Abstract
The propionate utilization operons of several bacteria differ from each other in the occurrence of two genes, acnD and prpF, in place of or in addition to the prpD gene encoding an Fe/S-independent 2-methylcitrate dehydratase enzyme. We cloned the acnD and prpF genes from two organisms, Shewanella oneidensis and Vibrio cholerae, and found that, together, the AcnD and PrpF proteins restored the ability of a prpD mutant strain of Salmonella enterica to grow on propionate as a source of carbon and energy. However, neither acnD nor prpF alone was able to substitute for prpD. The AcnD and PrpF proteins were isolated and biochemically analyzed. The AcnD protein required reconstitution of an Fe/S cluster for activity. All detectable AcnD activity was lost after incubation with iron-chelating agents, and no AcnD activity was observed after attempted reconstitution without iron. Nuclear magnetic resonance spectroscopy and in vitro activity assay data showed that AcnD dehydrated 2-methylcitrate and citrate to 2-methyl-cis-aconitate and cis-aconitate, respectively; AcnD also hydrated cis-aconitate. However, 2-methylisocitrate and isocitrate were not substrates for AcnD, indicating that AcnD only catalyzes the first half of the aconitase-like dehydration reactions. No aconitase-like activity was found for PrpF. It is hypothesized that, in vivo, PrpF is an accessory protein required to prevent oxidative damage of the Fe/S center of active AcnD enzyme or that it may be involved in synthesis or repair of the Fe/S cluster present in AcnD.
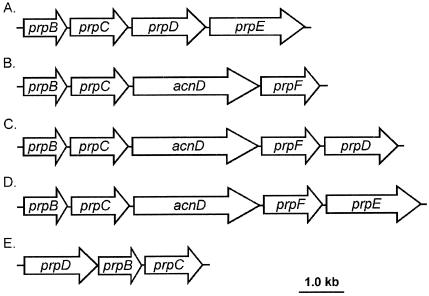
First demonstrated in Yarrowia lipolytica and several other filamentous fungi and yeast species (26, 32, 33), the 2-methylcitrate (2-MC) cycle was subsequently shown to occur in the bacteria Salmonella enterica serovar Typhimurium and Escherichia coli (20, 35). Since the finding that prokaryotes can also utilize the 2-MC cycle as their route of propionate catabolism, the 2-MC pathway was also demonstrated in other gram-negative bacteria such as Ralstonia eutropha (8) and Burkholderia sacchari (7) and in the gram-positive bacterium Corynebacterium glutamicium (10). Sequence analysis of the completed genomes of several other bacteria indicates that the 2-MC cycle may be widespread among bacteria; Vibrio cholerae, Shewanella oneidensis, Neisseria spp., and several Pseudomonas species contain propionate utilization (prp) operons (8, 17) (Fig. 1).
FIG. 1.
Structural variations on prp operons of several bacteria. (A) S. enterica serovar Typhimurium and E. coli (E. coli contains a 439-nt putative stem-loop region between prpB and prpC). (B) R. eutropha CH34, S. oneidensis MR-1, Neisseria meningitidis, Neisseria gonorrhea (the Neisseria spp. contain a 789-nt ORF of unknown function between prpC and acnD, which has been designated yfcA, and an ORF of 1,200 nt following prpF designated ackA that shows sequence similarity to propionate kinase [tdcD] of E. coli), and B. sacchari IPT101 (B. sacchari contains a putative ORF of 366 nt between acnD and prpF and two putative ORFs of 372 nt and 198 nt following prpF). (C) R. eutropha HF39, Bordetella pertussis, Pseudomonas aeruginosa, and Pseudomonas putida KT2440. (D) V. cholerae. (E) prpD2B2C2 operon of C. glutamicum. Putative regulators have been excluded from Fig. 1C and D, and the spaces between ORFs are not drawn to scale.
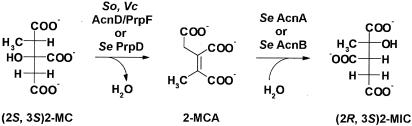
The prp operons of S. enterica and E. coli comprise four genes encoding structural proteins of the 2-MC cycle which have been characterized (Fig. 1A) as follows: prpB encodes 2-methylisocitrate (2-MIC) lyase (13, 14, 17), prpC encodes 2-MC synthase (17, 18, 35), prpD encodes 2-MC dehydratase (9, 17), and prpE encodes propionyl-coenzyme A synthetase (19). However, other prp operons have a gene organization that differs greatly from that of these two enterics (8, 17) (Fig. 1). Figure 1D shows an operon structure that contains two genes, acnD (17) and prpF, instead of prpD. In S. enterica, prpD encodes an Fe/S-independent 2-MC dehydratase that generates 2-methyl-cis-aconitate (2-MCA) from 2-MC but will not hydrate 2-MCA into 2-MIC (17). The hydration of 2-MCA is catalyzed by either aconitase AcnA or AcnB (Fig. 2) (17). All currently sequenced prp operons that contain an acnD ortholog also contain prpF and vice versa. The latter is an ortholog of open reading frame 5 (ORF5) of R. eutropa (8) and E. coli ybhH. The only work on acnD and prpF reported to date was performed in R. eutropha (8). The R. eutropha prp operon, represented in Fig. 1C, contains both the acnD and prpF genes and prpD. Brämer et al. recently reported that, in R. eutropha HF39, prpD was not required for a functional 2-MC cycle, but acnD and prpF (acnM and ORF5 in R. euthropha, respectively) functions were needed. However, efforts to demonstrate the conversion of 2-MC to 2-MIC by AcnM-enriched crude cell extracts were inconclusive (8).
FIG. 2.
Conversion of 2-MC to 2-MIC in bacteria. 2-MC and 2-MIC are drawn as Fischer projections; stereochemistry is based on intermediates of the E. coli 2-MC cycle (9). Se, S. enterica; Vc, V. cholerae; So, S. oneidensis.
In this paper we demonstrate that the acnD gene encodes an Fe/S-dependent 2-MC dehydratase enzyme that requires the prpF gene product to function in vivo. The acnD and prpF genes from V. cholerae and S. oneidensis were cloned independently or together and were used to compensate for the lack of the Fe/S-independent 2-MC dehydratase (PrpD) enzyme in S. enterica during growth on propionate. The AcnD and PrpF proteins were isolated. AcnD purified in the presence of air was inactive but was reactivated by protocols reported for the reactivation of aconitase (22). Reactivated AcnD had 2-MC dehydratase activity but no measurable 2-MIC dehydratase activity. Even though aconitase-like activity was not observed for PrpF in vitro, PrpF was required for the conversion of 2-MC into 2-MCA in vivo. Possible roles for the PrpF protein are discussed.
MATERIALS AND METHODS
Chemicals and culture media.
Cultures were maintained in Luria-Bertani (LB) broth and solid media. No-carbon E medium supplemented with MgSO4 (1 mM) and methionine (0.5 mM) was used as minimal medium (5, 11). Propionate and pyruvate were used at concentrations of 30 mM. Antibiotic concentrations in rich media were as follows (in μg/ml): ampicillin, 100; kanamycin, 25 (for plasmids, 50 μg/ml); tetracycline, 15; and chloramphenicol, 20. Bacterial strains harboring plasmids were grown in minimal media containing ampicillin (50 μg/ml) and kanamycin (30 μg/ml). Synthetic 2-MC was purchased from C/D/N Isotopes (Pointe-Claire, Quebec, Canada) as a mixture of stereoisomers; [2 -13C]propionate, 100% deuterium oxide (D2O), and tetramethylsilane were purchased from Cambridge Isotope Labs (Andover, Mass.). The 2-MIC was a gift from W. W. Cleland. All other chemicals were purchased from Sigma (St. Louis, Mo.) unless otherwise stated. A list of the strains and plasmids used and their genotypes is provided in Table 1.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| BL21(λDE3) | F−ampT hsdSB(rB− mB+) dcm gal λ(DE3) | New England Biolabs |
| DH5α/F′ | F′/endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15] | New England Biolabs |
| JE4570 | BL21(λDE3)/pPRP62 (S. enterica prpC+ in pET 15b bla+) | |
| JE4744 | BL21(λDE3)/pPRP67 (S. enterica prpD+ in pET15b bla+) | |
| S. enterica strains | ||
| TR6583b | metE205 ara-9 | K. Sanderson via J. Roth |
| Derivatives of TR6583c | ||
| JE3056 | prpB121::Tn10d(Tc) | 18 |
| JE3907 | prpC167 zai-6386::Tn10d(Tc)d | 18 |
| JE3909 | prpD169 zai-6386::Tn10d(Tc) | 18 |
| JE3946 | prpB195 zai-6386::Tn10d(Tc) | 18 |
| JE5993 | acnA2::cat+acnB3::kan+ | 17 |
| JE6501 | JE5993/pBAD30 (bla+) | |
| JE6502 | JE5993/pACN9 (S. enterica acnA+ in pBAD30 bla+) | |
| JE6503 | JE5993/pACN10 (S. enterica acnB+ in pBAD30 bla+) | |
| JE6504 | JE5993/pPRP121 (V. cholerae acnD+prpF+ in pBAD30 bla+) | |
| JE6506 | JE5993/pPRP123 (V. cholerae acnD+prpF+ in pBAD30 bla+) | |
| JE7235 | JE5993/pPRP138 (S. oneidensis acnD+ in pBAD30 bla+) | |
| JE7236 | JE5993/pPRP140 (S. oneidensis acnD+prpF+ in pBAD30 bla+) | |
| JE7357 | JE5993/pPRP163 (E. coli ybhJ+ in pBAD30 bla+) | |
| JE7590 | JE5993/pPRP166 (E. coli ybhHIJ+ in pBAD30 bla+) | |
| Plasmids | ||
| pBAD30 | ParaBAD expression vector, bla+ | 16 |
| pACN9 | S. enterica acnA+ in pBAD30 bla+ | 17 |
| pACN10 | S. enterica acnB+ in pBAD30 bla+ | 17 |
| pPRP12-5.4 | S. enterica prpBCD+ in pSU38 kan+ | 18 |
| pPRP21 | S. enterica prpB+ in pBAD30 bla+ | 18 |
| pPRP36 | S. enterica prpD+ in pBAD30 bla+ | 18 |
| pPRP35 | S. enterica prpC+ in pBAD30 bla+ | 18 |
| pPRP62 | S. enterica prpC+ in pET15b bla+ | 20 |
| pPRP67 | S. enterica prpD+ in pET 15b bla+ | 17 |
| pPRP121 | V. cholerae acnD+ in pBAD30 bla+ | |
| pPRP123 | V. cholerae acnD+prpF+ in pBAD30 bla+ | |
| pPRP138 | S. oneidensis acnD+ in pBAD30 bla+ | |
| pPRP140 | S. oneidensis acnD+prpF+ in pBAD30 bla+ | |
| pPRP141 | S. oneidensis prpB+prpC+acnD+prpF+ in pBAD30 bla+ | |
| pPRP149 | S. oneidensis prpB+ in pBAD18Kan kan+ | |
| pPRP150 | S. oneidensis prpC+ in pBAD18Kan kan+ | |
| pPRP151 | S. oneidensis prpB+prpC+ in pBAD 18Kan kan+ | |
| pPRP152 | S. oneidensis acnD+ in pTYB12 bla+ | |
| pPRP153 | S. oneidensis prpF+ in pBAD18Kan kan+ | |
| pPRP154 | E. coli ybhH+ in pBAD18Kan kan+ | |
| pPRP155 | V. cholerae prpF+ in pBAD18Kan kan+ | |
| pPRP156 | S. oneidensis prpF+ in pTYB12 bla+ | |
| pPRP163 | E. coli ybhJ+ in pBAD30 bla+ | |
| pPRP166 | E. coli ybhHIJ+ in pBAD30 bla+ |
All S. enterica strains are derivatives of the S. enterica serovar Typhimurium LT2 strain. Unless otherwise stated, strains and plasmids were constructed during the course of this work.
Formerly SA2979.
See Table 3 for additional strains (and their genotypes) used in this work.
Tn10d(Tc) is an abbreviation of Tn10DEL 16DEL17 (37).
Recombinant DNA techniques.
Restriction and modification enzymes were purchased from Promega (Madison, Wis.) unless otherwise stated and were used according to the manufacturer's specifications. All DNA manipulations were performed in E. coli strain DH5α/F′. Plasmids were transformed into S. enterica strains by a quick-electroporation method as follows. Strains were grown to approximately mid-exponential phase, and cells in 1.0 ml of culture were pelleted at 10,000 × g in a Microfuge 18 centrifuge (Beckman Coulter). Cells were washed once with 100 μl of cold H2O, resuspended in 100 μl of cold H2O, and allowed to equilibrate on ice for 5 min. Plasmids were electroporated into the competent cells with a Bio-Rad Gene Pulser (Hercules, Calif.), according to the manufacturer's recommendations.
Construction of plasmids.
Plasmids containing S. oneidensis or V. cholerae genes were constructed by PCR amplification of genomic DNA of S. oneidensis MR-1 (a gift from D. Saffarini, University of Wisconsin—Milwaukee) or V. cholerae N16961 (a gift from Ron Taylor, Dartmouth Medical School). PCRs typically contained the following in a 100-μl reaction mixture: 1.5 ng of genomic DNA, 50 pmol of each primer (IDTDNA, Coralville, Iowa), and deoxynucleoside triphosphate and HiFi DNA polymerase (Novagen, Madison, Wis.), each at a concentration of 0.2 μM, according to manufacturer's instructions. Reactions were performed under the following conditions: 35 cycles at 95°C for 30 s, at 50°C for 30 s, and at 72°C for 1 min per kb of target DNA. The PCR fragment was purified with a QIAquick PCR purification kit (QIAGEN, Chatsworth, Calif.). The methods of constructing the plasmids used are outlined in Table 2. The primer sequences used in plasmid constructions are available upon request.
TABLE 2.
Construction of plasmids used in this work
| Plasmid name | Vector backbone | Gene(s) cloned | Method of construction |
|---|---|---|---|
| pPRP117 | pDONR201a | V. cholerae acnD | Gateway BP reactionb |
| pPRP121 | pBAD30 | V. cholerae acnD | Gateway LR reaction |
| pPRP123 | pBAD30 | V. cholerae acnD prpF | KpnI, XbaI cloning |
| pPRP136 | pDONR201 | S. oneidensis acnDc | Gateway BP reaction |
| pPRP138 | pBAD30 | S. oneidensis acnD | Gateway LR reaction |
| pPRP137 | pDONR201 | S. oneidensis acnD prpF | Gateway BP reaction |
| pPRP140 | pBAD30 | S. oneidensis acnD prpF | Gateway LR reaction |
| pPRP141 | pBAD30 | S. oneidensis prpB prpC acnD prpF | SacI, XbaI cloning |
| pPRP149 | pBAD18-Kan | S. oneidensis prpB | SacI, XbaI cloning |
| pPRP150 | pBAD18-Kan | S. oneidensis prpC | SacI, XbaI cloning |
| pPRP151 | pBAD18-Kan | S. oneidensis prpB prpC | SacI, XbaI cloning |
| pPRP152 | pTYB12 | S. oneidensis prpF | BsmI, XhoI cloning |
| pPRP153 | pBAD18-Kan | S. oneidensis prpF | EcoRI, XbaI cloning |
| pPRP154 | pBAD18-Kan | E. colid ybhH | SacI, XbaI cloning |
| pPRP155 | pBAD18-Kan | V. cholerae prpF | KpnI, XbaI cloning |
| pPRP156 | pTYB12 | S. oneidensis acnD | BsmI, XhoI cloning |
| pPRP163 | pBAD30 | E. coli ybhJ | SacI, XbaI cloning |
| pPRP166 | pBAD30 | E. coli ybhH ybhl ybhJ | SacI, XbaI cloning |
pDONR201 was purchased from Invitrogen, Carlsbad, Calif.
Gateway BP and LR reactions are methods developed by Life Technologies, Invitrogen Corporation.
All S. oneidensis organisms are strain MR-1.
All E. coli organisms are strain MG1655.
Sequence verification of plasmid constructs.
All resulting plasmid constructs were sequenced to verify that no mutations were introduced into the genes of interest. PCR sequencing reactions were prepared with Big Dye (Biotechnology Center, University of Wisconsin—Madison). Reactions were purified by means of the CleanSEQ reaction clean-up protocol of Agencourt Bioscience Corporation (Beverly, Mass.) and sequenced at the Biotechnology Center. Searches for sequence similarity were performed by using the BLAST algorithm (1). Protein sequence alignments were conducted with the ClustalW multiple alignment tool (36).
Complementation analysis.
Plasmids were introduced into S. enterica strains as described above. The resulting strains were grown overnight in LB broth containing appropriate antibiotics. Four microliters of each overnight culture was used to inoculate 200 μl of fresh no-carbon E minimal medium supplemented with propionate (30 mM) and glycerol (1 mM) or acetate (30 mM), the appropriate antibiotic, and various amounts of l-(+)-arabinose (0, 100, or 500 μM). Medium was placed into the wells of a 96-well Falcon (Beckton Dickinson, Franklin Lakes, N.J.) microtiter dish, and the density of the cultures was monitored at 650 nm with a SpectraMAX Plus high-throughput spectrophotometer (Molecular Devices, Sunnyvale, Calif.). The plate chamber in the spectrometer was maintained at 37°C. Absorbance measurements were taken every 15 min for 72 h with agitation (for 780 s) between reads.
Anaerobic growth analysis.
For anaerobic growth experiments, LB plates containing 10 mM sodium tetrathionate as a terminal electron acceptor (27) were patched with the strains of interest and introduced into an anoxic environment with Gas-Pak jars (Bethesda Research Laboratories, Gaithersburg, Md.). Plates were incubated at 37°C for 24 h and then transferred into an anaerobic chamber, where they were replica printed onto minimal propionate medium plates supplemented with 10 mM tetrathionate and various concentrations of arabinose. Plates were incubated anoxically for 3 days at 37°C and growth was assessed.
Purification of Shewanella AcnD and PrpF proteins.
Plasmids pPRP152 (S. oneidensis prpF+) and pPRP156 (S. oneidensis acnD+) were introduced into E. coli BL21(λDE3) by the CaCl2 heat shock method described elsewhere (30). Cells (20 ml of an overnight culture) were inoculated into 2 liters of LB broth supplemented with 100 μg of ampicillin/ml and grown with shaking at 37°C. Cells were grown to an A600 of approximately 0.6, and the overproduction of proteins was induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cultures were harvested 3 h after induction for 10 min at 4°C and 11,700 × g. The cell pellets were frozen at −20°C for later use.
Cell pellets were resuspended in 25 ml of 20 mM (pH 7.5) 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (buffer A) (HEPES; Fisher Biotech, Itasca, Ill.), containing 100 mM KCl, 0.1% (vol/vol) Triton X-100, and 0.1 mM EDTA. The cell suspensions were broken at 104 kPa in a chilled French pressure cell. Cell debris was removed by centrifugation at 31,000 × g for 30 min at 4°C. Crude cell extracts were filtered through a 0.2-μm-pore-size filter and passed through a 5-ml column of chitin beads (New England Biolabs, Beverly, Mass.). The column was washed with buffer A according to the manufacturer's instructions and then quickly washed with 15 ml of buffer A containing 50 mM 1,4-dithio-dl-threitol (DTT; Promega, Madison, Wis.). The column flow was stopped, and the column was kept at 4°C for 96 h. Purified, untagged proteins were eluted off the column with buffer A containing 50 mM DTT and were visualized with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (23). Fractions containing protein were pooled and dialyzed overnight into 20 mM HEPES (pH 7.5), 100 mM KCl, 0.1 mM EDTA, and 5 mM DTT (buffer B). After dialysis for 2 h, the buffer was changed to buffer B lacking EDTA. The remaining part of the dialysis period was performed with buffer B containing 5% (vol/vol) glycerol. The protein was flash-frozen in liquid nitrogen and stored at −80°C.
Reactivation of purified proteins.
Purified AcnD was reactivated according to the method of Kennedy and Beinert (22). All reagents were prepared under strict anaerobic conditions (3, 15). Purified H6AcnA (17) was reactivated by the same procedure and was used as a positive control in the aconitase assays.
In vitro aconitase assays.
Aconitase assays were performed as described (17). Reaction mixtures contained 90 mM Tris (Tris-HCl) buffer (pH 8.0) and 20 mM citrate, isocitrate, 2-MC, 2-MIC, or 2 mM cis-aconitate in a 1-ml quartz cuvette. Reactivated enzyme was transferred to the cuvette with a 25-μl Hamilton syringe. No detectable loss of activity was observed during the duration of the assays (2 h). Reactions were monitored for 10 min at 240 nm in a Perkin-Elmer Lambda 40 spectrophotometer (Norwalk, Conn.) equipped with a circulating water bath, which maintained the temperature at 37°C. Specific activities were reported in micromoles per minute per milligram of protein and calculated from the extinction coefficients of 3,600 M−1 cm−1 for the cis-aconitate (21) and 4,500 M−1 cm−1 for the 2-methyl-cis-aconitate (2).
Requirement of an Fe/S cluster for AcnD activity.
To determine if iron was required for AcnD activity, AcnD was reactivated according to the protocol of Kennedy and Beinert (22) with iron excluded from the reactivation mixture. To determine the magnitude of inactivation of AcnD in the presence of iron-chelating agents, reactivated AcnD was incubated with EDTA and ferricyanide in the molar ratios of 1:50:20 (enzyme:EDTA:ferricyanide) as described by Kennedy and Beinert (22). Protein activity was assayed as described above.
H6PrpC, H6PrpD, and PrpE proteins.
The S. enterica H6PrpC and H6PrpD proteins were overproduced and purified as previously described (17, 20). The propionyl-coenzyme A synthetase (PrpE) enzyme was provided by V. J. Starai.
13C-NMR spectroscopy.
Peak assignments were compared to those previously reported (20). Glycerol was present in all protein samples and observed in the spectra due to the natural abundance of 13C (17). 13C-nuclear magnetic resonance (NMR) spectra were obtained at the National Magnetic Resonance Facility at the University of Wisconsin—Madison by means of a Bruker Instruments DMX-400 Avance console with a 9.4-T wide-bore magnet at 100.6 MHz.
In vitro enzymatic synthesis of [2-13C]MC.
[2-13C]MC was generated in vitro (17) in 0.5-ml reaction mixtures that contained potassium phosphate buffer (pH 7.5; 50 mM), ATP (2.5 mM), MgCl2 (5 mM), coenzyme A (2.5 mM), [2-13C]propionate (2.5 mM), oxaloacetate (5 mM), and homogeneous PrpE and H6PrpC proteins (75 μg each). To each sample, 0.1 ml of 100% D2O was added, and the reactions were transferred to 5-mm NMR tubes (Wilmad Glass, Buena, N.J.). A sealed tetramethylsilane capillary was added as an external reference.
Conversion of [2-13C]MC to [2-13C]MCA.
[2-13C]MC (∼2.5 mM) synthesized as described above was used as substrate for PrpF and reactivated AcnD or for H6PrpD. Reaction mixtures (0.5 ml) contained [2-13C]MC, H6PrpD, PrpF, or reactivated AcnD (25 μg each) or PrpF and reactivated AcnD (25 μg each). The reaction mixtures were allowed to incubate for 1 h at 37°C and were prepared for 13C-NMR analysis as described above.
Other procedures.
Protein concentrations were determined from a standard curve generated with bovine serum albumin by the method of Bradford (6) with the Bio-Rad protein reagent. Proteins were separated by SDS-12% PAGE and stained with Coomassie blue (28). Novagen Perfect Protein markers (Madison, Wis.) were used as standards for SDS-PAGE.
RESULTS
S. oneidensis and V. cholerae acnD and prpF gene product functions restore growth of a S. enterica prpD mutant strain on propionate.
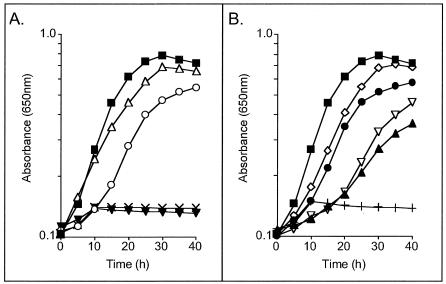
As pointed out above, prp loci from various prokaryotes contain two genes (acnD and prpF) in lieu of prpD (Fig. 1). The acnD and prpF genes from S. oneidensis and V. cholerae were cloned and used to determine whether they would compensate for the lack of PrpD function during growth of an S. enterica prpD mutant strain on propionate. The acnD and prpF genes were cloned into plasmids under the control of arabinose-inducible promoters either as a pair or individually on compatible plasmids. Plasmids carrying these genes were introduced into S. enterica strain JE3909 (prpD), and growth on propionate was assessed. Table 3 shows the doubling times of all strains tested. Both S. oneidensis acnD+prpF+ and V. cholerae acnD+prpF+ constructs complemented strain JE3909 (Fig. 3A). When the acnD and prpF genes were carried in separate plasmids, however, growth on propionate was observed only when both genes were present in the cell. In all cases when S. oneidensis and V. cholerae genes were mixed, strain JE3909 was able to grow on propionate (Fig. 3B). This result was not surprising considering that the S. oneidensis and V. cholerae AcnD and PrpF share 76 and 73% identity, respectively. These results indicate that both acnD and prpF are required to complement a prpD strain of S. enterica.
TABLE 3.
Heterologous complementation of S. enterica prp mutant strains
| Strain | Genotypea | Arabinose concentrationb (μM) | Doubling timec (h ± SD) |
|---|---|---|---|
| JE4175 | TR6583 (wild type)/pBAD30 | 500 | 7.3 ± 0.3 |
| JE6107 | JE3909/pPRP36 (S. enterica prpD+) | 500 | 7.6 ± 0.3 |
| JE7280 | JE3909/pPRP138 (S. oneidensis acnD+), pPRP153 (S. oneidensis prpF+) | 500 | 7.9 ± 0.3 |
| JE7232 | JE3909/pPRP140 (S. oneidensis acnD+prpF+) | 500 | 8.0 ± 0.5 |
| JE7227 | JE3909/pPRP141 (S. oneidensis prpB+prpC+acnD+prpF+) | 500 | 9.2 ± 1.0 |
| JE7282 | JE3909/pPRP138 (S. oneidensis acnD+), pPRP155 (V. cholerae prpF+) | 500 | 20.4 ± 1.2 |
| JE7283 | JE3909/pPRP121 (V. cholerae acnD+), pPRP153 (S. cholerae prpF+) | 100 | 11.0 ± 0.5 |
| JE7285 | JE3909/pPRP121 (V. cholerae acnD+), pPRP155 (V. cholerae prpF+) | 500 | 25.8 ± 2.0 |
| JE7234 | JE3909/pPRP123 (V. cholerae acnD+prpF+) | 100 | 13.0 ± 0.4 |
| JE6105 | JE3909/pBAD30 | 500 | NG |
| JE7238 | JE3056/pPRP12-5.4 (S. enterica prpB+prpC+prpD+) | 0 | 15.8 ± 0.5 |
| JE7274 | JE3056/pPRP151 (S. oneidensis prpB+prpC+), pPRP36 (S. oneidensis prpD+) | 100 | 12.7 ± 0.3 |
| JE7275 | JE3056/pPRP151 (S. oneidensis prpB+prpC+), pPRP140 (S. oneidensis acnD+prpF+) | 100 | 19.2 ± 0.4 |
| JE7277 | JE3056/pPRP151 (S. oneidensis prpB+prpC+), pPRP123 (V. cholerae acnD+prpF+) | 100 | 14.7 ± 0.5 |
| JE7239 | JE3056/pPRP141 (S. oneidensis prpB+prpC+acnD+prpF+) | 100 | 27.0 ± 1.4 |
| JE5585 | JE3056/pBAD30 | 500 | NG |
| JE7242 | JE3056/pPRP151 (S. oneidensis prpB+prpC+) | 500 | NG |
| JE5297 | JE3946/pPRP21 (S. oneidensis prpB+) | 500 | 9.7 ± 0.3 |
| JE7252 | JE3946/pPRP149 (S. oneidensis prpB+) | 100 | 11.3 ± 0.4 |
| JE7253 | JE3946/pPRP151 (S. oneidensis prpB+prpC+) | 100 | 10.5 ± 0.5 |
| JE5296 | JE3946/pBAD30 | 500 | NG |
| JE7270 | JE3907/pPRP35 (S. enterica prpC+) | 100 | 11.0 ± 0.2 |
| JE7254 | JE3907/pPRP150 (S. oneidensis prpC+) | 500 | 12.6 ± 0.3 |
| JE7255 | JE3907/pPRP151 (S. oneidensis prpB+prpC+) | 500 | 11.6 ± 0.4 |
| JE7251 | JE3907/pBAD30 | 500 | NG |
All plasmids except pPRP12-5.4 (S. enterica prpBCD+) contain arabinose-inducible promoters. Where two plasmids are indicated, they are of different incompatiblity groups. The following strains were used to construct the strains tested: JE3909 [metE205 ara-9 prpD169 zai-6386::Tn10d(Tc)], JE3907 [metE205 ara-9 prpC167 zai-6386::Tn10d(Tc)], JE3056 [metE205 ara-9 prpB121::Tn10d(Tc)], and JE3946 [metE205 ara-9 prpB195 zai-6386::Tn10d(Tc)].
Each construct was tested in media with 100 and 500 μM arabinose; the highest growth rate was included in this table.
JE7230 (JE3909/pPRP138 [S. oneidensis acnD+]), JE7271 (JE3909/pPRP153 [S. oneidensis prpF+]), JE7233 (JE3909/pPRP121 [V. cholerae acnD+]), JE7273 (JE3909/pPRP155 [V. cholerae prpF+]), JE7523 (JE3909/pPRP166 [E. coli ybhHIJ+]), JE7356 (JE3909/pPRP163 [E. coli. ybhJ+]), JE7272 (JE3909/pPRP154 [E. coli ybhH+]), or JE7359 (JE3909/pPRP154 [E. coli ybhH+], pPRP163 [E. coli ybhJ+]) did not grow in a propionate medium supplemented with 500 μM arabinose. NG, no growth observed (doubling times greater than 50 h).
FIG. 3.
Heterologous complementation studies. All curves shown are strain JE3909 (S. enterica prpD) with plasmids in trans. Filled squares, pPRP21 (S. enterica prpD+); open triangles, pPRP123 (V. cholerae acnD+ prpF+); open circles, pPRP140 (S. oneidensis acnD+ prpF+); filled inverted triangles, pPRP138 (S. oneidensis acnD+); X, pPRP153 (S. oneidensis prpF+); open diamonds, pPRP121 (V. cholerae acnD+) + pPRP153 (S. oneidensis prpF+); filled circles, pPRP138 (S. oneidensis acnD+) + pPRP153 (S. oneidensis prpF+); open inverted triangles, pPRP138 (S. oneidensis acnD+) + pPRP155 (V. cholerae prpF+); filled triangles, pPRP121 (V. cholerae acnD+) + pPRP155 (V. cholerae prpF+); plus signs, pPRP163 (E. coli ybhJ+) + pPRP154 (E. coli ybhH+).
To determine whether PrpF function was required along with AcnD function to complement an S. enterica prpD mutant grown on a propionate medium anaerobically, these strains were replica printed onto a propionate medium containing tetrathionate as a terminal electron acceptor. Only the positive control (prpD+) and strains containing both the acnD+ and prpF+ plasmids grew anaerobically on propionate. These data indicate that even under anaerobic conditions, both AcnD and PrpF functions are required to complement an S. enterica prpD mutant.
The possibility that the E. coli ybhJ and ybhH genes could restore the growth of strain JE3909 on propionate was also assessed. The E. coli YbhJ protein is an AcnA homolog that shares 22% identity (37% similarity) with S. oneidensis AcnD; the YbhH protein shares 34% identity (47% similarity) with S. oneidensis PrpF. The E. coli ybhJ and ybhH genes were cloned individually and as an operon (ybhHIJ) under the control of an arabinose-inducible promoter. None of the plasmids tested restored the growth of strain JE3909 on propionate (Table 3). Strain JE3909 failed to grow on propionate when E. coli ybhJ was introduced with S. oneidensis or V. cholerae prpF or when E. coli ybhH was added with S. oneidensis or V. cholerae acnD. These data indicated that the ybhHIJ operon of E. coli did not convert the 2-MC generated by S. enterica into 2-MCA or at least did not convert enough to complement the growth phenotype on propionate. The role of the ybhHIJ operon in E. coli remains unclear.
S. oneidensis PrpB (2-MIC lyase) and PrpC (2-MC synthase) restore growth of S. enterica prpB and prpC mutant strains on propionate.
It was of interest to determine whether S. oneidensis prpB (a 2-MIC lyase ortholog) and prpC (a 2-MC synthase ortholog) could complement S. enterica prpB or prpC mutant strains. S. oneidensis prpB and prpC were cloned as a pair into plasmid pBAD18Kan and introduced into strains JE3946 (prpB) and JE3907 (prpC) (Table 3). The resulting S. enterica strains (JE7253 and JE7255, respectively) grew on propionate, suggesting that the same stereoisomer of 2-MC was synthesized in S. enterica and S. oneidensis and that the PrpB enzymes from both organisms most likely use the same stereoisomer of 2-MIC. It has been shown that E. coli only synthesizes the 2S,3S isomer of 2-MC and generates the 2R,3S isomer of 2-MIC (9), and because S. enterica PrpB and PrpC are greater than 91 and 96% identical, respectively, to the corresponding proteins in E. coli, it is inferred that the same stereoisomers of the 2-MC cycle intermediates are produced in these organisms. The same results were obtained when S. oneidensis prpB or prpC were cloned individually and introduced into S. enterica JE3946 (JE7252) and JE3907 (JE7254), respectively (Table 3).
AcnD and PrpF do not substitute for TCA cycle aconitase.
It was also tested whether AcnD could substitute for S. enterica AcnA or AcnB during growth conditions that demanded a functional tricarboxylic acid (TCA) cycle. S. oneidensis or V. cholerae acnD individually or in combination with prpF were introduced into strain JE5993 (acnA acnB) (17). The resulting strains (JE6504, JE6506, JE7235, and JE7236) failed to grow on pyruvate and various concentrations of arabinose (as inducer). Only the control strains JE6502 (JE5993/pACN9 S. enterica acnA+) and JE6503 (JE5993/pACN10 S. enterica acnB+) grew on pyruvate under the conditions tested. Hence, it was concluded that the acnD and prpF genes cannot compensate for the lack of aconitase activity required for a functional TCA cycle of S. enterica, at least not to the level required for growth on pyruvate. No growth on pyruvate was observed when the E. coli ybhJ and ybhH genes or the complete ybhHIJ operon were introduced into strain JE5993.
Purification of AcnD and PrpF proteins.
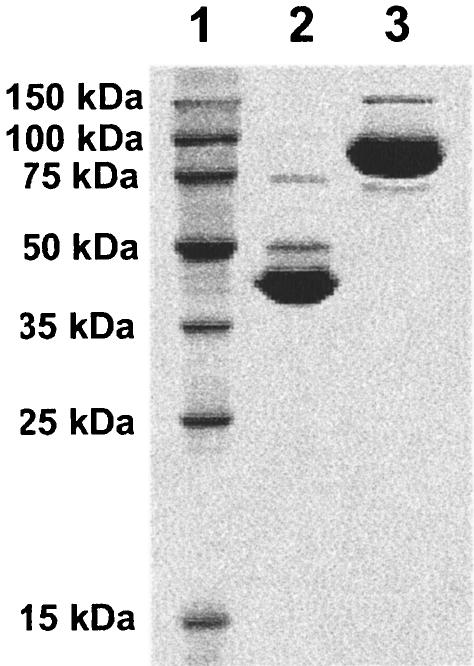
The putative biochemical activity of the AcnD and PrpF proteins was investigated in vitro. For this purpose, the S. oneidensis acnD and prpF genes were cloned and their products produced with an N-terminal chitin-binding tag (plasmids pPRP152 and pPRP156, respectively). Both proteins were purified by chitin affinity chromatography, and the tag was cleaved at the intein site according to the manufacturer's recommendations (New England Biolabs). Each protein was >95% pure as judged by scanning densitometry (Fig. 4). The relative mobility of both proteins was consistent with their predicted molecular masses, i.e., AcnD was observed at ∼94 kDa, and PrpF was observed at ∼42 kDa.
FIG. 4.
SDS-PAGE of purified S. oneidensis AcnD and PrpF proteins. Lane 1, MW standards (Novagen); lane 2, purified PrpF; lane 3, purified AcnD.
Enzymatic activity of AcnD.
Purified AcnD and H6AcnA (17) were reactivated as described in Materials and Methods and assayed spectrophotometrically for activity with various substrates (Table 4). AcnD used citrate, cis-aconitate, and 2-MC as substrates but not 2-MIC or isocitrate, indicating that, like PrpD (17), AcnD only catalyzes the first half of the aconitase-like reaction. The specific activity of AcnD for 2-MC was approximately 2.5-fold higher than that for citrate. The specific activity measured with 2-MC as substrate may be an underestimate of AcnD activity since commercially available 2-MC contained a mixture of stereoisomers, some of which may be inhibitory to the enzyme. On the other hand, AcnA used as positive control readily dehydrated 2-MIC, isocitrate, and citrate and hydrated cis-aconitate. In agreement with previous work, AcnA did not use 2-MC as substrate (17).
TABLE 4.
Specific activities of AcnD and AcnAa with different substrates
| Substrate | AcnD | AcnA |
|---|---|---|
| Citrate | 2.9 ± 0.5 | 26.5 ± 4.0 |
| cis-Aconitate | 5.0 ± 0.2 | 68.5 ± 7.4 |
| Isocitrate | NDb | 43.4 ± 3.1 |
| 2-MC | 7.8 ± 0.4 | ND |
| 2-MIC | ND | 12.7 ± 1.4 |
AcnD and AcnA were reconstituted as stated in Materials and Methods.
ND, no detectible activity observed (limits of detection were 0.5 μM/min for cis-aconitate conversion and 0.4 μM/min for 2-MCA formation) under the conditions tested.
AcnD is an Fe/S 2-MC dehydratase.
AcnD activity was only observed after anoxic reactivation with iron, sulfide, and reductant. When reactivation was attempted in the absence of iron, no enzymatic activity was observed. The activity of reconstituted AcnD was lost over time in the presence of air. Also, when ferricyanide and EDTA were added to the enzyme after reactivation, all detectable activity was lost within 15 min of incubation. The primary amino acid sequence of AcnD contains 22 of the 23 residues found at the active site of mitochondrial aconitase (mAcn), including the three cysteine residues that coordinate the 4Fe/4S cluster of mAcn (24). Taken together, these data indicated that in its active form AcnD contains an Fe/S center.
13C-NMR analysis of the AcnD reaction product.
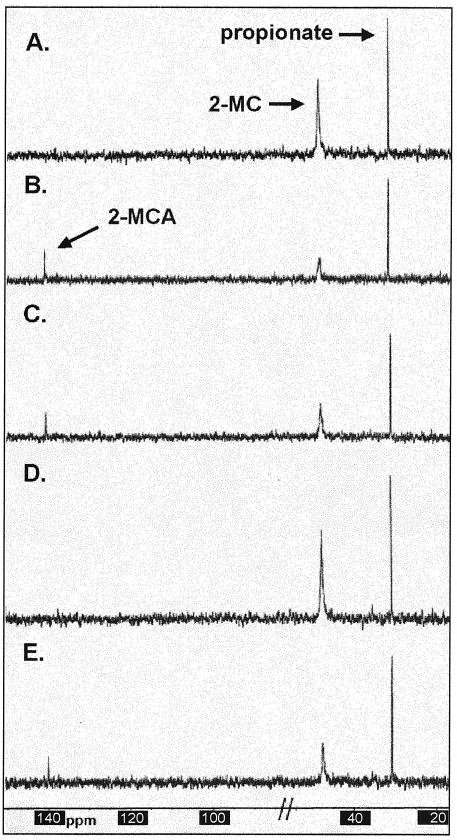
[2-13C]MC was synthesized in vitro as described (17). Reactivated AcnD was added to this reaction, incubated for 1 h, and analyzed by 13C-NMR spectroscopy. Peak assignments were based on those identified previously (17). Excluding two glycerol peaks (glycerol was added to the enzyme as cryoprotectant), only three peaks were observed: for [2-13C]propionate (30.8 ppm), [2-13C]MC (47.5 ppm), and [2-13C]MCA (141.5 ppm) (Fig. 5C). A positive control experiment for [2-13C]MCA production was performed with H6PrpD (Fig. 5B). The peaks observed with the H6PrpD-catalyzed reaction matched exactly the shifts of those seen with the AcnD reaction. The reaction mixture containing only [2-13C]MC had peaks at 30.8 and 47.5 ppm (Fig. 5A). These data indicated that both AcnD and PrpD had 2-MC dehydratase activity. To test whether the PrpF protein had aconitase-like enzymatic activity, PrpF was added to the [2-13C]MC reaction mixture and incubated for 1 h at 37°C. No peak shift or decrease in the 2-MC signal at 47.5 ppm was observed (Fig. 5D). PrpF was also added to the reaction mixture containing reactivated AcnD. No peak shift or decrease in the 2-MCA signal at 141.5 ppm was observed relative to the signal observed in the reaction mixture containing only AcnD (Fig. 5E). These results indicated that PrpF did not convert 2-MCA into 2-MIC or catalyze the conversion of 2-MC into 2-MCA under the assay conditions tested.
FIG. 5.
13C-NMR spectra of in vitro reactions. The composition of the reaction mixtures is described under Materials and Methods. Reaction A contained the S. enterica PrpE and PrpC enzymes; reaction B contained the S. enterica PrpE, PrpC, and PrpD enzymes; reaction C contained the S. enterica PrpE, PrpC, and reactivated S. oneidensis AcnD enzymes; reaction D contained the S. enterica PrpE and PrpC enzymes and the S. oneidensis PrpF protein; reaction E contained the S. enterica PrpE and PrpC enzymes and the S. oneidensis AcnD enzyme and PrpF protein. Chemical shifts expressed in parts per million are as follows: [2-13C]propionate 30.8 ppm; [2-13C]MC, 47.5 ppm; and [2-13C]MCA, 141.5 ppm. The portion of the spectra removed contained two glycerol peaks as previously described (17).
Probing for a role for the PrpF protein in propionate metabolism.
The possibility that the PrpF protein could catalyze the dehydration of citrate, isocitrate, 2-MC, or 2-MIC or the hydration of cis-aconitate was investigated. Even though PrpF does not contain an apparent Fe/S cluster binding motif, anoxic reconstitution of an Fe/S center was attempted. The PrpF protein with or without anoxic iron and sulfide reactivation did not have any detectable amounts of dehydratase or hydratase activities. PrpF was also added in twofold molar excess to AcnD in the cis-aconitate hydratase assay. No increase in the AcnD-catalyzed rate or in the overall conversion of substrate was observed.
We also tested whether AcnD required PrpF to catalyze the second half of the aconitase-like reaction, i.e., the conversion of 2-MCA to 2-MIC. PrpF was added to AcnD under anoxic and oxic conditions and was tested in the 2-MIC dehydratase assay. No 2-MIC dehydratase activity was observed.
We also looked into the possibility that PrpF could stabilize AcnD activity in the presence of oxygen. A twofold molar excess PrpF protein was added to anoxic AcnD. The mixture was incubated for 5 min before the seal was removed. Assays were performed, along with the control experiment with a reaction mixture that lacked PrpF protein. No significant differences in AcnD activity were observed over 2 h, and approximately 75% of AcnD (cis-aconitate hydratase) activity was lost over this period of time (data not shown). Work is currently being conducted to elucidate the role of PrpF in the 2-MC cycle.
DISCUSSION
This study established, both in vitro and in vivo, the biochemical activity of a new enzyme involved in the 2-MC cycle of several prokaryotes. The genes acnD and prpF of S. oneidensis and V. cholerae, when concurrently expressed, compensate for the lack of the Fe/S-independent PrpD enzyme in S. enterica prpD mutant strains during growth on propionate. The AcnD protein from S. oneidensis was isolated and shown to have a new activity for an enzyme containing an Fe/S center. AcnD catalyzes the dehydration of 2-MC and citrate but does not catalyze the dehydration of 2-MIC or isocitrate (Table 4). 13C-NMR spectroscopy of reactivated AcnD with [2-13C]MC revealed that AcnD could utilize the 2-MC generated by S. enterica PrpC and that the product of the AcnD reaction matched that produced by S. enterica PrpD (2-MCA) (Fig. 5). To our knowledge, this is the first report of an Fe/S-dependent 2-MC dehydratase.
To date, the only reported work on an AcnD homolog was performed with the acnM gene from R. eutropha (R. eutropha AcnM shares 83% identity with S. oneidensis AcnD) expressed in a crude extract system in E. coli. E. coli crude extracts containing R. eutropha AcnM protein were found to have cis-aconitate hydratase activity, but the data were inconclusive as to whether AcnM could dehydrate 2-MC. It was concluded, however, that AcnM may catalyze the hydration of 2-MCA into 2-MIC (8). In contrast, the data reported in this paper indicate that AcnD (and by extrapolation, AcnM) most likely does not catalyze the hydration of 2-MCA to 2-MIC because the enzyme will not dehydrate 2-MIC, and aconitases are known to catalyze freely reversible reactions. Additional support for this conclusion comes from NMR experiments where no evidence was obtained to indicate that active AcnD protein can convert 2-MC into 2-MIC. The only signal observed in the experiments was that of 2-MCA.
Studies on mammalian aconitases (mAcn) and AcnA and AcnB from both S. enterica and E. coli have demonstrated that these enzymes will not catalyze the dehydration of 2-MC; however, they will dehydrate 2-MIC and catalyze the full conversion of citrate into cis-aconitate into isocitrate (4, 9, 17, 29). The mechanism of aconitases is known to proceed by the binding of cis-aconitate in two ways to achieve the trans elimination or addition of water across the double bond (29). For this to occur, the substrate (cis-aconitate) must rotate 180°. The crystal structure of mAcn bound with 2-MIC allowed the prediction that if 2-MCA were rotated into the analogous 2-MC position, 2-MC would not be able to bind in the active site due to a steric clash of the methyl group with residue Asp165 (25). Interestingly, when aligned with mAcn, AcnD also contains this conserved aspartate residue, along with 21 of the other 22 active site residues of mAcn (24). It has been noted that all sequenced acnD homologs contain an Asn residue directly following one of the Cys residues that is likely to coordinate the Fe/S cluster, while in aconitases of the tricarboxylic acid cycle an Ile residue is found at this position (7). Experiments to test whether this residue plays a role in the substrate specificity of the AcnD enzyme are in progress.
It has been reported that horse heart aconitase can catalyze the formation of 2-MC and 2-MIC from 2-MCA (12). These data are in contradiction to the present information on aconitases, but the possibility exists that one enzyme may catalyze the complete conversion of 2-MC to 2-MIC. In Y. lipolytica, two enzymes, a 2-MC dehydratase and a 2-MIC dehydratase, have been implicated in its 2-MC cycle; both of these enzymes were found not to contain an Fe/S cluster or to be inactivated by iron-chelating agents (31, 34). To our knowledge, an Fe/S-independent 2-MIC dehydratase has not been identified in any other organism.
Possible roles for PrpF in propionate catabolism.
The possibility that PrpF may be an isomerase of one of the intermediates of the 2-MC cycle was considered. However, it is unlikely that PrpF is a 2-MIC isomerase because the prpB gene from S. oneidensis complemented a prpB mutant strain of S. enterica, and NMR and spectrophotometric data suggested that AcnD only catalyzed the conversion of (2S,3S)-MC into 2-MCA. It was also possible that PrpF could be a 2-MC isomerase. However, our data showed that the S. oneidensis prpC+ allele complemented an S. enterica prpC mutant strain, suggesting that the PrpC protein from S. oneidensis generated the same stereoisomer of 2-MC as that generated by the S. enterica PrpC enzyme. If the S. oneidensis PrpC protein generated a different 2-MC stereoisomer, it would be unlikely that the S. enterica PrpD enzyme would be able to use it as substrate. In support of this hypothesis, Brock et al. showed that the true substrate of the E. coli PrpD protein is most likely (2S,3S)-MC, and a 10-fold decrease in PrpD activity was observed when a racemic mixture of 2-MC stereoisomers was used (9). To further test these ideas, the stereochemical configuration of the reaction product of S. oneidensis PrpC must be determined.
Brämer et al. observed that PrpF was weakly similar (24%) to the pduG gene product of S. enterica, the proposed reactivation factor of diol dehydratase (7). Although this similarity is very weak, PrpF may be involved in AcnD Fe/S cluster formation or repair. As shown above, in vivo, PrpF must accompany AcnD to compensate for the lack of the PrpD enzyme during growth of a prpD mutant strain of S. enterica on propionate. If PrpF is required to stabilize AcnD in the presence of air, then growth under anoxic conditions should bypass the need for PrpF. However, PrpF was required even under anoxic growth conditions, suggesting that PrpF may not be needed to protect the AcnD Fe/S cluster from oxidation, but it could be involved in the formation, insertion, or stabilization of the Fe/S cluster. At present, the role of PrpF in propionate metabolism remains to be determined.
Bacteria use different strategies to convert 2-MC into 2-MIC.
It is interesting that the sequenced prp operons of several bacteria contain both prpD and the acnD/prpF genes. Since these gene products catalyze the same reaction, i.e., the conversion of 2-MC into 2-MCA, they would appear to perform redundant functions in these organisms. Why would these organisms employ this strategy? Perhaps PrpD, the Fe/S-independent 2-MC dehydratase, is needed by these organisms at times when oxygen levels are high in the cell and may be deleterious to AcnD. Then why do these organisms retain the acnD/prpF pair of genes? Maybe having both of these ways to convert 2-MC into 2-MCA allows for more efficient growth on carbon sources that require the 2-MC cycle as a route of metabolism. Or perhaps the acnD/prpF gene products do carry out the second half of the aconitase-like reaction and convert 2-MCA into 2-MIC, which may not have been apparent in vitro in this study, in addition to the AcnD 2-MC dehydratase activity observed. If these gene products only function to convert 2-MC into 2-MCA, then an enzyme outside of the prp operons, most likely one of the aconitases of the cell (most bacteria contain more than one aconitase), must catalyze the conversion of 2-MCA into 2-MIC. It has been described for Y. lipolytica that an Fe/S-independent enzyme catalyzes the conversion of 2-MCA into 2-MIC (34). Perhaps some bacteria that utilize the 2-MC cycle also contain a gene encoding an Fe/S-independent 2-MIC dehydratase. Elucidating the function of PrpF both in vivo and in vitro may provide the answers to some of these questions.
Acknowledgments
This work was supported by grant GM62203 from the NIGMS to J.C.E.-S. NMR spectroscopy studies were performed at the National Magnetic Resonance Facility at Madison, which is supported by the NIH Biomedical Technology Program (RR02301), with additional equipment funding from the University of Wisconsin, NSF Academic Infrastructure Program (BIR-9214394), NIH Shared Instrumentation Program (RR02781, RR08438), NSF Biological Instrumentation Program (DMB-8415048), and U.S. Department of Agriculture.
We thank A. R. Horswill for the AcnA protein, V. J. Starai for the PrpE protein, and W. W. Cleland for 2-MIC. We thank D. Saffarini and R. Taylor for their gifts of DNA.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, and E. W. Myers. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, H., H. Uchiyama, H. Umetsu, and T. Tabuchi. 1995. Isolation of 2-methylcitrate dehydratase, a new enzyme serving in the methylcitric acid cycle for propionate metabolism, from Yallowia lipolytica. Biosci. Biotechnol. Biochem. 59:1825-1828. [Google Scholar]
- 3.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beach, R. L., T. Aogaichi, and G. W. E. Plaut. 1977. Identification of d-threo-α-methylisocitrate as stereochemically specific substrate for bovine heart aconitase and inhibitor of TPN-linked isocitrate dehydrogenase. J. Biol. Chem. 252:2702-2709. [PubMed] [Google Scholar]
- 5.Berkowitz, D., J. M. Hushon, H. J. Whitfield, J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-255. [DOI] [PubMed] [Google Scholar]
- 7.Brämer, C. O., L. F. Silva, J. G. Gomez, H. Priefert, and A. Steinbüchel. 2002. Identification of the 2-methylcitrate pathway involved in the catabolism of propionate in the polyhydroxyalkanoate-producing strain Burkholderia sacchari IPT101(T) and analysis of a mutant accumulating a copolyester with higher 3-hydroxyvalerate content. Appl. Environ. Microbiol. 68:271-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brämer, C. O., and A. Steinbüchel. 2001. The methylcitric acid pathway in Ralstonia eutropha: new genes identified involved in propionate metabolism. Microbiology 147:2203-2214. [DOI] [PubMed] [Google Scholar]
- 9.Brock, M., C. Maerker, A. Schütz, U. Völker, and W. Buckel. 2002. Oxidation of propionate to pyruvate in Escherichia coli: involvement of methylcitrate dehydratase and aconitase. Eur. J. Biochem. 269:6184-6194. [DOI] [PubMed] [Google Scholar]
- 10.Claes, W. A., A. Pühler, and J. Kalinowski. 2002. Identification of two prpDBC gene clusters in Corynebacterium glutamicum and their involvement in propionate degradation via the 2-methylcitrate cycle. J. Bacteriol. 184:2728-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, R. W., D. Botstein, and J. R. Roth. 1980. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Gawron, O., and K. P. Mahajan. 1966. Alpha-methyl-cis-aconitic acid. Aconitase substrate. II. Substrate properties and aconitase mechanism. Biochemistry 5:2343-2350. [DOI] [PubMed] [Google Scholar]
- 13.Grimek, T. L., H. M. Holden, I. Rayment, and J. C. Escalante-Semerena. 2003. Residues C123 and D58 of the 2-methylisocitrate lyase (PrpB) enzyme of Salmonella enterica are essential for catalysis. J. Bacteriol. 185:4837-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimm, C., A. Evers, M. Brock, C. Maerker, G. Klebe, W. Buckel, and K. Reuter. 2003. Crystal structure of 2-methylisocitrate lyase (PrpB) from Escherichia coli and modelling of its ligand bound active centre. J. Mol. Biol. 328:609-621. [DOI] [PubMed] [Google Scholar]
- 15.Gunsalus, R. P., S. M. Tandon, and R. S. Wolfe. 1980. A procedure for anaerobic column chromatography employing an anaerobic Freter-type chamber. Anal. Biochem. 101:327-331. [DOI] [PubMed] [Google Scholar]
- 16.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horswill, A. R., and J. C. Escalante-Semerena. 2001. In vitro conversion of propionate to pyruvate by Salmonella enterica enzymes: 2-methylcitrate dehydratase (PrpD) and aconitase enzymes catalyze the conversion of 2-methylcitrate to 2-methylisocitrate. Biochemistry 40:4703-4713. [DOI] [PubMed] [Google Scholar]
- 18.Horswill, A. R., and J. C. Escalante-Semerena. 1997. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J. Bacteriol. 179:928-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horswill, A. R., and J. C. Escalante-Semerena. 1999. The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology 145:1381-1388. [DOI] [PubMed] [Google Scholar]
- 20.Horswill, A. R., and J. C. Escalante-Semerena. 1999. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J. Bacteriol. 181:5615-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy, M. C., M. H. Emptage, J. L. Dreyer, and H. Beinert. 1983. The role of iron in the activation-inactivation of aconitase. J. Biol. Chem. 258:11098-11105. [PubMed] [Google Scholar]
- 22.Kennedy, M. C., and H. Beinert. 1988. The state of cluster SH and S2− of aconitase during cluster interconversions and removal: a convenient preparation of apoenzyme. J. Biol. Chem. 263:8194-8198. [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage and structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Lauble, H., M. C. Kennedy, H. Beinert, and C. D. Stout. 1992. Crystal structures of aconitase with isocitrate and nitroisocitrate bound. Biochemistry 31:2735-2748. [DOI] [PubMed] [Google Scholar]
- 25.Lauble, H., and C. D. Stout. 1995. Steric and conformational features of the aconitase mechanism. Proteins 22:1-11. [DOI] [PubMed] [Google Scholar]
- 26.Miyakoshi, S., H. Uchiyama, T. Someya, T. Satoh, and T. Tabuchi. 1987. Distribution of the methylcitric acid cycle and β-oxidation for propionate catabolism in fungi. Agric. Biol. Chem. 51:2381-2387. [Google Scholar]
- 27.Rado, T. A., and J. A. Hoch. 1973. Phosphotransacetylase from Bacillus subtilis: purification and physiological studies. Biochim. Biophys. Acta 321:114-125. [DOI] [PubMed] [Google Scholar]
- 28.Sasse, J. 1991. Detection of proteins, p. 10.6.1-10.6.8. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. Wiley Interscience, New York, N.Y.
- 29.Schloss, J. V., M. H. Emptage, and W. W. Cleland. 1984. pH profiles and isotope effects for aconitase from Saccharomycopsis lipolytica, beef heart, and beef liver: α-methyl-cis-aconitate and threo-ds-α-methylisocitrate as substrates. Biochemistry 23:4572-4580. [DOI] [PubMed] [Google Scholar]
- 30.Seidman, C. G., K. Struhl, J. Sheen, and T. Jessen. 1997. Introduction of plasmid DNA into cells, p. 1.8.1-1.8.10. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. Wiley Interscience, New York, N.Y. [DOI] [PubMed]
- 31.Tabuchi, T., H. Aoki, H. Uchiyama, and T. Nakahara. 1981. 2-Methylcitrate dehydratase, a new enzyme functioning at the methylcitric acid cycle of propionate metabolism. Agric. Biol. Chem. 45:2823-2829. [Google Scholar]
- 32.Tabuchi, T., and S. Hara. 1974. Production of 2-methylcitric acid from n-parafins by mutants of Candida lipolytica. Agr. Biol. Chem. 38:1105-1106. [Google Scholar]
- 33.Tabuchi, T., and N. Serizawa. 1975. The production of 2-methylisocitric acid from odd-carbon n-alkanes by a mutant of Candida lipolytica. Agric. Biol. Chem. 39:1049-1054. [Google Scholar]
- 34.Tabuchi, T., H. Umetsu, H. Aoki, and H. Uchiyama. 1995. Characteristics of 2-methylisocitrate dehydratase, isolated from Yarrowia lipolytica, in comparison with aconitase. Biosci. Biotechnol. Biochem. 59:2013-2017. [Google Scholar]
- 35.Textor, S., V. F. Wendisch, A. A. De Graaf, U. Müller, M. I. Linder, D. Linder, and W. Buckel. 1997. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 168:428-436. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]