The aberrant-activation of tyrosine kinases represents an important oncogenic mechanism, and yet the majority of such events remain undiscovered. Here we describe a bead-based method capable of profiling tyrosine kinase phosphorylations in a multiplexed, high-throughput and low-cost manner. This approach allows for the discovery of tyrosine kinase-activating events, even when the DNA sequence is wild-type. In an effort to pilot the establishment of a tyrosine kinase activation catalog, we profiled 130 human cancer lines, and followed-up on the frequent SRC phosphorylation in glioblastoma. SRC was also activated in primary patient samples, and the SRC inhibitor dasatinib inhibited viability and cell-migration in vitro, and tumor-growth in vivo. Furthermore, while dasatinib inhibits many tyrosine kinases, the testing of dasatinib-resistant tyrosine kinase alleles showed that SRC is indeed the relevant target. These studies establish the power and feasibility of tyrosine-kinome-wide phosphorylation profiling, and point to SRC as a possible therapeutic target in glioblastoma.
All tumors, regardless of their cell of origin, arise through the aberrant activation of oncogenic pathways. Foremost among oncogenes discovered to date are the tyrosine kinases, which are receiving high priority in international efforts to systematically discover all recurrent genomic alterations in the cancer genome. While such cancer genomic efforts will certainly identify mutations in tyrosine kinase-encoding genes, there is mounting evidence of indirect, oncogenic activation of kinases. For example, the PDGF receptor beta is aberrantly activated through a chromosomal translocation that results in constitutive expression of its ligand, PDGFB 1. Similarly, hepatocyte growth factor (HGF) amplification has been proposed to lead to oncogenic activation of its receptor, MET 2. Such indirect mechanism of tyrosine kinase activation would not be detected by cancer genome resequencing approaches that focus on the kinases themselves. We therefore sought to develop a method capable of the systematic identification of activated tyrosine kinases, regardless of the mechanism of activation.
Because nearly all tyrosine kinases become tyrosine phosphorylated following activation, we asked whether profiling tyrosine kinase phosphorylation state might be used as a surrogate for aberrant kinase activity. First, we profiled tyrosine phosphorylation in 5 cell lines using phospho-tyrosine peptide immunoprecipitation followed by mass spectrometry (Figure 1a and Supplementary Table 1 online). Using this approach, we recovered known tyrosine phosphorylated tyrosine kinases and identities of specific pY-sites in these proteins (e.g. ABL1 in K562 harboring a BCR-ABL1 translocation) (Supplementary Table 1 online). However, this mass spectrometry-based approach required large numbers of cells (~ 108 per sample), many days of sample preparation, processing and instrument time per sample, and was therefore costly (~ $1,000 per sample). In addition, its sensitivity depends on the physical characteristics of the phospho-peptides. These attributes make the approach unattractive for the high-throughput tyrosine phosphorylation profiling that would be required to capture the diversity of tyrosine kinase activation events in human cancer.
Figure 1. Luminex immunosandwich assay and IP-MS approach.
(a) The left panel shows the Luminex immunosandwich assay. Antibody-coupled beads were incubated with cellular protein lysates to capture tyrosine kinases. A biotinylated phospho-tyrosine antibody and streptavidin R-phycoerythrin conjugate (SAPE) were added sequentially to bind to the phospho-tyrosine residues. The mixture was analyzed with a Luminex 100 instrument, whereby each microsphere is analyzed with two lasers -- one to detect the bead color and thereby the identity of the tyrosine kinase, and the other to detect the R-phycoerythrin signal reflecting the tyrosine phosphorylation levels on the bead. The right panel presents the IP-MS approach. Total protein lysates were subjected to enzymatic digestion. Subsequently, the digested lysates were immunoprecipitated with a cocktail of phospho-tyrosine antibodies to enrich for phospho-tyrosine containing peptides. Upon elution, the peptide mixture was analyzed with mass spectrometry to determine their sequences and phosphorylation sites. (b) Comparison between IP-MS and Luminex data on five cell lines. The tyrosine phosphorylations on tyrosine kinases were assayed on biological replicates with either Luminex immunosandwich assay or IP-MS approach as described. Van diagrams of pY-TKs are shown. Numbers of pY-TKs identified with each method alone or both methods are indicated in parentheses. (c) Luminex fluorescence intensities on selected positive control samples. Lysates were prepared from 293FT cells infected with either a control lentivirus (Ctrl) or virus expressing the tyrosine kinases specified in the figure. The samples were assayed on at least three independent days and the representative raw values are shown. Western analysis on the same samples used in the left panels with either a phospho-specific antibody against pY419SRC or pY530SRC. Due to the high sequence homology, pY419SRC antibody also recognizes the corresponding phosphorylation site in other family members. (d) Technical and biological variations of the Luminex assay. For technical variations (blue bars), protein lysates from 6 cell lines (one sample per line) were analyzed at least 8 times with Luminex assay. For biological variations (red bars), protein lysates from each cell line were harvested 12 times sequentially and analyzed with Luminex assay. Variations of active (dark blue or red) and inactive beads (light blue or red) were determined as described in Materials and Methods.
We therefore explored an alternative approach based on the multiplexed coupling of kinase-specific antibodies to polystyrene microspheres (beads). We used the approach to profile the tyrosine phosphorylation status of 62 of the 90 tyrosine kinases in the human genome (Figure 1a), including 4 pseudokinases (ERBB3, EPHB6, PTK7 and RYK) which lack intrinsic kinase activity. The method involves the coupling of tyrosine kinase-specific antibodies to polystyrene beads, with a different bead color representing each kinase. Whole protein lysates from cancer lines or patient samples are then mixed with the antibody-coupled beads in a multiplexed reaction, and then combined with a biotinylated anti-phosphotyrosine antibody that thus labels all tyrosine-phosphorylated tyrosine kinases in the mixture. The bead complexes are then detected on a Luminex flow-cytometer.
We assessed the performance characteristics of the method by subjecting 46 of the antibodies to further experimental measures of sensitivity and specificity, including the treatment of cells with kinase-activating ligands, blocking antibodies, or overexpression of tyrosine kinase-encoding expression constructs (see Supplementary Table 2-4 online). Of the 46 characterized kinases, 46 (100%) yielded the expected signals, although in some cases the testing of multiple antibodies was required to obtain a properly-functioning assay (Figure 1c, Supplementary Table 2-4 and Supplementary Fig. 1 online).
Additional experiments were carried out to determine the reproducibility of the method, addressing both technical and biological variations. The level of technical variability was very low, ranging from 3-11% (Figure 1d and Supplementary Table 5 online). As expected, variability increased when biological replicates were assessed (ranging from 9-35%, Figure 1d and Supplementary Table 5 online). Based on antibody-specific background levels, we established 3-fold over background to represent significant phosphorylation (Supplementary Fig. 2 online).
With a robust assay in hand, we next piloted the idea of generating a resource of tyrosine kinase phosphorylation data across a diversity of human cancers. Toward that end, we profiled 130 human cancer cell lines (Figure 2a and Supplementary Table 6 online; data available at www.broad.mit.edu/cancer/pubs/kinase_profile). As expected, most cell lines exhibited only a limited repertoire of tyrosine kinase tyrosine-phosphorylation events. Some kinases, in contrast (e.g. LCK, LYN, SRC, EGFR, FGFR3 and PTK2) showed a broad pattern of phosphorylation across the cell line panel. The method detected expected patterns of tyrosine phosphorylation, such as ABL1 phosphorylation in K562 (CML with BCR-ABL1 translocation) 3 and KIT phosphorylation in GIST882 cells (gastrointestinal stromal tumor with activating KIT mutation) 4. Other known examples include MET activation in SNU-5 (MET amplification) 5 and H2009 (constitutive MET phosphorylation) 6; EGFR in HCC827, HCC4006 (EGFR exon19del mutant) 7; ERBB3 in MDAMB453 (constitutive ERBB3 phosphorylation) 8; FGFR2 in KATOIII (FGFR2 amplification) 9; IGF1R in Huh7 (increased expression and secretion of IGF2 leads to constitutive IGF1R activation) 10. These results suggest this method is capable of identification of activated tyrosine kinases regardless of their mechanism of activation.
Figure 2. Luminex screen identifies activated tyrosine kinases in human cancer cell lines and primary glioblastomas.

(a) Luminex screen on human cancer cell lines. Total protein lysates were prepared from human cancer cell lines and assayed as described in the Experimental Procedures. The raw data were processed and positive signals were normalized to fold-over-backgrounds. The normalized readings were log-transformed and converted into a gct file and the heat map was generated using the GenePattern 3.0 software. (b) Luminex screen on human primary glioblastomas. Total protein lysates were prepared from freshly frozen patient samples as described in the Experimental Procedures. The data was acquired and processed same as in a). (c) Representative positive and negative pY419SRC IHC staining in primary GBM samples. Primary GBM tissue microarrays were staining as described in the Experimental Procedures. Normal brain samples on the same TMA consistently stained negative for pY419SRC (data not shown).
In its current configuration, the bead-based method assays 62 of the 90 human tyrosine kinases, requires limited numbers of cells (~105), is amenable to high-throughput studies performed in 96-well plates, is inexpensive (~$10 per assay), and exhibits sensitivity and specificity that is comparable to conventional immunoprecipitation-Western blotting approaches. The majority of tyrosine-phosphorylated tyrosine kinases identified by mass spectrometry were also identified by the bead assay, but some events were missed by one method or the other (Figure 1b). The low number of input cells (1,000-fold less than what is required for mass spectrometry), together with its low cost and high-throughput format may make the bead assay a favored method for screening applications. As with any antibody-based assay, however, the quality of the results is primarily determined by the specificity and sensitivity of the capture antibodies.
The bead-based assay should be distinguished from recent reports of reverse-phase protein microarrays 11. Such lysate arrays have the advantage of requiring only minute numbers of cells, but each analyte (e.g. kinase) must be studied one at a time (in contrast to the multiplexed assay described here), thereby substantially limiting throughput. In addition, lysates arrays require phospho-specific kinase antibodies, whereas our bead-based assay utilizes non-phospho-specific kinase antibodies with a secondary anti-phosphotyrosine antibody, thereby making the assay applicable to any kinase for which a primary antibody exists.
In an effort to demonstrate the feasibility of using the kinase profiling method for the discovery of new oncogenic, therapeutic targets in cancer, we followed up on one of our cell line screen findings – namely, the frequent activation of SRC in glioblastoma, a particularly devastating type of brain tumor (Figure 2a)12. SRC phosporylation was confirmed by immunoprecipitation and Western blotting (Supplementary Fig. 3a online), and documented that the phosphorylation signal derived from the SRC kinase domain (pY419, reflecting the active SRC conformation), as opposed to simply being part of a multi-protein complex containing other tyrosine-phosphorylated proteins (Figure 1c).
We next asked whether the SRC phosphorylation seen in GBM cell lines was also present in primary patient samples. We assessed this in two ways. First, we subjected 31 frozen GBM samples to kinase profiling using the Luminex assay. SRC phosphorylation was detected in 19 of 31 samples (61%) (Figure 2b and Supplementary Table 7 online). Of note, the assay also detected extensive EGFR phosphorylation as previously described 13, 14, and high level phosphorylation of MET in a sample harboring MET amplification 2. Second, we performed immunohistochemistry using a phospho-specific (pY419) SRC family kinase antibody on tissue microarrays derived from an independent collection of 33 GBM patients. Twenty-two of 33 samples were positive by IHC, consistent with the Luminex assay results (Figure 2c and Supplementary Table 8 online). Taken together, these results indicate that SRC activation is a common event in GBM. Importantly, however, SRC mutations in cancer are exceedingly rare. In particular, the initial Cancer Genome Atlas (TCGA) effort has yet to identify focal SRC amplification or somatic missense mutations15. This further emphasizes the value of functional profiling methods for the detection of kinase activation in the absence of kinase mutation.
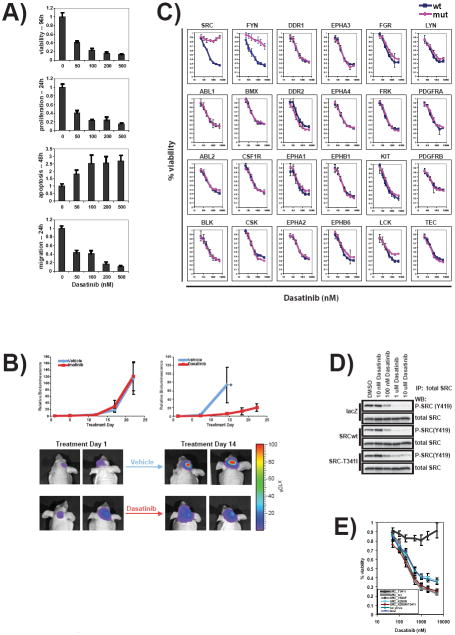
To determine whether inhibition of SRC might be of therapeutic value, we tested the effects of the SRC inhibitor dasatinib on a panel of 71 cancer cell lines. Dasatinib16 is an ATP-competitive inhibitor initially developed as an ABL inhibitor (for the treatment of chronic myeloid leukemia), but which is known to also potently inhibit SRC and other kinases. 30/71 cell lines exhibited an EC50 less than 1 μM using a viability assay (Figure 3a and Supplementary Table 9 online). Nearly all the cell lines tested displayed decreased BrdU incorporation upon dasatinib treatment, reflecting decreased proliferation (Figure 3a and Supplementary Table 9 online). Dasatinib also induced apoptosis in a subset of the cell lines tested (Figure 3a and Supplementary Table 9 online). Furthermore, dasatinib inhibited cell migration in a transmembrane assay, consistent with SRC’s documented role in cell motility 17. While the effects varied among cell lines (Figure 3a and Supplementary Table 9 online), dasatinib consistently inhibited GBM cell migration – even in cells where no growth inhibitory effect was observed, consistent with prior studies suggesting a possible role of SRC in the invasion of glioblastoma cells 18-20. The anti-GBM properties of dasatinib observed in vitro were next extended to a U87MG cell line-based orthotopic xenograft model of GBM that has been extensively validated 21. While treatment with neither vehicle nor the control inhibitor imatinib had any appreciable effect on in vivo tumor growth (Figure 3b), treatment with dasatinib resulted in significant attenuation of tumor growth (Figure 3b).
Figure 3. Dasatinib effectively blocked tumor progression in vitro and in vivo, and resistant mutants of SRC and FYN rescued dasatinib effects in glioma cells.
(a) Dasatinib diminished T98G glioma cell viability and migration in vitro. T98G cells were plated onto 96-well plates and treatment with either DMSO or dasatinib at concentrations specified in the figure. The cell viability was assayed with CellTiterGlo. Proliferation rates were assessed with BrdU incorporation at 24h. Apoptosis levels were analyzed with a Caspase3/7 activity assay at 48h. Cell migration through 0.45um membrane was measured at 24h post-treatment. Triplicate samples were assayed for each condition. The averages and standard deviations are presented. (b) Dasatinib effectively inhibited tumor growth in vivo. Mice with established intracranial U87 tumors were treated with imatinib at 75 mg/kg twice a day by oral gavage (n=6) or vehicle (n=5). Tumor burden was assessed by bioluminescence imaging and expressed relative to the start of treatment (Treatment Day 1). Data expressed as mean ± SD. A significant suppression of tumor growth was observed in mice treated with dasatinib at 10 mg/kg twice a day by oral gavage (n=5) by comparison to vehicle treated controls (n=5). Day 14 of treatment was the last imaging point with all vehicle treated animals. Student’s t-test p<0.05. Representative images of dasatinib and vehicle treated animals. Bioluminescence intensity is represented by the indicated false-color scale. (c) T98G cells were plated onto 96-well plates and infected with lentiviruses expressing the tyrosine kinases specified in the figure. 24 hours after infection, cells were treated with dasatinib ranging from 50nM to 5uM. Cell viability was assessed at 96h post-treatment. Triplicate samples were analyzed for each condition. The averages and standard deviations are shown. (d) Total protein lysates were prepared from T98G cells treated with dasatinib at concentrations specified in the figure. The lysates were immunoprecipitated with a total SRC antibody and blotted with either the pY419SRC antibody or a total SRC antibody. (e) Dasatinib-resistant mutant SRC but not other alleles confer resistance in glioma cells, indicating increased SRC activity alone does not confer dasatinib resistance. T98G cells were plated, infected and treated same as in figure 3. Besides the wild-type and drug resistant T341I mutant, cells were also infected with viruses expressing an activating allele (Y530F), a kinase dead allele (K598R) and a double mutant of K598R/T341I. Uninfected cells and lacZ virus infected cells were used as controls. Each condition was assayed in triplicates and the means and standard deviations are presented.
The experiments, while preliminary, further support the notion that the Luminex bead-discovered SRC activation represents a potential therapeutic target in GBM. Recent studies have indicated, however, that dasatinib is a potent inhibitor of many tyrosine kinases. Specifically, 20 tyrosine kinases have a Kd < 1nM in a biochemical assay of kinase inhibition 22. It is therefore plausible that the dasatinib effects observed in GBM might be attributable to tyrosine kinases other than SRC. To address this issue, we generated dasatinib-resistant alleles of candidate kinases and tested them for their ability to rescue the dasatinib effects observed in GBM cells. To make the dasatinib-resistant mutants, we leveraged the observation that all dasatinib-targeted kinases contain a conserved threonine (“gatekeepker”) in the kinase domain. This threonine, essential for ABL1 kinase sensitivity to imatinib and dasatinib, and a specific mutation (T315I) has been observed in CML patients who develop imatinib or dasatinib resistance 23, 24.
We therefore engineered the T315I gatekeeper mutation into each of 24 candidate dasatinib target kinases, and introduced either wild-type or mutant kinases into GBM cells by lentiviral transduction. Expression of the kinases was documented using the Luminex assay (Supplementary Table 4 online). As expected, none of the 24 wild-type kinases conferred resistance to dasatinib in vitro. (Figure 3c) Similarly, 22/24 of the mutant kinases failed to rescue dasatinib, indicating that they were not the relevant targets of dasatinib in this setting. In contrast, drug-resistant alleles of SRC and the SRC family member FYN rescued dasatinib effects (Figure 3c). We note, however, that whereas prominent phospho-SRC was detected by the Luminex assay and immunoblotting, phospho-FYN was not abundant in these cells (Figure 2a, Supplementary Fig. 3b and Supplementary Table 6 online). Of note, we also tested 30 different anti-SRC shRNA constructs, none of which resulted in >50% reduction in phospho-SRC in glioma cells (data not shown). This result likely reflects the cell’s enormous plasticity in upregulating residual kinase activity in the face of attempted down-regulation of a kinase on which the cell is dependent for survival.
The experiments described in this report demonstrate the feasibility and utility of a bead-based approach to profiling tyrosine kinase activity in cancer. In addition to recovering many of the known kinase activation events in cancer, the pilot screen identified and validated SRC as a potential new therapeutic target in GBM. Of note, a role for SRC was not suggested by recent DNA sequence or copy-number analysis15, but rather required protein phosphorylation data for its discovery. The availability of an orally-available, FDA-approved SRC inhibitor (dasatinib) should facilitate the rapid testing dasatinib in GBM clinical trials. Most importantly, however, the work described here establishes a general strategy for discovering activated tyrosine kinases in cancer, and suggests that large-scale efforts to generate comprehensive kinase activity profiles across the diversity of human cancers should be undertaken.
METHODS
Luminex immunosandwich assay
Individual bead-type of Luminex xMAP microspheres (Luminex Corporation, Austin, TX) were coupled separately to capture antibodies against total tyrosine kinases using the following procedure. 100μl Microspheres were washed with 100 μl ddH2O and resuspended in 80 μl 100mM NaH2PO4 pH 6.2. 10 μl 25mg/ml Sulfo-NHS (Pierce) and 10 μl 25mg/ml EDC (Pierce) were added and incubated at 25°C for 20min while shaking to activate the microspheres. The activated beads were washed with 250 μl 1x PBS pH7.4 twice and resuspended in 200 μl 1x PBS. 20 μg capture antibodies in 50 μl 1x PBS were added. The mixtures were incubated at 25°C for two hours while shaking in dark. The antibody-coupled beads were blocked with 500 μl PBS-TBN (0.02% Tween-20, 0.1% BSA and 0.05% NaN3 in 1x PBS pH7.4) at 25°C for 30 minutes while shaking. Finally, the microspheres were washed with 500 μl PBS-TBN twice and stored in 500 μl PBS-TBN at 4°C until use.
0.25 μl coupled microspheres per bead type were used for each sample. Microsphere mixture were transferred into a 96-well 1.2um filter plate (Millipore) and washed with 100 μl 1x PBS-1% BSA pH7.4 (Sigma) twice. 100 μl 1mg/ml protein lysates were added to each well and incubated at 4°C overnight while shaking. The microspheres were washed with 100μl 1x PBS-1% BSA pH7.4 twice. 50μl 4μg/ml biotin-4G10 (UpState) were added and incubated at 25°C for 30 minutes while shaking. The microspheres were then washed with 100μl 1x PBS-1% BSA pH7.4 twice and then incubated with 50μl 4μg/ml R-phycoerythrin streptavidin conjugates (Molecular Probes) at 25°C for 10 minutes. Finally, the samples were washed with 100μl 1x PBS-1% BSA pH7.4 twice and resuspended in 50μl 1x PBS-1% BSA pH7.4 for analysis.
The data were acquired with a Luminex 100 instrument (Luminex Corporation) according to manufacturer’s instructions. The background readings for each capture antibody were obtained using microspheres incubated with 1x cell lysis buffer (Cell Signaling Technology). Values were considered positive if they were 3-fold over the background and represented by log2-transformation of the folds over background. Negative values were threshold to -10. The preprocessed data were converted into .gct files and analyzed with GenePattern 3.0 (The Broad Institute).
Technical and Biological Variation
All analyses were based on median-based statistics. Variation was measured using the quartile coefficient of variation 25, defined as: (Q3-Q1)/(Q3+Q1), where Q1 is the lower quartile and Q3 is the upper quartile. To determine whether a TK was active or inactive in any given cell line, we evaluated its measured intensity against a reference as follows: 1) Measure the TK in a reference where it is inactive. Repeat the measurement n times and note the observed intensity; 2) Let max be the maximum of these n measurements in the reference; 3) For any new measurement of TK in any cell line, the TK is active if the new measurement exceeds max; the TK is otherwise inactive. With this procedure, the probability that a measurement greater than max could arise from an inactive TK depends on the value of n, and can be calculated using the binomial distribution 26.
IP-LC/MS/MS
The basics of the procedure is developed and described by Rush and colleagues 27. Briefly, 2x108 cells were lysed in 8M urea / 20 mM Hepes containing phosphatase inhibitors. Upon reduction of the disulfide bonds, the lysate was split into 2 aliquots and diluted with 20 mM Hepes to 2M or 0.5 M urea before digestion with trypsin or chymotrypsin, respectively. The total peptide mixtures were then desalted by Sep-Pak cartridge and resuspended in IP buffer, 50 mM MOPS/NaOH pH 7.2, 10 mM Na2PO4, 50 mM NaCl, along with 4 exogenous pTyr-containing peptides added as controls. IP was performed with a cocktail of 3 protein G agarose bead-bound phospho-tyrosine antibodies pY100 (Cell Signaling), 4G10 (UpState), and PY99 (Santa Cruz Biotechnology) to enrich for phosphotyrosine-containing peptides. Peptides captured by phospho-tyrosine antibodies were eluted under basic followed by acidic conditions. The IP eluates were analyzed by data-dependent LC/MS/MS using a Thermo LTQ-FT instrument. Each peptide’s site(s) of phosphorylation were uniquely mapped using the sequence information present in its ion trap MS/MS spectrum, unless it had several tyrosine residues and fragmented poorly. All MS and MS/MS data was processed using the Spectrum Mill software package (Agilent Technologies).
Dasatinib treatment, viability, proliferation, migration and apoptosis assays
Human glioblastoma T98G cells cultured in 96-well plates (1000 cells /well) were treated with different concentrations of dasatinib and a vehicle control (DMSO). Dasatinib16 was synthesized by the Chemical Biology Program at the Broad Institute. Treatments lengths varied from 24h for migration and proliferation assays to 48h for apoptosis and 96h for viability. Cell viability was determined by quantification of ATP present in culture using CellTiter-Glo (Promega). Cell proliferation was determined by incorporation of bromodeoxyuridine to actively proliferating cells and subsequent quantification of BrdU by immunohistochemistry (Calbiochem). The kit’s substrate solution was replaced with Glo Substrate (R&D) for higher sensitivity. CytoSelect™ Cell Migration Assay Kit (Cell Biolabs, INC.) was used to assess cell migration. Migratory cells were detected using CellTiter-Glo (Promega) instead of the dye provided with the kit. Apoptosis was determined using a luminescence assay that quantifies caspases 3 and 7 activities (Caspase Glo 3/7 Assay, Promega).
Orthotropic glioblastoma model
Orthotopic glioma xenografts were established as previously described28. Briefly, 50,000 U87-LucNeo cells were stereotactically implanted into the right brain of 6 week old male NCr nude mice using a 1:1 mix of propylene glycol and water as the vehicle. Bioluminescence imaging was performed 7 and 12 days after tumor implantation to identify mice with logarithmically growing tumors. Mice were divided into treatment groups with statistically equivalent mean bioluminescence. Treatments consisted of either imatinib at 75 mg/kg twice a day by oral gavage, dasatinib at 10 mg/kg twice a day by oral gavage, or vehicle. Tumor burden was assessed by bioluminescence imaging as previously described, and for each individual animal values were expressed relative to the bioluminescence at the start of treatment. All animal studies were performed under an IACUC approved protocol.
Overexpression vectors (cloning), transformations, transfections and infection
Cloning
Vectors containing tyrosine kinase ORFs were obtained from Harvard Institute of Proteomics or Dr. Mark Vidal at Dana-Farber Cancer Institute. The ORFs were amplified with primers listed in Supplementary Table 10 and inserted into pENTR/D-TOPO (Invitrogen). Site-directed mutagenesis was performed with the primers listed in Supplementary Table 10 according to manufacturer’s recommendations (Stratagene). Both wild-type and mutant constructs were sequence verified. LR recombination reaction into pLenti6/UbC/V5-DEST vector was done according to Invitrogen’s guideline. The inserts were verified again with sequencing.
Transfections
293-FT producer cells (Invitrogen) were transfected using ViraPower™ Lentiviral Expression Systems (Invitrogen) according to manufacturer’s protocol. Virus was harvested at 48h and 72h. Infections. Cells were plated in 96-well plates (1000 cells/well) and infected with 10 uL of wild type and mutant TK lentivirus. Infections were done in triplicates. On day 2 cells were treated with various concentrations of dasatinib. Viability was assessed at 96h after dasatinib treatment.
Supplementary Material
Acknowledgments
We would like to thank Drs. Levi Garraway and Johnathan Fletcher for cell lines. We thank Julie Dang for her technical support on the pY419SRC IHC experiment. JD is supported by a fellowship from the Leukemia and Lymphoma Society and The Irving Family. IKM is funded by the Brain Tumor Funders’ Collaborative. The project has been funded in part with funds from the National Cancer Institute’s Initiative for Chemical Genetics, National Institutes of Health, under Contract No. N01-CO-12400, and via the NCI’s Integrative Cancer Biology Program.
Footnotes
Authors contributions J.D. developed the Luminex immunosandwich assay, profiled cell lines and primary gliomas with the assay, performed the in vitro experiments on SRC and dasatinib, prepared samples for IP-MS analysis, and contributed to the planning and writing of the manuscript. P.B. contributed to sample preparation for the Luminex assay and IP-MS analysis, Luminex assay antibody validation, in vitro experiments on SRC and dasatinib, and writing the manuscript. K.R.C. and S.A.C. performed mass spectrometric analysis. D.M. performed the variation analysis on the Luminex assay. S.P.F. and M.L. performed IHC on tissue microarrays and analyzed the staining results. R.B. provided the genomic data on the primary glioma samples. M.B. contributed to the cell line screen with the Luminex assay. B.J. contributed to the antibody validation and writing of the manuscript. X.P.P. and H.H. provided the expression constructs for antibody validation. R.L.M. and T.A.L. synthesized dasatinib for this study. L.M.L., P.N. and I.K.M. provided the frozen primary glioma specimens and corresponding genomic data. D.N.L. provided the primary glioma tissue microarrays. A.L.K. performed the animal experiments with dasatinib. T.R.G. planned and designed the project and contributed to the planning and writing of the manuscript.
Note: Supplementary information is available on the Nature Biotechnology website.
The authors report no conflicts of interest relevant to this work.
References
- 1.Simon MP, et al. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet. 1997;15:95–98. doi: 10.1038/ng0197-95. [DOI] [PubMed] [Google Scholar]
- 2.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heisterkamp N, Stam K, Groffen J, de Klein A, Grosveld G. Structural organization of the bcr gene and its role in the Ph’ translocation. Nature. 1985;315:758–761. doi: 10.1038/315758a0. [DOI] [PubMed] [Google Scholar]
- 4.Tuveson DA, et al. STI571 inactivation of the gastrointestinal stromal tumor c-KIT oncoprotein: biological and clinical implications. Oncogene. 2001;20:5054–5058. doi: 10.1038/sj.onc.1204704. [DOI] [PubMed] [Google Scholar]
- 5.Smolen GA, et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci U S A. 2006;103:2316–2321. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura Y, et al. Constitutive activation of c-Met is correlated with c-Met overexpression and dependent on cell-matrix adhesion in lung adenocarcinoma cell lines. Cancer science. 2008;99:14–22. doi: 10.1111/j.1349-7006.2007.00640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 8.Kita YA, et al. NDF/heregulin stimulates the phosphorylation of Her3/erbB3. FEBS letters. 1994;349:139–143. doi: 10.1016/0014-5793(94)00644-x. [DOI] [PubMed] [Google Scholar]
- 9.Ueda T, et al. Deletion of the carboxyl-terminal exons of K-sam/FGFR2 by short homology-mediated recombination, generating preferential expression of specific messenger RNAs. Cancer research. 1999;59:6080–6086. [PubMed] [Google Scholar]
- 10.Desbois-Mouthon C, et al. Dysregulation of glycogen synthase kinase-3beta signaling in hepatocellular carcinoma cells. Hepatology (Baltimore, Md. 2002;36:1528–1536. doi: 10.1053/jhep.2002.37192. [DOI] [PubMed] [Google Scholar]
- 11.Tibes R, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular cancer therapeutics. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 12.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 13.Libermann TA, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 14.Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombardo LJ, et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2- hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. Journal of medicinal chemistry. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 17.Parsons SJ, Parsons JT. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–7909. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- 18.Angers-Loustau A, Hering R, Werbowetski TE, Kaplan DR, Del Maestro RF. SRC regulates actin dynamics and invasion of malignant glial cells in three dimensions. Mol Cancer Res. 2004;2:595–605. [PubMed] [Google Scholar]
- 19.Lund CV, et al. Reduced glioma infiltration in Src-deficient mice. Journal of neurooncology. 2006;78:19–29. doi: 10.1007/s11060-005-9068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park CM, et al. Ionizing radiation enhances matrix metalloproteinase-2 secretion and invasion of glioma cells through Src/epidermal growth factor receptor-mediated p38/Akt and phosphatidylinositol 3-kinase/Akt signaling pathways. Cancer research. 2006;66:8511–8519. doi: 10.1158/0008-5472.CAN-05-4340. [DOI] [PubMed] [Google Scholar]
- 21.Phuong LK, et al. Use of a vaccine strain of measles virus genetically engineered to produce carcinoembryonic antigen as a novel therapeutic agent against glioblastoma multiforme. Cancer research. 2003;63:2462–2469. [PubMed] [Google Scholar]
- 22.Carter TA, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci U S A. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 24.Shah NP, et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 25.Feinstein AR. Principles of Medical Statistics. Chapman & Hall / CRC; 2002. [Google Scholar]
- 26.Efron B. Nonparametric Standard Errors and Confidence Intervals. The Canadian Journal of Statistics. 1981;9:139–158. [Google Scholar]
- 27.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 28.Rubin JB, et al. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc Natl Acad Sci U S A. 2003;100:13513–13518. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.