sigB operon of B. cereus ATCC 14579 and a predicted novel regulator of σB activity (RsbY), which is located directly downstream of the sigB operon. σB was activated under several stress conditions, particularly during heat shock but also during other stresses, such as osmotic upshock and ethanol exposure. No correlation between intracellular ATP levels and σB activation was found, indicating that σB activation is not triggered by energy depletion. We mapped two σB-dependent promoters in the σB operon, which revealed the transcriptional organization of the σB operon in B. cereus. Finally, a sigB null mutant exhibited impaired survival at 50°C after preadaptation to 42°C compared to the survival of the parent strain. This indicates that σB plays a role in the adaptive response of B. cereus during heat stress.
MATERIALS AND METHODS
Bacterial strains, culture media, growth conditions, and plasmids.
B. cereus ATCC 14579 was cultured in brain heart infusion (BHI) medium at 30°C with aeration at 200 rpm. All Escherichia coli strains were cultured in Luria broth at 37°C (40). E. coli DH5α (40) was used as a general-purpose cloning host. E. coli BL21-Codonplus-(DE3)-RIL (Stratagene, La Jolla, Calif.) was used as the host for SigB overproduction. E. coli HB101/pRK24 (44) was used as the donor host in conjugation experiments. The antibiotics used were ampicillin at a concentration of 50 μg/ml, kanamycin at a concentration of 70 μg/ml, erythromycin at a concentration of 150 μg/ml (for E. coli) or 5 μg/ml (for B. cereus), spectinomycin at a concentration of 100 μg/ml, and polymyxin B at a concentration of 50 μg/ml for counterselection against E. coli upon conjugation. The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant properties | Source or reference |
|---|---|---|
| pGEM-T | PCR cloning vector, Ampr | Promega |
| pET28-b | E. coli overexpression vector, Kanr | Novagen |
| pMT01 | pET28-b derivative containing sigB under control of the T7 RNA polymerase promoter | This study |
| pUC18ERY | Ampr Eryr | 45 |
| pATΔS28 | tra+ conjugative suicide vector for B. cereus group, Spcr | 32 |
| pATΔS28ery | pATΔS28 derivative containing erythromycin resistance cassette from pUC18ERY, Spcr Eryr | This study |
| pATΔS28eryBY | pATΔS28ery derivative containing 1.1-kb downstream flanking region of sigB, Spcr Eryr | This study |
| pATΔsigB | pATΔS28ery derivative containing 1.1-kb upstream and downstream flanking regions of sigB, Spcr Eryr | This study |
DNA manipulation and sequencing.
Plasmid DNA was purified with a Qiaprep Spin Miniprep kit (Westburg, Leusden, The Netherlands). B. cereus chromosomal DNA was isolated by using a Wizard genomic DNA purification kit (Promega, Madison, Wis.) according to the manufacturer's instructions. DNA sequencing was performed with an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, Calif.) and a DYEnamic ET terminator cycle sequencing kit (Amersham Biosciences, Roosendaal, The Netherlands). Restriction endonucleases and DNA ligase (MBI Fermentas GmbH, St. Leon-Rot, Germany) were used according to the manufacturer's instructions. PCR experiments for cloning DNA into vectors were performed with Pwo polymerase (Roche Diagnostics, Almere, The Netherlands). The oligonucleotides used are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| OEcSigBF | GCAGCCATGGTGGAAATCCAATCTCAACCT |
| OEcSigBR | GCAGCTCGAGTGTATCTAAAAATGCGGCTTG |
| EryCasF | GCGATCTAGAGTCCGCAAAAGAAAAACG |
| EryCasR | GCGAGGATCCCATACCTAATAATTTATCTAC |
| FlSigbupF | CACGTCTAGACTTACGACTTGCCTTGGTTC |
| FlSigbupR | CCCTTCTAGACCTGCGCTTCATCACATTGG |
| FlSigbdownF | GCGACCCGGGGGTTAGGTATTTCACAAATG |
| FlSigbdownR | GCGAGAATTCTTTAATTCCGATTTCAAGCG |
| PrRsbVF | AAATGATGGAGGTTATACG |
| PrRsbVR | TAATATTTCTGTTAACCCTG |
| PrOrf4F | TTTAGCAGGAGAATACTCAG |
| PrOrf4R | AACTCTGTCATATTTAATTTCG |
| PERsbV | AATCTACGTTATGAAAATCTA |
| PEOrf4 | TGTCCTTGTTCATCACTAAT |
| SeqRsbV | GGAATGATTACGGGAAAAGACT |
| SeqOrf4 | AATGAAAATTCCTGCAAAGG |
Introduced restriction sites are underlined.
The B. cereus ATCC 14579 genome sequence database was used throughout this study (http://www.integratedgenomics.com). Comparisons with the B. anthracis Ames genome and the unfinished B. cereus ATCC 10897 genome were made by using BLAST with microbial genomes at http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/genom_table_cgi. Comparisons with the incomplete B. thuringiensis subsp. israelensis ATCC 35646 genome were made at ERGO Light (http: //www.ergo-light.com). The free energy of stem-loop structures was calculated by using the Mfold server at http://mfold.burnet.edu.au.
Overexpression and purification of σB in E. coli and generation of polyclonal antibodies.
sigB was amplified from B. cereus chromosomal DNA by PCR by using primers OEcSigBF and OEcSigBR, which introduced an NcoI site and an XhoI site, respectively. The PCR product was cloned into pET28-b (Novagen, Madison, Wis.), and the resulting vector (pMT01) was transformed into E. coli BL21-Codonplus-(DE3)-RIL. To produce SigB, a 200-ml culture of this strain was grown at 37°C in Luria broth with kanamycin. When the cells reached the mid-logarithmic phase (optical density at 600 nm [OD600], ∼0.5), isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and incubation was continued for 2 h. Cells were then harvested, resuspended in lysis buffer (50 mM HEPES-NaOH [pH 7.5], 0.5 M NaCl, 1 mM dithiothreitol, 5 mM Pefabloc, 0.5 mg of lysozyme per ml), and incubated for 30 min at 4°C. Cells were then lysed by addition of Triton X-100 to a final concentration of 1% and subsequent sonication. The extract was then treated with DNase I (20 μg/ml) for 1 h at 37°C. The insoluble fraction containing the inclusion bodies was centrifuged (30,000 × g, 20 min, 4°C) and washed twice with phosphate-buffered saline with 1% Triton X-100. The washed pellet was resuspended in 20 ml of binding buffer (20 mM sodium phosphate buffer [pH 7.4], 8 M urea, 0.5 M NaCl, 10 mM imidazole, 1 mM dithiothreitol, 5 mM Pefabloc, 10% glycerol) and kept at 4°C for 1 h. After removal of debris by centrifugation, an aliquot of the supernatant, corresponding to 12 mg of protein, was loaded on a 1-ml Hi-Trap chelating HP Ni2+ column (Amersham Biosciences). The column was then washed with 10-ml portions of binding buffer containing decreasing concentrations of urea (8, 6, 4, 2, 1, and 0 M), which allowed on-column refolding of the protein. The protein was then eluted with an imidazole gradient (20 to 500 mM) and dialyzed against dialysis buffer (50 mM sodium phosphate buffer [pH 7.8], 0.3 M NaCl, 50% glycerol). Protein purity was assessed by Coomassie blue staining of a sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel. The protein concentration was measured by the bicinchoninic acid assay.
Rabbit serum containing anti-σB antibodies was prepared by Eurogentec S.A. (Herstal, Belgium) according to the company's standard protocol. Two rabbits were used to raise antibodies against σB. At the start of the protocol 100 μg of purified σB was mixed with Freund's adjuvant and injected intradermally. After 14, 28, and 56 days booster injections consisting of 100 μg of antigen mixed with incomplete Freund's adjuvant were administered. Finally, the animals were bled at day 66. The serum of the animal with the highest antibody titer was used for all experiments.
Protein extraction and immunoblotting techniques.
Total cellular protein was extracted by bead beating as described previously (33). The protein concentration was determined by the bicinchoninic acid assay. Samples containing 40 μg of protein were loaded on two SDS-PAGE gels. One of the gels was used in Western blotting experiments, and the other gel was stained with Coomassie blue and visually inspected to confirm that equal amounts of protein were loaded.
Proteins were separated by SDS-PAGE by using 15% polyacrylamide gels and a Criterion II vertical electrophoresis system (Bio-Rad, Richmond, Calif.). Bio-Rad's broad-range prestained SDS-PAGE standards were used as molecular weight markers. After electrophoresis, proteins were electroblotted at 100 V onto nitrocellulose membranes. After the membranes were blocked by incubation with TBS (20 mM Tris-HCl [pH 7.5], 500 mM NaCl) with 0.1% sodium caseinate, the blots were incubated with TBS-0.05% Tween 20 supplemented with 2,000-fold-diluted rabbit immune serum containing the polyclonal anti-σB antibodies. Immunocomplexes were incubated with goat anti-rabbit peroxidase (Bio-Rad) and were visualized with 3,3′-diaminobenzidine tetrahydrochloride. The signal intensities of the Western blots were quantified by using the Gel-Pro Analyzer software package (Media Cybernetics, Silver Spring, Md.).
Determination of the intracellular ATP pool.
ATP was measured by the firefly luciferase assay by using the the LuminATE luciferin-luciferase reagent (Celsis, Landgraaf, The Netherlands). Cells were lysed by adding 1 ml of culture to 2 ml of absolute ethanol that was prechilled to −20°C. The suspension was incubated for 10 min at −20°C before the luciferase reaction results were determined with a Lumac biocounter M2500. To correct for ATP present in the culture broth, 1-ml aliquots of the cultures were centrifuged (12,000 × g, 1 min), and each supernatant was analyzed to determine the ATP concentration as described above.
Construction of a sigB null mutant.
First, an erythromycin resistance cassette was amplified from pUC18ERY with primers EryCasF and EryCasR. After digestion with XbaI and BamHI, the erythromycin resistance cassette was cloned into pATΔS28, resulting in pATΔS28ery. Subsequently, a 1.2-kb downstream flanking region of sigB, which contained 82 bp of the 3′ end of sigB, was amplified by PCR with primers FlSigbdownF and FlSigbdownR and, after digestion with XmaI and EcoRI, inserted into pATΔS28ery, resulting in pATΔS28eryBY. Finally, a 1.2-kb upstream flanking region of sigB, which contained 96 bp of the 5′ end of sigB, was amplified by PCR with primers FlSigbupF and FlSigbupR and, after digestion with XbaI, inserted into pATΔS28eryBY, resulting in pATΔsigB. The orientation of the inserts in the vector was checked by restriction analysis and sequencing. The vector was then transformed into E. coli HB101/pRK24, and the resulting strain was used in conjugation experiments with B. cereus. Conjugation was carried out by using the standard protocol for conjugative plasmid transfer from E. coli to gram-positive bacteria (4). Transconjugants were selected for erythromycin resistance and screened for spectinomycin sensitivity. PCR and Southern analysis confirmed that the strains selected harbored the deleted allele of sigB and that the erythromycin resistance cassette had recombined into the chromosome through a double-crossover event rather than integration of the entire plasmid (data not shown). The B. cereus sigB null mutant was designated B. cereus FM1400.
Isolation of total RNA, Northern blotting, and primer extension.
Total RNA was isolated from B. cereus by using Trizol (Invitrogen, Breda, The Netherlands). After precipitation of the nucleic acid, the residual DNA was removed with 10 U of RNase-free DNase I (Roche). After phenol-chloroform extraction and precipitation, the RNA was quantitated by measuring the OD260. All RNA samples had an OD260/OD280 ratio of ≥1.9.
RNA was separated on a 1.2% agarose-0.66 M formaldehyde-morpholinepropanesulfonic acid (MOPS) gel, which was electrophoresed at 40 V (constant voltage) and blotted onto Zeta-Probe membranes (Bio-Rad). Blots were hybridized and washed according to the manufacturer's instructions. Internal PCR fragments of rsbV (generated with primers PrRsbVF and PrRsbVR, resulting in a 270-bp product) and orf4 (generated with primers PrOrf4F and PrOrf4R, resulting in a 302-bp product) were used as probes. Gel-purified probes were radiolabeled with [α-32P]dATP by nick translation. After hybridization and washing, the blots were exposed to a PhosphorImager screen. After exposure for 16 to 72 h, the screen was scanned with a Storm 840 system (Amersham Biosciences). A 0.24- to 9.5-kb RNA ladder (Invitrogen) was used to determine the transcript sizes.
Primer extension analysis was performed as described by Kuipers et al. (24). The oligonucleotides used were PERsbV and PEOrf4, which are complementary to rsbV and orf4, respectively. Four picomoles of primer was added to 50 μg of RNA in reaction buffer containing 10 nmol of dCTP, 10 nmol of dGTP, 10 nmol of dTTP, and 3.3 nmol of [α-32P]dATP. cDNA was synthesized by addition of 200 U of Superscript II RNase H− reverse transcriptase (Invitrogen) and incubation for 10 min at 42°C, followed by addition of 10 nmol of cold dATP and incubation for 10 min at 42°C; the final volume of the reaction mixture was 20 μl. After the enzyme was inactivated by heating the preparation for 15 min at 70°C, 12 μl of formamide loading dye (95% formamide, 18 mM EDTA, 0.025% SDS, xylene cyanol, bromophenol blue) was added. After the samples were denatured by heating them at 80°C for 10 min, 5-μl aliquots were analyzed on a 7 M urea-8% PAGE sequencing gel prior to visualization by autoradiography. Sequence ladders were obtained by cloning template DNA (generated with PERsbV and SeqRsbV, resulting in a 941-bp product, and with PEOrf4 and SeqOrf4, resulting in a 1,429-bp product) into pGEM-T (Promega) and performing radioactive sequencing with a T7 DNA polymerase sequencing kit (USB, Cleveland, Ohio) with the same primers that were used for the primer extension reaction.
Assay for virulence factors and heat resistance of B. cereus cells.
Protease, lecithinase, and hemolytic activities were assayed on BHI agar plates supplemented with 5% milk, 5% egg yolk, and 5% sterile sheep blood (Biotrading, Mijdrecht, The Netherlands), respectively. Two microliters of an overnight culture of B. cereus was spotted on each plate. The plates were examined after 24 h of incubation at 30 and 37°C. The presence of enterotoxins in the supernatants of overnight cultures of B. cereus was determined with a Tecra BDE kit (Tecra Diagnostics, Frenchs Forest, Australia), which detects the NheA subunit of the nonhemolytic enterotoxin Nhe.
The heat resistance of vegetative B. cereus cells was assayed as described previously (33). Briefly, cells from a culture in the mid-logarithmic growth phase were exposed to a lethal temperature (50°C) with or without preexposure to 42°C for 30 min. Survival at 50°C was determined by plating appropriate dilutions on BHI agar plates, followed by overnight incubation at 30°C. For all heat exposures, three independent experiments were performed, and samples were plated in duplicate for each time point.
RESULTS
Sequence analysis of the σB gene cluster in B. cereus ATCC 14579.
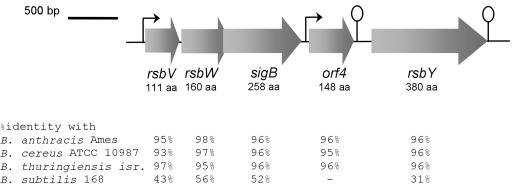
A gene cluster encoding σB and its regulators was identified in the recently completed genome sequence of B. cereus ATCC 14579 (20) (Fig. 1). This gene cluster consisted of five open reading frames, which encode regulators of σB activity, the σB structural gene, and a protein with an unknown function. The products of all these open reading frames exhibit high amino acid identity with homologues encoded in the recently completed B. anthracis Ames genome (39), the unfinished B. cereus ATCC 10897 genome, and the unfinished B. thuringiensis subsp. israelensis ATCC 35646 genome, highlighting the close relationship among the different members of the B. cereus group. All of the open reading frames, except orf4, also have homologues in B. subtilis (26) with amino acid identities between 31 and 56% (Fig. 1).
FIG. 1.
Diagram of the organization of the sigB gene cluster of B. cereus ATCC 14579. The large arrows represent open reading frames and indicate their orientations and sizes. The code numbers of these open reading frames in the B. cereus genome database (20) are RZC05131, RZC05126, RZC05124, RZC05127, and RZC05128. The predicted sizes of the encoded proteins (in amino acids [aa]) are also indicated. Predicted terminators downstream of orf4 and rsbY are indicated by stem-loop structures. σB-dependent promoters identified in this study are indicated by small arrows. The levels of amino acid identity with homologues of the open reading frames in B. anthracis Ames (39), B. cereus ATCC 10987 (http://www.tigr.org), B. thuringiensis subsp. israelensis ATCC 35646 (http://www.ergo-light.com), and B. subtilis 168 (26) are indicated at the bottom. Note that in B. subtilis 168 the gene encoding the RsbY homologue (rsbP) is not located directly downstream of the sigB operon like rsbY in the B. cereus group.
The first open reading frame of the gene cluster is rsbV, which encodes a 111-amino-acid protein with the predicted function of an anti-sigma factor antagonist. The next open reading frame is rsbW, which is predicted to encode an 160-amino-acid protein that can function as an anti-sigma factor of σB. The sigB gene overlaps rsbW for 12 codons and codes for a protein consisting of 258 amino acids. The overlap between rsbW and sigB is conserved in B. subtilis (13 codons overlap), L. monocytogenes (13 codons overlap), and S. aureus (8 codons overlap) (1, 21, 25, 50). In B. subtilis these genes have been shown to be translationally coupled, ensuring that equimolar amounts of σB and its cognate anti-sigma factor are present (3). The fourth open reading frame was designated orf4 and could code for a 148-amino-acid protein. The BLAST hit in the GenBank database with the highest significance for the gene encoding this protein is the B. anthracis homologue (96% identity), which is also situated directly downstream of sigB. Orf4 from B. cereus is distantly related to bacterioferritins and Dps-like DNA binding proteins, as previously reported for Orf4 of B. anthracis (9). A homologue with an unknown function from the recently discovered and sequenced organism Oceanobacillus iheyensis strain HTE831 (29, 43) is also relatively closely related to Orf4, with a level of identity of 63%. This homologue is not part of the σB operon of O. iheyensis. Directly downstream of orf4 a stem-loop structure was identified, which may function as a terminator. The free energy of formation of this structure is −9.4 kcal/mol.
The last open reading frame in the sigB gene cluster is rsbY. The 380-amino-acid protein that could be encoded by this open reading frame contains a C-terminal PP2C serine phosphatase domain and an N-terminal response regulator receiver domain homologous to CheY. The latter domain retained the highly conserved and functionally important residues equivalent to D12, D13, the phosphorylation site D57, T87, and K109 of CheY (49). Previously, rsbY was not identified in B. anthracis (9), presumably because an incomplete version of the B. anthracis genome sequence was used. A new search of the genomes available for the B. cereus group revealed that in all cases rsbY is present and located directly downstream of orf4. The closest well-described relative of RsbY is RsbP in B. subtilis. RsbP is also a two-domain protein; the N-terminal domain is a sensor PAS domain, while the C-terminal domain is a PP2C serine phosphatase domain. RsbP is activated upon energy stress and is then able to dephosphorylate RsbV, leading to σB activation (46). Directly downstream of rsbY a strong stem-loop structure with a calculated free energy of formation of −20.6 kcal/mol was identified.
The genome of B. cereus ATCC 14579 was also searched for homologues of other regulators (RsbQ, -R, -S, -T, -U, and -X) of the environmental stress and energy stress pathway of σB activation in B. subtilis. RsbU exhibited some homology with the C-terminal part of RsbY and with stage II sporulation protein E, but the other σB regulators of B. subtilis have no apparent homologues in B. cereus ATCC 14579.
Overexpression of sigB in E. coli, purification of σB, and generation of anti-σB antibodies.
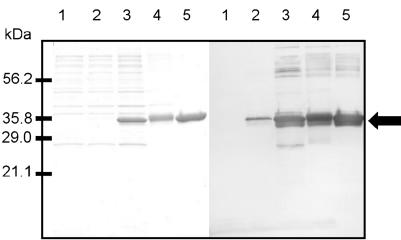
To analyze the role of σB in the stress response of B. cereus, we raised polyclonal antibodies against σB to determine intracellular σB levels under a variety of stress conditions. To obtain sufficient amounts of antigen for the immunization protocol, σB was overexpressed in E. coli. The sigB gene was cloned into the overexpression vector pET28-b, creating a fusion with six C-terminal histidine residues. Subsequently, σB was purified by Ni2+ affinity chromatography. In the standard overexpression host E. coli strain BL21λDE3(pLysS), only very limited production of σB could be obtained (data not shown), presumably because of the differences in codon usage between E. coli and B. cereus. In the codon bias-adjusted BL21 derivative E. coli BL21-Codonplus-(DE3)-RIL, σB production was dramatically increased. Purification of σB resulted in a >95% pure protein as judged on a Coomassie blue-stained SDS-PAGE gel. The purified protein was then used for generation of polyclonal antibodies against σB (Fig. 2). Even when the histidine tag was taken into account, σB migrated as a slightly larger protein in the SDS-PAGE gel than predicted on the basis of its predicted molecular mass (29 kDa). This is a property of many sigma factors because of highly positive and negative charge clusters in the proteins (6, 13, 15, 42). The antiserum that was raised against σB reacted specifically with purified σB, although some cross-reaction was also seen with larger proteins in the purified σB sample (Fig. 2).
FIG. 2.
Overproduction and purification of σB. (Left panel) SDS-PAGE of protein extracts from E. coli BL21-Codonplus-(DE3)-RIL carrying either pET28-b or pMT01. (Right panel) Immunodetection of σB with anti-σB antiserum. Ten micrograms of protein was loaded for each sample. Lane 1, crude protein extract from E. coli carrying pET28-b; lane 2, crude protein extract from E. coli carrying pMT01; lane 3, crude protein extract from E. coli carrying pMT01 after induction with 1 mM IPTG for 2 h; lane 4, inclusion bodies isolated from E. coli carrying pMT01 after induction with 1 mM IPTG for 2 h; lane 5, representative fraction of purified σB after elution from an Ni2+ affinity column. The arrow indicates the position of the overproduced σB protein.
Activation of σB under stress conditions.
To characterize the σB response of B. cereus under stress conditions, σB levels in total protein extracts from stressed B. cereus cells were determined by Western blotting by using the anti-σB antiserum. The antiserum reacted strongly with a protein band at 34 kDa, which corresponded to the expected migration of native σB in B. cereus. This band was absent when protein extracts from the B. cereus sigB null mutant (see below) were studied, confirming that it was indeed σB.
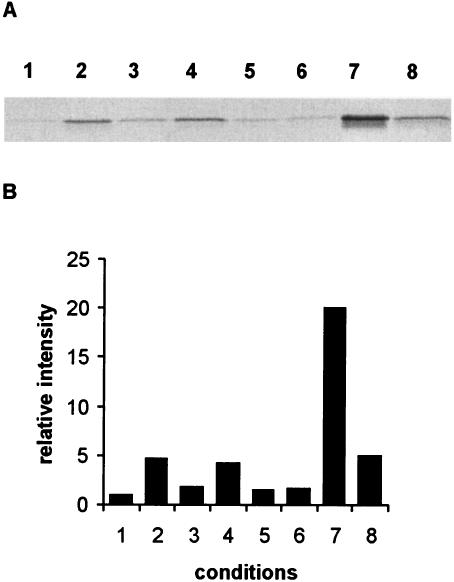
We found that several types of stress can lead to activation of σB (Fig. 3A). In cells taken from a culture in the mid-logarithmic growth phase (OD600, 0.4 to 0.5), low levels of σB were present, but upon exposure to stress conditions, the σB levels rose rapidly. To quantify activation of σB under stress conditions, we determined the signal intensities of the σB bands (Fig. 3B). The greatest effect was observed after a heat shock at 42°C. Densitometric analysis of the σB band on the Western blot revealed that there was 20.1-fold activation of σB. In an overnight culture, which had been in the stationary phase for several hours, σB was found to be expressed at levels that were 5.0 fold higher than the levels in cells in the mid-logarithmic growth phase. Addition of 4% ethanol or 2.5% NaCl or an acid shock at pH 5.2 led to 4.6-, 4.2-, and 1.8-fold induction of σB, respectively. A limited effect on σB levels was observed after oxidative stress induced by addition of 50 μM H2O2 or the thiol-specific oxidizing agent diamide at a concentration of 1 mM (1.5- and 1.6-fold σB activation, respectively).
FIG. 3.
Stress-induced activation of σB in B. cereus. (A) Cellular σB levels upon exposure to stress. Protein extracts from mid-logarithmic-phase B. cereus cells (lane 1) and stressed B. cereus cells were prepared as described in Materials and Methods. B. cereus cells in the mid-logarithmic growth phase were exposed to 4% (vol/vol) ethanol (lane 2), pH 5.2 (the pH was adjusted by addition of HCl) (lane 3), 2.5% (wt/vol) NaCl (lane 4), 50 μM H2O2 (lane 5), 1 mM diamide (lane 6), and 42°C for 30 min (lane 7). Proteins were also extracted from an overnight culture (lane 8). Forty micrograms of protein of each sample was loaded on the SDS-PAGE gel. Immunoblotting was performed with the sample by using the σB antiserum. (B) Relative amounts of σB. The signal intensities from the Western blot shown in panel A were quantified by using the Gel-Pro Analyzer software package (Media Cybernetics). The value for the mid-logarithmic-phase culture was defined as 1.
Energy stress is not an important trigger for activation of σB.
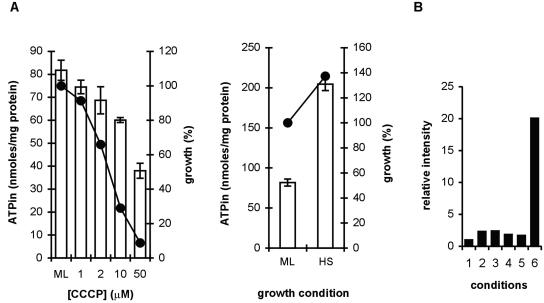
In B. subtilis, the energy stress pathway of σB activation is controlled by RsbP, which responds to a decrease in the size of the intracellular ATP pool (46, 48). Because of the presence of the RsbP homologue RsbY in the proteins encoded by the B. cereus sigB gene cluster, we decided to test if a reduction in the intracellular ATP level would also result in σB activation in B. cereus. The intracellular ATP pool was depleted by using increasing concentrations of carbonyl cyanide m-chlorophenylhydrazone (CCCP), which is an agent that uncouples oxidative phosphorylation. A 30-min exposure to CCCP resulted in a decrease in the intracellular ATP concentration and inhibition of growth (Fig. 4A). A limited σB-activating effect (≤2.5-fold induction on the protein level) was observed in the cultures exposed to CCCP (Fig. 4B). During a heat shock from 30 to 42°C for 30 min, the intracellular ATP concentration rose more than twofold and the σB level increased 20.1-fold. These results indicate that the σB response in B. cereus is not triggered by a drop in the intracellular ATP concentration, as in B. subtilis (48), but seems to occur solely under environmental stress conditions.
FIG. 4.
Effects of CCCP on growth, the intracellular concentration of ATP, and σB expression of B. cereus. (A) Growth (circles) and intracellular ATP concentration (bars) in B. cereus cells from a mid-logarithmic-phase culture (ML) after exposure to 1, 2, 10, and 50 μM CCCP (left panel) and after a heat shock (HS) at 42°C for 30 min (right panel). Growth of the culture was monitored by determining the increase in the OD600 during the 30 min of exposure to CCCP or 42°C. The increase in the OD600 over 30 min for an untreated culture was defined as 100%, and growth in the CCCP-treated and heat-shocked cultures was related to this value. Intracellular ATP concentrations were determined by the firefly luciferase assay as described in Materials and Methods. (B) Relative levels of σB in CCCP-treated B. cereus cells (lanes 1 to 5 contained 0, 1, 2, 10, and 50 μM CCCP, respectively) and heat-shocked B. cereus cells (lane 6). The cellular levels of σB were estimated by immunoblotting with anti-σB antiserum and quantification of the signal by the Gel-Pro Analyzer software package as described in the text. Forty micrograms of protein from each sample was loaded on the SDS-PAGE gel used for Western blotting.
Transcriptional analysis of the sigB operon.
To study transcriptional regulation of the sigB operon and the physiological role of σB in the resistance of B. cereus to stress, a sigB null mutant (B. cereus FM1400) was constructed, in which the sigB gene was disrupted by an erythromycin resistance cassette. PCR and Southern blotting analysis showed that the erythromycin resistance cassette had correctly integrated into the sigB gene (data not shown).
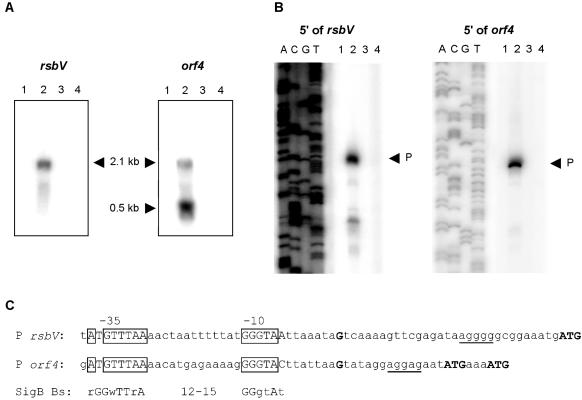
Transcription of the sigB operon in the sigB null mutant and its parent strain was studied. The activation of σB under stress conditions was confirmed by Northern blot experiments performed with RNA isolated from cultures of B. cereus FM1400 and the parent strain in the mid-logarithmic growth phase and after 10 min of exposure to 42°C (Fig. 5A). In the sigB null mutant, no transcription of the sigB operon was observed under both these conditions. In wild-type cells in the mid-logarithmic growth phase no transcription of the sigB operon was observed, results which corresponded to the barely detectable σB levels in the Western blot experiments. In the RNA isolated from wild-type cells after 10 min of exposure to 42°C, a 2.1-kb mRNA transcript hybridized with the rsbV probe, corresponding to a transcript covering the rsbV-rsbW-sigB-orf4 region. This transcript could also be visualized after the blot was probed with DNA probes specific for rsbW and sigB (data not shown). These results demonstrate that the sigB operon is autoregulated by σB. When an orf4-specific probe was used, the 2.1-kb transcript was also identified after a heat shock, but in addition a strong band at 0.5 kb was also observed. This transcript could not be visualized in the RNA samples from B. cereus FM1400, which indicates that the transcript is σB dependent.
FIG. 5.
Analysis of transcription of the sigB operon in B. cereus. (A) Northern blot analysis of transcription of the sigB operon. Total RNA was extracted from B. cereus ATCC 14579 and B. cereus FM1400 cells during mid-logarithmic growth in BHI medium (lanes 1 and 3, respectively) and after a 10-min exposure to 42°C (lanes 2 and 4, respectively). 32P-labeled internal PCR products of rsbV and orf4 were used as probes. Hybridization of the probe with target RNA was visualized by exposure to a Phosphoscreen and scanning with a Storm scanner. Transcript sizes are indicated by arrowheads. (B) Primer extension analysis of promoters 5′ of rsbV and orf4. For all reactions 50 μg of RNA was used. Total RNA was extracted from B. cereus ATCC 14579 and B. cereus FM1400 cells during logarithmic growth in BHI medium (lanes 1 and 3, respectively) and after a 10-min exposure to 42°C (lanes 2 and 4, respectively). Mapped transcriptional start sites are indicated by arrowheads. Lanes A, C, G, and T contained the corresponding sequencing ladders for localization of the transcripts. (C) Promoter sequence alignment for the σB-dependent promoters 5′ of rsbV and orf4. The positions of identified −35 and −10 regions are indicated. The nucleotides in boxes in the −35 and −10 regions fit the σB promoter consensus sequence of B. subtilis (18). Uppercase letters in the B. subtilis σB promoter consensus sequence indicate highly conserved residues, and lowercase letters indicate less conserved residues (R = A or G, W = A or T). Transcriptional start sites and ATG start codons are indicated by boldface type. Putative Shine-Dalgarno sequences are underlined.
On the basis of the results obtained in the Northern blot experiments, we performed primer extension analysis and mapped two σB-dependent promoters in the sigB operon (Fig. 5B and C). These promoters are located upstream of rsbV and orf4, and both of them are silent during mid-logarithmic growth but are activated upon a heat shock. In B. cereus FM1400 these promoters are silent under both conditions, showing that they are σB dependent. The promoter upstream of rsbV has −35 and −10 sequences of ATGTTTAA and GGGTAA, respectively, with a spacing of 13 nucleotides. This promoter sequence closely resembles the consensus sequence of σB promoters in B. subtilis. For this organism, the consensus sequences for the −35 and −10 regions have been described as rGGwTTrA and GGgtAt, respectively (uppercase letters indicate highly conserved residues, and lowercase letters indicate less conserved residues; R = A or G, W = A or T), which are separated by 12 to 15 nucleotides (18). The promoter 5′ of orf4 was also shown to be activated only in the wild-type strain upon heat shock. The −35 and −10 regions of this promoter are ATGTTTAA and GGGTAC, respectively, which are separated by 13 nucleotides. This means that the −35 and −10 regions of this promoter are practically identical to the regions of the promoter 5′ of rsbV; the only difference is the last residue of the −10 region (C instead of A).
Role of σB in production of virulence factors and heat resistance of vegetative cells.
We did not observe any radical difference between the phenotypes of the B. cereus sigB null mutant and its parent strain. The growth rate at 30°C of the sigB null mutant in BHI broth did not differ from the growth rate of the parent strain, and cultures of both strains reached the same cell density in the stationary phase. We assayed protease, lecithinase, and hemolytic activities and the production of nonhemolytic enterotoxin by both wild-type and sigB null mutant cells and found no significant differences between the strains in any of the assays. Hence, σB does not play a role in the production of virulence factors in B. cereus.
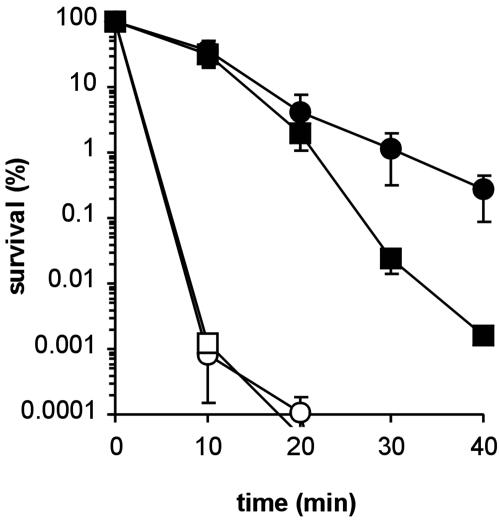
To examine the role of σB in the adaptive response of B. cereus, we focused on the thermotolerance of vegetative cells. Previously, it has been shown that mid-logarithmic cells of B. cereus are very sensitive to a high temperature (50°C) but that they can be protected by preadaptation to a permissive temperature, 42°C (33). We showed that upon heat shock from 30 to 42°C the σB levels increased substantially (Fig. 3), suggesting that σB may play a role in survival of the cells at high temperatures. Therefore, we tested the thermotolerance of B. cereus FM1400 and its parent strain in the mid-logarithmic growth phase with and without preadaptation to 42°C (Fig. 6). In the nonadapted cultures there was no significant difference between the levels of survival of the two strains; both died rapidly at 50°C, and the viable counts were around or below the detection limit after 20 min of incubation at 50°C. After preadaptation to 42°C, the wild-type cells showed dramatically increased survival at 50°C. The sigB null mutant was also protected by preadaptation at 42°C, but it was clearly protected less than the wild-type cells, which resulted in >100-fold-lower survival after 40 min at 50°C compared to the survival of the parent strain. These results demonstrate that in B. cereus σB is involved in the protective response of vegetative cells against high temperature.
FIG. 6.
Survival of of B. cereus ATCC 14579 cells (circles) and B. cereus FM1400 cells (squares) in the mid-logarithmic growth phase at 30°C (open symbols) and after pretreatment at 42°C for 30 min before exposure to 50°C (solid symbols). The averages of three independent experiments are shown. The error bars indicate standard deviations.
DISCUSSION
In this paper we describe the transcriptional organization and expression of the sigB operon of B. cereus and provide data concerning activation of σB upon exposure to stress. In this study we also found that σB is involved in the protective heat stress response. Our results establish a starting point for further studies of the role of σB in the stress response of the food pathogen B. cereus and related members of the B. cereus group. We specifically studied regulation of σB activity, and hence we can compare and contrast the B. cereus σB response with the responses of other gram-positive bacteria.
Several stresses activate σB in B. cereus. A heat shock from 30 to 42°C leads to 20.1-fold activation of σB and is by far the most powerful trigger leading to σB activation in B. cereus. This is in agreement with a previous study, in which several stress-induced proteins of B. cereus were identified by two-dimensional gel electrophoresis (33). One of the proteins identified was the anti-sigma factor antagonist RsbV, which was upregulated >20-fold after a heat shock. The data obtained suggested that σB is activated upon heat shock. In this study, we obtained experimental evidence that σB is indeed activated upon heat shock. Furthermore, we found that σB is also activated under other stress conditions, albeit at a level that is an order of magnitude less than the level during a heat shock, which may explain why the effects were not observed in a previous proteomic study by our group (33).
In B. subtilis, activation of σB in response to heat stress has been well documented (16, 19, 36, 37) and seems to be signaled through the environmental stress pathway (48). It has been proposed that in B. subtilis structural changes in the ribosome during stress could lead to induction of this pathway (51). Conceivably, a similar mechanism could also be involved in triggering σB activation in B. cereus. Energy stress can be ruled out as an important factor in σB activation in B. cereus, because (i) decreasing the intracellular ATP concentration results in limited activation of σB (≤2.5-fold) and (ii) during a heat shock at 42°C the intracellular concentration of ATP increases but, nevertheless, σB is strongly activated.
Analysis of the transcriptional organization of the σB operon revealed that this operon is transcribed as a 2.1-kb transcript encompassing rsbV, rsbW, sigB, and orf4. orf4 is also under control of an additional σB-dependent promoter, and this makes orf4 a member of the σB regulon in B. cereus. The exact role of Orf4 in the stress response of B. cereus and other members of the B. cereus group is still unknown. This protein may function as a bacterioferritin, as predicted on the basis of its distant homologues, as proposed previously for Orf4 in B. anthracis (9), but experimental data are needed to verify this.
The sigB operon ends directly downstream of orf4, because the transcripts starting upstream of rsbV and orf4 both end at this site, which suggests that the stem-loop structure downstream of orf4 functions as a terminator. Downstream of this structure we identified an open reading frame, rsbY. The encoded protein is annotated as a σB regulatory protein. The rsbY gene was found to be present directly downstream of the σB operon in all members of the B. cereus group whose genome sequences are available. The position of RsbY in the sigB operon and its domain structure strongly suggest that it can function like the PP2C serine phosphatase proteins RsbU and RsbP in B. subtilis, which play crucial regulatory roles in activation of σB in that organism (36).
In other gram-positive bacteria, σB has not been studied as extensively as it has been in B. subtilis, but there are some intriguing similarities and differences between the σB responses of these organisms and the σB response of B. cereus. In L. monocytogenes, σB is also activated in response to different stresses, but in this organism osmotic shock is the most powerful trigger. Heat shock is also an important activating factor, and other stresses, like ethanol stress and acid shock, result in significantly less marked activation of σB (1). In S. aureus, a thorough examination of the stresses which activate σB has not been performed, but the available data suggest that heat shock is the most potent inducer in this organism, while ethanol shock has a limited effect (25). Of all the sigB operons in gram-positive bacteria, the pattern of activation in the S. aureus operon seems to be the most comparable to the pattern in the B. cereus operon. Interestingly, in S. aureus, σB seems to be regulated by three Rsb proteins, just as it is in B. cereus. An important difference between S. aureus and B. cereus, however, is the fact that the phosphatase containing the PP2C phosphatase domain is a single domain protein (RsbU) in S. aureus (25, 50). This again underlines the uniqueness of σB in the B. cereus group; both the stresses that activate σB and the organization of the operon are markedly different from the stresses that activate σB and the organization of the operon in other gram-positive bacteria studied thus far. In B. cereus σB also plays a role in protecting the cells against high temperature. In B. subtilis (47) and S. aureus (5), σB is also involved in the protective heat stress response, but an L. monocytogenes sigB null mutant was not more heat sensitive than its parent (7). This inconsistency in the phenotypes of sigB null mutants may be caused by differences in the set of σB-regulated genes in the various organisms.
The observation that σB does not play a role in the production of virulence factors and the nonhemolytic enterotoxin Nhe was not unexpected, since in the B. cereus group the production of these factors is governed by the pleiotropic regulator PlcR and σB seems not to be involved in transcription of the plcR gene (27, 28). However, a sigB null mutant may show a weakened stress response. This may indirectly decrease its virulence by impeding the growth or survival of the organism in food or the host, analogous to the role of the stress response in the virulence of the food pathogens L. monocytogenes and Salmonella spp. (10).
The fact that σB is activated when B. cereus is exposed to several different stress conditions and the observation that σB plays a role in survival during exposure to high temperature can have significant consequences for the control of B. cereus. B. cereus is a pathogen whose responses to different environmental situations can lead to cross-protection against normally lethal conditions (23, 33). The data presented here prove that σB is activated under several conditions and that σB has an important role in the adaptive response of B. cereus. Activation of σB may lead to increased survival of B. cereus during food processing and thus to increased risk of food poisoning outbreaks. In future research, the role of σB in the stress response of B. cereus will be elucidated further, and studies will focus on the σB regulon and the pathway leading to σB activation in B. cereus.
Acknowledgments
Integrated Genomics (Chicago, Ill.) is acknowledged for use of the B. cereus genome sequence database and for release of the preliminary B. thuringiensis subsp. israelensis ATCC 35646 genome sequence. We thank Nuno Miguel Sampaio Osório for performing the plate assays for B. cereus virulence factors and Agnes Fouet for supplying pATΔS28 and E. coli HB101/pRK24.
REFERENCES
- 1.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beecher, D. J., T. W. Olsen, E. B. Somers, and A. C. Wong. 2000. Evidence for contribution of tripartite hemolysin BL, phosphatidylcholine-preferring phospholipase C, and collagenase to virulence of Bacillus cereus endophthalmitis. Infect. Immun. 68:5269-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, A. K., and W. G. Haldenwang. 1993. Regulation of σB levels and activity in Bacillus subtilis. J. Bacteriol. 175:2347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bron, S. 1990. Plasmids, p. 75-174. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, Great Britain.
- 5.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deora, R., T. Tseng, and T. K. Misra. 1997. Alternative transcription factor σSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 179:6355-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouet, A., O. Namy, and G. Lambert. 2000. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 182:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gahan, C. G., and C. Hill. 1999. The relationship between acid stress response and virulence in Salmonella typhimurium and Listeria monocytogenes. Int. J. Food Microbiol. 50:93-100. [DOI] [PubMed] [Google Scholar]
- 11.Gaur, A. H., C. C. Patrick, J. A. McCullers, P. M. Flynn, T. A. Pearson, B. I. Razzouk, S. J. Thompson, and J. L. Shenep. 2001. Bacillus cereus bacteremia and meningitis in immunocompromised children. Clin. Infect. Dis. 32:1456-1462. [DOI] [PubMed] [Google Scholar]
- 12.Gaur, A. H., and J. L. Shenep. 2001. The expanding spectrum of disease caused by Bacillus cereus. Pediatr. Infect. Dis. J. 20:533-534. [DOI] [PubMed] [Google Scholar]
- 13.Gitt, M. A., L. F. Wang, and R. H. Doi. 1985. A strong sequence homology exists between the major RNA polymerase sigma factors of Bacillus subtilis and Escherichia coli. J. Biol. Chem. 260:7178-7185. [PubMed] [Google Scholar]
- 14.Granum, P. E. 2001. Bacillus cereus, p. 327-336. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. American Society for Microbiology, Washington, D.C.
- 15.Haldenwang, W. G., and R. Losick. 1980. Novel RNA polymerase sigma factor from Bacillus subtilis. Proc. Natl. Acad. Sci. 77:7000-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hecker, M., and S. Engelmann. 2000. Proteomics, DNA arrays and the analysis of still unknown regulons and unknown proteins of Bacillus subtilis and pathogenic gram-positive bacteria. Int. J. Med. Microbiol. 290:123-134. [DOI] [PubMed] [Google Scholar]
- 17.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A. B. Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmann, J. D., and C. P. J. Moran. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 19.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, E. S. Dusko, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 21.Kalman, S., M. L. Duncan, S. M. Thomas, and C. W. Price. 1990. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J. Bacteriol. 172:5575-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keim, P., and K. L. Smith. 2002. Bacillus anthracis evolution and epidemiology. Curr. Top. Microbiol. Immunol. 271:21-32. [DOI] [PubMed] [Google Scholar]
- 23.Kotiranta, A., K. Lounatmaa, and M. Haapasalo. 2000. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2:189-198. [DOI] [PubMed] [Google Scholar]
- 24.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 25.Kullik, I., and P. Giachino. 1997. The alternative sigma factor σB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167:151-159. [DOI] [PubMed] [Google Scholar]
- 26.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 27.Lereclus, D., H. Agaisse, M. Gominet, S. Salamitou, and V. Sanchis. 1996. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J. Bacteriol. 178:2749-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lereclus, D., H. Agaisse, C. Grandvalet, S. Salamitou, and M. Gominet. 2000. Regulation of toxin and virulence gene transcription in Bacillus thuringiensis. Int. J. Med. Microbiol. 290:295-299. [DOI] [PubMed] [Google Scholar]
- 29.Lu, J., Y. Nogi, and H. Takami. 2001. Oceanobacillus iheyensis gen. nov., sp. nov., a deep-sea extremely halotolerant and alkaliphilic species isolated from a depth of 1050 m on the Iheya Ridge. FEMS Microbiol Lett. 205:291-297. [DOI] [PubMed] [Google Scholar]
- 30.Lund, T., M. L. De Buyser, and P. E. Granum. 2000. A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 38:254-261. [DOI] [PubMed] [Google Scholar]
- 31.McKillip, J. L. 2000. Prevalence and expression of enterotoxins in Bacillus cereus and other Bacillus spp., a literature review. Antonie Leeuwenhoek 77:393-399. [DOI] [PubMed] [Google Scholar]
- 32.Namy, O., M. Mock, and A. Fouet. 1999. Co-existence of clpB and clpC in the Bacillaceae. FEMS Microbiol. Lett. 173:297-302. [DOI] [PubMed] [Google Scholar]
- 33.Periago, P. M., W. van Schaik, T. Abee, and J. A. Wouters. 2002. Identification of proteins involved in the heat stress response of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 68:3486-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria, p. 179-197. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress response. ASM Press, Washigton, D.C.
- 36.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 37.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 38.Radnedge, L., P. G. Agron, K. K. Hill, P. J. Jackson, L. O. Ticknor, P. Keim, and G. L. Andersen. 2003. Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 69:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strickland, M. S., N. E. Thompson, and R. R. Burgess. 1988. Structure and function of the σ70 subunit of Escherichia coli RNA polymerase. Monoclonal antibodies: localization of epitopes by peptide mapping and effects on transcription. Biochemistry 27:5755-5762. [DOI] [PubMed] [Google Scholar]
- 43.Takami, H., Y. Takaki, and I. Uchiyama. 2002. Genome sequence of Oceanobacillus iheyensis isolated from the Iheya Ridge and its unexpected adaptive capabilities to extreme environments. Nucleic Acids Res. 30:3927-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. An integrative vector exploiting the transposition properties of Tn1545 for insertional mutagenesis and cloning of genes from gram-positive bacteria. Gene 106:21-27. [DOI] [PubMed] [Google Scholar]
- 45.Van Kranenburg, R., J. D. Marugg, I. I. van Swam, N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 46.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 47.Voelker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volz, K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741-11753. [DOI] [PubMed] [Google Scholar]
- 50.Wu, S., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, S., J. M. Scott, and W. G. Haldenwang. 2001. Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor σB. J. Bacteriol. 183:2316-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]