Abstract
The Pseudomonas aeruginosa plasmid pKLC102 coexists as a plasmid and a genome island in clone C strains. Whereas the related plasmid pKLK106 reversibly recombines with P. aeruginosa clone K chromosomes at one of the two tRNALys genes, pKLC102 is incorporated into the tRNALys gene only close to the pilA locus. Targeting of the other tRNALys copy in the chromosome is blocked by a 23,395-bp mosaic of truncated PAO open reading frames, transposons, and pKLC102 homologs. Annotation and phylogenetic analysis of the large 103,532-bp pKLC102 sequence revealed that pKLC102 is a hybrid of plasmid and phage origin. The plasmid lineage conferred oriV and genes for replication, partitioning, and conjugation, including a pil cluster encoding type IV thin sex pili and an 8,524-bp chvB glucan synthetase gene that is known to be a major determinant for host tropism and virulence. The phage lineage conferred integrase, att, and a syntenic set of conserved hypothetical genes also observed in the tRNAGly-associated genome islands of P. aeruginosa clone C chromosomes. In subgroup C isolates from patients with cystic fibrosis, pKLC102 was irreversibly fixed into the chromosome by the insertion of the large 23,061-bp class I transposon TNCP23, which is a composite of plasmid, integron, and IS6100 elements. Intramolecular transposition of a copy of IS6100 led to chromosomal inversions and disruption of plasmid synteny. The case of pKLC102 in P. aeruginosa clone C documents the intraclonal evolution of a genome island from a mobile ancestor via a reversibly integrated state to irreversible incorporation and dissipation in the chromosome.
Genome diversity in bacteria is caused by sequence diversity in coding and noncoding regions, genome islands, and islets in the chromosome and mobile genetic elements, such as plasmids, phages, and transposons (3, 56). Comparative intraspecies mapping and sequencing has uncovered an abundance of genome islands in numerous taxa, particularly among the gram-negative proteobacteria, with the pathogenicity islands in enterobacteria being the most thoroughly investigated examples (30, 31). Pathogenicity islands were typically found to be integrated into tRNA genes of the host chromosome. The evolution of genome islands has mainly been deduced from indirect evidence gained from the comparison of clonal lineages, compositional analysis of global genome features, and/or reconstruction of the evolutionary tree (19, 31), but reports in which the transition from a mobile element to a chromosomally integrated genome island could be directly documented by isolates retrieved from the natural habitats are scarce (8, 49, 50, 63).
Our group studies genome diversity in the γ-proteobacterium Pseudomonas aeruginosa. This ubiquitous and metabolically versatile microorganism (57) is characterized by a core genome with conserved synteny of genes and a low average nucleotide substitution rate of 0.5% (33, 35, 51, 66, 72). Only 2.5% of the coding sequences (CDS) exhibit significantly higher sequence diversity (66). Clone- or strain-specific genome islands and genome islets define the variable part of the chromosome, which results in variations of genome size between 5.2 and 7 Mbp (53, 62). Four genome islands have so far been sequenced (4, 40, 41). They all encode phenotypic traits that are absent in the completely sequenced reference strain, PAO1 (68). In the two cases analyzed in the major P. aeruginosa clone C (55), the genome island had been incorporated into tRNA genes (40). The tRNAGly-associated genome islands PAGI-2(C) and PAGI-3(SG) show a global structure similar to that of the 105-kb self-transmissible clc element of Pseudomonas putida, which is the only known genome island in the genus Pseudomonas that can be mobilized and laterally transferred to other strains, even across species and genus barriers (50, 63, 67). The site-specific integrative recombination between the clc element's attachment site (attP) and the chromosomal attachment site at the 3′end of the tRNAGly gene is accomplished by an integrase that is highly homologous to those encoded by PAGI-2(C) and PAGI-3(SG) (40, 50, 63).
PAGI-2(C) is located in a so-called hypervariable region close to the lipH locus. The other two hypervariable regions in the P. aeruginosa chromosome with pronounced genomic variability reside in the vicinity of the pilA and phnAB loci (33, 51, 53). tRNALys genes were identified as the hot spots for the integration and excision of DNA in these regions (36). The large plasmid pKLK106 sequentially recombined with either of the two tRNALys genes in P. aeruginosa clone K strains, giving rise to reversible rearrangements of a 106-kb genome island in sequential isolates. In clone C strains, the plasmid pKLC102 was reversibly incorporated into the tRNALys gene of the pilA region. Clone C isolates from the environment and most disease habitats harbored both the free plasmid and the chromosomally integrated pKLC102, whereas isolates from the lungs of patients with cystic fibrosis (CF) carried no episomal forms (53). Physical mapping revealed that one subgroup of clone C strains from CF lungs had captured additional DNA in pKLC102, which induced large chromosomal inversions in the progeny (39, 53).
The two related plasmids pKLK106 and pKLC102 are one of the very few cases known in which mobile DNA coexists as a free plasmid and a genome island in a bacterial cell. Hence, first we sequenced this connecting link between the plasmid and the island in order to resolve the features that allow this dual lifestyle and to get a clue to the impact of this extra DNA on the phenotype of the host. The clone C plasmid pKLC102 was selected for sequencing (Table 1) because clone C is a major clone of the present P. aeruginosa population in environmental and disease habitats, and hence, its genome organization has been studied in detail (53, 55, 62). Twenty-one clone C chromosomes have been mapped, two of which were chosen for the sequencing of the genome islands PAGI-2(C) and PAGI-3(SG) in the lipH hypervariable region (40). Second, the organization of the phn region and the makeup of pKLK106 and pKLC102 were compared in order to address the issue of why the clone K plasmid sequentially recombines with both tRNALys genes whereas the target site in the phn region is not accessible to the clone C plasmid. Third, the type of genetic element of the DNA inserted into the chromosomally integrated pKLC102 of subgroup C strains was identified by sequencing. All data were compiled to trace the evolution of the P. aeruginosa clone C chromosome. Annotation revealed that pKLC102 was assembled from a phage lineage and a plasmid lineage that endowed this hybrid with the uncommon flexibility to exist as a conjugative plasmid and a genome island. In other words, these peculiar features make pKLC102 a physically existing piece of evidence for the evolution of a genome island from mobile ancestors.
TABLE 1.
Comparison of general features of sequenced gene islands and PAO1 genome
| Genomic region (reference) | Size (bp) | GC content (%) | Coding sequence (%) | No. of CDS | No. of CDS per 10 kb |
|---|---|---|---|---|---|
| pKLC102 | 103,532 | 60.9 | 86.3 | 105 | 10.1 |
| TNCP23 | 23,061 | 63.1 | 90.7 | 29 | 12.6 |
| PAGI-4(C) | 23,395 | 56.0 | 86.6 | 24 | 10.3 |
| PAGI-2(C) (40) | 104,955 | 64.7 | 90.4 | 113 | 10.7 |
| PAO1 genome (68) | 6,624,403 | 66.6 | 89.4 | 5,570 | 8.9 |
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The selected P. aeruginosa clone K and C strains (55) C, C17, K, K1, and K2 were isolated from the lungs of patients with CF (36, 53). Clone C strain SG17 M was recovered from the aquatic environment (55). The bacteria were routinely grown in Luria-Bertani medium (59) at 37°C.
DNA techniques.
DNA manipulations were done by standard procedures (5). A genomewide cosmid library was constructed according to the protocols of Wenzel and Herrmann (71) as described previously (40). Small-scale isolations of cosmid DNA were performed by using QIAprep spin miniprep kits (Qiagen); larger amounts of cosmid DNA were purified using QIAtip100 columns or QIAtip500 and the large-construct kit (Qiagen) according to the instructions of the supplier. The high-molecular-mass plasmids pKLK106 and pKLC102 were prepared on a large scale by modified alkaline lysis (5, 36).
Southern hybridization.
For colony blots, cell suspensions were inoculated on Hybond N+ membranes (Amersham) by using a 96-needle replication device and were grown on 2YT-amp plates. Alternatively, colony lifts were performed directly from agar plates onto Hybond N+ membranes. The cells were lysed, and the DNA was fixed (71). Blotting of chromosomal or cosmid DNA digested with appropriate restriction enzymes to nylon membranes, the hybridization procedure, and immunological detection of probe signals were performed according to previously described protocols (52). For the screening of the library, probes were prepared from purified plasmid DNA, from the SpeI fragment SpAB-specific clone 2A (54), or from gel-purified restriction fragments of plasmids and cosmids by using a digoxigenin labeling kit (Roche) (52).
Construction of a pKLC102 tiling path in the strain C chromosome.
The pKSCC cosmid library was screened with plasmid pKLC102 and clone 2A as probes. Thirty-seven probe-reactive cosmids were digested with BamHI or EcoRI plus HindIII and separated by agarose gel electrophoresis. Comparison of the gel-separated restriction fragment pattern with the restriction maps of pKLC102 and of clone C strains C and SG17 M in this chromosomal region identified the recombination point for chromosomal integration on the plasmid restriction fragment BmQ (53) and the integration of a further large 23-kb DNA segment on BmG. The cosmids were ordered by Southern hybridization of restricted pKSCC cosmids with BamHI fragments of pKLC102. The cosmids pKSCC785, -187, -050, and -867 represented the contig of minimal overlap and hence were selected for sequencing. The remaining large 2.6-kb gap between cosmids pKSCC187 and -050 (Fig. 1), reaching from fragments BmY1 to BmO (53), was closed by recombinant PCR using GoldStar DNA polymerase (Eurogentech).
FIG. 1.
(a) Restriction map of plasmid pKLC102 (inner circle, EcoRI; outer circle, PvuII). The recombination site for chromosomal integration and the position of the insertion of integron TNCP23 in strain C are indicated. The thick arcs represent the tiling path of cosmids and gap-spanning PCR products utilized for sequencing pKLC102 DNA in the strain C chromosome. The darkly shaded area is absent in pKLK106 (see panel b). (b) Comparative restriction analysis of pKLC102 and pKLK106. (I) Separated HindIII, EcoRI, and PvuII restriction digests of cosmids pKSCC785 (lanes 1), pKSCC187 (lanes 2), pKSCC050 (lanes 3), and pKSCC867 (lanes 4). PCR, gap-spanning PCR product (undigested); λ, BstEII digest of λ DNA used as a size standard. (II) Southern blot of gel I, hybridized with plasmid pKLK106. The letters in gel I indicate bands with no or lower-than-expected hybridization signals due to DNA that is not represented in the pKLK106 probe. P, PAO1 DNA flanking the inserted pKLC102 in strain C; V, vector DNA; T, integron TNCP23; C (circled), pKLC102-specific DNA absent in pKLK106.
Identification of a PAGI-4(C) covering cosmid.
Several pKSCC-cosmids hybridized with the pKLC102 fragment BmQ (53), although their inserts were mapped to another chromosomal region (fragment SpB on SpeI-restricted pulsed-field gel electrophoresis-separated chromosomal DNA) (62). In one of these cosmids, pKSCC673, the cross-hybridizing DNA was assigned to a 5.4-kb EcoRI fragment. Sequencing of this fragment revealed that a tRNALys gene separates BmQ-homologous DNA from DNA nonhomologous to BmQ but with a high degree of similarity to PAO sequence. Next, the EcoRI-HindIII double-digestion fragment patterns of pKSCC673 and the other BmQ-cross-hybridizing cosmids from this region were compared. The insert ends of the two cosmids with the most divergent EcoRI-HindIII fingerprints were sequenced, using a T3 (5′-AATTAACCCTCACTAAAGGG) primer and a T7 (5′-CATAATACGACTCACTATAGGG) primer. Cosmid pKSCC260 was chosen for sequencing, because PAO1-like DNA from the oprL-phnAB region was identified at both insert ends, suggesting that pKSCC260 spans the whole PAGI-4(C) gene island.
Sequencing.
The ends of cosmid inserts (500 to 800 bp) were determined by single reads of one strand using T3 or T7 primers. Inserts of cosmids pKSCC785, -187, -050, -867, and -260 were completely sequenced by random sequencing of small-insert plasmid libraries (1.0 to 2.5 kb). After assembly, the sequence gaps were closed by editing the ends of sequence traces and/or primer walking on plasmid clones, and physical gaps were closed by combinatorial PCR followed by sequencing of the PCR product. The final sequences had an accuracy of >99.99%.
Annotation.
Putative open reading frames (ORFs) were identified by using a dictionary-driven gene-finding program (64; http://cbcsrv.watson.ibm.com/tgi.html) and by GeneMark and GeneMark.HMM programs (6, 44). Predicted ORFs were reviewed individually for the assignment of the start codon based on additional contextual information, such as the proximity of ribosome-binding sequence motifs and alignments with known proteins retrieved by BLAST search (2). tRNA genes were identified by the program tRNA-Scan SE (43). Public databases were searched for similar sequences with the BLASTN, BLASTX, and BLASTP/PSI- and PHI-BLAST algorithms. Sequence comparisons with the P. aeruginosa PAO1 genome (68) were retrieved from the website of the Pseudomonas Genome Project (http://www.pseudomonas.com). The sequences were scanned for palindromes, tandems, and signal sequences using programs available at http://bioweb.pasteur.fr/. The features of the predicted proteins were examined by the programs Pfam (http://www.sanger.ac.uk/Software/Pfam/search.shtml), Block Searcher (http://blocks.fhcrc.org/blocks/blocks_search.html), COGnitor (http://www.ncbi.nih.gov/COG/xognitor.html), “DAS” Transmembrane Prediction server (16), and SOSUI (http://sosui.proteome.bio.tuat.ac.jp/cgi-bin/sosui.cgi?/sosui_submit.html). Secondary DNA-RNA structure was analyzed by a Greedy algorithm with an energy threshold of −10 kcal using the programs GeneBee, available at http://www.genebee.msu.su/genebee.html (11), and Mfold, available at http://www.bioinfo.rpi.edu/applications/mfold/ (60). The program BioEdit version 5.0.9 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) was used for storing sequences in a database, pairwise comparison, alignment, and phylogenetic tree design. GC contents and GC skew were calculated with in-house programs. Restriction maps were constructed with the program Webcutter version 2.0, written by Max Heiman (available at http://www.firstmarket.com/cutter/cut2.html).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in the GenBank database {accession numbers AY257538 [pKLC102], AY257539 [TNCP23], and AY258138 [PAGI-4(C)]}.
RESULTS AND DISCUSSION
Comparison of plasmids pKLC102 and pKLK106.
Plasmids pKLC102 from P. aeruginosa clone C strains and pKLK106 from P. aeruginosa clone K strains were predicted to be highly similar (36). Both plasmids are ∼100 kb in size, integrate into the chromosome at the 3′ ends of tRNALys genes (the att site), and exhibit virtually identical BamHI/SpeI restriction maps. Map differences were evident in only three regions. In order to differentiate pKLK106-homologous segments from nonhomologous sequence in the pKLC102 region of the clone C chromosome, a tiling path represented by the gel-separated HindIII-, EcoRI-, or PvuII-restricted cosmids pKSCC785, pKSCC187, pKSCC050, and pKSCC867 and a gap-spanning PCR product (see Material and Methods) was hybridized with pKLK106 (Fig. 1B). The comparison of the gel (Fig. 1B, left) and the blot (Fig. 1B, right) uncovered strong hybridization signals for almost all restriction fragments derived from the episomal plasmids, indicating that pKLC102 is composed of >97% sequence that is homologous with pKLK106.
Restriction fragments (Fig. 1B, left) with no or weak hybridization signals (Fig. 1B, right) represent cosmid-vector, transposon TNCP23 (in pKSCC187 [see below]), PAO1 DNA (in pKSCC785 and pKSCC867), or apparently pKLC102-specific DNA that is absent in pKLK106 (Fig. 1b). All pKLC102-specific DNA was assigned within or adjacent to fragment PvP (Fig. 1a). PvP is the only part of pKLC102 in which the ORFs exhibit the highest number of BLAST hits with P. aeruginosa PAO1 sequence (see Table 3 and Fig. 3). The CDS CP84, CP85, and CP86 are homologous to PA2566, PA2565, and PA2564, respectively, and are flanked by two 239-bp direct repeats upstream of CP84 and downstream of CP86. Hence, this stretch of sequence has the characteristics of a “mobile cassette” that was probably incorporated into the plasmid after the divergence of pKLC102 and pKLK106 from a common ancestor. Besides CP84 to CP86, no further segments that did not hybridize with pKLK106 were detected in plasmid pKLC102. These data confirm the prediction that clone K and clone C strains harbor almost identical plasmids.
TABLE 3.
Annotation of all ORFs located within pKLC102
| Gene identification | Coordinates
|
Direction | Length (aa)a | GC content (%) | Gene name | Annotation result and homologous gene products | BLAST search resultb | E value of best BLAST hit | |
|---|---|---|---|---|---|---|---|---|---|
| Left | Right | ||||||||
| CP1 | 113 | 997 | → | 294 | 60.23 | soj | Similar to chromosome partitioning-related protein (Xylella fastidiosa 9a5c) | NP_299073 | 6e-67 |
| CP2 | 999 | 1724 | → | 241 | 59.78 | Similar to hypothetical protein from Pseudomonas resinovorans | NP_758706 | 2e-25 | |
| CP3 | 1.721 | 2.218 | → | 165 | 62.25 | Hypothetical protein similar to PA2226 (P. aeruginosa) | NP_250916 | 5E-27 | |
| CP4 | 2.224 | 2.970 | → | 248 | 61.85 | Hypothetical protein similar to unknown protein encoded by gene Z2097 within prophage CP-933O (E. coli O157:H7 EDL933) | NP_287555 | 6E-10 | |
| CP5 | 2.967 | 3.653 | → | 228 | 64.05 | Hypothetical protein similar to Magn3145 (Magnetospirillum magnetotacticum) | ZP_00050489 | 1E-10 | |
| CP6 | 3.653 | 4.354 | → | 233 | 64.53 | Hypothetical protein similar to an intragenic region upstream of Psyr3996 (P. syringae pv. syringae B728a) | NZ_AABP02000009 | 2E-61 | |
| CP7 | 4.351 | 5.058 | → | 235 | 58.62 | Hypothetical protein homologous to EsV-1-119 ORF119 (Ectocarpus siliculosus virus) | NP_077604 | 0.039 | |
| CP8 | 5.042 | 5.260 | → | 72 | 57.08 | No significant similarity | |||
| CP9 | 5.323 | 6.600 | → | 425 | 61.11 | dnaB | Replicative DNA helicase PA4931 (dnaB) (P. aeruginosa) | NP_253618 | E-122 |
| CP10 | 7.039 | 7.395 | ← | 118 | 55.74 | No significant similarity | |||
| CP11 | 7.838 | 8.365 | → | 175 | 61.74 | No significant similarity | |||
| CP12 | 8.362 | 8.619 | → | 85 | 60.08 | No significant similarity | |||
| CP13 | 8.612 | 9.109 | → | 165 | 62.85 | No significant similarity | |||
| CP14 | 9.102 | 9.335 | → | 77 | 66.24 | Conserved hypothetical protein similar to Bcep0486 (B. fungorum) | ZP_00027718 | 1E-06 | |
| CP15 | 9.335 | 10.351 | → | 338 | 64.01 | Conserved hypothetical protein homologous to PA3849 (P. aeruginosa) and to a nucleoid associated protein (E. coli O157:H7) | NP_252538 | E-175 | |
| CP16 | 10.348 | 10.602 | → | 84 | 58.43 | Putative DNA-binding protein homologous to PA3385 (AlgZ) (P. aeruginosa) | AAD55364 | 0.028 | |
| CP17 | 10.599 | 12.332 | → | 577 | 58.36 | Conserved hypothetical protein similar to ORF SG102 (P. aeruginosa); ParB-like nuclease domain | AAN62323 | E-104 | |
| CP18 | 12.360 | 13.115 | → | 251 | 59.13 | Hypothetical protein Bcep0488 (B. fungorum) | ZP_00027720 | 1E-30 | |
| CP19 | 15.766 | 16.494 | → | 242 | 63.24 | Conserved hypothetical protein similar to ORF C104 (P. aeruginosa) | AAN62197 | 4E-64 | |
| CP20 | 16.500 | 17.048 | → | 182 | 60.84 | Conserved hypothetical protein similar to ORF C103 (P. aeruginosa) | AAN62196 | 1E-24 | |
| CP21 | 17.095 | 17.934 | → | 279 | 60.83 | Putative antirepressor homologous to sb41 (Salmonella enterica serovar Typhimurium phage ST64B) | NP_700414 | 6E-23 | |
| CP22 | 17.964 | 18.452 | → | 162 | 62.17 | ssb | Putative single-stranded DNA-binding protein homologous to ssb (P. aeruginosa); single-strand binding conserved domain | AAN62318 | 5E-30 |
| CP23 | 18.871 | 19.152 | ← | 93 | 53.90 | No significant similarity | |||
| CP24 | 19.289 | 19.549 | ← | 86 | 52.87 | No significant similarity | |||
| CP25 | 19.571 | 19.981 | ← | 136 | 60.83 | Putative nucleic acid binding protein; contains PIN conserved domain (similarity with an analogous gene from Salmonella enterica serovar Typhimurium LT2) | NP_461950 | 2E-51 | |
| CP26 | 19.981 | 20.211 | ← | 76 | 57.57 | Putative virulence-associated protein similar to cytoplasmic protein from Salmonella enterica serovar Typhimurium LT2; VagC conserved domain | NP_490591 | 2E-20 | |
| CP27 | 20.467 | 22.386 | → | 639 | 62.19 | topA | Topoisomerase I; similar to XFa0003 (X. fastidiosa) | NP_061659 | E-148 |
| CP28 | 22.694 | 22.903 | → | 69 | 51.90 | cspA | Cold acclimation protein similar to PA3266 (CspA) (P. aeruginosa) | NP_251956 | 2E-18 |
| CP29 | 23.124 | 25.013 | → | 629 | 60.37 | No significant similarity | |||
| CP30 | 25.010 | 26.983 | → | 657 | 64.18 | Putative helicase similar to Psyr4003 (P. syringae pv. syringae); SNF2 family N-terminal domain; DEAD-like helicase superfamily; helicase superfamily C-terminal domain | ZP_00127683 | 0.0 | |
| CP31 | 27.105 | 27.887 | → | 260 | 46.23 | Putative ABC transporter similar to mlr7981 (Mesorhizobium loti); ATP-binding protein | BAB53638 | 9E-85 | |
| CP32 | 27.920 | 29.173 | → | 417 | 41.55 | Hypothetical protein similar to SMa2241 (S. meliloti plasmid pSymA) | NP_436454 | E-126 | |
| CP33 | 29.347 | 30.471 | → | 374 | 65.16 | pilL | PilL (Yersinia pseudotuberculosis) | AAL05416 | 3E-36/PICK> |
| CP34 | 30.471 | 32.180 | → | 569 | 63.86 | pilN | PilN (E. coli) | AAL05518 | 8E-56 |
| CP35 | 32.298 | 33.509 | → | 403 | 64.03 | pilO | PilO (Salmonella enterica serovar Typhi) | AAF14815 | 5E-13 |
| CP36 | 33.499 | 34.032 | → | 177 | 70.97 | pilP | PilP (Salmonella enterica serovar Typhimurium plasmid R64) | BAA77976 | 2E-04 |
| CP37 | 34.041 | 35.621 | → | 526 | 63.25 | pilQ | PilQ (S. enterica subsp. enterica serovar Dublin) | AAF81213 | 7E-74 |
| CP38 | 35.621 | 36.700 | → | 359 | 63.80 | pilR | PilR (Salmonella enterica serovar Typhi) | AAC98886 | 2E-40 |
| CP39 | 36.779 | 37.252 | → | 157 | 60.13 | pilS | PilS; prepilin similar to hypothetical protein from P. syringae pv. syringae B728a and to type IV prepilin (Y. pseudotuberculosis) | ZP_00127690 | 3E-18 |
| CP40 | 37.249 | 38.190 | → | 313 | 66.03 | pilU | PilU; prepilin peptidase (P. aeruginosa) | S54702 | 1E-15 |
| CP41 | 38.195 | 39.523 | → | 442 | 63.81 | pilV | PilV; pilus tip adhesin (E. coli) | CAA44099 | 6E-48 |
| CP42 | 39.552 | 39.989 | → | 145 | 68.26 | pilM | PilM (Salmonella enterica serovar Typhimurium plasmid R64) | BAA77973 | 1.1 |
| CP43 | 40.911 | 41.300 | → | 129 | 60.00 | No significant similarity | |||
| CP44 | 41.373 | 41.552 | → | 59 | 53.33 | No significant similarity | |||
| CP45 | 41.554 | 41.688 | → | 44 | 58.52 | No significant similarity | |||
| CP46 | 42.012 | 42.491 | → | 159 | 57.29 | Conserved hypothetical protein (P. fluorescens) | ZP_00087894 | 4E-46 | |
| CP47 | 42.861 | 43.058 | → | 65 | 53.54 | Conserved hypothetical protein (S. enterica subsp. enterica serovar Typhi) | NP_458626 | 1E-10 | |
| CP48 | 43.163 | 43.441 | → | 92 | 50.18 | No significant similarity | |||
| CP49 | 43.544 | 44.251 | → | 235 | 52.68 | Conserved hypothetical protein similar to Pflu5179 (P. fluorescens) | ZP_00087892 | 1E-41 | |
| CP50 | 44.487 | 44.837 | → | 116 | 63.25 | No significant similarity | |||
| CP51 | 44.894 | 45.685 | → | 263 | 61.62 | Conserved hypothetical protein similar to XF1760 (X. fastidiosa) | NP_299049 | 2E-42 | |
| CP52 | 45.788 | 46.150 | → | 120 | 58.13 | Conserved hypothetical protein similar to ORF C77 (P. aeruginosa) | AAN62171 | 9E-21 | |
| CP53 | 46.218 | 46.472 | → | 84 | 61.18 | Conserved hypothetical protein similar to XAC2240 (Xanthomonas axonopodis pv. citri) | NP_642557 | 2E-08 | |
| CP54 | 46.564 | 47.169 | → | 201 | 64.03 | Conserved hypothetical protein similar to XAC2241 (X. axonopodis pv. citri 306) | NP_642558 | 1E-50 | |
| CP55 | 47.199 | 48.644 | → | 481 | 63.69 | Hypothetical protein similar to Bcep3609 (B. fungorum), | ZP_00030785 | E-133 | |
| CP56 | 48.743 | 50.998 | → | 751 | 63.25 | Putative helicase similar to hypothetical protein Psyr4040 (P. syringae pv. syringae); SNF2 family N-terminal domain; DEAD-like helicase superfamily; helicase superfamily C-terminal domain | ZP_00127717 | 0.0 | |
| CP57 | 51.579 | 52.364 | ← | 261 | 55.47 | Putative polyketide synthase similar to XF2135 (X. fastidiosa); HCCA isomerase conserved domain | NP_299414 | 8E-23 | |
| CP58 | 52.303 | 52.644 | ← | 113 | 58.48 | Conserved hypothetical protein similar to Bcep7601 (B. fungorum); carboxymuconolactone decarboxylase conserved domain | ZP_00034706 | 2E-15 | |
| CP59 | 52.739 | 53.299 | ← | 186 | 54.55 | Putative TetR family transcription regulator protein similar to ORF43 (P. putida plasmid pWWO); TetR conserved domain | NP_542832 | 3E-17 | |
| CP60 | 53.362 | 54.147 | ← | 261 | 58.78 | Putative reductase protein similar to ORF44 (P. putida plasmid pWWO); short-chain dehydrogenase conserved domain | NP_542833 | 3E-76 | |
| CP61 | 54.303 | 54.716 | ← | 137 | 59.66 | Putative TetR family transcription regulator protein similar to MA2493 (Methanosarcina acetivorans) | NP_617399 | 0.12 | |
| CP62a | 54.709 | 55.560 | ← | 283 | 57.51 | Phage integrase similar to hypothetical protein Reut5633 (R. metallidurans); phage integrase domain | ZP_00026607 | 1E-23 | |
| CP62b | 54.904 | 55.407 | → | 167 | 59.52 | No significant similarity; inner ORF of the phage integrase CP62a | |||
| CP63 | 56.331 | 57.023 | → | 230 | 63.78 | Conserved hypothetical protein similar to Psyr4041 (P. syringae pv. syringae) | ZP_00127718 | 2E-26 | |
| CP64 | 57.034 | 57.789 | → | 251 | 65.21 | Conserved hypothetical protein similar to Psyr4042 (P. syringae pv. syringae) | ZP_00127719 | 5E-53 | |
| CP65 | 57.774 | 58.355 | → | 193 | 65.64 | Hypothetical protein similar to ORF SG70 (P. aeruginosa) | AAN62161 | 5E-35 | |
| CP66 | 58.352 | 58.852 | → | 166 | 66.27 | Hypothetical protein similar to ORF SG69 (P. aeruginosa) | AAN62291 | 7E-31 | |
| CP67 | 59.134 | 61.365 | → | 743 | 65.10 | Putative conjugation protein TraG similar to hypothetical protein Psyr4044 (P. syringae pv. syringae); TraG/TraD family conserved domain | ZP_00127721 | 0.0 | |
| CP68 | 61.365 | 62.111 | → | 248 | 61.18 | Conserved hypothetical protein similar to ORF SG67 (P. aeruginosa) | AAN62289 | 5E-52 | |
| CP69 | 62.122 | 63.603 | → | 493 | 59.85 | Putative helicase similar to hypothetical protein Psyr4046 (P. syringae pv. syringae); UvrD helicase conserved domain | ZP_00127723 | 0.0 | |
| CP70 | 63.735 | 64.844 | → | 369 | 46.58 | Hypothetical protein similar to a probable RND efflux transporter mll1426 (M. loti) | NP_103018 | 4.4 | |
| CP71 | 64.889 | 65.200 | ← | 103 | 49.68 | Conserved hypothetical protein similar to PA0713 (P. aeruginosa) | NP_249404 | 5E-16 | |
| CP72 | 65.373 | 65.672 | → | 99 | 57.00 | No significant similarity | |||
| CP73 | 65.883 | 66.104 | → | 73 | 67.57 | Hypothetical protein similar to Avin1322 (A. vinelandii) | ZP_00089650 | 3E-24 | |
| CP74 | 66.107 | 66.340 | → | 77 | 64.53 | Hypothetical protein similar to Avin1321 (A. vinelandii) | ZP_00089649 | 2E-8 | |
| CP75 | 66.358 | 66.714 | → | 118 | 57.98 | Hypothetical protein similar to Avin1320 (A. vinelandii) | ZP_00089648 | 3E-15 | |
| CP76 | 66.725 | 67.111 | → | 128 | 66.93 | Hypothetical protein similar to Avin1319 (A. vinelandii) | ZP_00089647 | 1E-32 | |
| CP77 | 67.108 | 67.767 | → | 219 | 61.06 | Hypothetical protein similar to Avin1318 (A. vinelandii) | ZP_00089646 | 4E-78 | |
| CP78 | 67.764 | 68.648 | → | 294 | 66.33 | Hypothetical protein similar to Avin1317 (A. vinelandii) | ZP_00089645 | 5E-88 | |
| CP79 | 68.710 | 70.137 | → | 475 | 64.15 | Hypothetical protein similar to Avin1316 (A. vinelandii) | ZP_00089644 | E-149 | |
| CP80 | 70.211 | 70.558 | → | 115 | 64.94 | Hypothetical protein similar to Avin1315 (A. vinelandii) | ZP_00089643 | 1E-47 | |
| CP81 | 70.558 | 73.500 | → | 980 | 63.00 | Hypothetical protein similar to Avin1314 (A. vinelandii) | ZP_00089642 | 0.0 | |
| CP82 | 73.497 | 73.781 | → | 94 | 58.25 | Hypothetical protein similar to Avin1313 (A. vinelandii) | ZP_00089641 | 2E-19 | |
| CP83 | 73.778 | 74.434 | → | 218 | 62.25 | Conserved hypothetical outer membrane protein similar to PA0982 (P. aeruginosa) | NP_249673 | 3E-87 | |
| CP84 | 75.208 | 76.395 | → | 395 | 62.88 | Putative flavoprotein reductase similar to hypothetical protein PA2566 (P. aeruginosa) | NP_251256 | 0.0 | |
| CP85 | 76.418 | 76.840 | → | 140 | 66.43 | Hypothetical protein similar to PA2565 (P. aeruginosa) | NP_251255 | 5E-25 | |
| CP86 | 76.830 | 77.696 | → | 288 | 63.78 | cheR | CheR; trans-aconitate 2-methyltransferase similar to PA2564 (P. aeruginosa) | NP_251254 | E-126 |
| CP87 | 77.834 | 78.265 | → | 143 | 61.81 | Conserved hypothetical protein similar to Avin1308 (A. vinelandii) | ZP_00089637 | 6E-20 | |
| CP88 | 78.265 | 79.203 | → | 312 | 64.86 | Hypothetical protein similar to Avin1307 (A. vinelandii) | ZP-00089636 | E-123 | |
| CP89 | 79.038 | 80.603 | → | 521 | 64.56 | Putative integral membrane protein similar to ORF C42 (P. aeruginosa) | AAN62272 | E-122 | |
| CP90 | 80.603 | 80.950 | → | 115 | 62.64 | Hypothetical protein similar to Avin1305 (A. vinelandii) | ZP_00089634 | 0.001 | |
| CP91 | 80.947 | 82.482 | → | 511 | 58.72 | Hypothetical protein similar to Avin1304 (A. vinelandii) | ZP_00089633 | 0.0 | |
| CP92 | 82.924 | 83.256 | → | 110 | 60.61 | Hypothetical protein (P. fluorescens); conserved domain of NikR transcriptional regulator | ZP_00086716 | 8E-16 | |
| CP93 | 83.260 | 83.610 | → | 116 | 62.68 | Conserved hypothetical protein similar to AGR_C_2415 (A. tumefaciens strain C58; Cereon); ParE conserved domain of plasmid stabilization system protein | G97518 | 5E-15 | |
| CP94 | 83.988 | 92.501 | ← | 2,837 | 65.32 | chvB | Beta (1->2) glucan biosynthesis protein similar to ChvB (A. tumefaciens) | NP_533395 | 0.0 |
| CP95 | 93.343 | 93.567 | → | 74 | 61.33 | Hypothetical protein similar to Pflu4565 (P. fluorescens) | ZP_00087289 | 0.13 | |
| CP96 | 93.782 | 94.501 | → | 239 | 61.25 | Transcriptional regulator-related protein similar to SO2551 (Shewanella oneidensis); helix-turn-helix and cAMP regulatory protein conserved domains | NP_718137 | 5E-78 | |
| CP97 | 94.572 | 94.934 | → | 120 | 60.33 | No significant similarity | |||
| CP98 | 94.852 | 95.532 | → | 226 | 64.02 | Hypothetical protein similar to XF0240 (X. fastidiosa) | NP_297533 | 7.1 | |
| CP99 | 95.641 | 97.659 | → | 672 | 62.51 | Fusion protein; putative fatty acid synthase similar to cyclopropane-fatty-acyl-2 phospholipid synthase mll4091 (M. loti) | BAB50831 | 3E-67 | |
| CP100 | 97.815 | 99.008 | ← | 397 | 61.81 | Probable fatty acid desaturase similar to PA0286 (P. aeruginosa) | AAG03675 | E-124 | |
| CP101 | 99.396 | 99.752 | → | 118 | 45.94 | Hypothetical protein similar to Avin0596 (A. vinelandii) | ZP_00088936 | 3E-21 | |
| CP102 | 99.964 | 101.883 | → | 639 | 58.70 | Conserved hypothetical protein; putative conjugative relaxase Tral; similar to hypothetical protein Avin0927 (A. vinelandii) | ZP_00089260 | 0.0 | |
| CP103a | 101.880 | 103.163 | → | 427 | 60.00 | xerC | Phage integrase similar to XerC (P. syringae) | AAM77365 | E-101 |
| CP103b | 101.970 | 102.476 | ← | 168 | 60.36 | No significant similarity; inner ORF of phage integrase CP103a | |||
aa, amino acids.
GenBank accession number.
FIG. 3.
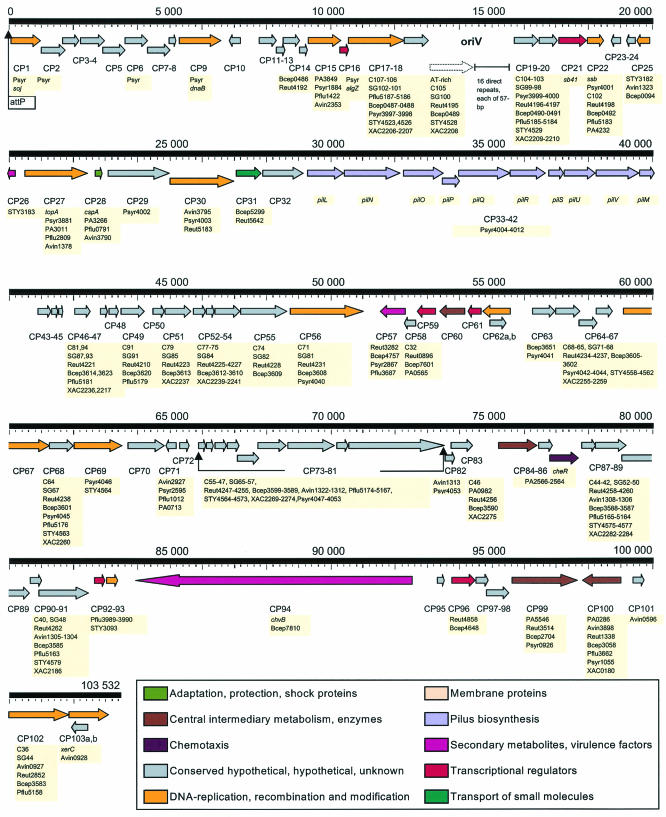
Gene map of pKLC102. The map is calibrated to the chromosomal integration attP site, marked by a flag. The leading strand was defined by colinearity with the P. aeruginosa PAO1 genome sequence. Predicted coding regions are shown by arrows indicating the direction of transcription. The genes are color coded according to their functional categories, as shown in the legend below the map. All genes carry identification numbers according to the CDS numbering in Table 3. Homologs in other microorganisms retrieved by a BLASTP search and identified gene names are highlighted beneath the corresponding CDS. oriV is the predicted origin of replication. The putative CDS within the origin of replication is shown by a dotted arrow. The syntenic CDS CP73 to CP81 that were subjected to cladistic analysis (Fig. 5) are marked by bent arrows.
Genome island PAGI-4(C) is at tRNALys(1) close to oprL-phnAB.
In clone K strains, plasmid pKLK106 can reversibly integrate into the two chromosomal copies of the tRNALys gene (36). Clone C strains incorporate plasmid pKLC102 into only one of these sites (36), whereas the tRNALys(1) gene in the oprL-phnAB region is not used for plasmid insertion. Sequencing of the cosmid pKSCC260, comprising DNA adjacent to this tRNALys(1) gene, revealed that another block of DNA had inserted at this point in strain C DNA.
In strain PAO1, the tRNALys(1) gene is located between CDS PA0976 and PA0977 (68). The 8.9-kb DNA block 3′ of tRNALys from PA0977 to PA0987 represents a nonconserved insertion that terminates with 22 duplicated base pairs of the 3′ end of the tRNALys(1) gene, presumably the former attP site of the integrated element. This 8.9-kb block of PAO-specific DNA is absent in clone K strains harboring PA0988 as their first PAO homolog downstream of tRNALys(1) (36).
The sequence annotation of pKSCC260 revealed that in strain C, a large 23.4-kb gene island called PAGI-4(C) is integrated at this tRNALys(1) site (Table 2 and Fig. 2). PAGI-4(C) substitutes PA0977 for PA0994, and correspondingly the chaperone-usher cupC cluster (PA0992-PA0994) (70) is missing in strain C. PAGI-4(C) apparently consists of two blocks of non-PAO sequence, each flanked by short stretches of PAO-homologous sequence. The first 370 bp downstream of the tRNALys(1) gene show 92% identity with the PAO sequence. The CL1 gene is a truncated homolog of PA0977; a frameshift mutation gives rise to a stop codon 48 nucleotides prior to the 3′ end of PA0977. Another stretch of 832 bp in the middle of PAGI-4(C) is 95% identical with the PAO1 sequence and contains the PA0980 homolog, CL11, and the initial 57% of the sequence of PA0981.
TABLE 2.
Annotation of ORFs located within PAGI-4(C) in P. aeruginosa strain C
| Gene identification | Coordinates
|
Direction | Length (aa)a | GC content (%) | Gene name | Annotation result and homologous gene productsb | Best BLAST hitc | E value of best BLAST hit | |
|---|---|---|---|---|---|---|---|---|---|
| Left | Right | ||||||||
| CL1 | 125 | 253 | ← | 42 | 54.3 | Hypothetical protein; homologous to part of PA0977; probably truncated ORF | AAG04366 | 1E-12 | |
| CL2a | 371 | 1651 | ← | 426 | 60.3 | xerC | Put. integrase, phage-like; highly similar to XerC integrase from pKLC102 | AY257538 | 0.0 |
| CL2b | 1055 | 1744 | → | 229 | 60.0 | Put. excision regulator; “inner ORF”; similar to CP103b from pKLC102 | AY257538 | 0.0 | |
| CL3 | 1648 | 3474 | ← | 608 | 59.0 | Conserved hypothetical protein; homology to CP102 from pKLC102 | AY257538 | 0.0 | |
| CL4 | 3898 | 4248 | ← | 116 | 61.0 | Conserved hypothetical protein; homology to CP93 from pKLC102; similar to ParE domain (plasmid stabilization system protein) | AY257538 | 1E-51 | |
| CL5 | 4252 | 4584 | ← | 110 | 62.5 | Conserved hypothetical protein; homology to CP92 from pKLC102; similar to DNA-binding domain of transcriptional regulators | AY257538 | 3E-52 | |
| CL6 | 5013 | 6524 | ← | 503 | 58.9 | Conserved hypothetical protein; homology to CP91 from pKLC102 | AY257538 | 0.0 | |
| CL7 | 6521 | 6868 | ← | 115 | 62.9 | Hypothetical protein; homology to CP90 from pKLC102 | AY257538 | 2E-57 | |
| CL8 | 6868 | 8250 | ← | 460 | 64.6 | Conserved hypothetical protein; homology to CP89 from pKLC102 | AY257538 | 0.0 | |
| CL9 | 8274 | 9212 | ← | 312 | 65.8 | Conserved hypothetical protein; homology to CP88 from pKLC102 | AY257538 | 0.0 | |
| CL10 | 9212 | 9643 | ← | 143 | 62.5 | Conserved hypothetical protein; homology to CP87 from pKLC102 | AY257538 | 2E-69 | |
| CL11 | 10022 | 10306 | → | 94 | 47.4 | Hypothetical protein; highly homologous to PA0980 | AAG04369 | 4E-43 | |
| CL12 | 10830 | 13844 | → | 1,004 | 55.9 | tnpA | Put. transposase; similar to transposase from Tn4652 (P. putida) | NP_758719 | 0.0 |
| CL13 | 13841 | 14203 | → | 120 | 56.8 | tnpC | Put. transposase regulator-repressor; tnpC located next to transposase in Tn4652 | NP_542894 | 1E-39 |
| CL14 | 14373 | 14837 | ← | 154 | 41.3 | Put. acetyltransferase; plasmid-encoded homolog Nostoc sp. | NP_490276 | 2E-18 | |
| CL15 | 14845 | 15750 | ← | 301 | 48.5 | Put. membrane protein; up to 10 TM domains predicted | TP_00083447 | 1E-02 | |
| CL16 | 15747 | 16475 | ← | 242 | 42.3 | Hypothetical protein | NP_442107 | 2E-16 | |
| CL17 | 16491 | 17900 | ← | 469 | 45.4 | Put. homospermidine synthase | ZP_700003187 | 1E-121 | |
| CL18 | 18312 | 19628 | ← | 438 | 56.3 | Conserved hypothetical protein | NP_758589 | 0.0 | |
| CL19 | 19650 | 20393 | ← | 247 | 60.6 | Put. ABC transporter subunit; domains like in bacteriophage protein gp37 or DNA repair photolyase SplB (partial) | NP_758590 | 1E-128 | |
| CL20 | 20423 | 21394 | ← | 323 | 59.9 | tnpS | Cointegrate resolution protein S; homologous to tnpS from Tn4652 (P. putida) | NP_758591 | 1E-153 |
| CL21 | 21575 | 22576 | → | 333 | 63.0 | tnpT | Put. cointegrate resolution protein T; homologous to tnpT from Tn4652 (P. putida) | NP_758592 | 2E-66 |
| CL22 | 22615 | 22836 | → | 73 | 60.4 | Unknown; no significant similarity | |||
| CL23 | 22869 | 23171 | ← | 100 | 55.8 | Hypothetical protein; BLAST hit for conserved domain protein | AAN70967 | 4E-23 | |
aa, amino acids.
Put., putative.
GenBank accession number.
FIG. 2.
Map of tRNALys-phnAB regions of strain K, PAO1, and C chromosomes. The tRNALys sites are indicated by thick black bars. In clone K strains, the tRNALys site can be used for reversible integration of plasmid pKLK106 (green triangle) (36). PAO1 carries an additional block (light gray triangle) at this site, comprising CDS PA0977 to PA0987. Strain C carries the gene island PAGI-4(C) at this position. Base pair counting starts after tRNALys. Two small segments (dark gray) with ORFs PA0977 and PA0980 are homologous to PAO1 sequence; two larger areas (yellow and orange) are C specific. The blue arrows show PAO1 CDS and their counterparts in K and C; the yellow and orange arrows represent C-specific CDS in PAGI-4(C). The blue boxes represent truncated PAO1 CDS in strain C.
The subsequent 12.7-kb DNA segment flanked by truncated versions of PA0981 and PA0994 (Fig. 2) encodes the typical elements of a transposon similar to Tn4652 from P. putida (34): the transposase gene tnpA (CL12), the transposase regulator gene tnpC (CL13), and the cointegrate resolution protein genes tnpS (CL20) and tnpT (CL21). With regard to the remaining eight CDS, a metabolic function could be ascribed only to CL14, CL15, CL17, and CL19 (Table 2).
The other block of novel DNA between CL1 and CL11 consists of 9.5 kb of non-PAO-homologous sequence. CL2a is predicted to encode a XerC-like integrase (23) (Table 2). All CDS of the CL2a-CL10 block have homologs in plasmid pKLC102, with conserved synteny and 87 to 99% amino acid sequence identity (CP103a, CP102, and CP93-CP87) (Table 3). CL10, adjacent to PA0980, is homologous to CP87 in pKLC102. The CP87-CP86 sequence contig in pKLC102 contains the 239-bp direct repeat (see above), and we noted that the repeat is 90% conserved in the CL11-CL10 contig in PAGI-4(C) (nucleotide identity at 216 of 239 positions). Moreover, the first 68 bp of the repeat (88% sequence identity) occur once in the PAO1 chromosome close to PA0980, in the intergenic region between PA0981 and PA0982 (Fig. 2). Shared sequence is known to trigger incorporation of donor into recipient DNA (22), and correspondingly, the direct repeat could have been involved in the evolution of the present PAGI-4(C) from an ancestor.
PAGI-4(C) was probably generated by at least two independent recombination events at a transposition close to the tRNALys(1) recognition site. The 9.5-kb part adjacent to the tRNALys(1) gene is homologous not only with sequences of the chromosomal and episomal versions of pKLC102 in clone C but also with the tRNAGly-associated gene island PAGI-2(C) (Table 2). The >95% sequence identity of the 9.5-kb stretch of DNA with parts of pKLC102 suggests the following scenario. An ancestor C strain, like the present clone K strains, was reversibly harboring a pKLC102-like plasmid at this site. When the 239-bp direct repeat was captured by the plasmid, a short stretch of sequence matched with the intergenic sequence between PA0981 and PA0982 located just five genes downstream of the att site in the tRNALys gene (Fig. 2). A similar situation is encountered in the tRNAGly-associated gene islands PAGI-2(C) and PAGI-3(SG) of clone C strains (40), in which another stretch of the direct repeat (positions 158 to 177) is found close to the attB sequence at the end of the island. We consider this coincidence to be relevant, because no further hits of sequences matching the direct repeat were retrieved from the databases. Thus, we propose that additional matching sequence in the vicinity of the att integration signal at the 3′ end of the tRNA gene could stabilize the maintenance of a genome island in the chromosome. However, in the case of the ancestor clone C strain, the acquisition of direct-repeat sequence may also have predisposed it to secondary changes, such as the truncation of the plasmid and the integration of the additional transposon. This proposal is substantiated by the fact that the clone K strains, which reversibly integrate pKLK106 at the tRNALys(1) site, do not harbor the repeat sequence in the chromosome (no PA0981-PA0982).
Sequence of pKLC102 at tRNALys(2), close to the pil region.
The organization of predicted CDS within the large 103,532-bp plasmid pKLC102 is displayed in Fig. 3. The annotation (Table 3) revealed 105 CDS, in two of which a smaller CDS resided in a larger CDS on the opposite strand (CP62a and -b and CP103a and -b).
Plasmid replication and recombination genes.
Of 105 identified CDS, 60 were classified as hypothetical or of unknown origin. Many of these hypothetical genes have DNA replication, recombination, and modification genes as neighbors (Fig. 3). Syntenic sets of homologous genes were identified in other plasmids and gene islands among gram-negative bacteria, including PAGI-2(C) and PAGI-3(SG) of P. aeruginosa clone C (40) (see Fig. 5). These genes may play a role in plasmid maintenance or horizontal gene transfer. At least 18 identified genes of pKLC102 are involved in plasmid conjugation, recombination, and repair, among them genes for two phage integrases (CP62a and CP103a), soj (encoding a chromosome-partitioning protein; CP1), genes for four helicases (CP9, CP30, CP56, and CP69), ssb (encoding a single-strand binding protein; CP22), the topoisomerase gene topA (CP27), and traG and traI (encoding conjugative proteins; CP67 and CP102).
FIG. 5.
Circular domain similarity plot. The inner and outer circles represent 50 and 100% similarities, respectively. Plasmid coordinates are shown along the outer circle.
The reversible chromosomal integration of pKLC102 at tRNALys(2) probably occurs by a phage-like mechanism under the control of the phage integrase XerC (CP103a) (23, 36, 50). Site-specific recombination takes place between the plasmid attachment site (attP) and the chromosomal attachment site (attB) at the 3′ end of the tRNALys(2) gene. attP is located 370 bp upstream from xerC and 68 bp downstream from soj. After integration of the plasmid into the chromosome, xerC and soj become the outermost genes of the gene island.
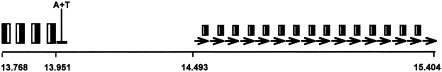
The region between CP18 and CP19 was recognized as the possible origin of replication, oriV, of pKLC102 (Fig. 3). Sixteen highly conserved 57-bp direct repeats constitute the right part of oriV (Fig. 4). All repeats except the last terminate with the 19-bp palindrome 5′-GTGGTGCCACTGGCACCAC (complementary sequence underlined), similar to synchrons of the Pseudomonas fluorescens plasmid pL6.5 (AJ250853) (P. Herbelin, unpublished data). The highly conserved nonpalindromic part of the repeats (38 bp) may serve as replication protein binding sites; however, their sequence is not similar to those of iterons of experimentally characterized oriVs of plasmids (20). In the left part of oriV (Fig. 4), an A+T-rich region is preceded by four palindromes, GAGTTCGGATGCCGAACTC, with the first loop inverted with respect to the others. A similar organization of the oriV locus, albeit shorter at the right side with only four repeats, was found in the intergenic region between Psyr3998 and Psyr3999 in the Pseudomonas syringae pv. syringae B728a genome. The oriV locus of pKLC102 is flanked by genes that are typically found in the ori regions of plasmids, such as dnaB (CP9), ssb (CP22), and topA (CP27). The episomal pKLC102 is probably replicated by the strand displacement mechanism (20, 28), because (i) no turning point indicative of the terminus of replication was detected by GC skew and (ii) in silico analysis of secondary DNA structure by the energy-optimized Greedy algorithm (11) predicted thermodynamically stable hairpins at the ori locus, which is typical for this mode of replication.
FIG. 4.
Structure of the origin of replication of pKLC102. Identical sequences are indicated by the sizes of the symbols. Adjacent solid and open boxes represent palindromes; the arrows indicate the sequences of 16 consecutive direct repeats. The A+T-rich region is indicated by a horizontal black bar.
Phenotypic traits.
Plasmid pKLC102 confers two major phenotypic traits on strain C, a pilin gene cluster (pilI to pilM) and the large 8,514-bp chvB gene (CP94), which most likely encodes a cyclic β-(1,2)-glucan synthetase. Cyclic β-glucan is a constitutive component of the periplasm of Agrobacterium tumefaciens, Sinorhizobium meliloti, and Brucella abortus. Knockouts of the glucan synthetase abrogate nodule formation by rhizobia (14, 26), gall tumor induction by Agrobacterium (21), and intracellular multiplication of Brucella in mammalian host cells (10). The pKLC102 gene exhibits the highest homology to the A. tumefaciens and Burkholderia fungorum chvB genes. Since the glucan synthetase is essential for the interaction of the prokaryote with its eukaryotic host (18), it is tempting to assume that chvB is a major determinant for host tropism and/or virulence of the respective strain.
A putative operon of 10 genes from CP33 to CP42 is similar in size, sequence, and gene arrangement to the pil operon of the Escherichia coli IncI plasmid R64 (73) and of the major pathogenicity island of Salmonella enterica serovar Typhi (75). In both cases, these pil operons encode type IV thin sex pili (42). The closest homolog of the pKLC102 pil operon was found to be a functionally uncharacterized operon in the P. syringae pv. syringae B728a genome, with the level of identity ranging from 29 to 47%. The CP39 gene product is homologous to the prepilin PilS, which is processed prior to assembly by pilU (CP40), which removes the N-terminal leader peptide. The adhesin at the pilus tip is encoded by pilV (CP41). In contrast to enterobacterial pil operons, in which the terminal pilV gene is followed by shufflon sequences (38), and the site-specific recombinase gene rci, the pil operons of pKLC102 and P. syringae terminate with pilM (CP42) and do not contain any recombination genes. The genetic organization of the pil operon in pKLC102 is appropriate for mating but lacks the option to evade the eukaryotic host immune response as it has evolved in enterobacteria. The transport of plasmid DNA through the sex pili requires coupling and pilot proteins (42). A putative FtsK coupling protein and the pilot protein (encoded by the conjugative relaxase gene traI) were identified as being encoded by CP81 and CP102, respectively. The activity of the FtsK proteins is controlled by a XerC integrase (1) represented by CP103a in pKLC102. Hence, the plasmid contains all of the genes that are necessary for conjugation. This pil operon is unrelated in sequence and genetic organization to the pil clusters of the P. aeruginosa chromosome that confer twitching motility and type II secretion (46), which corroborates the conclusion that pKLC102 encodes conjugative sex pili.
Besides chvB and the pil cluster, annotation provided no unequivocal clues about the additional extra metabolic features that pKLC102 confers on its host strain. Two genes (CP99 and CP100) encode novel fatty acid synthases. A putative chemotaxis operon (CP84 to CP86) and a cold adaptation protein (encoded by CP28) may provide further options for the response to environmental signals, and a polyketide synthase (encoded by CP57) and a protein with a VagC domain (encoded by CP26) are putative virulence-associated proteins. Moreover, an Arc repressor (encoded by CP16), a phage antirepressor (encoded by CP21) (17), and four putative transcription regulators (encoded by CP59, CP61, CP92, and CP96) were identified.
Origin, source, and horizontal gene transfer.
According to sequence database comparisons, plasmid pKLC102 shares DNA with numerous proteobacteria, of which P. aeruginosa PAO1 contributed only a minor part (the gene cassette PA2566-PA2564 [see above]) (Fig. 5). The genetic repertoire of pKLC102 was predominantly assembled from two lineages. One part exhibits strong homology with gene islands in the P. syringae pv. syringae B728a and enterobacterial genomes. This DNA block includes oriV, the pil cluster, and conjugative elements, which points to the inheritance of these genes from a common ancestral plasmid (Fig. 5). The other major DNA block is homologous to several tRNA-integrated genome islands, of which 35 CDS distributed on six segments are similar to CDS in the clone C islands PAGI-2(C) and PAGI-3(SG) (40) and genome islands of other proteobacteria (Fig. 5). To explore the phylogenetic relationships in more detail, the longest conserved gene contig (CP73 to CP81) of the six segments was selected for cladistic comparison. pKLC102 of strain C was found to segregate with other tRNALys-associated gene islands found in Azotobacter vinelandii and P. fluorescens, whereas PAGI-2(C) of strain C was more closely related to other tRNAGly-associated gene islands of Burkholderia fungorum and Ralstonia metallidurans.
In summary, pKLC102 is composed of a mosaic of blocks of diverse origin. The orthologs and paralogs with the highest sequence similarities were typically identified in A. vinelandii, P. syringae, P. fluorescens, and Burkholderia spp., all of which are associated with plants, particularly with the rhizosphere. Hence, pKLC102 most likely evolved in plant-associated microbial communities.
Integrases.
pKLC102 recombines within the 3′ end of the tRNALys(2) structural gene in the chromosome. tRNA genes are typical integration sites for phages, but not for plasmids (12). Annotation revealed that integration and excision are probably mediated by the phage tyrosine integrase XerC (encoded by CP103a) (Fig. 3 and 6). CP103a shows 60 and 55% amino acid identity with the xerC genes Avin0928 and AAM77365 detected by BLAST in the A. vinelandii and P. syringae strain BR2R genomes, suggesting that these three XerC integrases have a common chromosomal target site. Tyrosine integrases are a family of site-specific recombinases found in bacteria, plasmids, and bacteriophages (1, 9, 15, 25, 32, 47, 56). The conserved C-terminal protein domains cleave and religate the DNA; thus, a covalent intermediate is formed between DNA and the tyrosine in the active site of the integrase (7, 37, 61). The nonconserved N-terminal domains possess high-affinity DNA binding sites and act as context-sensitive modulators of enzyme activity.
FIG. 6.
Inner opposite CDS of XerC integrases CL2ab, CP103ab, and Avin0928. The integrase genes and putative traI genes CL3, CP102, and Avin0927 located upstream of the integrases are shown by open arrows. The integration attachment sites downstream of the integrases are indicated by solid boxes. Identified inner ORFs (putative excisionases) are depicted by shaded arrows. The boxed sequences indicate the putative termination loops following the inner ORFs in pKLC102 and A. vinelandii.
Within CP103a, a second divergently transcribed ORF, CP103b, was identified (Fig. 6). The gene prediction programs revealed comparably high likelihood coefficents for both ORFs to encode a protein, implying that the two ORFs are not artifacts but two transcribed CDS. Moreover, the A. vinelandii homolog Avin0928 is strikingly similar to CP103 in its makeup (Fig. 6), i.e., an inner ORF resides within a larger ORF and the ribosome binding sites, start codon, and stem-loop terminator for the transcription of the inner ORF are located at corresponding positions.
The opposite activities of an integrase to catalyze both integration into and the excision from the chromosome are regulated by an excisionase (13, 23). In enterobacteria, the integrase and excisionase are encoded by adjacent int and xis genes that may partially overlap, as is the case for the E. coli phage λ (7, 23, 37, 61). Hence, a complete overlap of the two genes is reasonable. Accordingly, the outer ORF, CP103a, and the inner ORF, CP103b, were annotated as int and xis; thus, the gene product of the latter, like its weak homolog Cox of phage P2 (58, 74), may function not only as an excisionase but also as a trancription regulator for proteins that mobilize the gene island.
The int locus should play a key role in the chromosomal incorporation and mobilization of pKLC102. In the case of the clc element (50, 63), which so far is the only experimentally characterized gene island in Pseudomonas, the presence of int was necessary and sufficient for integration into and mobilization from the chromosome. In order to execute these opposite activities through one locus, a complex genetic structure is instrumental in expressing just one activity at a time. The divergent transcription of the same sequence observed in CP103 and Avin0928 (Fig. 6) is a mechanism of genetic control to meet this requirement.
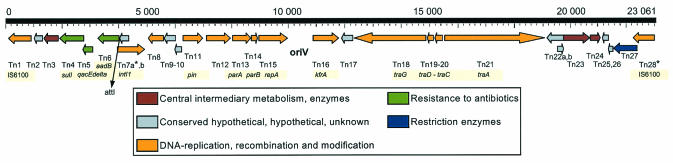
Integron TNCP23 within pKLC102 of subgroup C chromosomes.
A large 23,061-bp class I transposon (Table 4) inserted into an AT-rich region of pKLC102. This transposon, called TNCP23, was found only in clone C chromosomes of subgroup C (39, 53). TNCP23 is flanked at both ends by the insertion sequence (IS) element IS6100 (65). TNCP23 integrated upstream of the pil operon at position 28,440 of pKLC102 (Fig. 1); thus, the last 8 nucleotides 5′ of the breakpoint (positions 28,433 to 28,440) were duplicated so that the 17-mer inverted repeats at the termini of IS6100 are flanked on both sides by the direct repeat 5′-TTCCGAAC. Hence, the sequences spanning the integration point read 5′-TTCCGAACGGCTCTGTTGCAAAAAT at the right end and 5′-ATCTTTGCAACAGAGCCTTCCGAAC at the left end. Inspection of the adjacent plasmid sequence did not disclose any known recombination signals, such as direct or inverted repeats; however, the breakpoint is located approximately in the middle of a 2-kb region with a GC content (42%) significantly lower than the average GC content (60.9%) of pKLC102 (Table 1). The lower thermodynamic stability of base pairs in AT-rich regions may have facilitated the targeting of the transposon to this site.
TABLE 4.
Annotation of all ORFs located within integron TNCP23
| Gene identification | Coordinates
|
Direction | Length (aa)a | GC content (%) | Gene name | Annotation result and homologous gene products | BLAST search resultb | E value of best BLAST hit | |
|---|---|---|---|---|---|---|---|---|---|
| Left | Right | ||||||||
| TNCP1 | 63 | 857 | ← | 264 | 61.01 | tnp | Transposase; similar to Tnp from S. enterica subsp. enterica serovar Typhimurium; IS6100 element | AAG03007 | E-112 |
| TNCP2 | 1.023 | 1.310 | ← | 95 | 64.31 | Hypothetical protein similar to ORF6 (P. aeruginosa) | AAK96399 | 8E-33 | |
| TNCP3 | 1.334 | 1.834 | ← | 166 | 66.27 | Acetyltransferase similar to ORF5 (P. aeruginosa) | CAA11478 | 1E-90 | |
| TNCP4 | 1.962 | 2.801 | ← | 279 | 61.57 | sulI | Dihydropteroate synthase Sul1 (P. aeruginosa); sulfonamide resistance protein | AAA25859 | E-143 |
| TNCP5 | 2.795 | 3.142 | ← | 115 | 50.00 | qacEde | Ethidium bromide resistance protein | AAC44316 | 1E-42 |
| lta | QacEdelta1 (P. aeruginosa) | ||||||||
| TNCP6 | 3.299 | 4.048 | ← | 249 | 51.76 | aadB | Aminoglycoside adenylyltransferase AadB (P. aeruginosa); streptothricin-spectinomycin resistance protein | AAD39934 | 7E-97 |
| TNCP7a | 3.978 | 4.937 | → | 319 | 61.25 | int1 | Phage integrase similar to Int from P. aeruginosa | AAA25857 | E-154 |
| TNCP7b | 4.068 | 4.376 | ← | 102 | 62.35 | Hypothetical protein similar to ORF2 (E. coli) | B26839 | 3E-47 | |
| TNCP8 | 5.054 | 5.596 | → | 180 | 63.17 | ISxac3 transposase similar to XACa0010 (Xanthomonas axonopodis pv. citri) | NP_644696 | 1E-18 | |
| TNCP9 | 5.602 | 5.997 | → | 131 | 62.37 | Conserved hypothetical protein similar to XCC1632 (Xanthomonas campestris pv. campestris) | NP_637002 | 4E-40 | |
| TNCP10 | 5.994 | 6.245 | ← | 83 | 61.51 | Conserved hypothetical protein similar to XCC1631 (X. campestris pv. campestris) | NP_637001 | 1E-35 | |
| TNCP11 | 6.310 | 6.993 | → | 227 | 64.77 | Putative invertase/recombinase (X. axonopodis) | NP_644692 | 2E-75 | |
| TNCP12 | 7.113 | 7.964 | → | 283 | 69.25 | Putative resolvase similar to pVS1 resolvase | AAD19676 | 2E-80 | |
| TNCP13 | 8.045 | 8.677 | → | 210 | 63.98 | parA | ParA partitioning protein (Serratia marcescens) | BAB71950 | 9E-92 |
| TNCP14 | 8.698 | 8.919 | → | 73 | 60.36 | parB | Putative ParB plasmid stabilization protein (Pseudomonas alcaligenes) | AAD40335 | 0.006 |
| TNCP15 | 8.972 | 9.985 | → | 337 | 63.91 | repA | RepA replication protein (X. axonopodis pv. citri) | NP_642756 | E-110 |
| TNCP16 | 10.900 | 11.790 | → | 296 | 70.82 | kfrA | KfrA protein (X. axonopodis pv. citri) | NP_644728 | 2E-12 |
| TNCP17 | 11.964 | 12.356 | ← | 130 | 69.47 | No significant similarity | |||
| TNCP18 | 12.367 | 14.889 | ← | 840 | 25.68 | traG | Plasmid transfer factor protein similar to TraG (M. loti) | BAB54919 | 8E-36 |
| TNCP19 | 14.924 | 15.145 | ← | 73 | 59.46 | traD | Putative conjugative protein similar to TraD (Rhizobium sp.) | NP_443830 | 0.22 |
| TNCP20 | 15.168 | 15.425 | ← | 85 | 55.81 | traC | Conjugal transfer protein similar to TraC (Brucella suis 1330) | AE014536 | 1E-12 |
| TNCP21 | 15.442 | 18.963 | → | 1,173 | 18.26 | traA | Conjugal transfer protein similar to TraA (M. loti); MobA/MobL family conserved domain | BAB52121 | E-132 |
| TNCP22a | 19.145 | 19.717 | ← | 190 | 63.70 | Similar to truncated N-terminal conserved domain of phage integrases | |||
| TNCP22b | 19.511 | 19.729 | → | 72 | 63.01 | No significant similarity, overlapping by TNCP22a | |||
| TNCP23 | 19.716 | 20.651 | → | 311 | 68.27 | Serine protease similar to Ycg4E (C. glutamicum) | AAG00285 | 4E-74 | |
| TNCP24 | 20.747 | 21.085 | → | 112 | 66.67 | Hypothetical protein similar to coenzyme F390 synthetase XF1916 (Xylella fastidiosa) | NP_299202 | 0.16 | |
| TNCP25 | 21.148 | 21.354 | ← | 68 | 66.18 | Hypothetical protein similar to XfasO0625 (X. fastidiosa) | ZP_00040826 | E-6 | |
| TNCP26 | 21.351 | 21.524 | ← | 57 | 71.84 | No significant similarity | |||
| TNCP27 | 21.521 | 22.357 | ← | 278 | 63.08 | Putative type II restriction enzyme similar to XfasA1931 (X. fastidiosa Dixon) | ZP_00039987 | E-104 | |
| TNCP28 | 22.284 | 23.030 | ← | 248 | 60.78 | tnp | Truncated transposase; similar to Tnp from S. enterica subsp. enterica serovar Typhimurium IS6100 element | AAG03007 | E-112 |
aa, amino acids.
GenBank accession number.
TNCP23 has a complex structure (Fig. 7). The two large 880-bp IS6100 elements (65) are each flanked by two 17-bp inverted repeats, and each encodes a transposase, one of which is inactivated by a frameshift mutation. tnp of the left copy IS6100-L is intact and exhibits 100% identity to tnpA on Tn610 from Mycobacterium fortuitum (65). IS6100-L represents the 3′ end of a type In7 class I integron (25, 48) (Fig. 7). The large 4,871-bp integron possesses the typical recombination signal attI; the three conserved genes qacΔ1, sulI, and orf5i (TNCP3) (Table 4); and an integrase gene, int1; the last, however, is truncated by 203 bp at the 5′ end and therefore is probably nonfunctional. As in pKLC102, a divergently transcribed xis gene was identified within int1 (TNCP7a and -b). A gene cassette with an aadB gene (TNCP6) encoding an aminoglycoside-adenyltransferase for gentamicin and tobramycin is inserted into attI. Integrons of similar structure are known from the P. aeruginosa plasmid R1033 (accession no. U12338) and the Corynebacterium glutamicum plasmid pCG4 (48), but the deletion in int1 at the 5′ end has so far not been reported.
FIG. 7.
Gene map of TNCP23. The map is calibrated to the site of integration into the chromosome of strain C. The leading strand was defined by colinearity with the P. aeruginosa PAO1 genome sequence. Predicted coding regions are shown by arrows indicating the direction of transcription. The genes are color coded according to their functional categories as shown in the legend below the map. All of the genes carry identification numbers according to the CDS numbering in Table 4, but the abbreviation Tn was used instead of TNCP due to space limitations. Gene names are highlighted beneath the corresponding CDS. oriV is the predicted origin of replication.
Strain C was isolated from the airways of a CF patient who regularly received tobramycin for the treatment of her P. aeruginosa infection. The aadB gene confers resistance to aminoglycosides, which may have selected for the incorporation of the transposon into the chromosomally integrated pKLC102. In contrast, the transposon genes outside the integron confer no apparent advantage on the bacterial host. About 50% of the genes are homologs of typical plasmid genes, such as traACDG, needed for conjugation; repA, oriV, and kfrA, needed for replication; and pin, parAB, and repA, needed for the distribution and maintenance of plasmid DNA during cell division. In summary, TNCP23 is a mosaic of IS elements, integron, and plasmid. One tnp gene and the int1 gene were probably inactivated by a frameshift mutation and deletion, respectively. The IS6100-L element with the intact tnp proved to be the starting point for chromosomal inversions (Fig. 8) (39).
FIG. 8.
Evolution of P. aeruginosa strains linked to plasmid DNA. (a) Reversible integration of plasmid DNA into two possible sites of clone K strains. (b) Different forms of plasmid DNA in clone C strains. In subgroup SG17M, pKLC102 is found episomally and integrated into the genome at tRNALys(2). Strain C5 apparently lost the pKLC102 DNA, while strain C2 harbors only the integrated form. In subgroup C, the integron TNCP23 inserted into chromosomally integrated pKLC102. Free plasmid is not detectable in subgroup C strains, indicating that TNCP23 prevented mobilization. TNCP23 is flanked by copies of IS6100. Intramolecular transposition of the left copy of IS6100-L is coupled with an inversion of the chromosomal region between the transposed copy and IS6100-L in some strains of subgroup C. For these strains C8, C9, C10, and C19, the tRNALys(1) area is not shown.
A copy of IS6100-L, called IS6100-t, inserted into several P. aeruginosa clone C subgroup C chromosomes, causing the region between the transposed copy and IS6100-L to be inverted. These large inversions were detected exclusively in sequential clone C isolates from CF lung infections (53). The inversion breakpoints in strains C9, C10, and C19 were identified as wbpM, pilB, and mutS, which gave rise to O-antigen deficiency, loss of twitching motility, and hypermutability, respectively (39). The action of IS6100-L on clone C strains was thus twofold: after capture by integration, the established additional copy, IS6100-t, reorganized the chromosome by large inversions and disrupted genes that are typically inactivated during the adaptation of P. aeruginosa to the CF lung habitat (see the original article by Kresse et al. [39] for more details).
Genome evolution in P. aeruginosa clone C.
The related clones C and K are among the major clones of the present P. aeruginosa population (36, 55). The abundance of several hundred C and K isolates in our collection of >3,000 strains from clinical and environmental habitats made it possible to evaluate intraclonal genome diversity by physical mapping and sequencing and, as shown here, to trace the underlying genome rearrangements. The P. aeruginosa clones K and C are thus among the first examples for which bacterial genome evolution could be documented by analyzing related isolates retrieved from their natural habitats.
pKLK106 and pKLC102 are highly homologous plasmids. pKLK106 reversibly recombines with clone K chromosomes at one of the two tRNALys genes (Fig. 8). In all investigated clone K strains, both episomal and chromosomal copies were detected. During the propagation of single colonies on agar plates in vitro, progeny that had retargeted pKLK106 into the other tRNALys locus were regularly observed, indicating that pKLK106 is mobilized and reintegrated into the clone K chromosome at high frequency.
Plasmid pKLC102 could recombine with the tRNALys (2) gene only close to the pilA locus, because the other site was blocked by PAGI-4(C). The only extra DNA of pKLC102 that is absent in pKLK106 is a P. aeruginosa operon flanked by direct repeats which match with PAO chromosomal sequence in the vicinity of tRNALys(1), which is present in C but not in K chromosomes. Repeats and tRNALys(1) encompass a 9.5-kb block which is found again with conserved synteny and >90% sequence homology in pKLC102. We assume that the proximity of two targeting signals in cis initiated complex genome rearrangements which led to the irreversible incorporation of one small part of a pKLC102 ancestor next to the tRNALys(1) gene.
All investigated clone C isolates from aquatic habitats and the hospital environment harbored chromosomal and episomal copies of pKLC102. However, many isolates from CF lungs contain either no (C5) or only chromosomally integrated (C2) pKLC102 (Fig. 8). The latter scenario is typical for a genome island (29, 30, 31). Of the four subgroups of clone C (53), subgroup C is exclusively represented by CF lung isolates and differs from the other three groups by the insertion of the class I composite transposon TNCP23 into chromosomally integrated pKLC102, which may have been acquired because of the aadB gene conferring gentamicin resistance (Fig. 8). P. aeruginosa converges in CF lungs to a common phenotype characterized by the decreased production of membrane components, cellular appendages, and secreted factors (45, 69). This phenotypic signature was partially gained in subgroup C strains by TNCP23-mediated chromosome remodeling. Intramolecular transposition of the active IS6100 element of TNCP23 led to large chromosomal inversions, which disrupted genes that are typically inactivated during the adaptation of P. aeruginosa to the atypical habitat of CF lungs (Fig. 8). In parallel, the integrity of pKLC102 was destroyed. The two attachment sites were separated, so that the genetic content of pKLC102 was irreversibly fixed in the chromosome. In summary, Fig. 8 portrays the evolution of a plasmid from a mobile genetic element to an irreversibly fixed genome island that finally was disrupted and distributed among separate chromosomal regions. It should be noted that the increasing complexity of genome organization caused by insertion, transposition, and inversion was accompanied by mutation, deletion, and/or duplication of sequence close to the breakpoint.
Horizontally acquired elements, such as prophages, plasmids, and genome islands, have been detected in numerous completely sequenced bacterial genomes (3, 19, 24, 29-31) based on sequence homology, phylogenetic profiling, the presence of diagnostic genes (for example, int, tnp, ori, and tra), and/or global criteria, such as atypical GC content, codon usage, or oligonucleotide frequency bias. However, with the exception of the spread of resistance determinants, most in silico findings are not backed up by knowledge about the original donors and recipients and the underlying mode of transmission. pKLC102 is one of the rare examples for which the causative action on genome evolution can be demonstrated. pKLC102 coexists in the episomal and chromosomal states and recombines with and mobilizes from the chromosome at high frequency, even in the absence of any apparent stress stimuli. Annotation and phylogenetic analyses point to the possible origin of this double role of plasmid and genome island. The closest homologs of pKLC102 are plasmids and phage-type genome islands (Fig. 5). The plasmid lineage conferred genes for replication, partitioning, and conjugation, and the phage lineage conferred integrase, att, and the syntenic set of conserved hypothetical genes also observed in the tRNAGly-associated gene islands on clone C chromosomes (40). Interestingly, the closest neighbors of the phage lineage inhabit the rhizosphere, while the closest neighbors of the plasmid lineage colonize the phyllosphere (Fig. 5). Hence, pKLC102 probably emerged in a plant habitat from a phage lineage and a plasmid lineage that endowed this hybrid with the uncommon flexibility to exist as a conjugative plasmid and as a genome island.
Genome islands adapt over time to the taxospecies-specific signature of the core genome (29, 30, 31). pKLC102 escaped this adaptation. Its tetranucleotide frequency bias defines a lineage that is separate from those of the completely sequenced P. aeruginosa, P. putida, and P. syringae genomes (data not shown). Moreover, the genetic repertoire of pKLC102 includes mainly genes for its own maintenance and propagation. Even the putative virulence gene chvB may primarily facilitate the spread of the plasmid; its impact on the pathogenicity and fitness of the host bacterium may be just an implicit secondary effect. In conclusion, pKLC102 exhibits typical features of a selfish genetic element, and this is probably a major reason why it coexists in most isolates from environmental and disease habitats as both a plasmid and a genome island.
Acknowledgments
We thank Christian Weinel for the provision of software and Helmut Hilbert and Andre Bahr for DNA sequencing.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) (Tu40-5, Schwerpunktprogramm “Ökologie bakterieller Krankheitserreger-molekulare und evolutionäre Aspekte”). J.K. and O.R. are members of the DFG-sponsored Europäisches Graduiertenkolleg “Pseudomonas: Pathogenicity and Biotechnology.”
REFERENCES
- 1.Abremski, K., and S. Gottesman. 1981. Site-specific recombination. Xis-independent excisive recombination of bacteriophage lambda. J. Mol. Biol. 153:67-78. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arber, W. 2000. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 24:1-7. [DOI] [PubMed] [Google Scholar]
- 4.Arora, S. K., M. Bangera, S. Lory, and R. Ramphal. 2001. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc. Natl. Acad. Sci. USA 98:9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. Wiley, New York, N.Y.
- 6.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Better, M., C. Lu, R. C. Williams, and H. Echols. 1982. Site-specific DNA condensation and pairing mediated by the Int protein of bacteriophage λ. Proc. Natl. Acad. Sci. USA 79:5837-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briones, G., N. I. de Iannino, M. Roset, A. Vigliocco, P. S. Paulo, and R. A. Ugalde. 2001. Brucella abortus cyclic β-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect. Immun. 69:4528-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodsky, L. I., V. V. Ivanov, Y. L. Kalaydzidis, A. M. Leontovich, V. K. Nikolaev, S. I. Feranchuk, and V. A. Drachev. 1995. GeneBee-NET: internet-based server for analyzing biopolymer structure. Biochemistry (Moscow) 60:923-928. [PubMed] [Google Scholar]
- 12.Campbell, A. M. 1992. Chromosomal insertion sites for phages and plasmids. J. Bacteriol. 174:7495-7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrol, D., M. A. Kehoe, D. Cavanagh, and D. C. Coleman. 1995. Novel organization of the site-specific integration and excision recombination functions of the Staphylococcus aureus serotype F virulence-converting phages φ13 and φ42. Mol. Microbiol. 16:877-893. [DOI] [PubMed] [Google Scholar]
- 14.Castro, O. A., A. Zorreguieta, V. Ielmini, G. Vega, and L. Ielpi. 1996. Cyclic β-(1,2)-glucan synthesis in Rhizobiaceae: roles of the 319-kilodalton protein intermediate. J. Bacteriol. 178:6043-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collis, C. M., M.-J. Kim, S. R. Partridge, H. W. Stokes, and R. M. Hall. 2002. Characterization of the class 3 integron and the site-specific recombination system it determines. J. Bacteriol. 184:3017-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in procariotic membrane proteins: the Dense Alignment Surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 17.Davis, B. M., H. H. Kimsey, A. V. Kane, and M. K. Waldor. 2002. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 15:4240-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Iannino, N. I., G. Briones, M. Tolmasky, and R. A. Ugalde. 1998. Molecular cloning and characterization of cgs, the Brucella abortus cyclic β-(1-2) glucan synthetase gene: genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J. Bacteriol. 180:4392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Cruz, F., and J. Davies. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8:128-133. [DOI] [PubMed] [Google Scholar]
- 20.Del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglas, C. J., W. Halperin, and E. W. Nester. 1982. Agrobacterium tumefaciens mutants affected in attachment to plant cells. J. Bacteriol. 152:1265-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 23.Echols, H., and G. Guarneros. 1983. Control of integration and excision, p. 75-92. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Eisen, J. A. 2000. Assessing evolutionary relationships among microbes from whole-genome analysis. Curr. Opin. Microbiol. 3:475-480. [DOI] [PubMed] [Google Scholar]
- 25.Fluit, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 26.Geremia, R. A., S. Cavaignac, A. Zorreguieta, N. Toro, J. Olivares, and R. A. Ugalde. 1987. A Rhizobium meliloti mutant that forms ineffective pseudonodules in alfalfa produces exopolysaccharide but fails to form β(1-2)glucan. J. Bacteriol. 169:880-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberg, J. B., and D. E. Ohman. 1984. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J. Bacteriol. 158:1115-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gophna, U., A. Parket, J. Hacker, and E. Z. Ron. 2003. A novel ColV plasmid encoding type IV pili. Microbiology 149:177-184. [DOI] [PubMed] [Google Scholar]
- 29.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity—a Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 31.Hacker, J., and J. B. Kaper (ed.). 2002. Pathogenicity islands and the evolution of pathogenic microbes. Curr. Top. Microbiol. Immunol. 264:1-211. [PubMed] [Google Scholar]
- 32.Hansson, K., L. Sundström, A. Pelletier, and P. H. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heuer, T., C. Bürger, G. Maaβ, and B. Tümmler. 1998. Cloning of prokaryotic genomes in yeast artificial chromosomes: application to the population genetics of Pseudomonas aeruginosa. Electrophoresis 19:486-494. [DOI] [PubMed] [Google Scholar]
- 34.Horak, R., and M. Kivisaar. 1998. Expression of the transposase gene tnpA of Tn4652 is positively affected by integration host factor. J. Bacteriol. 180:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiewitz, C., and B. Tümmler. 2000. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J. Bacteriol. 182:3125-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiewitz, C., K. Larbig, J. Klockgether, C. Weinel, and B. Tümmler. 2000. Monitoring genome evolution ex vivo: reversible chromosomal integration of a 106 kb plasmid at two tRNALys gene loci in sequential Pseudomonas aeruginosa airway isolates. Microbiology 146:2365-2373. [DOI] [PubMed] [Google Scholar]
- 37.Kim, S.-H., and A. Landy. 1992. Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR. Science 276:198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komano, T. 1999. Shufflons: multiple inversion systems and integrons. Annu. Rev. Genet. 33:171-191. [DOI] [PubMed] [Google Scholar]
- 39.Kresse, A. U., S. D. Dinesh, K. Larbig, and U. Römling. 2003. Impact of large chromosomal inversions on the adaptation and evolution of Pseudomonas aeruginosa chronically colonizing cystic fibrosis lungs. Mol. Microbiol. 47:145-158. [DOI] [PubMed] [Google Scholar]
- 40.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H.-J. Fritz, and B. Tümmler. 2002. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang, X., X.-Q. T. Pham, M. V. Olson, and S. Lory. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 43.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukashin, A., and M. Borodovsky. 1998. GeneMark: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 47.Murata, T., M. Ohnishi, T. Ara, J. Kaneko, C.-G. Han, Y. F. Li, K. Takashima, H. Nojima, K. Nakayama, A. Kaji, Y. Kamio, T. Miki, H. Mori, E. Ohtsubo, Y. Terawaki, and T. Hayashi. 2002. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J. Bacteriol. 184:3194-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nesvera, J., J. Hochmannova, and M. Patek. 1998. An integron of class 1 is present on the plasmid pCG4 from gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol. Lett. 169:391-395. [DOI] [PubMed] [Google Scholar]
- 49.Osborn, A. M., and D. Böltner. 2002. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid 48:202-212. [DOI] [PubMed] [Google Scholar]
- 50.Ravatn, R., S. Studer, A. J. B. Zehnder, and J. R. van der Meer. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Römling, U., J. Greipel, and B. Tümmler. 1995. Gradient of genomic diversity in the Pseudomonas aeruginosa chromosome. Mol. Microbiol. 17:323-332. [DOI] [PubMed] [Google Scholar]
- 52.Römling, U., T. Heuer, and B. Tümmler. 1994. Bacterial genome analysis by pulsed field gel electrophoresis techniques. Adv. Electrophoresis 7:353-406. [Google Scholar]
- 53.Römling, U., K. D. Schmidt, and B. Tümmler. 1997. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J. Mol. Biol. 271:386-404. [DOI] [PubMed] [Google Scholar]
- 54.Römling, U., and B. Tümmler. 1992. Comparative mapping of the Pseudomonas aeruginosa PAO genome with rare-cutter linking clones or two-dimensional pulsed-field gel electrophoresis protocols. Electrophoresis 14:283-289. [DOI] [PubMed] [Google Scholar]
- 55.Römling, U., J. Wingender, H. Müller, and B. Tümmler. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roy, P. H. 1999. Horizontal transfer of genes in bacteria. Microbiol. Today 26:168-170. [Google Scholar]
- 57.Sabath, C. D. (ed.). 1980. Pseudomonas aeruginosa: the organism, diseases it causes, and their treatment. Hans Huber Publishers, Berne, Switzerland.
- 58.Saha, S., E. Haggård-Ljungquist, and K. Nordström. 1987. The cox protein of bacteriophage P2 inhibits the formation of the repressor protein and autoregulates the early operon. EMBO J. 6:3191-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 60.SantaLucia, J. J. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 95:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarkar, D., M. Radman-Livaja, and A. Landy. 2001. The small DNA binding domain of λ integrase is a context-sensitive modulator of recombinase functions. EMBO J. 20:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt, K. D., B. Tümmler, and U. Römling. 1996. Comparative mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 178:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sentchilo, V., A. J. Zehnder, and J. R. van der Meer. 2003. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. strain B13. Mol. Microbiol. 49:93-104. [DOI] [PubMed] [Google Scholar]
- 64.Shibuya, T., and I. Rigoutsos. 2002. Dictionary-driven prokaryotic gene finding. Nucleic Acids Res. 30:2710-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith, B., and P. Dyson. 1995. Inducible transposition in Streptomyces lividans of insertion sequence IS6100 from Mycobacterium fortuitum. Mol. Microbiol. 18:933-941. [DOI] [PubMed] [Google Scholar]
- 66.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Springael, D., K. Peys, A. Ryngaert, S. V. Roy, L. Hooyberghs, R. Ravatn, M. Heyndrickx, J. R. Meer, C. Vandecasteele, M. Mergeay, and L. Diels. 2002. Community shifts in a seeded 3-chlorobenzoate degrading membrane biofilm reactor: indications for involvement of in situ horizontal transfer of the clc-element from inoculum to contaminant bacteria. Environ. Microbiol. 4:70-80. [DOI] [PubMed] [Google Scholar]
- 68.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 69.Tümmler, B., and C. Kiewitz. 1999. Cystic fibrosis: an inherited susceptibility to bacterial respiratory infections. Mol. Med. Today 5:351-358. [DOI] [PubMed] [Google Scholar]
- 70.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wenzel, R., and R. Herrmann. 1996. Cosmid cloning with small genomes, p. 197-222. In B. Birren and E. Lai (ed.), Nonmammalian genomic analysis: a practical guide. Academic Press, San Diego, Calif.
- 72.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoshida, T., S.-R. Kim, and T. Komano. 1999. Twelve pil genes are required for biogenesis of the R64 thin pilus. J. Bacteriol. 181:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu., A., and E. Haggård-Ljungquist. 1993. The Cox protein is a modulator of directionality in bacteriophage P2 site-specific recombination. J. Bacteriol. 175:7848-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, X.-L., I. S. M. Tsui, C. M. C. Yip, A. W. Y. Fung, D. K.-H. Wong, X. Dai, Y. Yang, J. Hackett, and C. Morris. 2000. Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 68:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]