Abstract
The clinical and research value of human embryonic stem cells (hESC) depends upon maintaining their epigenetically naïve, fully undifferentiated state. Inactivation of one X chromosome in each cell of mammalian female embryos is a paradigm for one of the earliest steps in cell specialization through formation of facultative heterochromatin. Mouse ES cells are derived from the inner cell mass (ICM) of blastocyst stage embryos prior to X-inactivation, and cultured murine ES cells initiate this process only upon differentiation. Less is known about human X-inactivation during early development. To identify a human ES cell model for X-inactivation and study differences in the epigenetic state of hESC lines, we investigated X-inactivation in all growth competent, karyotypically normal, NIH approved, female hESC lines and several sublines. In the vast majority of undifferentiated cultures of nine lines examined, essentially all cells exhibit hallmarks of X-inactivation. However, subcultures of any hESC line can vary in X-inactivation status, comprising distinct sublines. Importantly, we identified rare sublines that have not yet inactivated Xi and retain competence to undergo X-inactivation upon differentiation. Other sublines exhibit defects in counting or maintenance of XIST expression on Xi. The few hESC sublines identified that have not yet inactivated Xi may reflect the earlier epigenetic state of the human ICM and represent the most promising source of NIH hESC for study of human X-inactivation. The many epigenetic anomalies seen indicate that maintenance of fully unspecialized cells, which have not formed Xi facultative heterochromatin, is a delicate epigenetic balance difficult to maintain in culture.
The potential of human embryonic stem cells (hESC) in cell replacement therapies depends upon stable maintenance in culture of the epigenetic competence of developmentally naïve cells. Inactivation of one X chromosome in mammalian females is a prominent example of early epigenetic regulation and “commitment” to cell-specific facultative heterochromatin which is then propagated to all subsequent cell generations. Embryonic stem (ES) cells are derived from the inner cell mass (ICM) of blastocyst stage embryos. During mouse development, random X-inactivation has not yet occurred in the ICM, and it has been well established that undifferentiated mouse ES cells do not display an inactive X (reviewed in Chow et al., 2005; Heard and Disteche, 2006). As uncommitted ICM or ES cells begin to differentiate, a large non-coding RNA from the X-linked Xist gene accumulates across the one X-chromosome chosen at random for inactivation in a given cell, while the other Xist allele is silenced by methylation (reviewed in Alexander and Panning, 2005). Thus, only after 1–2 days of ES cell differentiation does a large, Xist RNA interphase territory accumulate over the future Xi and initiate the inactivation process over a period of several days (reviewed in Heard and Disteche, 2006). This coating of the chromosome by human/mouse XIST/Xist RNA initiates a cascade of histone modifications that culminates in the silent state of Xi facultative heterochromatin (reviewed in Chang et al., 2006). Once established, the same XIST RNA coated heterochromatic chromosome is maintained in subsequent cell generations, such that the embryo is henceforth mosaic with regard to paternal or maternal X-linked gene expression.
To date, studies of the initiation of mammalian X-inactivation have been largely restricted to mouse, and any studies involving human X-inactivation have been very limited and restricted to somewhat abnormal systems, such as embryonal carcinoma (EC) cells or inducible transgenes in somatic cells (see Discussion Section). Previous reports indicated that two of the eleven NIH approved human female ES lines did not initiate X-inactivation upon differentiation, as expected from studies in mouse (Enver et al., 2005; Hoffman et al., 2005). One line (H7) never initiated X-inactivation even when differentiated for many weeks, whereas another (H9) exhibited apparently premature X-inactivation while maintained as an undifferentiated culture (Hoffman et al., 2005). However another study, focusing on using gene trapping as a tool for genome annotation, suggested that in H9 cells X-inactivation occurred after differentiation, as expected (Dhara and Benvenisty, 2004). These various findings imply that there are differences in the epigenetic status and X-inactivation competence of not only hESC lines but potentially sublines grown in different labs. Thus identifying particular lines or subcultures of the available NIH approved female lines that maintain this delicate epigenetic balance will be very valuable for the X-inactivation field, but may also suggest a source for possibly more epigenetically “naive” ES cells.
It is well established in mouse embryogenesis or ES cells, that cells of the ICM contain two active X-chromosomes for a short window of time before they initiate random X-inactivation as they begin to differentiate to specific lineages (Okamoto et al., 2004; Heard and Disteche, 2006). Not surprisingly, little is known about the timing of X-inactivation in the human ICM (Daniels et al., 1997; Ray et al., 1997), such that it is not clear when this takes place in human female embryos. Examining the X-inactivation status of female hESC lines may give insight into this question. Therefore, we investigated the available NIH-approved female hESC lines, as well as several sublines from multiple sources.
Materials and Methods
hES cell culture and characterization
Slides of hESC cultures were grown and fixed in the different labs that maintained the lines/sublines (U. Washington hESC core facility and Technion in Israel) and characterized them for karyotypic stability and pluripotency. One H9 subculture was characterized in the Stein Lab (U. Massachusetts hESC Core Facility), but briefly grown, differentiated and fixed in the Lawrence Lab. All hESC lines were grown on coverslips, on either a feeder layer of MEFs or matrigel and prepared for analysis without disruption of colonies. Ware Lab (hESC Core Facility for the University of Washington): Human ES cell lines (HSF6, BG03, ES01, ES02, ES03, H7, and H9) were cultured as had been previously described (Ware et al., 2005). Itskovitz-Eldor Lab (Technion, Haifa, Israel): hES cell lines I-4 and I-3 were cultured as previously described (Amit et al., 2000). Stein and Lawrence Labs: The H9 hESC line was maintained as recommended by the supplier (WiCell, Madison, WI) and as previously described (Becker et al., 2006). Due to space constraints, all protocols are briefly outlined in Supporting Information.
Differentiation
Rare differentiated cells found among the feeders functioned as internal differentiation controls on the same slide as undifferentiated colonies, and were discriminated by morphology and lack of alkaline phosphatase (Fig. 1A) (Vector labs, Burlingame, CA), TRA-160 and TRA-181 (Supplemental Fig. 2C–D). Longer term differentiated cells (7–14 days) were also stained with AP, TRA-160, and TRA-181, as well as Pan-cytokeratin (Supplemental Fig. 2E–G) to assess differentiation status of cells/colonies scored. Human cells can be distinguished from mouse feeders by the DAPI dense chromocenters in mouse nuclei and the presence of human Cot-1 RNA (See Figs. 1A and 2).
Fig. 1.
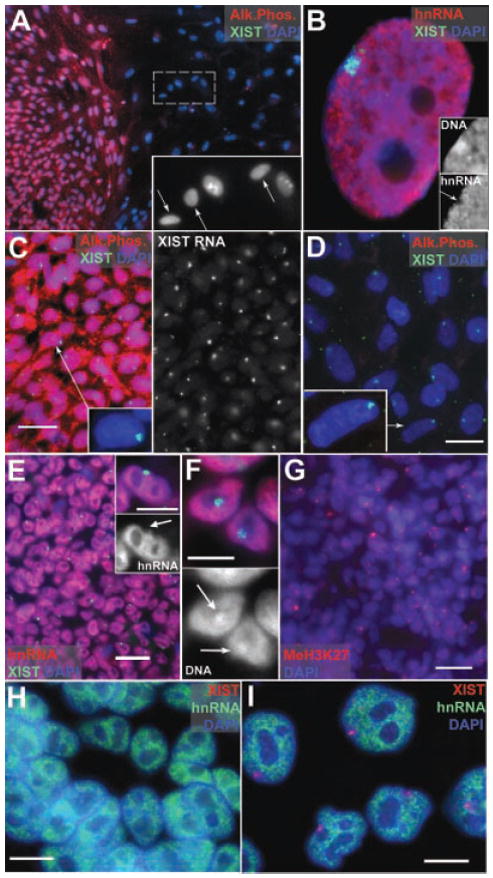
A: Undifferentiated hESC (H9) colony shows AP staining. The large differentiated nuclei among the feeders (arrows insert) do not. B: Xi in female WI38, show silencing (arrow insert), and Barr Body (insert). C: Undifferentiated hESC (ES01) with AP and XIST RA territories. D: Differentiated ES01 do not label with AP. E: Undifferentiated hESCs show a “hole” (insert, arrow) where hnRNA transcription is repressed. F: Undifferentiated hESCs have Barr Bodies (arrows DAPI channel). G: Undifferentiated colonies with MeH3K27 (red). H: Undifferentiated H9+X with no XIST RNA territories. I: Differentiated nuclei show only a single XIST territory, and hnRNA transcriptional inhibition. Scale bars: C,D,E,G,H,I = 10 μm, F and insert E = 5 μm.
Fig. 2.
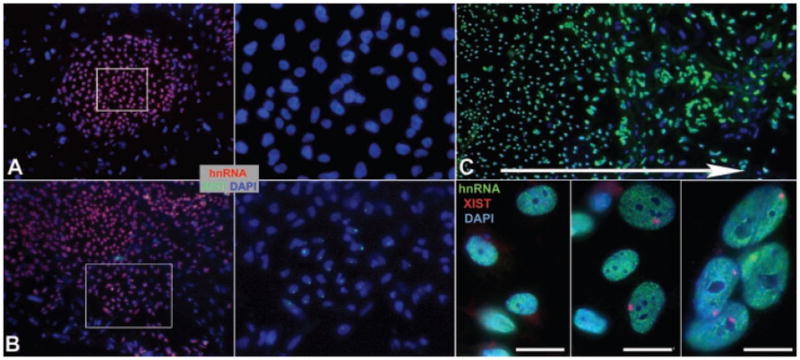
A: Undifferentiated H9 with no XIST territories. B: Undifferentiated H9 with a patch of XIST positive cells (cells in box enlarged at right for both). C: Differentiated H9 show increased XIST singals (red) with differentiation. Close-ups of cells from center (left), inner edge (middle) and outer edge (right) of differentiating colony in the large low-mag image. Arrow indicates the direction of increased differentiation, and increased number of XIST positive cells. Scale bars are 10 μM.
Longer term differentiated cultures—Lawrence Lab
EBs were cultured in differentiation medium (DMEM, 20% FBS, penicillin, and streptomycin) for 4 days in suspension, then were re-plated at low density onto matrigel coated coverslips for 14–16 days (for subsequent IHC and FISH analyses). Ware Lab protocol: hESC were plated onto coverslips covered with MEF feeder cells at 104 cells per cm2 in differentiation medium (DMEM, 20% FBS, penicillin, and streptomycin), and allowed to spontaneously differentiate for 7–14 days before cell fixation.
Cell preparation and in situ hybridization
All labs used our standard paraformaldehyde fixation protocol as previously described (Lawrence et al., 1989; Clemson et al., 1996; Tam et al., 2004) and briefly summarized in the Supporting Information. Probes were nick translated with either biotin or digoxigenin-dUTP (Roche, Basel, Switzerland). RNA hybridization was performed as described previously (Clemson et al., 1996; Tam et al., 2004). XIST was detected using XIST G1A: a ~10 kb genomic plasmid spanning from intron 4 to the 3′ end of the XIST gene, and collagen was detected using a full length (46 kb) genomic human Collagen |α| gene probe (gift from Rudolph Jaenisch, MIT). Human Cot-1 DNA (Roche) was nick translated and used to detect hnRNA transcription in non-denatured cells as previously published (Hall et al., 2002). The Barr body was identified using DAPI DNA staining (0.1 μg/ml for 4′). For simultaneous labeling of XIST RNA with antibody or alkaline phosphatase (Vector labs, VC-SK-5100) staining was performed first followed by a 5 min fix in 4% paraformaldehyde and XIST RNA hybridization. Antibodies used were: TRA-160 (Chemicon/Millipore, Bedford, MA), TRA-181 (Chemicon), H3H27me (Abcam, Cambridge, UK), H4K20me (Abcam), Pan-cytokeratin (Sigma, St. Louis, MO) and FK2 (BIOMOL/Affinity, Plymouth Meeting, PA).
Image analysis
Images presented are single plane images obtained using AxioVision LE 4.4 with either an Axiovert 200 or an Axiophot microscope (Carl Zeiss, Thornwood, NY) equipped with Chroma 83000 multi-bandpass dichroic and emission filter sets (Brattleboro, VT), set up in a wheel to prevent optical shift. Images were captured with an Orca-ER camera (Hamamatsu, Bridgewater, NJ) or a cooled charge-coupled device (CCD) camera (200 series, Photometrics, Inc., Tucson, AZ). HESC cells were scored in an unbiased blind study. Cell percentages are derived from 100 cell counts in random fields or among random colonies on a slide. For low frequency events 200 cells were counted. The sample sizes for percent of colonies in a given culture affected are listed on Table 1.
TABLE 1.
Summary of XIST territories in undifferentiated hESC cultures
| hESC line (Lab and passage #) | Colonies counted | XIST (+) colonies (%) | XIST (±) colonies (%) | XIST (−) colonies (%) |
|---|---|---|---|---|
| ES01—Ware (P63) | 16a | 100 | ||
| ES02—Ware (P37) (manual) | 15a | 100 | ||
| ES02—Ware (P59) (dispase) | 51 | 96 | 2 | 2 |
| ES03—Ware (P87) | 9a | 100 | ||
| ES03—Ware (P88) | 6a | 100 | ||
| BG03—Ware (P47) | 41 | 96 | 2 | 2 |
| HSF6—Ware (P54) | 35 | 100 | ||
| H9—Ware (P31) | 40 | 12 | 13 | 75 |
| H9—Ware (P35) | 72 | 21 | 13 | 64 |
| H9+X—Ware (P44) | 51 | 4 | 96 | |
| H9+X—Ware (P46) | 41 | 46 | 28 | 26 |
| TE03—Technion (P30) | 5a | 40 | 60 | |
| TE04—Technion (P54) | 3a | 33 | 66 | |
| H9—Stein (P48) | 50 | 100 | ||
| H7—Ware (P41) | 30 | 100 |
hESC lines and lab that supplied them, with passage numbers and sample size. The breakdown of the percentage of colonies on the undifferentiated slide is shown under each category. Colonies in the category XIST (+) are considered to show “precocious inactivation,” where >90% of the cells in the colony contain an XIST RNA territory. Although most colonies in the XIST (−) category contained no cells with XIST territories, the few colonies with <5% XIST positive cells were also included. XIST (±) are those colonies with a mixed population of cells, showing between 10% and 80% of the cells in the colony with XIST RNA territories. These slides contained recently passaged hESC colonies, which consisted of usually between 100 and 300 cells. More than one passage (ES03, UW-H9, UW-H9+X) or culture technique (ES02) were assessed.
The entire slide was scanned and all available colonies on the slide were scored.
Results
There are only 11 NIH approved female hESC lines available and of these the SA02 line has an altered karyotype (47XX+13) and the ES06 line proved very difficult for us to grow. Therefore, we examined the remaining nine hESC lines available from UCSF (HSF6), BresaGen, Inc. (Thebarton, Australia) (BG03), Cellartis (Dundee, UK) (SA02), WiCell (H9 and H7), ES Cell International (Helios, Singapore) (ES01, ES02, ES03, and ES06), and Technion (TE03 and TE04). In most cases, undifferentiated (or differentiated) cultures were prepared directly in the core facility/company lab that was routinely growing, maintaining and characterizing that line/subline. Importantly, this avoided alterations in the epigenetic status of cells during live cell transfer, freeze/thaw selection or changes in culture conditions between labs, and also allowed us to sample sublines and cultures maintained and characterized as undifferentiated directly in multiple established hESC labs. Usually fixed slides of hESC cultures were obtained directly from the labs maintaining these lines/sublines (Materials and Methods Section). All lines were assessed for X-inactivation status by identical procedures in one laboratory (Lawrence Lab). Care was taken to examine cells in colonies prepared without disruption (see Materials and Methods Section), in order to preserve valuable information in the native cell and colony morphology. The various lines were maintained and characterized by routine procedures in each core facility (Materials and Methods Section), and slides were stained in parallel for TRA-160, TRA-181, and Alkaline Phosphatase (AP) ES markers and Pan-cytokeratin (Pan-CK) for verification of differentiation status.
Most undifferentiated cultures of NIH female HESC lines contain an inactive X
X-inactivation status was based on the presence of an XIST RNA territory, transcriptional silencing (Cot-1 assay), histone H3K27 and/or H4K20 modifications, and the DAPI-dense Barr body. The undifferentiated samples prepared by the three different labs were grown under conditions which were shown to maintain pluripotency in each lab (Materials and Methods Section). The vast majority of colonies in these samples exhibited the characteristic well defined undifferentiated morphology (clear colony edges with no invasion of the feeders) of small tightly packed cells with no cytoplasm (5 μM diameter nuclei), and bright staining for AP (78%), TRA-160 (79%), and TRA-181 (87%) (Fig. 1A,C and Supplemental Fig. 2). This contrasts with the occasional spontaneously differentiated cells often found among the feeders or at colony edges (Fig. 1A(insert),D), which appear similar to the large cells growing as monolayers on slides allowed to differentiated for 7–14 days. These differentiated cells have large nuclei (≥10 μM diameter) and cytoplasms, with distinctive morphologies, and do not generally stain for the three ES markers, but show abundant cells positive for Pan-CK (Supplemental Fig. 2).
In over half of the undifferentiated hESC lines examined (5 of 9 lines), essentially all cells and colonies on the slide (96–100%) showed unmistakable XIST RNA territories (Fig. 1C) (Table 1). This was not due to occasional differentiating cells or colonies, since it was uniformly present in all cells in all colonies, the vast majority of which showed a clear undifferentiated phenotype. XIST RNA was localized to one site in a large well defined territory (not a small transcription focus) located at the perimeter of the nucleus (inserts Fig. 1C,D), indistinguishable from the Xi in normal somatic cells (Fig. 1B). In some lines (Table 1) we tested more than one passage and for the ES02 line we examined two different culture conditions, dispase (P59) versus mechanical expansion (P37-38); yet all showed identical Xi status.
Importantly, these hESC cultures also show transcriptional silencing of the Xi, as in normal female somatic cells (Fig. 1B) (Hall et al., 2002), where absence of hnRNA over the XIST painted chromosome indicates transcriptional repression as shown in Figure 1E (arrow). Additionally, methylation of histone H3 K27 (a hallmark of the Xi) and a condensed Barr Body was visible in all cells in all colonies on the slide as shown in Figure 1F(arrows), G. Since essentially all human cells on the slide showed X-inactivation hallmarks, and well over three quarters of the colonies were clearly AP positive, this indicates that, similar to the previously studied H9 culture (Hoffman et al., 2005), over half of the NIH approved female lines had undergone X-inactivation prior to differentiation (see Table 2). Since this was not expected from studying mouse embryos and ES cells, we refer to X-inactivation prior to differentiation as “precocious” inactivation.
TABLE 2.
Summary of hESC X-inactivation status broken down by XIST RNA phenotype
| Precocious X-inactivation | |
| H9—Carpenter (P61–63)a | Hoffman et al. (2005) |
| H7—Andrews (P39)a | Enver et al. (2005) |
| ES01—Ware (P63) | |
| ES02—Ware (P37–59) | |
| ES03—Ware (P87–88) | |
| BG03—Ware (P47) | |
| HSF6—Ware (P54) | |
| Mostly non-precocious colonies (but initiation is possible) | |
| H9—Benvenisty (P45–48)a | Dhara and Benvenisty (2004 |
| H9—Ware (P31–32) | |
| H9 + X—Ware (P44–46) | |
| TE03—Technion (P30) | |
| TE04—Technion (P54) | |
| Demonstrated to initiate Xi upon differentiation | |
| H9—Benvenisty (P45–48)a | Dhara and Benvenisty (2004 |
| H9—Ware (passage: low 30 s) | |
| H9 + X—Ware (passage: low 40 s) | |
| Never inactivates an X-chr | |
| H7—Andrews (P110)a | Enver et al. (2005) |
| H7—Carpenter (P48-72)a | Hoffman et al. (2005) |
| H7—Ware (P41) | |
| Loss of XIST expression after precocious inactivation | |
| H9—Stein (P48) | |
| X-trisomy with only a single Xi | |
| H9 + X—Ware (P44–46) | |
hESC lines grouped by phenotype, the lab it was grown in, and the passages assessed. Seven NIH approved hESC lines show precocious X-inactivation. Seven sublines have low levels of precocious inactivation, but having some colonies with Xi suggests they are capable of inactivation (unlike UW-H7 and UM-H9). Three H9 sublines have been demonstrated to initiate X-inactivation upon differentiation. One subline (UM-H9) may have lost XIST expression following precocious inactivation, and one line (H7) never initiates inactivation upon differentiation.
Sublines from previous literature; Hoffman et al. (2005), Dhara and Benvenisty (2004), Enver et al. (2005).
Some hESC sublines retain cells that have not yet formed the Xi and retain competence to initiate X-inactivation
The differences in Xi-status between the previously published results on the H9 line grown in two different labs (Dhara and Benvenisty, 2004; Hoffman et al., 2005) suggests there may be epigenetically distinct sublines of a given hESC line. Thus we examined more sources and subcultures of H9 cells, to determine if there are sublines that remain “uncommitted” with regard to X-inactivation, and then inactivate Xi during early differentiation. This would also bear on the important question of whether the precocious X-inactivation seen in the above five hESC lines, and in our previous results on H9 (Hoffman et al., 2005), was present from the earliest isolation of the ES cells from the ICM (an thus potentially a property of the embryo), or whether it is an epigenetic property that commonly arises in cultured hESC lines. Where possible we tested cultures that were relatively “low passage,” as contrasted with the 100 passages (year long culture) used to identify “adapted” cultures by Enver et al. (2005), and screened multiple undifferentiated subcultures of H9 from the Stein lab (UM-H9) and the Ware lab (UW-H9).
Importantly, the University of Washington H9 culture (UW-H9) did not exhibit XIST territories in most colonies while undifferentiated, indicating that most of cells had not precociously inactivated Xi (Fig. 1H). The vast majority of colonies (96%) did not contain any cells with XIST RNA territories, and the remaining 4% of colonies contained only a small number (~10%) of cells with XIST RNA territories. However, karyological analysis discovered that this line carried X-chromosome trisomy (47, XXX) in 5 of 5 chromosome spreads (the rest of the karyotype was normal), which we later verified by DNA FISH (see below and Fig. 3D). Therefore we acquired an earlier UW-H9 freeze (P32) with a normal diploid karyotype for further assessment. Like UW-H9+X, this diploid culture contained many colonies (78%) that did not show any XIST RNA territories (Fig. 2A). There was also significant mosaicism among the remaining colonies, with all of the cells in 12% of the colonies containing an Xi while the other 10% of colonies were variable, showing XIST positive cells ranging from 10% to 58% of the colony (Fig. 2B). Thus, this karyotypically normal subculture still maintained most colonies lacking XIST RNA territories.
Fig. 3.
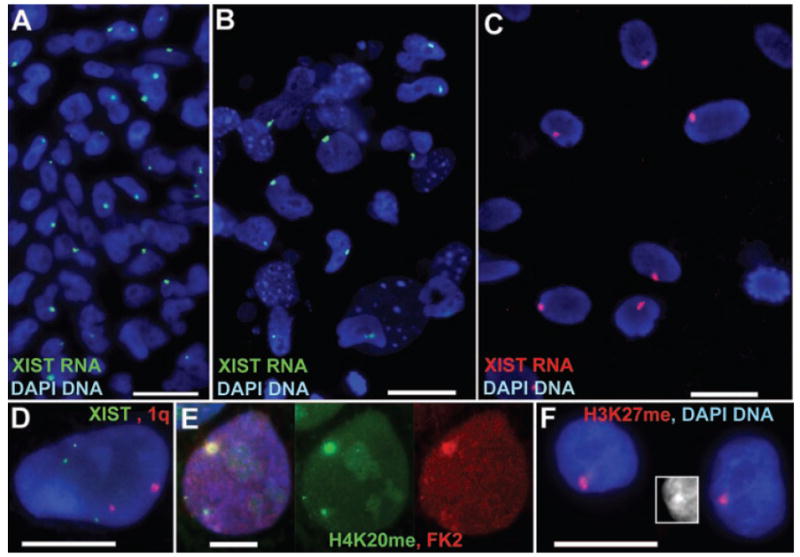
A: Undifferentiated, (B) Differentiated H9+X cells on same slide and (C) 14 days differentiated H9+X hESC with only one XIST territory. D: Two 1q singals (red) and three XIST genes (green) confirm the trisomy X in H9+X line. The MA-H9 Xi lacks XIST RNA but has methyl-H3K27 and ubiquitin (E) monomethyl-H4K20 (F), and a Barr Body (F-insert).
Because the first UW-H9+X culture contained a small number (<2%) of large cells scattered among the feeders, that stained negative for AP (see Fig. 1A insert), and did have XIST RNA territories, it suggested that this subline may inactivate upon differentiation (Fig. 1I). To assess the potential of both the diploid and trisomic H9 sublines to inactivate upon differentiation, we obtained additional undifferentiated samples of both cultures (UW-H9 (P31–35) and UW-H9+X (P45)) from the UWash core facility, and parallel 7–14 days differentiated slides. Undifferentiated cultures had abundant colonies (~78%) staining brightly with AP, TRA-160, and TRA-181 in parallel samples. When only undifferentiated AP positive colonies were scored for the presence of XIST RNA territories, most colonies showed no X-inactivation in any cells of the colony, with some variability between freeze thaws of each H9 subline (e.g., UW-H9: ~60–80%). The remaining colonies showed a range of precocious inactivation where most had less than 50%, and some had 100% of the cells labeled. However when the cell numbers are pooled across the 20–30 colonies scored on each slide, only ~16% of the AP (+) cells showed precocious inactivation and ~84% did not. This supports that these two H9 sublines tend to maintain two active X-chromosomes while undifferentiated.
On the other hand, both differentiated cultures clearly showed increased indicators of X-inactivation, with a greater frequency of cells with Xi and robust XIST RNA territories. The majority of cells covering the differentiated slide were large in size, compared to ES cells, generally grew as a monolayer, and did not stain with the three ES markers (TRA-160, TRA-181, and AP). There were usually around 6–10 small regions across the slide that contained more densely packed cells and were positive for the ES markers, suggesting some small regions on the slide remained undifferentiated. The epithelial differentiation marker pancytokeratin was also abundant on many of the large monolayer cells (~80% of cells) (Supplemental Fig. 2E–G). Only AP-negative, largely Pan-CK positive (info taken from parallel slides), large cells, growing in monolayer were scored for XIST RNA territories, and 50–80 cells were scored for each of 10 different regions across the slide, to get a sampling of Xi status across the differentiated population. For the UW-H9 subline, 100% of the regions sampled contained cells with XIST RNA territories, compared to 40% of the colonies on the undifferentiated slide. Half the regions scored contained over 65% of the cells with XIST territories, and only one region had less than 10% of the population with XIST RNA territories. Undifferentiated colonies on the slide generally did not show Xi, while the larger differentiated cells growing away from these undifferentiated regions often contained XIST RNA coated Xi (Fig. 2C). The difference in the number of cells with XIST territories between undifferentiated (~15%) and 7–14 days randomly differentiated slides (~60%), suggests that X-inactivation in these cultures was progressing with cellular differentiation. In addition, these sublines (UW-H9 and UW-H9+X) support that cells initially derived from the ICM were likely still “epigenetically naïve” with respect to X-inactivation, and that the presence of the Xi in other H9 sublines (Hoffman et al., 2005), (and likely the other five hESC lines above), arose in culture.
We were also able to obtain a few slides of undifferentiated cultures for the two NIH approved female hESC lines, TE03 (P30) and TE04 (P54), from Technion in Israel. Interestingly, these cultures looked similar to the UW-H9 subculture, and contained substantial undifferentiated ES colonies lacking XIST expression (TE04=66%, TE03=60%), although precocious Xi was also apparent in the remaining colonies. These findings indicate that these lines, like UW-H9, may also be a source of subcultures that may remain epigenetically naïve with respect to X-inactivation.
Specific hESC sublines exhibit interesting anomalies in X-inactivation
While UW-H9+X contained colonies that initiated X-inactivation during differentiation, our initial observations suggested an anomaly in the number of Xi in these XXX cells. Since normally two X chromosomes would be inactivated in trisomy X, we anticipated two XIST RNA territories, but found ~92% of the cells with XIST RNA territories had only one (Fig. 1I), irrespective of whether this appeared to be the precocious Xi of an undifferentiated cell in an ESC colony (100%) (Fig. 3A), or the Xi of the larger, more differentiated cells scattered among the feeders (95%) (Fig. 3B). We verified the X-trisomy directly by DNA FISH, which showed three XIST alleles in nuclei of 10 out of 10 colonies on the slide (Fig. 3D); in contrast, the presence of only two 1q signals in the same cells confirmed X-trisomy, not triploidy. Even after allowing (P45) H9+X cultures to spontaneously differentiate for 7–14 days, the vast majority of differentiated cells contained only one Xi (95%) (Fig. 3C). Although two Xi were occasionally observed (5%), these were in unusually large nuclei (well over 40 μM) which were very likely polyploid. To further support this, cells with two Xi appeared at essentially the same frequency on the diploid UW-H9 differentiated slides (9%), which would not be expected to have two Xi unless they had become polyploid. This suggests that most cells in the UW-H9+X cells may exhibit an X chromosome counting anomaly and inactivate only one of the three X chromosomes.
Another anomaly seen in some lines was the absence of any inactive X after differentiation, or the loss of XIST RNA signal after precocious inactivation had apparently occurred. We and others had previously published that some H7 samples did not contain any Xi after differentiation, despite containing two active Xs (Enver et al., 2005; Hoffman et al., 2005). Here we examined another undifferentiated subline of H7 (P41) (verified to have two X chromosomes in the Ware lab), and found similar results (100% of colonies had no XIST territories). To verify that this low passage UW-H7 line fails to inactivate upon differentiation similar to our previous observation (Hoffman et al., 2005), we spontaneously differentiated this line for 1 week before re-analysis. The large UW-H7 cells growing in monolayer on the slide did not stain for the three ES markers (AP, TRA-160, and TRA-181), and were positive for Pan-cytokeratin (all cells in seven different differentiated regions were Pan-CK positive), but lacked XIST RNA expression (100%), or H3K27me (100%).
Finally, another low passage (P48) subculture of H9 acquired from the Stein Lab (MA-H9) never showed XIST territories even following 16 days of differentiation (100% of the cells lacked XIST territories in five separate experiments), which was very unusual for H9, based on analysis of several other H9 sources (above and in previous literature). Although we repeated the XIST RNA hybridization several times, with parallel positive controls, we never detected XIST expression, yet we found other hallmarks of Xi facultative heterochromatin: methylated H3K27 (95% of cells), mono-methyl H4K20 (65% of cells) and ubiquitination (40% of cells) (Fig. 3E,F) (Kohlmaier et al., 2004; Rougeulle et al., 2004; Smith et al., 2004). Over 35% of the cells showed enrichment of both H4K20 and ubiquitin on what appeared to be the large DAPI bright “Barr body” localized at the nuclear periphery (insert Fig. 3F), suggesting that precocious XIST RNA expression had been shut down, although several hallmarks of the Xi remain in the absence of the RNA.
Discussion
Human embryonic stem cells are a unique and critical resource to study epigenetics during early human development and have enormous clinical potential, but the varying epigenetic status of these different hESC lines may have a great impact on their future utility (Humpherys et al., 2001; Li et al., 2005; Mitalipov, 2006; Allegrucci et al., 2007). X-inactivation represents the early formation of facultative heterochromatin, that distinguishes one cell from another. From mouse it is widely believed that this commitment occurs with the onset of the earliest lineage commitments. Thus, a cell that has not already formed the cell specific and chromosome wide facultative heterochromatin of the Xi can be reasonably considered more “epigenetically naive” than one that has taken this step, even if both populations express similar ES markers.
The ability to study the human X-inactivation process requires the availability of female hES cell lines that are capable of initiating X-inactivation in culture. The only other systems available to study X-inactivation in human cells are: karyotypically abnormal male embryonal carcinoma (EC) cells (e.g., Looijenga et al., 1997; Chow et al., 2003), mouse/human somatic cell hybrids induced to express XIST (e.g., Clemson et al., 1998; Anderson and Brown, 2002), or transgenic somatic human cancer cell lines or mouse ES lines (e.g. Migeon et al., 1999, 2001; Chow et al., 2002; Hall et al., 2002). For the mouse, it took the derivation of many female mouse ES lines before a few were found that maintained two X-chromosomes and initiated X-inactivation appropriately (Zvetkova et al., 2005). Our findings indicate that this may also be the case for the human ES cell system. Here we find that subcultures of the majority of NIH approved female hES lines we have screened have undergone precocious X-inactivation while in an undifferentiated state.
Importantly, however, through detailed analysis of “sublines” of H9, we were able to identify two H9 sublines, one karyotypically normal (UW-H9), and one trisomy X (UW-H9+X), that retained most cells which had not yet inactivated their Xi, and which largely did so upon differentiation. We suggest that this delicate epigenetic balance is unstable and that there is a propensity for hESC maintained in culture to undergo “precocious” or “premature” X-inactivation. A recent report used PCR to examine the expression of many genes including XIST in a large number of hESC lines, but did not assess the X-inactivation status of these lines (Adewumi et al., 2007). Few of the lines examined were NIH approved, and their competence to initiate X-inactivation upon differentiation was not studied. In addition, the detection of XIST expression in undifferentiated ES03 and ES02 cells is consistent with our demonstration of precocious inactivation. Adewumi et al. (2007) also found that undifferentiated BG03 and HES01 had low XIST, similar to male cells, but it remains to be determined if these cells are capable of initiating inactivation. Additionally, as further demonstrated in our study, this status can vary significantly between sublines of the same line. Thus, to effectively use any NIH approved sublines to study human X-inactivation, and to propagate the most “epigenetically naïve” lines available, it will be valuable to expand and preserve sublines that retain competence to initiate X-inactivation upon differentiation.
Interestingly, these findings bear on the timing of the human X-inactivation process, which cannot be assumed to be the same as in the mouse, where neither the ICM of the blastocyst, nor the ES cells derived from it, show random X-inactivation. While the presence of Xi in so many apparently undifferentiated hESC cultures might suggest that random Xi occurs earlier in the human ICM at the blastocyst stage of normal (or IVF) human embryogenesis, we pursued detailed analysis of sublines of one line in attempt to determine if more epigenetically naïve cells were present in the original line established from the ICM. Our findings of certain subcultures that are a “tableau rasseau” with respect to Xi suggests that at least for the H9 isolate most likely follows a mouse-like pattern, and that the “precocious” inactivation is likely due to adaptation to culture. This could also be true for the other lines as well.
It is possible that in human ES cells, as in mouse, there may be selection against the presence of two active X chromosomes. Some evidence in mouse indicates that this is due to abnormal methylation levels (Zvetkova et al., 2005). It may be very difficult and unnatural to induce ES cells of the blastocyst ICM to propagate indefinitely with two active X-chromosomes, which they were never biologically programmed to do. In fact, there is a very small window in mammalian development in which the two X-chromosomes remain active. Prior to the mouse blastocyst stage (<day 3.5), the paternal X remains inactive due to parental imprinting, and after this stage (>day 5.5) random X-inactivation has taken place, thus, leaving just 2 days during which two X-chromosomes may be active (Heard and Disteche, 2006). However, in contrast to many female mouse ES lines, human cells do not eliminate an X-chromosome during propagation, but tend to inactivate it. Since human X-chromosomes contain numerous genes that escape inactivation and are required in two doses, human XX ES cells may need to find other ways to eliminate the presumed growth suppression associated with having two X-chromosomes active during ESC propagation. Our data and others suggest that the most common route is for hESC lines to initiate inactivation early.
In this screen we also found three very interesting hESC phenotypes which may provide valuable systems to study certain aspects of X-inactivation. The UW-H9 line often acquires an additional X-chromosome (UW-H9+X) (Ware et al., 2005), but we find that only one of the three X-chromosomes is inactivated in this subline whereas we saw two Xi in our previously described H9 trisomy-X line (Hoffman et al., 2005). While we have not investigated the precise mechanism to account for this, this UW-H9 line could potentially provide a valuable model for defective X-chromosome “counting.”
Here we have surveyed the female human embryonic cell lines that were already established and approved for NIH funding when federal funding of hESC research was first authorized in August of 2001. While undifferentiated cultures of most sublines already contain an Xi (Enver et al., 2005; Hoffman et al., 2005; Allegrucci et al., 2007), scrutiny of numerous sources of one line (H9) identified some sublines for which most cells “appropriately” initiate inactivation upon differentiation. These H9 sublines are thus the most promising source for cells that retain competence to initiate X-inactivation, and potentially other developmental commitments. As cultures of two hES lines from Technion also showed a mosaic X-inactivation phenotype, we believe it likely that careful analysis and cultivation of these and other hES lines, may identify additional sublines that retain X-inactivation competence. While this NIH-funded study does not address whether non-NIH approved hES lines show a similar diversity of phenotypes, our prior collaborative study with the Carpenter lab (Hoffman et al., 2005) and a recent broad survey of gene expression by the International Stem Cell Initiative (Adewumi et al., 2007) suggest that this may also appear to be the case for non-NIH approved lines as well (also see note below). The very act of establishing and forcing the expansion of “non-differentiated” hES cells may change the epigenetic properties present in the native ICM, including a strong propensity to inactivate one of the two X chromosomes. Consistent with our findings of differences between lines, other studies have shown distinct gene expression profiles between lines and cultures that do not seem to be explained by differentiation status (Doherty et al., 2000; Bibikova et al., 2006). Although a recent report did not directly determine if hESC lines displaying precocious X-inactivation also exhibited abnormal status for other imprinted genes (Adewumi et al., 2007), it may be that in future efforts to establish developmentally naïve cells, either from ICM or from reversing the programming of more advanced cells, X-inactivation status may provide a valuable tool to evaluate the epigenetic status and competence of those cells.
Note Added in Proof
Silva et al. have a similar study, predominantly in non NIH-approved human ESC lines, in press in PNAS [Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X chromosome inactivation and epigenetic fluidity in human embryonic stem cells. 2008. PNAS (in press)]. This study also finds a variety of epigenetic anomalies in Xi in the hESC lines studied, as well as some lines that initiate X-inactivation in most cells upon differentiation. Thus, the epigenetic anomalies seen here are not exclusive to the NIH approved lines, but are a broader property of cultured hESC.
Supplementary Material
Acknowledgments
This research was supported by NIH grants GM53234 (Lawrence) and GM032010 (Stein) including supplements for human embryonic stem cell research, and partly supported by NIH grants GM069983-01 (Ware) and R24RR18405 (Itskovitz-Eldor). We thank Kohava Shariki for her technical assistance to the Itskovitz-Eldor lab.
Footnotes
This article includes Supplementary Material available from the authors upon request or via the Internet at http://www.interscience.wiley.com/jpages/0021-9541/suppmat.
Literature Cited
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, Blum B, Brooking J, Chen KG, Choo AB, Churchill GA, Corbel M, Damjanov I, Draper JS, Dvorak P, Emanuelsson K, Fleck RA, Ford A, Gertow K, Gertsenstein M, Gokhale PJ, Hamilton RS, Hampl A, Healy LE, Hovatta O, Hyllner J, Imreh MP, Itskovitz-Eldor J, Jackson J, Johnson JL, Jones M, Kee K, King BL, Knowles BB, Lako M, Lebrin F, Mallon BS, Manning D, Mayshar Y, McKay RD, Michalska AE, Mikkola M, Mileikovsky M, Minger SL, Moore HD, Mummery CL, Nagy A, Nakatsuji N, O’Brien CM, Oh SK, Olsson C, Otonkoski T, Park KY, Passier R, Patel H, Patel M, Pedersen R, Pera MF, Piekarczyk MS, Pera RA, Reubinoff BE, Robins AJ, Rossant J, Rugg-Gunn P, Schulz TC, Semb H, Sherrer ES, Siemen H, Stacey GN, Stojkovic M, Suemori H, Szatkiewicz J, Turetsky T, Tuuri T, van den Brink S, Vintersten K, Vuoristo S, Ward D, Weaver TA, Young LA, Zhang W. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Alexander MK, Panning B. Counting chromosomes: Not as easy as 1, 2, 3. Curr Biol. 2005;15:R834–R836. doi: 10.1016/j.cub.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Allegrucci C, Wu YZ, Thurston A, Denning CN, Priddle H, Mummery CL, Ward-van Oostwaard D, Andrews PW, Stojkovic M, Smith N, Parkin T, Edmondson Jones M, Warren G, Yu L, Brena RM, Plass C, Young LE. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16:1253–1268. doi: 10.1093/hmg/ddm074. [DOI] [PubMed] [Google Scholar]
- Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J, Thomson JA. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- Anderson CL, Brown CJ. Variability of X chromosome inactivation: effect on levels of TIMP1 RNA and role of DNA methylation. Hum Genet. 2002;110:271–278. doi: 10.1007/s00439-002-0676-8. [DOI] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, Gonzalez R, Crook J, Davidson B, Schulz TC, Robins A, Khanna A, Sartipy P, Hyllner J, Vanguri P, Savant-Bhonsale S, Smith AK, Chakravarti A, Maitra A, Rao M, Barker DL, Loring JF, Fan JB. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Tucker T, Thorogood NP, Brown CJ. Mechanisms of X-chromosome inactivation. Front Biosci. 2006;11:852–866. doi: 10.2741/1842. [DOI] [PubMed] [Google Scholar]
- Chow J, Hall LL, Lawrence JB, Brown CJ. Ectopic XIST transcripts in human somatic cells show variable expression and localization. Cytogene Genome Res. 2002;99:92–98. doi: 10.1159/000071579. [DOI] [PubMed] [Google Scholar]
- Chow J, Hall LL, Clemson C, Lawrence JB, Brown CJ. Characterization of expression at the human XIST locus in somatic, embryonal carcinoma and transgenic cell lines. Genomics. 2003;82:309–322. doi: 10.1016/s0888-7543(03)00170-8. [DOI] [PubMed] [Google Scholar]
- Chow JC, Yen Z, Ziesche SM, Brown CJ. Silencing of the mammalian X chromosome. Annu Rev Genomics Hum Genet. 2005;6:69–92. doi: 10.1146/annurev.genom.6.080604.162350. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Chow JC, Brown CJ, Lawrence JB. Stabilization and localization of Xist RNA are controlled by separate mechanisms and are not sufficient for X inactivation. J Cell Biol. 1998;142:13–23. doi: 10.1083/jcb.142.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, Zuccotti M, Kinis T, Serhal P, Monk M. XIST expression in human oocytes and preimplantation embryos. Am J Hum Genet. 1997;61:33–39. doi: 10.1086/513892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhara SK, Benvenisty N. Gene trap as a tool for genome annotation and analysis of X chromosome inactivation in human embryonic stem cells. Nucleic Acids Res. 2004;32:3995–4002. doi: 10.1093/nar/gkh746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- Enver T, Soneji S, Joshi C, Brown J, Iborra F, Orntoft T, Thykjaer T, Maltby E, Smith K, Dawud RA, Jones M, Matin M, Gokhale P, Draper J, Andrews PW. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14:3129–3140. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci USA. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation inmammals: Fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Hoffman LM, Hall L, Batten JL, Young H, Pardasani D, Baetge EE, Lawrence J, Carpenter MK. X-inactivation status varies in human embryonic stem cell lines. Stem Cells. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM, III, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Li T, Vu TH, Ulaner GA, Littman E, Ling JQ, Chen HL, Hu JF, Behr B, Giudice L, Hoffman AR. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol Hum Reprod. 2005;11:631–640. doi: 10.1093/molehr/gah230. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, van Gurp RJ, Verkerk AJ, Oosterhuis JW. X inactivation in human testicular tumors. XIST expression and androgen receptor methylation status. Am J Pathol. 1997;151:581–590. [PMC free article] [PubMed] [Google Scholar]
- Migeon BR, Kazi E, Haisley-Royster C, Hu J, Reeves R, Call L, Lawler A, Moore CS, Morrison H, Jeppesen P. Human X inactivation center induces random X chromosome inactivation in male transgenic mice. Genomics. 1999;59:113–121. doi: 10.1006/geno.1999.5861. [DOI] [PubMed] [Google Scholar]
- Migeon BR, Winter H, Kazi E, Chowdhury AK, Hughes A, Haisley-Royster C, Morrison H, Jeppesen P. Low-copy-number human transgene is recognized as an X inactivation center in mouse ES cells, but fails to induce cis-inactivation in chimeric mice. Genomics. 2001;71:156–162. doi: 10.1006/geno.2000.6421. [DOI] [PubMed] [Google Scholar]
- Mitalipov SM. Genomic imprinting in primate embryos and embryonic stem cells. Reprod Fertil Dev. 2006;18:817–821. doi: 10.1071/rd06112. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Ray PF, Winston RM, Handyside AH. XIST expression from the maternal X chromosome in human male preimplantation embryos at the blastocyst stage. Hum Mol Genet. 1997;6:1323–1327. doi: 10.1093/hmg/6.8.1323. [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, Avner P, Heard E. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Byron M, Clemson CM, Lawrence JB. Ubiquitinated proteins including uH2A on the human and mouse inactive X chromosome: Enrichment in gene rich bands. Chromosoma. 2004;113:324–335. doi: 10.1007/s00412-004-0325-1. [DOI] [PubMed] [Google Scholar]
- Tam R, Smith KP, Lawrence JB. The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatic unlike most human telomeres. J Cell Biol. 2004;167:269–279. doi: 10.1083/jcb.200403128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Nelson AM, Blau CA. Controlled-rate freezing of human ES cells. Biotechniques. 2005;38:879–880. 882–883. doi: 10.2144/05386ST01. [DOI] [PubMed] [Google Scholar]
- Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


