Abstract
Mercury resistance mediated by mercuric reductase (MerA) is widespread among bacteria and operates under the control of MerR. MerR represents a unique class of transcription factors that exert both positive and negative regulation on gene expression. Archaea and bacteria are prokaryotes, yet little is known about the biological role of mercury in archaea or whether a resistance mechanism occurs in these organisms. The archaeon Sulfolobus solfataricus was sensitive to mercuric chloride, and low-level adaptive resistance could be induced by metal preconditioning. Protein phylogenetic analysis of open reading frames SSO2689 and SSO2688 clarified their identity as orthologs of MerA and MerR. Northern analysis established that merA transcription responded to mercury challenge, since mRNA levels were transiently induced and, when normalized to 7S RNA, approximated values for other highly expressed transcripts. Primer extension analysis of merA mRNA predicted a noncanonical TATA box with nonstandard transcription start site spacing. The functional roles of merA and merR were clarified further by gene disruption. The merA mutant exhibited mercury sensitivity relative to wild type and was defective in elemental mercury volatilization, while the merR mutant was mercury resistant. Northern analysis of the merR mutant revealed merA transcription was constitutive and that transcript abundance was at maximum levels. These findings constitute the first report of an archaeal heavy metal resistance system; however, unlike bacteria the level of resistance is much lower. The archaeal system employs a divergent MerR protein that acts only as a negative transcriptional regulator of merA expression.
The element mercury is a toxic heavy metal that occurs naturally in several forms, including elemental (Hg0), ionized (inorganic salts Hg2+ and Hg+), organic (typically alkylated), or sulfidic (cinnabar). Mercury use is widespread, particularly in the production of gold, vaccines, antimicrobials, amalgams, and electronics. Mercuric chloride (HgCl2) is most often used in experimental studies because it is soluble and poisonous. Mercury is a redox-active transition metal in both biotic and abiotic environments. In vivo, mercury plays a critical role in modulating cellular redox status by depleting antioxidant pools (16). Both ionic and organic mercury form covalent bonds with sulfur atoms in cysteine residues of target proteins.
Bacteria respond to mercury exposure using several strategies. While mechanisms involving tolerance occur (33, 34, 58), enzymatic reduction of mercuric ion to elemental mercury-catalyzed by-products of the mer operon is the only resistance mechanism that has been described (reviewed in references 4, 30, 31, and 50). The mer operon (merTPCAD) encodes a group of proteins involved in the detection, transport, and reduction of mercury. The NADPH-dependent enzyme, mercuric reductase (MerA), transfers two electrons to mercuric ion, Hg2+, reducing it to elemental mercury Hg0. Elemental mercury is volatile and is released from the cell. Mercuric ion is scavenged from the environment through the action of the periplasmic protein, MerP, which binds Hg2+ and transfers it to the membrane protein MerT. MerT transports mercuric ion into the cytoplasm. Additional mer genes occur, notably merB, an organomercurial lyase. Inclusion of merB in the mer operon results in a so-called broad-spectrum mercury resistance.
Expression of the mer operon by the activity of its major promoter, PT, is controlled by MerR, a dual-function transcriptional regulatory protein that remains bound to PT at an inverted repeat sequence called merO (51). In the absence of mercury, MerR binds and bends the DNA and then attracts RNA polymerase to the operator and holds it there in an inactive state and represses transcription (2, 3). In the presence of mercury, the MerR homodimer undergoes a conformational change that underwinds the DNA, creating optimal PT topology by rotating the −10 region 30° closer to the helix face of the −35 region, thereby facilitating access to it by prebound RNA polymerase, which subsequently activates mer operon transcription. MerO is located in the intergenic region between the divergently transcribed merR gene and the mer operon, where it simultaneously exerts control over both transcription units. MerR consists of an N-terminal helix-turn-helix domain coupled to a C-terminal mercuric ion binding domain containing three active cysteines. MerR constitutes a distinct class of transcriptional regulatory factors as yet not reported in either archaea or eukaryotes (11).
Archaea and bacteria are both prokaryotes (56, 57). Archaea however, use simplified versions of several eukaryotic-like subcellular processes, including transcription (5, 8). The regulation of gene expression is an active area of research in archaea because of this evolutionary overlap and the attraction of a less complex experimental system. Examples of this overlap include homologous promoter structure (21, 41), orthologs of TATA binding protein (29, 37, 48), TFIIB (referred to as TFB in archaea [18, 38, 39]), TFIIEα (TFEα [6, 22]), TFIIS (TFS [25]), and the 12-subunit RNA polymerase II (28). They appear, however, to lack homologs of TFIIA, TFIIF, and TFIIH. In the crenarchaeal subdivision of the archaea, RNA polymerase is recruited by TATA binding protein and TFB to an octameric TATA box (YTTTTAAA [40]) and 5′-flanking TFB recognition element (BRE) hexamer having a consensus sequence of RNWAAW (where R is a purine, N is any base, and W is A or T [8]). The midpoint of the TATA box is located 26 nucleotides from the start point of transcription (41). Conservation of a eukaryotic-type transcription apparatus has notable consequences for the mechanism of regulation of gene expression in archaeal prokaryotes. Consequently, gene regulatory studies in these organisms are expanding and include efforts on negative (7, 12, 13, 54) and positive (32, 36) transcriptional mechanisms of control.
Sulfolobus solfataricus is a hyperthermophile and a member of the crenarchaeal subdivision of the archaea. This organism is found in acidic geothermal pools, while in the laboratory it grows chemoheterotrophically on reduced carbon compounds at an optimal temperature of 80°C (14, 19, 23, 24). Hot springs are typically rich in heavy metals, but little is known about the interaction between these elements and resident archaea. To investigate this relationship, S. solfataricus was used because it grows aerobically in defined media, has a sequenced genome (49), and offers powerful experimental genetic techniques (27, 59).
MATERIALS AND METHODS
Archaeal strains, cultivation, and construction.
Archaeal strains, plasmids, and primers used in this work are indicated in Table 1. S. solfataricus strain 98/2 and mutant derivatives were grown at 80°C in batch culture as described previously (9, 60). A defined minimal medium consisted of the basal salts of Allen (1) as modified by Brock (10), supplemented either with 0.2% (wt/vol) sucrose (SM) or lactose (LM) as the sole carbon and energy source. Recovery of transformed cells employed a rich medium (RM) supplemented with 0.2% (wt/vol) tryptone. Growth was monitored at a wavelength of 540 nm using a Cary 50 Bio, UV-visible spectrophotometer (Varian). A solid medium was prepared using 0.6% (wt/vol) gelrite gellan gum (Kelco) and 8.0 mM magnesium chloride. Mercury-containing plates were formed by mixing mercuric chloride dissolved in double-distilled water with liquid plate medium just prior to pouring. Plates were incubated at 80°C in plastic containers with sufficient hydration to prevent desiccation.
TABLE 1.
Archaeal strains, plasmids, and primers
| Strain, plasmid, or primer | Genotype or sequence | Source or derivation |
|---|---|---|
| Strains | ||
| PBL2000 | Wild-type S. solfataricus strain 98/2 | 44 |
| PBL2002 | lacS::IS1217 | 59 |
| PBL2020 | merA::lacS | PBL2002 by targeted recombination |
| PBL2025 | del(SSO3004-3050) | PBL2000 spontaneous deletion |
| PBL2026 | merR::lacS del(SSO3004-3050) | PBL2025 by targeted recombination |
| Plasmids | ||
| pUC19 | bla | New England Biolabs |
| pMerA1 | merA | This work |
| pMerAS1 | merA::lacS | This work |
| pMerRS1 | merR::lacS | This work |
| pMerA10 | merA (nt 133 to 801) | This work |
| Primers | ||
| MerA-F | 5′ GTTCCATCGAAGAGAATGTCTAGAATAGG 3′ | |
| MerA-R | 5′ TGCTGCATGCAAATTTAAATCTACATTGG 3′ | |
| MerA2-F | 5′ ATAGGAATTCCAATTCATTTGTAAGAGGCT 3′ | |
| MerA2-R | 5′ CGTAGGATCCCTATACACAACAACTCATTTACTAACGTC 3′ | |
| MerR-L-BamHI-F | 5′ ATGCCGCGGATCCATCTTGTGAAAATTAAGGATGCGAT 3′ | |
| MerR-L-BamHI-R | 5′ ATGCCGCGGATCCTCTTGAAAGGCTTGGAAAAATTCTG 3′ | |
| MerR-OL-MfeI-F | 5′ AAGTGTGGAGCCTCTTACAAATCAATTGGAATCACTATTCTCTGCCCTAG 3′ | |
| MerR-OL-MfeI-R | 5′ CTAGGGCAGAGAATAGTGATTCCAATTGATTTGTAAGAGGCTCCACACTT 3′ | |
| LacS-MfeI-F | 5′ AGTCAGCAATTGAATACTAGGAGGAGTAGCAT 3′ | |
| LacS-MfeI-R | 5′ CTGACTCAATTGAGTATTAAATCTAAATGAC 3′ | |
| MerAp | 5′ CGAGTTCATTTGCCCTAATTAACG 3′ |
A Jerome 431X mercury vapor analyzer (Arizona Instrument) was used to measure volatilization of elemental mercury (Hg0). Cultures were grown in minimal medium to a cell density of 108/ml in glass screw-cap Erlenmeyer flasks (250 ml) fitted with neoprene O-rings, and mercuric chloride was added to the cultures to a final concentration of 0.3 μM. Cultures were then incubated for 4 h at 80°C and cooled for 1 h at room temperature. After cooling, flask lids were removed and mercury content was measured in units with the Jerome analyzer intake tube centered at a distance of 12 to 13 mm above the flask opening. The intake was 750 ml/min (12.5 ml/s), and the duration of the reading was 13 s for a total volume of 163 ml of air per reading. Ambient air was sampled to ensure a zero baseline reading. Detergent-treated cell extracts were prepared by addition of N-laurylsarcosine at 2.5% (wt/vol) prior to inoculation.
Strain PBL2002 (lacS::IS1217) was used to create the merA disruption mutant and is a spontaneous derivative of wild-type S. solfataricus (PBL2000) with an insertion of IS1217 at position 1242 in lacS (59). Strain PBL2025 was used to create the merR disruption mutant and is deleted for lacS and flanking genes. PBL2025 was selected from a collection of spontaneous lacS mutants (24) because it harbors a 58-kb deletion spanning open reading frames SSO3004 to SSO3050 as annotated by She and coworkers (49). Both strains are unable to utilize β-linked disaccharides as a sole carbon and energy source, and reintroduction of lacS integrated at other chromosomal loci is necessary and sufficient to restore this ability. Transformation procedures were as described previously, using electroporation to mobilize DNA into target cells (59). Following electroporation, cells were subcultured into RM for 8 h to allow recovery and then subcultured again into LM to enrich for chromosomal recombinants. Following appearance of turbidity, dilutions of the cultures were prepared and spread on RM plates to allow colony formation. Chromosomal recombinants resulting from insertion of the lacS gene were detected by spraying plates with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside solution to identify Lac+ colonies. These isolates were then subcultured into RM medium and analyzed in greater detail. Disruption of merA was conducted as described previously for other genes (59), while construction of the merR mutant employed an alternative strategy with a new strain to increase the incidence of recombination at the targeted locus.
Primers used for PCR of merA were forward primer MerA2-F and reverse primer MerA2-R. MerA2-F starts 325 bp upstream relative to the merA start codon, and MerA2-R starts 1,362 bp downstream relative to the merA start codon, beginning with the merA stop codon. MerA2-F encodes an added EcoRI site, and MerA2-R encodes an added XbaI site. PCR primers for lacS were forward primer LacS-MfeI-F and reverse primer LacS-MfeI-R, and both encode added MfeI sites. LacS-MfeI-F starts 170 bp 5′ to the lacS start codon, while LacS-MfeI-R starts 165 bp 3′ to the lacS stop codon. Plasmid pMerA1 was constructed by insertion of an EcoRI-XbaI-digested PCR merA amplicon produced with primers MerA2-F and MerA2-R and cloned into the EcoRI- XbaI sites of pUC19. Plasmid pMerASI was constructed by insertion of an MfeI-digested PCR lacS amplicon into the MfeI site of pMerA1 in the reverse orientation relative to merA. PCR and restriction analysis were used to verify the identity of the merA recombinant strain. Amplification of the wild-type undisrupted merA locus using primers MerA2-F and MerA2-R produced a single fragment of 1.69 kb and two fragments of 1.26 and 0.43 kb after digestion with MfeI. Amplification of the disrupted merA locus in strain PBL2020 produced a single fragment of 3.62 kb and three fragments following digestion representing the 5′ and 3′ ends of merA and the 1.93-kb lacS insert.
The MfeI site located in merR of pMerRS1 was created by overlap extension PCR (26) with primers MerR-OL-MfeI-F and MerR-OL-MfeI-R. The 5′ end of MerR-OL-MfeI-F begins 4 bp upstream of the merR start codon and is complementary to MerR-OL-MfeI-R. PCR of the modified merR allele, wild-type allele, and lacS-disrupted merR allele for verification of recombinant identity employed primers MerR-L-BamHI-F and MerR-L-BamHI-R. The 5′ start of MerR-L-BamHI-F is located 547 bp upstream of the merR start codon. The 3′ end of MerR-L-BamHI-R is located 893 bp downstream of the merR start codon. PCR and restriction analysis were used to verify the identity of the merR recombinant strain. Amplification of wild-type merR and flanking regions using primers MerR-L-BamHI-F and MerR-L-BamHI-R produced a single band of 1.44 kb that was cut into two bands by digestion with MfeI. Amplification of the disrupted merR locus in strain PBL2026 produced a single band of 3.37 kb, approximately 1.9 kb larger than that observed with the undisrupted locus due to the presence of the inserted copy of lacS. This fragment was cut by MfeI into three fragments of 0.57, 0.85, and 1.93 kb that represented the 5′ and 3′ ends of merR and the lacS insert, respectively.
Molecular biology methods.
DNA cloning, PCR, and plasmid transformation of Escherichia coli were performed as described elsewhere (24, 43). DNA sequencing was as described previously (45). DNA and RNA concentrations were measured using either a DyNA Quant 200 fluorometer (Hoefer) or a UV-visible Genesys 2 spectrophotometer (Spectronics). All manipulations of RNA were as described previously (9, 23). Protein concentrations were measured using a bicinchoninic acid protein assay reagent kit (Pierce). Unless otherwise indicated, all chemicals were obtained from common chemical suppliers.
Northern blot analysis.
RNA extraction and Northern hybridization using antisense riboprobes were performed as described previously (9, 23). RNAs were detected by autoradiography on X-Omat AR film (Kodak). Digital images were acquired using a GDS7800 gel documentation system (UVP). Scanning densitometry of the images was performed using GelBase-Pro software (UVP). The 7S RNA probe was prepared as described elsewhere (9). The merA probe was prepared by PCR amplification using chromosomal DNA and primers MerA-F and MerA-R, which were complementary to positions 133 to 161 and 773 to 801, respectively, in the merA coding region. The 640-bp fragment was cloned at the XbaI and SphI sites of pT7T3/18U (Pharmacia). In riboprobe synthesis merA was linearized using SmaI, and T3 RNA polymerase was used for transcription to produce a 32P-labeled antisense RNA.
Primer extension analysis.
The merA transcript was subjected to primer extension using primer MerAp, which is complementary to positions 47 to 70 downstream from the merA start codon. The primer extension oligonucleotide was labeled at the 5′ end with [γ-32P]ATP using T4 kinase (NEB) as described previously (9, 53). The labeling reaction was terminated by EDTA addition followed by heating at 65°C. The labeled primer was purified using a Sep-pak cartridge (Waters), dried, and resuspended in 10 μl of 10 mM Tris-Cl (pH 8.0), 1 mM EDTA. A typical reaction yielded 10 μl of 106-cpm/μl labeled oligo, and 1 μl of this oligo was used for each reverse transcription reaction. Reverse transcription was as described elsewhere (9, 53) with modifications. Samples of total RNA (20 μg) were hybridized with the labeled primer in 150 mM MgCl2, 10 mM Tris-Cl (pH 8.3), and 1 mM EDTA, heated at 65°C for 90 min, and cooled to allow primer annealing. The mixture was adjusted to 20 mM Tris-Cl (pH 8.3), 10 mM MgCl2, 0.5 mM dithiothreitol, 0.15 mg of actinomycin D/ml, and 0.15 mM deoxynucleoside triphosphates, and 5 U of avian myeloblastosis virus reverse transcriptase (Pharmacia) was added. The reaction was incubated for 1 h at 42°C and terminated by addition of 17.5 ng of salmon sperm DNA/ml, 14 ng of RNase A/ml followed by incubation for 15 min at 37°C. The reaction was extracted with phenol-chloroform (1:1), and primer-extended DNA was recovered by ethanol precipitation, dried, and resuspended in the stop solution of the T7 Sequenase version 2.0 DNA sequencing kit (Amersham). The primer-extension primer also was used to generate the sequencing ladder for mapping the start site of transcription of merAp. The template used to generate the DNA sequencing ladder for merAp primer extension mapping was plasmid pMerA10. DNA sequencing reaction products were separated on preequilibrated 8% (wt/vol) denaturing polyacrylamide sequencing gels as described previously (45).
Bioinformatic analysis.
Sequences used for phylogenetic analysis were derived by BLAST queries against the NCBI database using S. solfataricus P2 open reading frames SSO2689 and SSO2688. CLUSTAL W (52) was used to create multiple sequence alignments. PHYLIP version 3.57c (20) and the distance method of analysis were used for phylogenetic studies. SEQBOOT was used to generate 100 bootstrapped data sets, distance matrices were determined using PROTDIST and the Dayhoff PAM matrix option, unrooted trees were inferred by neighbor-joining analysis using NEIGHBOR, CONSENSE was used to identify the most likely tree, and FITCH was used to create branch lengths proportional to distance values. Nearly full-length sequences were used for both the MerA phylogenetic tree (433 residues) and MerR phylogenetic tree (104 residues). BOXSHADE version 3.21 was used to create boxshade diagrams. Sequences employed for the MerA tree were the following: Acinetobacter calcoaceticus (Q52109), Pseudomonas aeruginosa Tn501 (RDPSHA); Shigella flexneri R100 (P08332), Thiobacillus ferrooxidans (P17239), Shewanella putrefaciens (Q54465), Bacillus sp. strain RC 067 (P16171), Clostridium butyricum (T44505), Staphylococcus aureus pI258 (P08663), S. solfataricus P2 (NP_344015), Thermoplasma acidophylum (CAC12462), Thermoplasma volcanium (NP_110770), Aeropyrum pernix (B72625), Halobacterium sp. strain NRC-1 (AAG18773), and mouse transcription factor SOX 10 (Q04888). Sequences employed for the MerR tree were E. coli Tn21 (AAC33922), Pseudomonas fluorescens Tn501 (P06688), E. coli pDU1358 (A33858), S. aureus p1258 (P22874), T. ferrooxidans (P22896), Archaeoglobus fulgidus (AAB90568), S. solfataricus P2 (AAK42804), Streptomyces lividans (P30346), and Halobacterium salinarum (AAG18773).
RESULTS
Adaptive response to mercury challenge.
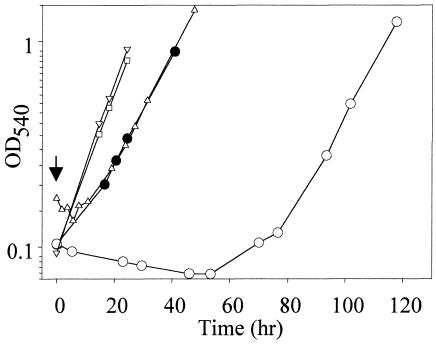
The minimal growth-inhibitory concentration of mercuric chloride for S. solfataricus that was effective in a defined liquid medium (SM) was determined in batch culture (Fig. 1). Growth was unaffected following addition of the metal at a final concentration of 0.3 μM, but at 0.5 μM a transient growth lag was apparent that was followed by resumption of a normal growth rate. Addition of a yet-higher metal concentration (1.5 μM) terminated growth, and recovery was apparent only after prolonged incubation. The rate of response to addition of an inhibitory metal concentration was quite rapid and evident within minutes. In a complex liquid medium (RM), the MIC was 2.5 μM and higher than that in the defined medium, due to thiol titration by complex medium components.
FIG. 1.
Adaptive resistance to mercuric chloride. Wild-type S. solfataricus was grown in SM and challenged with mercuric chloride. Open symbols, unadapted cultures; closed symbols, adapted culture; inverted triangles, untreated control. Unadapted cells challenged with 0.3 (squares), 0.5 (triangles), or 1.5 μM (open circles). The arrow indicates the time of addition of mercuric chloride.
While these data indicate the organism could overcome growth-inhibitory levels of this heavy metal, they do not assess the existence of an adaptive response. To test whether S. solfataricus exhibits an adaptive response to mercuric chloride, cultures were pretreated by addition of a concentration of this metal that was transiently growth inhibitory (0.5 μM) and then challenged using a dose (1.5 μM) that blocked growth of untreated cells and could only be overcome with prolonged incubation (Fig. 1). The growth of mercury-adapted cells was insensitive to addition of the higher challenge dose, since no growth lag was apparent. This indicates that S. solfataricus harbors an adaptive mechanism to detoxify mercuric chloride.
Protein phylogeny of MerA and MerR.
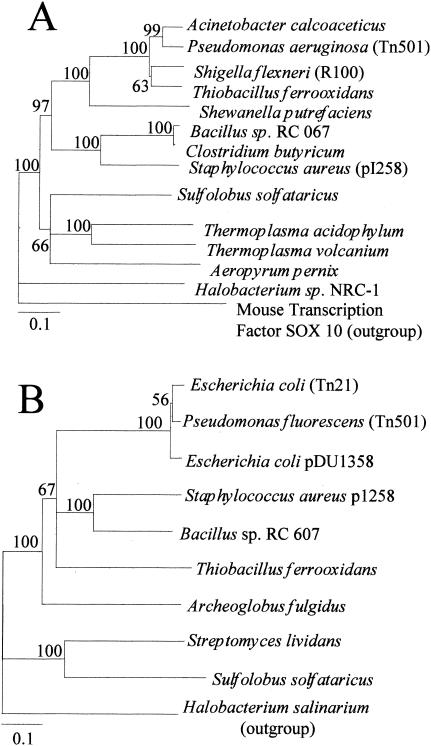
Several mer genes have been annotated in the S. solfataricus strain P2 genome, including a putative mercuric reductase (merA; SSO2689) and a mercury transcriptional regulatory protein (merR; SSO2688 [49]). The merR and merA genes are arranged in opposite directions, and their open reading frames are separated by a 300-nucleotide (nt) sequence (Fig. 2). Immediately downstream of merA is an open reading frame designated SSO2690 of unknown function separated from merA by 141 nt. BLAST analysis indicated that the S. solfataricus MerA protein exhibited significant homology to biochemically validated bacterial MerA proteins. The best match was to the S. aureus protein, with 40% amino acid identity over the length of the protein. Since MerA orthologs have been annotated in other archaeal genomes, a protein phylogenetic analysis of these and selected bacterial sequences was conducted to ascertain the relationship between these proteins (Fig. 3A). A consensus neighbor-joining distance tree with robust topology indicated the existence of two distinct clades. One clade comprised the bacterial MerA proteins, while the other contained crenarchaeal MerA sequences. The halobacterial NRC1 protein fell outside the crenarchaeal clade despite the use of an unrelated sequence as an outgroup. Unlike merA homologs, merR homologs are only rarely evident in archaeal genomes. To assess the significance of the putative MerR protein in S. solfataricus, a similar protein phylogenetic procedure was employed (Fig. 3B). In this case, a small clade containing Sulfolobus and Streptomyces lividans MerR representatives was evident and distinct from all other bacterial MerR sequences.
FIG. 2.
S. solfataricus mer locus. Lengths are in nucleotides.
FIG. 3.
Protein phylogenies of MerA and MerR. (A) MerA tree; (B) MerR tree. Neighbor-joining distance trees are based on comparison of near-full-length protein sequences of 422 residues for MerA and 104 residues for MerR. Distances are indicated by the bar in the lower left corner and represent 10 substitutions per 100 residues. Percent occurrence among 100 trees is given for all nodes. Accession numbers are indicated in the text.
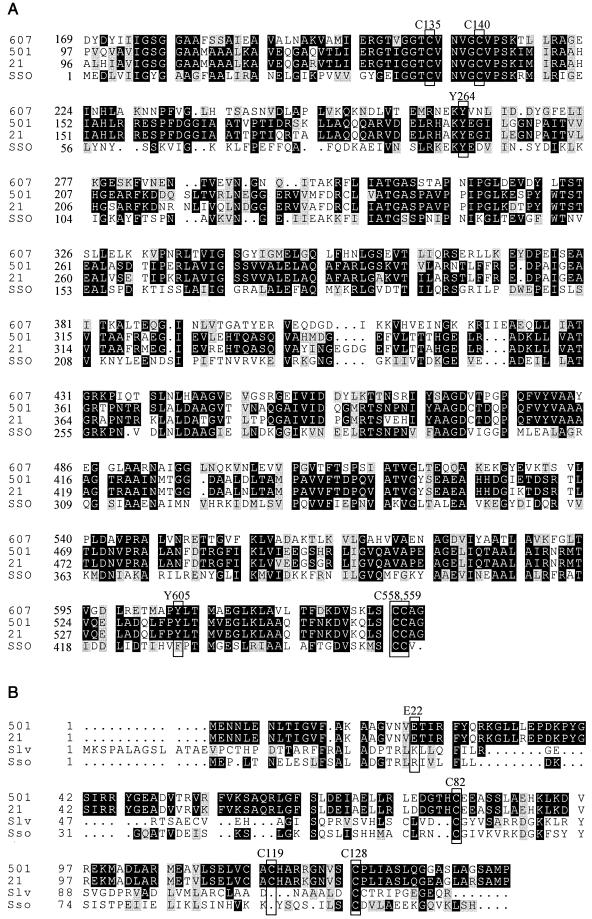
A multiple sequence alignment comparing MerA proteins from Bacillus sp. strain RC601, Tn501, Tn21, and S. solfataricus is also shown (Fig. 4A). The catalytically active cysteines (C135, C140, C558, and C559 [15]) and one of two tyrosines (Y264 [42]) are present. In addition, the S. solfataricus protein lacks the N-terminal extension commonly found in bacterial MerA proteins and present in the bacterial sequences used in this alignment (4). A multiple sequence alignment of MerR proteins from Tn501, Tn21, S. lividans, and S. solfataricus is also shown (Fig. 4B). The S. solfataricus MerR homolog exhibits several key differences from most of its bacterial counterparts. While it has a conserved N-terminal DNA binding motif, the sequence of this domain is divergent and it lacks a critical glutamic acid residue (E22) required for DNA binding of the MerR protein from transposon Tn501 (46). The S. solfataricus MerR homolog is one-third shorter in length than most of its bacterial counterparts and contains only two of the three catalytically active cysteines (C82 and C128) found in Tn501 and Tn21 MerR proteins (4). The third active cysteine (C119) is missing.
FIG.4.
Multiple sequence alignments of MerA and MerR. Sequence conservation is indicated by boxshading. (A) MerA alignment. Multiple sequence alignment is shown for Bacillus sp. strain RC607 (607), Tn501 (501), Tn21 (21), and S. solfataricus (Sso) MerA proteins. Numbering refers to the position of the protein relative to its amino-terminal end. Conserved catalytically active residues are outlined in boxes and labeled. (B) MerR alignment. Multiple sequence alignment is shown for Tn501 (501), Tn21 (21), and S. lividans (Slv) and S. solfataricus (Sso) MerR proteins. The conserved glutamate required for DNA binding and the catalytically active cysteines are outlined in boxes and labeled.
Transcriptional regulation of merA expression by mercury.
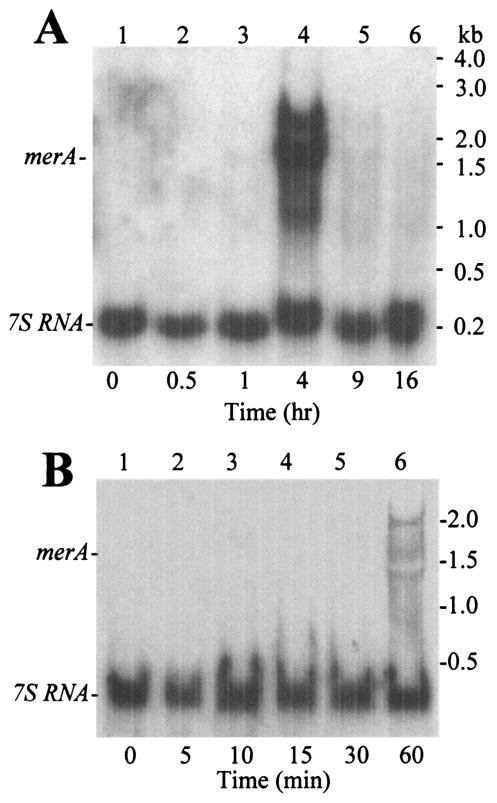
Northern blot analysis of merA was conducted to test if expression of this gene was influenced by mercuric chloride challenge (Fig. 5). Batch cultures grown in a defined minimal SM were treated with 0.3 μM mercuric chloride, and samples were removed for analysis at times thereafter. The signal recognition particle 7S RNA was used to standardize mRNA band intensity as described previously (9). Blots were probed simultaneously with antisense merA and 7S RNA riboprobes. Levels of merA mRNA were undetectable before mercuric chloride addition and for a short period immediately following addition. After 4 h, however, several abundant transcripts became evident. The smaller of the two transcripts (approximately 1.5 kb) was of sufficient size to encode merA, while the larger transcript (approximately 2.0 kb) was sufficient in size to encode both merA and the adjacent gene, SSO2690. After 9 h of exposure these mRNAs were no longer detected. Several smaller RNAs representing possible mRNA degradation products were also evident. Analysis of transcript induction using samples from earlier times and with longer autoradiographic exposure indicated merA transcripts could be detected within 1 h of mercuric chloride addition. Fully induced merA mRNA levels normalized to 7S RNA levels in the same samples and expressed as a percentage of that amount averaged 119.9% ± 84.9% (mean ± standard deviation). Transcript abundances for other highly expressed S. solfataricus genes expressed in an identical manner were as follows: lacS, 45.2% ± 14.4%; tfb-1, 67.2% ± 16.6%; malA, 47.8% ± 24.3%; sod, 88.3% ± 40.0%; glnA-1, 45.0% ± 12.1%; and dhg-1, 88.4% ± 14.9% (9). In all cases the number of samples examined was at least three, and the variation between triplicate measurements is indicated. This comparison indicates merA transcript abundance was within the range of these other mRNAs and, therefore, that merAp is a reasonably strong promoter.
FIG. 5.
Northern analysis of merA following mercuric chloride challenge. Cells in exponential phase were treated with 0.3 mM mercuric chloride, and RNA was extracted at the times indicated beneath the figures for analysis. Blots were probed simultaneously using merA and 7S RNA riboprobes. The data in panels A and B were prepared from independent experiments.
Primer extension analysis of merAp.
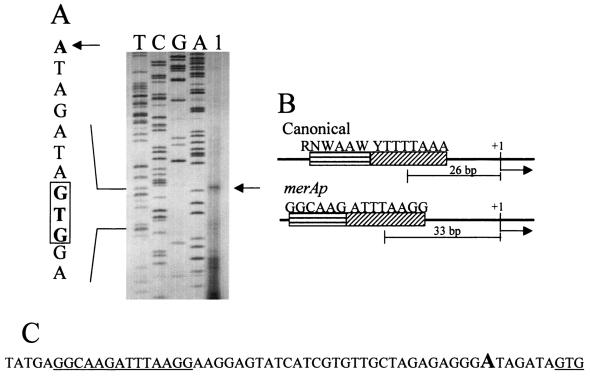
To investigate the basis for merA regulation by mercuric chloride challenge, primer extension analysis was conducted to help identify the merA promoter (merAp). RNA was extracted from cells 4 h after treatment with 0.3 μM mercuric chloride to ensure the presence of merA transcript. The start point occurred at an A residue 7 nt upstream of the start codon (GTG) of merA (Fig. 6A). There is no TATA box or BRE sequence apparent at the expected position 26 nt from the transcription start site (Fig. 6B); however, a putative TATA box and BRE are evident 5 nt upstream of this location. The midpoint of this putative TATA box (ATTTAAGG) is located 33 nt 5′ to the start point of transcription and is flanked on the 5′ side by a putative BRE sequence (GGCAAG) (Fig. 6C).
FIG. 6.
Primer extension analysis and DNA sequence of merAp. (A) Primer extension analysis of RNA prepared from mercuric chloride-treated cells. The sequencing ladder is on the left, and the extension reaction is in lane 1. The start codon is boxed. The location of the start site is indicated by the arrow. (B) Location and composition of the TATA box and BRE for canonical and merAp promoters. (C) DNA sequence of merAp with underlines indicating the positions of the BRE, TATA box, and start codon. The large A indicates the transcription start site.
Construction and analysis of the merA disruption mutant.
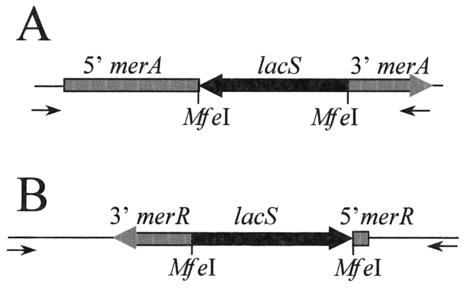
The role of merA in the adaptive resistance response to mercuric chloride challenge was investigated by creating a mutant strain encoding a loss-of-function mutation in this gene. The merA disruption mutant was created by targeted recombination, placing a copy of the β-glycosidase gene (lacS) in the middle of the target gene in the chromosome (Fig. 7A) using a previously developed procedure (59). The lacS gene, including its promoter, and 169 and 157 bp of 5′ and 3′ flanking DNA sequence, respectively, were inserted into a unique MfeI site in merA located 932 bp downstream from the merA start codon in a reverse orientation relative to merA.
FIG. 7.
Disruption of merA and merR. Schematic representations of the disrupted loci indicating the location of the disrupting copy of lacS (black region) in the target genes (grey regions). The direction of transcription is indicated by the arrows; merA (A) and merR (B) are divergently transcribed. The primers used in the analysis of the disrupted and wild-type alleles are indicated beneath the schematic.
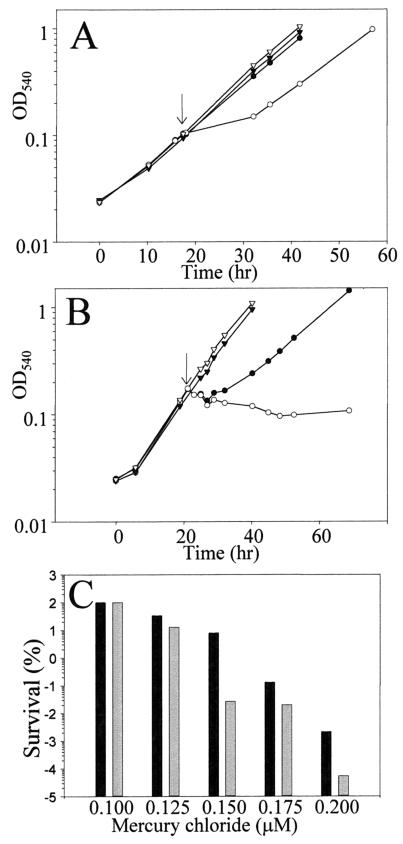
To assess the physiological consequence of merA disruption, the response of the merA mutant strain to mercuric chloride challenge was compared to that of the otherwise isogenic wild-type strain (Fig. 8). Both strains were grown in a liquid sucrose minimal medium. At a cell density of 108 cells/ml, 0.3 μM mercuric chloride was added to each culture. Cultures of both strains with no added mercury were included as controls (Fig. 8A). Growth of the wild-type strain was unaffected by mercury addition, while the mutant strain exhibited a reduced rate of growth for nearly 12 h followed by a resumption of the previous growth rate. In response to a higher concentration of mercuric chloride (0.5 μM), the wild-type strain exhibited a lag followed by a reduced rate of growth, while the merA disruption mutant discontinued growth altogether (Fig. 8B). The efficiency of plating (EOP) of both strains was examined on RM plates over a range of concentrations of added mercuric chloride. The EOP of the mutant relative to that of the wild-type strain was most impaired at 0.15 μM mercuric chloride, reaching only 0.34% of the wild-type value (Fig. 8C). At concentrations below and above this amount the differential between the strains was less pronounced, but in all cases the disruption mutant exhibited greater sensitivity than the wild-type strain.
FIG. 8.
Response of the merA disruption mutant to mercuric chloride. (A and B) Response of the wild type (closed symbols) and merA disruption mutant (open symbols) to mercuric chloride challenge during growth in SM liquid medium. (A) 0.3 μM mercuric chloride challenge (circles); (B) 0.5 μM mercuric chloride challenge (circles). Inverted triangles (A and B), no addition. The arrow indicates the time of addition of mercuric chloride. (C) EOP of the wild type (filled bars) and the merA disruption mutant (grey bars) on RM plates containing mercuric chloride. Values are a percentage of the EOP observed with no added mercuric chloride. Data are averages from duplicate plates.
Production of elemental mercury was assessed as an additional indication of the role of the merA gene in mercury resistance. Mercuric reductase catalyzes the reduction of mercuric ion to elemental mercury. Since elemental mercury is volatile, it can accumulate in the headspace of a culture flask. Detection of this form of the metal was accomplished using a Jerome meter, which measures conductivity changes produced by the interaction of mercury with a gold film sensor. Cultures of both strains were cultivated in a liquid SM, and 0.3 μM mercuric chloride was added. After 4 h of additional incubation, the flasks were allowed to cool and the concentration of volatile mercury trapped in the headspace was measured. Levels of mercury (in nanograms) were as follows: wild type, 62.8 ± 11.9; merA mutant, 11.8 ± 3.8; no inoculum, 41 ± 1.9; detergent-treated wild type, <0.4. The wild-type strain produced sixfold more elemental mercury than the mutant. A control sample consisting of the minimal medium with mercuric chloride but without added cells also produced detectable mercury, though at considerably lower levels. Since the headspace of the mutant culture also contained detectable mercury, an effort was made to identify its source. Continued release of low levels of mercury from the mutant strain together with its ability to withstand exposure to lower challenge doses (Fig. 8C) suggested additional pathways for mercury reduction that could be present in this strain. To test this possibility, the ability of the mutant strain to undergo an adaptive response to mercuric chloride challenge was examined as described above for the wild-type strain (Fig. 1); however, no adaptation was observed. Since cell extracts prepared by detergent solubilization of the cell envelope incubated in the same medium with added mercuric chloride produced no measurable mercury, it seems plausible that reduced intracellular thiol compounds were responsible for residual mercury reduction in the mutant strain.
Construction and analysis of the merR disruption mutant.
Disruption of the merR gene used a strategy similar to that employed for merA disruption, but the alternative strain, PBL2025, replaced the use of PBL2002. PBL2025 harbors a large chromosomal deletion spanning lacS and flanking regions and increases the frequency of recovery of recombinants at the target locus by avoiding those occurring at the lacS locus. The deletion in this strain was spontaneous and was recovered using a screen for isolates that had lost lacS (22). PCR was used to determine the extent of the deleted region. Open reading frames that could be amplified from this strain included SSO3000, -3002, -3003, -3051, and -3052, while those that could not be amplified included SSO3004, -3006, -3017, -3019, -3032, -3036, -3048, -3049, and -3050. DNA sequence analysis indicated the presence of an IS1173 inserted in SSO3052 at the 3′ end. These results showed that the deleted region extends from SSO3004 through SSO3050. The suicide plasmid pMerS1 was transformed into PBL2025 by electroporation, and chromosomal recombinants were recovered by selection for lactose utilization as described previously (59). Since lacS was deleted, recombinants could only arise by homologous recombination between the chromosomal and plasmid-encoded copies of merR. The lacS gene was inserted in a reverse orientation relative to merR at an artificial MfeI site that converted a G into a C at nt 19 relative to the merR start codon (Fig. 7B).
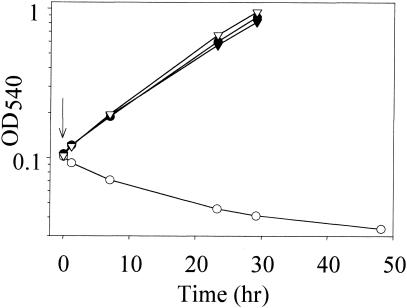
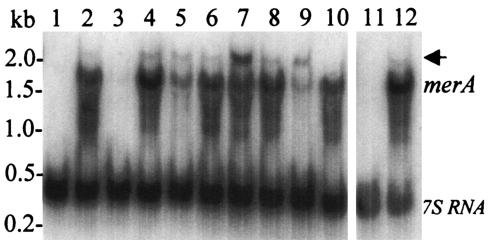
The merR disruption mutant exhibited increased resistance to mercuric chloride (Fig. 9). The adaptive resistance response of wild-type cells enabled them to grow without a lag when challenged with doses of mercuric chloride that were otherwise growth inhibitory. The merR disruption mutant was insensitive to addition of 0.75 μM mercuric chloride, while this same dose blocked growth of the wild-type strain. Since mercuric reductase appears necessary for mercuric chloride resistance in wild-type cells, the resistance of the previously unconditioned merR disruption mutant suggested merA expression might be constitutive. Northern analysis of merA levels in the merR mutant and its parental strain were examined to determine the correlation between transcript abundance and mercury resistance (Fig. 10). Mercuric chloride (0.3 μM) was added to both cultures, and RNA samples were removed at the indicated times. Unlike the inducible pattern of merA expression observed in the wild-type strain, in the merR disruption mutant the merA transcript was produced constitutively, and this expression was independent of mercury challenge. The ratio of the levels of merA transcript relative to those of the 7S RNA control in the merR disruption strain were similar to those observed for the wild-type strain at peak levels of merA expression. These results indicated that merR is necessary for the inducible pattern of merA expression evident in wild-type cells. They also indicate that MerR must be absent for constitutive expression of merA.
FIG. 9.
Response of the merR disruption mutant to mercuric chloride. Cells were grown in SM liquid medium and challenged with 0.75 μM mercuric chloride (arrow). Cultures were merR disruption mutant (triangles) or wild type (circles). Open symbols, untreated cultures; closed symbols, treated cultures.
FIG. 10.
Northern analysis of merA in the merR disruption mutant following mercuric chloride challenge. Levels of merA in the merR disruption mutant and the wild type (PBL2025) were determined in response to mercuric chloride challenge (0.3 μM). Lanes 1, 3, 5, 7, 9, and 11, wild type; lanes 2, 4, 6, 8, and 10, merR disruption mutant. Sample times and lane numbers were as follows: lanes 1 and 2, 0 h; lanes 3 and 4, 0.5 h; lanes 5 and 6, 1 h; lanes 7 and 8, 2 h; lanes 9 and 10, 4 h; lanes 11 and 12, 9 h. The positions of the merA and 7S RNA are indicated. A larger transcript possibly encoding merA and SSO2690 is indicated by the arrow.
DISCUSSION
These studies investigated the relevance of MerA and MerR homologs to the sensitivity of the archaeon S. solfataricus to challenge by the heavy metal mercury. To explain the observed results, the following model is proposed. MerR regulates merA transcription, acting in a negative fashion on merAp activity. Mercuric ion relieves this effect by interacting with MerR. If MerR binds merAp, then mercuric ion interaction may stimulate MerR release. However, merAp structure appears insufficient to explain the merA expression pattern observed in the merR disruption mutant strain, because merAp is unlikely to constitute a strong promoter. Though the merAp promoter has a putative TATA box and a consensus BRE, the presence of a G at the 3′ end of another archaeal TATA hexamer reduced promoter strength by 75% (21). In addition, merAp exhibits nonstandard spacing between the TATA box and the start point of transcription. The consensus for this distance in archaeal promoters is 26 nt measured from the midpoint of the octameric TATA box to the start point of transcription (21, 40, 41). In merAp, this distance is 33 nt and would thus rotate the TATA box around the DNA helix relative to the start point of transcription. Consequently, constitutive expression of merAp should require the action of some additional factor to overcome this topological constraint on promoter recognition by general transcription factors and RNA polymerase.
The pattern of merA expression observed in the merR disruption mutant also indicated that MerR is only negatively acting. This is unlike the case with most bacterial MerR proteins, and the only exception yet reported is the MerR protein of S. lividans (47). Interestingly, protein phylogenetic analysis of the S. solfataricus MerR protein revealed that it occurred in a clade with the S. lividans protein and separately from other bacterial MerR proteins. This finding suggests that these proteins have a common evolutionary origin. The absence of the third catalytically active cysteine residue (C119) in the S. solfataricus MerR protein does not block its ability to act as a negative regulator. In addition, the absence of the conserved glutamate (E22) in the N-terminal domain of bacterial MerR proteins, which is required for DNA binding, again appears unnecessary for the action of the S. solfataricus MerR protein as a negative regulatory element for merA transcription.
Restoration of repression of merA expression observed in wild-type S. solfataricus strains following mercury challenge occurred prior to the onset of significant cell division. Disappearance of merA transcript must therefore require that MerR repression be reinstated to prevent new synthesis and that merA transcript be actively degraded. The stability of mRNAs in this organism has been examined (9). In all cases the rate of turnover was low, indicating that mRNA degradation proceeded at a slow rate, and in the case of merA transcript a half-life of less than an hour seemed necessary.
The level of mercuric chloride resistance observed in S. solfataricus containing a functional mercuric reductase is surprisingly low relative to that observed in bacteria. Mercury-resistant bacteria exhibit tolerance to mercuric chloride concentrations 20- to 40-fold greater (17, 55) than that reported here for S. solfataricus. Though the archaeal merA is chromosomally encoded and bacterial mer operons are typically plasmid encoded, R factors are low copy, and merA gene dosage appears unrelated to the level of mercury resistance (35). Consequently, the difference observed between this archaeon and bacteria could be explained by proposing that S. solfataricus MerA is a relatively inefficient enzyme, or that there is some other fundamental difference between bacteria and archaea controlling the biological activity of this heavy metal. Homologs of MerA are evident in many archaeal genomes. Proof of the importance of this gene in S. solfataricus indicates that other archaeal homologs may indeed play functional roles. Since merR homologs rarely accompany these merA sequences, it will be of interest to understand how these merA homologs are regulated and what factors might control their expression.
Construction of the merR disruption mutant employed a new strategy involving the use of a host strain with the disrupting marker gene, lacS, deleted. This approach simplified recovery of recombinant isolates by preventing occurrence of those resulting from recombination at the chromosomal copy of lacS as observed previously (59). The present system should be suitable to allow identification of cis-acting sequences required for merR function as well as to address questions concerning merAp promoter activity. Further advances in genetic strategies for the manipulation of this organism will facilitate studies on mechanisms involving gene regulation of the archaeal transcription apparatus and provide a complement to biochemical approaches.
Acknowledgments
This research was supported by National Science Foundation grants MCB-0085216 and MCB-0235167.
We thank Anne Summers for advice on early stages of the research.
REFERENCES
- 1.Allen, M. B. 1959. Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch. Mikrobiol. 32:270-277. [DOI] [PubMed] [Google Scholar]
- 2.Ansari, A. Z., M. L. Chael, and T. V. O'Halloran. 1992. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature 355:87-89. [DOI] [PubMed] [Google Scholar]
- 3.Ansari, A. Z., J. E. Bradner, and T. V. O'Halloran. 1995. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374:371-375. [DOI] [PubMed] [Google Scholar]
- 4.Barkay, T., S. M. Miller, and A. O. Summers. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355-384. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, P., S. A. Qureshi, and S. P. Jackson. 1995. Transcription: new insights from studies on Archaea. Trends Genet. 11:279-283. [DOI] [PubMed] [Google Scholar]
- 6.Bell, S. D., A. B. Brinkman, J. van der Oost, and S. P. Jackson. 2001. The archaeal TFIIEα homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep. 2:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell, S. D., S. S. Cairns, R. L. Robson, and S. P. Jackson. 1999. Transcriptional regulation of an archaeal operon in vivo and in vitro. Mol. Cell 4:971-982. [DOI] [PubMed] [Google Scholar]
- 8.Bell, S. D., P. L. Kosa, P. B. Sigler, and S. P. Jackson. 1999. Orientation of the transcription preinitiation complex in archaea. Proc. Natl. Acad. Sci. USA 96:13662-13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bini, E., V. Dikshit, K. Dirksen, M. Drozda, and P. Blum. 2002. Stability of mRNA in hyperthermophilic archaea. RNA 8:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brock, T. D., K. M. Brock, R. T. Belly, and R. L. Weiss. 1972. Sulfolobus: a genus of sulfur oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84:54-68. [DOI] [PubMed] [Google Scholar]
- 11.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 12.Cohen-Kupiec, R., C. Blank, and J. A. Leigh. 1997. Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc. Natl. Acad. Sci. USA 94:1316-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlke, I., and M. Thomm. 2002. A pyrococcus homolog of the leucine-responsive regulatory protein, LrpA, inhibits transcription by abrogating RNA polymerase recruitment. Nucleic Acids Res. 30:701-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Rosa, M., A. Gambacorta, and J. D. Bu'lock. 1975. Extremely thermophilic acidophilic bacteria convergent with Sulfolobus acidocaldarius. J. Gen. Microbiol. 86:156-164. [DOI] [PubMed] [Google Scholar]
- 15.Distefano, M. D., M. J. Moore, and C. T. Walsh. 1990. Active site of mercuric reductase resides at the subunit interface and requires Cys135 and Cys140 from one subunit and Cys558 and Cys559 from the adjacent subunit: evidence from in vivo and in vitro heterodimer formation. Biochemistry 29:2703-2713. [DOI] [PubMed] [Google Scholar]
- 16.Ercal, N., H. Gurer-Orhan, and N. Aykin-Burns. 2001. Toxic metals and oxidative stress. Part I: mechanism involved in metal induced oxidative damage. Curr. Top. Med. Chem. 1:529-539. [DOI] [PubMed] [Google Scholar]
- 17.Foster, T. J., H. Hakahara, A. A. Weiss, and S. Silver. 1979. Transposon A-generated mutations in the mercuric resistance genes of plasmid R100-1. J. Bacteriol. 140:167-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gohl, H. P., B. Grondahl, and M. Thomm. 1995. Promoter recognition in archaea is mediated by transcription factors: identification of transcription factor aTFB from Methanococcus thermolithotrophicus as archaeal TATA-binding protein. Nucleic Acids Res. 23:3837-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grogan, D. W. 1989. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 171:6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felsenstein, J. 1989. PHYLIP phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 21.Hain, J., W.-D. Reiter, U. Hudepohl, and W. Zillig. 1992. Elements of an archaeal promoter defined by mutational analysis. Nucleic Acids Res. 20:5423-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanzelka, B. L., T. J. Darcy, and J. N. Reeve. 2001. TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEα. J. Bacteriol. 183:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haseltine, C., R. Montalvo-Rodriguez, E. Bini, A. Carl, and P. Blum. 1999. Coordinate transcriptional control in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 181:3920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haseltine, C., R. Montalvo-Rodriguez, A. Carl, E. Bini, and P. Blum. 1999. Extragenic pleiotropic mutations that repress glycosyl hydrolase expression in the hyperthermophilic archaeon Sulfolobus solfataricus. Genetics 152:1353-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hausner, W., U. Lange, and M. Musfeldt. 2000. Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J. Biol. Chem. 275:12393-12399. [DOI] [PubMed] [Google Scholar]
- 26.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonuscheit, M., E. Martusewitsch, K. M. Stedman, and C. Schleper. 2003. A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol. Microbiol. 48:1241-1252. [DOI] [PubMed] [Google Scholar]
- 28.Klenk, H.-P., P. Palm, F. Lottspeich, and W. Zillig. 1992. Component H of the DNA-dependent RNA polymerase of archaea is homologous to a subunit shared by the three eucaryal nuclear RNA polymerases. Proc. Natl. Acad. Sci. USA 89:407-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsh, T. L., C. I. Reich, R. B. Whitelock, and G. J. Olsen. 1994. Transcription factor IID in the Archaea: sequences in the Thermococus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc. Natl. Acad. Sci. USA 91:4180-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 31.Osborn, A. M., K. D. Bruce, P. Strike, and D. A Ritchie. 1997. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 19:239-262. [DOI] [PubMed] [Google Scholar]
- 32.Ouhammouch, M., R. E. Dewhurst, W. Hausner, M. Thomm, and E. P. Geiduschek. 2003. Activation of archaeal transcription by recruitment of the TATA-binding protein. Proc. Natl. Acad. Sci. USA 100:5097-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pak, K.-R., and R. Bartha. 1998. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Appl. Environ. Microbiol. 64:1987-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan-Hou, J. S. K., and N. Imura. 1981. Role of hydrogen sulphide in mercury resistance determined by plasmid of Clostridium cochlearium T2. Arch. Microbiol. 129:49-52. [DOI] [PubMed] [Google Scholar]
- 35.Philippidis, G. P., L. H. Malmberg, W. S. Hu, and J. L. Schottel. 1991. Effect of gene amplification on mercuric ion reduction activity of Escherichia coli. Appl. Environ. Microbiol. 57:3558-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plosser, P., and F. Pfeifer. 2002. A bZIP protein from halophilic archaea: structural features and dimer formation of cGvpE from Halobacterium salinarum. Mol. Microbiol. 45:511-520. [DOI] [PubMed] [Google Scholar]
- 37.Qureshi, S. A., P. Baumann, T. Rowlands, B. Khoo, and S. P. Jackson. 1995. Cloning and functional analysis of the TATA binding protein from Sulfolobus shibatae. Nucleic Acids Res. 23:1775-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qureshi, S. A., B. Khoo, P. Baumann, and S. P. Jackson. 1995. Molecular cloning of the transcription factor TFIIB homolog from Sulfolobus shibatae. Proc. Natl. Acad. Sci. USA 92:6077-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qureshi, S. A., and S. P. Jackson. 1998. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol. Cell 1:389-400. [DOI] [PubMed] [Google Scholar]
- 40.Reeve, J. N. 2003. Archaeal chromatin and transcription. Mol. Microbiol. 48:587-598. [DOI] [PubMed] [Google Scholar]
- 41.Reiter, W.-D., U. Hudepohl, and W. Zillig. 1990. Mutational analysis of an archaebacterial promoter: essential role of a TATA box for transcription efficiency and start-site selection in vitro. Proc. Natl. Acad. Sci. USA 87:9509-9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rennex, D., R. T. Cummings, M. Pickett, C. T. Walsh, and M. Bradley. 1993. Role of tyrosine residues in Hg(II) detoxification by mercuric reductase from Bacillus sp. strain RC607. Biochemistry 32:7475-7478. [DOI] [PubMed] [Google Scholar]
- 43.Rockabrand, D., K. Livers, T. Austin, R. Kaiser, D. Jensen, R. Burgess, and P. Blum. 1998. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J. Bacteriol. 180:846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rolfsmeier, M., and P. Blum. 1995. Purification and characterization of a maltase from the extremely thermophilic crenarchaeote Sulfolobus solfataricus. J. Bacteriol. 177:482-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rolfsmeier, M., C. Haseltine, E. Bini, A. Clark, and P. Blum. 1998. Molecular characterization of the α-glucosidase gene (malA) from the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 180:1287-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross, W., S. J. Park, and A. O. Summers. 1989. Genetic analysis of transcriptional activation and repression in the Tn21 mer operon. J. Bacteriol. 171:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rother, D., R. Mattes, and J. Altenbuchner. 1999. Purification and characterization of MerR, the regulator of the broad-spectrum mercury resistance genes in Streptomyces lividans 1326. Mol. Gen. Genet. 262:154-162. [DOI] [PubMed] [Google Scholar]
- 48.Rowlands, T., P. Baumann, and S. P. Jackson. 1994. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science 264:1326-1329. [DOI] [PubMed] [Google Scholar]
- 49.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeote Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silver, S., and L. T. Phung. 1996. Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50:753-789. [DOI] [PubMed] [Google Scholar]
- 51.Summers, A. O. 1992. Untwist and shout: a heavy metal-responsive transcriptional regulator. J. Bacteriol. 174:3097-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Triezenberg, S. J. 1992. Preparation and analysis of RNA, p. 4.8.1-4.8.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley, New York, N.Y.
- 54.Vierke, G., A. Engelmann, C. Hebbeln, and M. Thomm. 2003. A novel archaeal transcriptional regulator of heat shock response. J. Biol. Chem. 278:18-26. [DOI] [PubMed] [Google Scholar]
- 55.Wireman, J., C. A. Liebert, T. Smith, and A. O. Summers. 1997. Association of mercury resistance with antibiotic resistance in the gram-negative fecal bacteria of primates. Appl. Environ. Microbiol. 63:4494-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74:5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woese, C., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains archaea, bacteria and eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wood, J. M., F. S. Kennedy, and C. G. Rosen. 1968. Synthesis of methyl-mercury compounds by extracts of a methanogenic bacterium. Nature 220:173-174. [DOI] [PubMed] [Google Scholar]
- 59.Worthington, P., V. Hoang, F. Perez-Pomarez, and P. Blum. 2003. Targeted disruption of the α-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 185:482-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Worthington, P., P. Blum, F. Perez-Pomares, and T. Elthon. 2003. Large-scale cultivation of acidophilic hyperthermophiles for recovery of secreted proteins. Appl. Environ. Microbiol. 69:252-257. [DOI] [PMC free article] [PubMed] [Google Scholar]