Abstract
The opportunistic pathogen Burkholderia cenocepacia produces the yellow-green fluorescent siderophore, pyochelin. To isolate mutants which do not produce this siderophore, we mutagenized B. cenocepacia with the transposon mini-Tn5Tp. Two nonfluorescent mutants were identified which were unable to produce pyochelin. In both mutants, the transposon had integrated into a gene encoding an orthologue of CysW, a component of the sulfate/thiosulfate transporter. The cysW gene was located within a putative operon encoding other components of the transporter and a polypeptide exhibiting high homology to the LysR-type regulators CysB and Cbl. Sulfate uptake assays confirmed that both mutants were defective in sulfate transport. Growth in the presence of cysteine, but not methionine, restored the ability of the mutants to produce pyochelin, suggesting that the failure to produce the siderophore was the result of a depleted intracellular pool of cysteine, a biosynthetic precursor of pyochelin. Consistent with this, the wild-type strain did not produce pyochelin when grown in the presence of lower concentrations of sulfate that still supported efficient growth. We also showed that whereas methionine and certain organosulfonates can serve as sole sulfur sources for this bacterium, they do not facilitate pyochelin biosynthesis. These observations suggest that, under conditions of sulfur depletion, cysteine cannot be spared for production of pyochelin even under iron starvation conditions.
The genus Burkholderia includes a number of gram-negative bacterial species that are pathogenic to animals and/or plants. One group, the B. cepacia complex (BCC), is noted for the ability to cause opportunistic infections in humans, particularly in patients with cystic fibrosis (CF) (33). In addition, some members of the BCC have attracted interest as antagonists of soilborne plant pathogens and/or as plant growth-promoting agents and are therefore under development as biocontrol agents. Furthermore, their exceptional metabolic diversity has stimulated interest in their use as agents of bioremediation (6). The BCC contains at least nine closely related but genetically distinct species (or genomovars), of which members of genomovar III (recently designated B. cenocepacia) are the most prevalent in CF infections (6, 35, 63). The complete genome sequence has recently been determined for a B. cenocepacia strain (http://www.sanger.ac.uk/Projects/B_cenocepacia/).
B. cenocepacia (and some pseudomonads) produces the yellow-green fluorescent siderophore pyochelin as a mechanism for acquiring iron from the host (9, 10, 55, 56, 57). Pyochelin is a low-affinity but active tridentate siderophore which binds iron with a stoichiometry of two pyochelin molecules per Fe(III) ion (7, 8). This siderophore exists as two stereoisomers in nature, pyochelin I and pyochelin II, and can also chelate Zn(II), Cu(II), Co(II), Mo(VI), and Ni(II) (2, 45, 67). Pyochelin is biosynthesized from salicylate by the successive addition and cyclization of two molecules of cysteine (41, 42, 44). Salicylate also forms complexes with iron and was identified as a siderophore in some members of the BCC and some pseudomonads (38, 58, 68). In the first step in pyochelin biosynthesis, condensation of salicylate with a molecule of l-cysteine, followed by cyclization of the cysteine moiety to a thiazoline ring, results in formation of an intermediate, dihydroaeruginoic acid (Dha) (2, 3, 42, 50). Condensation and cyclization of a second molecule of l-cysteine generates the second thiazoline ring, which is subsequently reduced and methylated (50).
The genes required for the biosynthesis of salicylate from chorismate, and for the biosynthesis of pyochelin from salicylate, have been identified in Pseudomonas aeruginosa (43, 44, 49). These include pchA and pchB, which are required for conversion of chorismate into salicylate via isochorismate (49), and pchE and pchF, which encode nonribosomal peptide synthetases that are required for the sequential formation of Dha and pyochelin (44). Regulation of these genes in P. aeruginosa requires the Fur protein, a repressor of transcription of the pchDCBA and pchEF operons (44, 50), and PchR, which activates the fptA gene encoding the ferri-pyochelin receptor (18). As part of our investigation into iron regulated gene expression in B. cenocepacia we have used several approaches to identify genes involved in pyochelin biosynthesis. Here we describe a mutagenesis approach that resulted in the isolation of mutants of B. cenocepacia deficient in the production of pyochelin which were affected in sulfate transport. Our results show that pyochelin production in this organism is crucially dependent upon the availability of sulfur as well as iron.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
B. cenocepacia 715j is an isolate from a CF patient (10, 31, 36) and was maintained on M9 minimal salts agar containing glucose (0.5%) as the carbon source (5). To adjust the sulfate content of this medium, the concentration of MgSO4 (present at 1 mM in standard M9 medium) was reduced as required, and the balance of magnesium ions was maintained at 1 mM by the addition of the appropriate amount of MgCl2. Sulfur-limited solid medium was made by solidifying sulfate-free M9-glucose medium with 0.6% molecular-biology-grade agarose (Gibco-BRL). Cysteine or methionine was added to 100 μg/ml to analyze the effect of each on pyochelin production. For analyzing their efficiency as sulfur sources, cysteine, methionine, ethanesulfonate, and taurine (2-aminoethanesulfonate) were used at 500 μM. Sulfite and sulfide were provided as sulfur sources in the form of ethanesulfonate and potassium thiocyanate, respectively. Where indicated, a mixture of 18 amino acids (excluding cysteine and methionine) was added to liquid medium to a final concentration of 20 μg/ml with respect to each amino acid. For measurement of growth rates in the presence of different sulfur sources, cells from overnight cultures were collected by centrifugation, the cell pellet was resuspended in 0.5 vol of 0.85% saline, and the cell suspension was used to inoculate fresh M9-glucose medium (50 ml) at a 200-fold dilution. Cultures were grown in flasks at 37°C with vigorous shaking.
Transposon mutagenesis.
Mini-Tn5Tp is a derivative of mini-Tn5Cm (11, 12) containing the dfr (Tpr) gene from p34E-Tp (15) and will be described elsewhere (C. A. Lowe, K. Agnoli, K. L. Farmer, and M. S. Thomas, unpublished data). This transposon was maintained on plasmid pUT (12, 19). pUTmini-Tn5Tp was introduced into B. cenocepacia 715j by conjugal transfer using the Escherichia coli strain BW19851 (37) as the donor as described previously (12, 19), and transposon mutants (arising at a frequency of ∼1.25 × 10−4 per recipient) were selected on M9 minimal agar containing glucose (0.5%), trimethoprim (40 μg/ml), and kanamycin (25 μg/ml). Following incubation at 37°C for 3 days, candidate pyochelin-deficient mutants were identified by screening colonies for the absence of yellow-green fluorescence on a long-wave UV transilluminator.
Analysis of pyochelin production.
The analysis of pyochelin production in liquid cultures was based on the method of Sokol (55) and Visca et al. (67). Cells were cultured in M9-glucose minimal medium (10 to 30 ml) at 37°C with aeration for 24 to 48 h. The bacteria were collected by centrifugation, and the spent culture supernatants were filtered, acidified, and extracted with 0.4 volume of ethyl acetate. Pyochelin was concentrated by evaporating the organic phase to dryness and resuspending the residue in 50 to 100 μl of methanol. Pyochelin was analyzed by chromatography on a thin (0.2-mm) layer of silica gel 60 (Merck) with chloroform-acetic acid-ethanol (90:5:2.5 [vol/vol]). After developing the plate, two yellow-green fluorescent bands corresponding to two pyochelin stereoisomers, pyochelin I and II (Rf, 0.35 and 0.37, respectively), and a blue fluorescent band corresponding to salicylate (Rf, 0.74) were visualized under UV light (2, 45). In some cases, the precursor of pyochelin, Dha, was also present as a yellow-green fluorescent band migrating between pyochelin and salicylate (50). Production of pyochelin by bacteria grown on M9 minimal agar was evident by the yellow-green fluorescence produced by colonies after 2 to 3 days of incubation at 37°C. This occurred on standard M9 medium and did not require addition of an iron chelator.
Identification of transposon insertion sites.
B. cenocepacia genomic DNA was purified using the Genomix DNA extraction kit (Talent), digested overnight with appropriate restriction enzymes, fractionated in an 0.8% agarose gel, and transferred to Hybond-N+ nylon membrane (Amersham Pharmacia Biotech) by Southern blotting (47), following which hybridization was carried out under normal-stringency conditions using probe DNA labeled by the ECL Direct Nucleic Acid Labeling and Detection System (Amersham Pharmacia Biotech). The probe DNA used contained the dfr gene from p34E-Tp. Genomic DNA fragments corresponding in size to the fragment hybridizing to the probe were eluted from gel slices using the freeze-thaw method of Seth (51) or the QIAquick Gel Extraction kit (Qiagen), ligated to plasmid pHG165 (60) or pUC19 (72), and used to transform E. coli NM522 (17) or MC1061 (4). Selection of transformants was made on Iso-Sensitest agar (Oxoid, Basingstoke, United Kingdom) containing ampicillin (100 μg/ml) and trimethoprim (20 μg/ml).
Sulfate transport assays.
Cultures of cells were grown overnight at 37°C in 20 ml of M9-glucose minimal medium supplemented with 1 mM djenkolic acid and 1 mM MgCl2 (in place of MgSO4). A further 20 ml of the same medium was added, and incubation continued until the culture reached an A600 of 0.5 to 0.8, whereupon the cells were collected by centrifugation and resuspended in 0.5 ml of the same medium. A 0.1-ml aliquot of cell suspension was mixed with 2 ml of the same medium containing chloramphenicol (50 μg/ml) and then transferred to an oxygen electrode. After the addition of 1 μCi of sodium [35S]sulfate (1,050 to 1,600 Ci/mmol; New England Nuclear), 200-μl amounts were removed at regular intervals and mixed with 5 ml of stop buffer (M9-glucose minimal medium containing 2 mM MgSO4 and 2 mM sodium thiosulfate). Radiolabeled cells were filtered through 0.45-μm-pore-size nitrocellulose filters, washed with 5 ml of stop buffer, dried, and counted.
Nucleotide sequence accession number.
The GenBank accession number for the sequence reported in this paper is AF374458.
RESULTS
Isolation of B. cenocepacia mutants which fail to produce pyochelin.
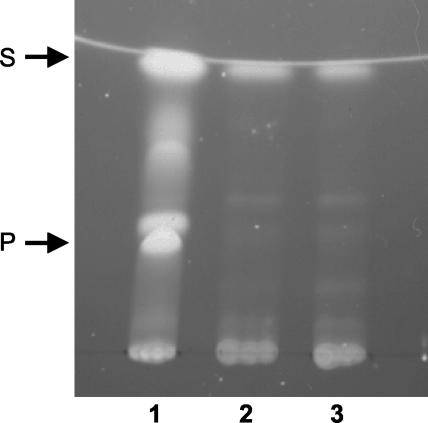
Tn5 derivatives have been shown to be useful for genetic analysis of members of the genus Burkholderia (1, 13, 14). Due to the high intrinsic resistance of B. cenocepacia clinical isolates to most of the antibiotics commonly used to select for plasmids and transposons, we employed a mini-Tn5 derivative, mini-Tn5Tp, which contains a trimethoprim resistance cassette (11; C. A. Lowe, K. Agnoli, K. L. Farmer, and M. S. Thomas, unpublished data) to generate mutants of B. cenocepacia 715j deficient in pyochelin production (Pch−). As this strain has been shown to produce two siderophores, ornibactin and pyochelin, Pch− mutants were identified by screening for transconjugant colonies which failed to exhibit the characteristic yellow-green fluorescence associated with the production of pyochelin on iron-limited M9 minimal agar. Of a total of 15,000 trimethoprim-resistant mutants, arising from three independent experiments, two were identified in which the yellow-green fluorescence was completely absent. Pyochelin was not detectable in culture supernatants from either of these two mutants, KLF2 and KLF3, following growth for up to 72 h in minimal medium (Fig. 1). Production of the pyochelin precursor, salicylic acid, was also reduced but not abolished (Fig. 1). However, production of the other siderophore biosynthesized by 715j, ornibactin (10, 39, 59), remained unaffected in the mutants, as judged by isoelectric focusing followed by chrome azurol S overlay analysis of culture supernatants (26) (results not shown).
FIG. 1.
Analysis of pyochelin and salicylate production by KLF2 and KLF3. Bacteria were grown with aeration in M9-glucose minimal medium for 24 h at 37°C. Pyochelin and salicylate were extracted from culture supernatants with ethyl acetate, resuspended in methanol following evaporation of ethyl acetate, and subjected to thin-layer chromatography in silica gel. Siderophores were visualized under UV light. Lane 1, 715j; lane 2, KLF2; lane 3, KLF3. Abbreviations: P, pyochelin (stereoisomers I and II); S, salicylate.
Genomic loci of mini-transposons in pyochelin-deficient mutants.
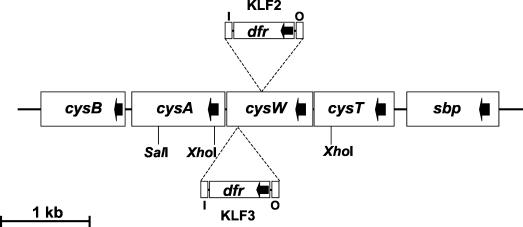
KLF2 and KLF3 chromosomal DNA was digested with the restriction enzymes PvuII and XhoI (which do not cut within mini-Tn5Tp) and SalI (which cuts adjacent to the O end of the transposon) and probed with a DNA fragment containing the dfr (Tpr) gene following Southern blotting. The probed digests revealed that in both mutants mini-Tn5Tp had inserted into chromosomal PvuII and XhoI fragments of the same size (12.0 and 2.0 kb, respectively) (results not shown). The probed SalI digests gave rise to hybridizable fragments of 1.8 and 1.6 kb for KLF2 and KLF3, respectively (results not shown), indicating that although the insertion sites of the mini-transposons in the two mutants may have been at the same genomic locus, they were nonidentical. The chromosomal XhoI and SalI fragments containing the integrated mini-Tn5Tp in KLF2 were cloned into plasmid pHG165, giving rise to pKF001 and pKF002, respectively. The complete sequence of the cloned B. cenocepacia DNA flanking the transposon on both plasmids was determined (total of 1.926 kb of contiguous sequence). In addition, the SalI fragment containing the mini-Tn5Tp integrated in KLF3 was cloned into plasmid pUC19, and the sequence of the genomic DNA adjacent to the I end of the transposon was determined. Database searches and alignment tools revealed that, in both mutants, mini-Tn5Tp had inserted at separate sites (250 bp apart) within a gene encoding a putative orthologue of the E. coli CysW protein (48% amino acid identity) (Fig. 2). The translated product of B. cenocepacia cysW exhibits stronger homology to the putative CysW proteins from members of the α (Mesorhizobium loti and Caulobacter crescentus; 70 and 61% identity, respectively) and β (Neisseria meningitidis; 59% identity) subdivisions of the Proteobacteriaceae, as well as CysW of P. aeruginosa (60% identity) (23, 40, 61, 62). Putative orthologues of the E. coli cysT and cysA genes were identified upstream and downstream, respectively, from B. cenocepacia cysW and were present in the same orientation. The cysW translation initiation codon overlaps the termination codon for cysT translation, and the cysA translation initiation codon is located only 13 bp downstream from the cysW termination codon (not shown), suggesting that these three genes form part of a single operon. In E. coli, CysT, CysW, and CysA form the cytoplasmic membrane component of an ATP-binding cassette (ABC) transporter required for sulfate and thiosulfate import (the sulfate/thiosulfate permease), and the respective genes are organized similarly as part of the cysPTWA operon, where cysP encodes a periplasmic thiosulfate binding protein (20, 28, 53, 54). However, examination of the region around cysW of B. cenocepacia, using data from the B. cenocepacia genome sequencing project, revealed that upstream of cysT and in the same orientation is a gene encoding a putative orthologue of the E. coli sulfate binding protein (Sbp), the periplasmic component of the sulfate permease (Fig. 2) (28). Downstream of cysA is an open reading frame encoding a polypeptide that exhibits high homology to the two LysR-type regulators of sulfur assimilation in E. coli, CysB and Cbl (22, 27, 34, 65).
FIG. 2.
Organization of the cysA locus of B. cenocepacia. Sites of insertion of mini-Tn5Tp giving rise to KLF2 and KLF3 are shown. Cleavage sites for restriction endonucleases used to clone cysA operon DNA from the mutants are also shown. The direction of transcription of the cysA operon genes and the dfr gene in mini-Tn5Tp are indicated by arrows. I and O represent the I and O ends of mini-Tn5Tp. The transcription terminators present in the progenitor of mini-Tn5Tp (mini-Tn5Cm) were removed along with the Cmr cassette during construction of mini-Tn5Tp (11; C. A. Lowe, K. Agnoli, K. L. Farmer, and M. S. Thomas, unpublished data).
Transposon insertion mutants KLF2 and KLF3 are defective in sulfate uptake.
To examine whether the transposon insertions in KLF2 and KLF3 had resulted in impaired sulfate transport, we carried out sulfate uptake assays. Our results showed that whereas the wild-type strain, 715j, imports sulfate at a rate of 11.8 fmol/min/mg of cell protein, the rate of accumulation of sulfate by the mutants is 0.25 and 0.24 fmol/min/mg of cell protein for KLF2 and KLF3, respectively; i.e., the mutants accumulate sulfate at only 2% of the rate of the parent strain.
Growth of KLF2 is impaired under conditions of sulfate depletion, but not under conditions of sulfite or sulfide depletion.
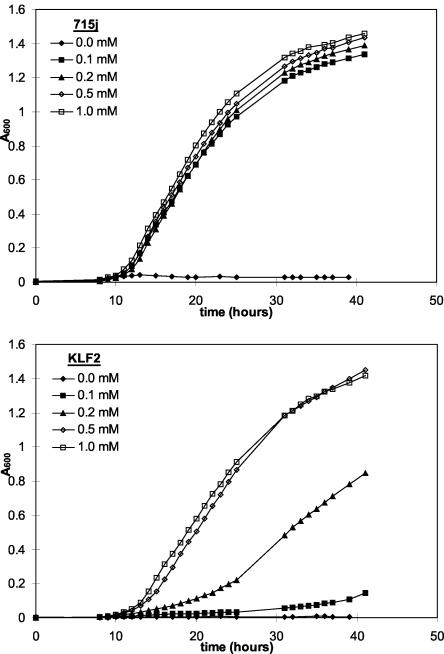
Despite the large decrease in the rate of sulfate uptake by KLF2 and KLF3, both mutants appeared to grow normally on M9-glucose minimal agar, the medium on which these mutants were originally selected. Standard M9 medium contains 1 mM sulfate, added in the form of MgSO4. Therefore, we examined the effect of decreasing the concentration of sulfate on the growth of the mutant strains. The wild-type strain grew normally on plates containing sulfate concentrations as low as 0.1 mM, whereas growth of both mutant strains was retarded at a concentration of 0.5 mM sulfate and inhibited in the presence of 0.1 mM sulfate (results not shown). The effect of sulfate limitation on the growth of KLF2 was also monitored in liquid medium. The growth rate and growth yield of the wild-type strain were not significantly affected by decreasing the exogenous sulfate concentration from 1.0 to 0.1 mM (Fig. 3). However, although growth of KLF2 occurred at the same rate as growth of the wild-type parent in medium containing ≥0.5 mM sulfate, it was impaired in the presence of 0.2 mM sulfate and severely retarded in the presence of 0.1 mM sulfate (Fig. 3). Similar experiments were performed in which the sole sulfur source present was ethanesulfonate or thiocyanate, which enter the sulfur assimilatory pathway at the level of sulfite or sulfide, respectively, and therefore do not require the sulfate transport system (24, 25, 28). These experiments showed that KLF2 grows at the same rate as the wild-type strain at all concentrations of ethanesulfonate and thiocyanate tested (0.1 to 1.0 mM) (results not shown). These results suggest that the defect in sulfur acquisition is restricted to the sulfate uptake system and does not affect assimilatory steps subsequent to the reduction of sulfate to sulfite (the first step in the assimilation of sulfur from inorganic sulfate).
FIG. 3.
Effect of sulfate concentration on the growth of 715j and KLF2. Cells were grown at 37°C with aeration in M9-glucose minimal medium containing various concentrations of sulfate (provided as MgSO4) as indicated. The total concentration of magnesium ions was maintained at 1 mM, with the balance being provided in the form of MgCl2.
KLF2 and KLF3 produce pyochelin in the presence of exogenous cysteine.
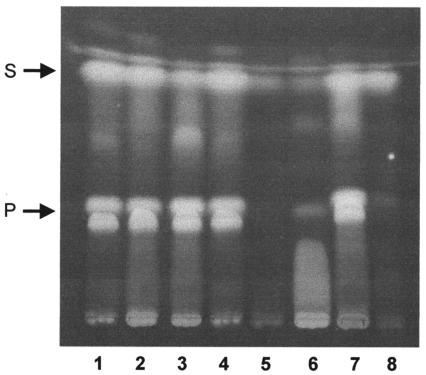
Pyochelin contains two sulfur atoms, each derived from separate cysteine moieties (42). To test the hypothesis that the defect in sulfate uptake in the mutants restricts pyochelin biosynthesis by limiting the intracellular sulfur (i.e., cysteine) pool, we analyzed the ability of KLF2 to produce pyochelin during growth in medium containing l-cysteine. The results show that the presence of cysteine in the medium restored the ability of this strain to produce normal amounts of pyochelin and salicylate (Fig. 4). On the other hand, addition of methionine did not facilitate the production of pyochelin by KLF2, although it did appear to stimulate increased salicylate production. Cysteine had no significant stimulatory effect on pyochelin production by the wild-type strain.
FIG. 4.
Effect of cysteine and methionine on production of pyochelin and salicylate by 715j and KLF2. Cells were grown in M9-glucose minimal medium (containing 1 mM magnesium sulfate) at 37°C with aeration for 24 h. Medium contained no additions, or an 18-amino-acid mixture (18AA; concentration of each amino acid, 20 μg/ml) with or without cysteine or methionine (100 μg/ml each). Pyochelin and salicylate were extracted from culture supernatants as described above and analyzed by thin-layer chromatography. Lanes 1 to 4, 715j grown in medium with no supplements, 18AA, 18AA plus cysteine, or 18AA plus methionine, respectively; lanes 5 to 8, KLF2 grown in medium with no supplements, 18AA, 18AA plus cysteine, or 18AA plus methionine, respectively. Abbreviations: P, pyochelin (stereoisomers I [migrating as a doublet] and II); S, salicylate.
We could also assess the ability of exogenous cysteine or methionine to suppress the defect in pyochelin production by examining colonies, grown under iron-limited conditions, for the restoration of yellow-green fluorescence. Colonies of both KLF2 and KLF3 exhibited the characteristic yellow-green fluorescence associated with pyochelin on standard M9 medium containing cysteine at a concentration of 100 μg/ml but not on medium containing cysteine at 20 μg/ml or methionine at 100 μg/ml (results not shown).
B. cenocepacia does not produce pyochelin under conditions of low sulfate availability.
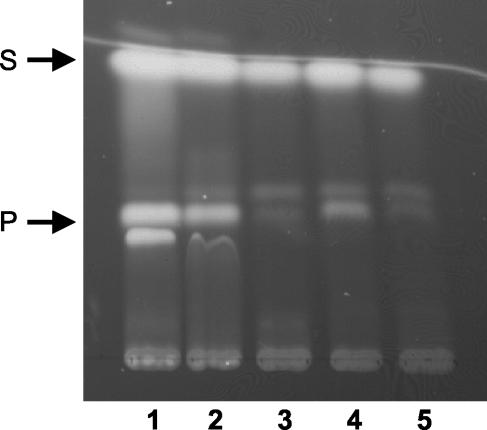
The observation that sulfate concentrations which support normal growth of a B. cenocepacia cysW mutant do not result in production of the siderophore pyochelin under iron-limiting conditions implies that the synthesis of certain secondary metabolites containing sulfur may be particularly sensitive to sulfur availability. To test this, we examined the effect of inorganic sulfate concentration on the ability of the wild-type B. cenocepacia strain, 715j, to produce pyochelin. We found that halving the normal concentration of sulfate in M9 minimal medium (i.e., 0.5 mM sulfate) led to a large decrease (∼70%) in the amount of pyochelin produced by this strain, with little or no change in the amount of salicylic acid produced (Fig. 5). Pyochelin production was essentially abolished when the mutant was grown in the presence of 0.25 mM sulfate. Salicylate production was slightly reduced, but not abolished, when cells were grown in the presence of ≤0.25 mM sulfate. Thus, in the absence of alternative sources of sulfur, optimal production of pyochelin by B. cenocepacia requires extracellular concentrations of sulfate far greater than is required to sustain normal growth under laboratory conditions.
FIG. 5.
Effect of sulfate concentration on production of pyochelin by 715j. Strain 715j was grown at 37°C with aeration for 24 h in M9-glucose minimal medium containing various concentrations of sulfate (provided as MgSO4). The total concentration of magnesium ions was maintained at 1 mM, with the balance being provided in the form of MgCl2. Pyochelin and salicylate were extracted from culture supernatants as described above and analyzed by thin-layer chromatography. Lanes 1 to 5, siderophores produced by bacteria growing in medium containing 1.0, 0.5, 0.25, 0.1, and 0.05 mM sulfate, respectively. Abbreviations: P, pyochelin (stereoisomers I and II); S, salicylate.
As expected, colonies of 715j were nonfluorescent when grown on M9 agar devoid of added sulfate (the ability of solidified medium to support bacterial growth is largely the result of sulfate residues in the agar, estimated to be ∼0.015% [28]). Addition of cysteine at 1,000 μM (but not at 200 μM) to these plates facilitated the production of the yellow-green fluorescence associated with pyochelin production, whereas addition of methionine (750 μM) did not restore pyochelin production (results not shown).
Methionine can serve as a sole source of sulfur for growth of B. cenocepacia but not for pyochelin biosynthesis.
One reason for the failure of methionine supplementation to compensate for the defect in pyochelin biosynthesis under conditions of sulfur limitation is that methionine cannot be used as a sulfur source by this organism. To test this possibility, we grew strain 715j in M9-glucose minimal medium containing different sulfur sources at a concentration of 500 μM. We found that the growth rate of this strain in medium containing methionine is similar to the growth rate in the presence of cysteine or inorganic sulfate (which are nearly identical). The cysW mutant strains also grew as efficiently as the wild-type strain with methionine as the sole sulfur source (data not shown). The growth rate of the wild-type strain in the presence of either of the alkyl sulfonates, ethanesulfonate or taurine, was also similar to that of cells grown in the presence of inorganic sulfate (data not shown). 715j formed colonies of similar size on M9-glucose agarose in the presence of each of these sulfur sources, but whereas colonies produced a strong yellow-green fluorescence in the presence of cysteine or sulfate as the sole sulfur source, they did not fluorescence on medium containing methionine and were only weakly fluorescent in the presence of the alkanesulfonates (results not shown). These results indicate that methionine, ethanesulfonate, or taurine can act as an efficient source of sulfur for B. cenocepacia.
DISCUSSION
We have identified two pyochelin-deficient mutants of B. cenocepacia in which transposon insertions have disrupted an operon encoding the CysT, CysW, and CysA proteins. In E. coli these proteins comprise the membrane component of an ABC transporter for the import of sulfate and thiosulfate (the sulfate/thiosulfate permease) (28, 53). ABC transporters are comprised of two ATP-binding domains (also known as ABC domains), which are located on the inner face of the cytoplasmic membrane in bacteria, and two channel-forming transmembrane domains, each of which has six segments that span the cytoplasmic membrane (32, 48). Their role is to couple the energy of ATP hydrolysis to the movement of substrates across membranes. In the E. coli sulfate/thiosulfate permease, a CysA homodimer provides the two ATP-binding domains, while the two transmembrane domains are provided separately by CysT and CysW (25, 28, 32). Accordingly, the CysW protein of B. cenocepacia is also homologous to the CysT protein of E. coli (31% identity) and other bacteria (not shown). The ATP-binding subunit consensus region of bacterial ABC transporters comprises a segment of approximately 215 residues near the N terminus of the polypeptide and is followed by a variable C-terminal region (48). The consensus region contains a number of strongly conserved motifs, all of which are found within the N-terminal 264 residues of B. cenocepacia CysA. These include the Walker box motifs, which are proposed to form the ATP binding pocket and are present in many other ATP-binding proteins (69); the linker peptide, which is unique to and diagnostic of the ABC transport family (signature sequence); and a recently recognized switch region (32, 48).
It is intriguing that the transposon insertions in cysW did not result in a cysteine-requiring auxotrophy. In organisms as diverse as E. coli and Mycobacterium tuberculosis, mutations in the cysTWA gene cluster confer a requirement for cysteine (28, 71). Our results show that sulfate uptake by either of the B. cenocepacia cysW mutants is ∼50-fold less efficient than that by the wild-type strain. However, in the presence of ≥0.5 mM sulfate, this rate of transport is able to sustain normal growth of the mutants, although the intracellular sulfur pool must clearly be depleted. A TBLASTN search of the B. cenocepacia genome sequence using CysW as the query did not reveal additional copies of cysW-like genes. Therefore, we may be able to explain the low level of sulfate uptake by the mutants by proposing that the CysT gene product can partially substitute for CysW. Furthermore, the orientation of the inserted transposon in both mutants is such that the strong dfr gene promoter (15, 30) is driving transcription of cysA (and cysB), as there is no transcription terminator within mini-Tn5Tp. This hypothesis may explain why only mutations in cysW were obtained (and not in cysT or cysA).
In both E. coli and P. aeruginosa the internalized sulfate is activated and reduced to sulfite and then to sulfide, which is subsequently transferred either to O-acetylserine to produce cysteine or, in an alternative pathway occurring in P. aeruginosa, to O-succinylhomoserine to generate homocysteine (24, 28, 66). In both species, most other forms of exogenous sulfur, such as organic sulfonates and sulfate esters, feed into this pathway and are also assimilated into cysteine and homocysteine (24), although thiosulfate reacts directly with O-acetylserine to give S-sulfocysteine, catalyzed by the cysM gene product (52). The biosynthesis of pyochelin in P. aeruginosa (and presumably in B. cenocepacia) essentially involves the successive condensation and cyclization of two molecules of cysteine with salicylic acid (42). Thus, the biosynthesis of cysteine can be viewed as an upstream extension of the pyochelin biosynthetic pathway. Our observations suggest that the biosynthesis of pyochelin in B. cenocepacia is particularly sensitive to the availability of assimilatable sulfur, most probably due to knock-on effects on the intracellular cysteine pool, and may indicate another level of regulation of pyochelin biosynthesis in this organism in addition to regulation by iron (16, 68). Therefore, it is likely that a regulatory hierarchy exists whereby, under conditions of sulfur limitation, bacteria channel the available sulfur into essential metabolites, with the consequential curtailment of the biosynthesis of nonessential secondary metabolites. Our results also reinforce the idea that the ability to synthesize a secondary metabolite under laboratory conditions should not necessarily be taken as evidence for the production of this compound in the natural environment of the organism.
One important difference between the sulfur assimilatory pathways of E. coli and P. aeruginosa is the fact that methionine can be converted to cysteine via the reverse transsulfuration pathway in P. aeruginosa but not in E. coli. Thus, methionine can be efficiently used as the sole sulfur source by P. aeruginosa, whereas for E. coli, which can only use the sulfur moiety of methionine as a sulfur source by an alternative pathway, growth is poor with methionine as the sulfur source (21, 66). The inability of exogenous methionine to facilitate pyochelin biosynthesis in B. cenocepacia cysW mutants suggested that, as in E. coli, methionine may not be efficiently converted into cysteine in this bacterium. However, growth rate measurements indicated that B. cenocepacia utilizes methionine as efficiently as cysteine as a sulfur source for the biosynthesis of central metabolites necessary for growth. Nevertheless, it is possible that the sulfur pool is sufficiently restricted during growth on methionine so as to mitigate against the biosynthesis of certain secondary metabolites, including pyochelin. Another possible reason for the different efficiencies of pyochelin production observed in the presence of different sulfur sources is that certain sulfur sources may exert a negative regulatory effect on pyochelin biosynthesis.
Surprisingly, we were only able to isolate two mutants out of ∼15,000 in which pyochelin was not produced under the conditions used for the screen. Curiously, neither of the transposon insertion events giving rise to the pyochelin-deficient phenotype had occurred within genes specific to the biosynthesis of this siderophore. There are two possibilities which might explain this observation: either there is genetic redundancy with respect to pyochelin biosynthetic genes in the B. cenocepacia strain we used for these experiments or the screen adopted for the identification of pyochelin-deficient mutants was too stringent. Although members of the BCC are renowned for their large genome size (29, 46, 64), we believe the first possibility to be less likely, as this would involve duplication of a large repertoire of genes (43). Furthermore, in the genome sequence of B. cenocepacia strain J2315, only single copies of genes homologous to those required for pyochelin biosynthesis in P. aeruginosa appear to be present. Members of the BCC are noted for the large number of secondary metabolites they produce, some of which also contain sulfur (70). In our screen to identify Pch− mutants, we investigated only mutants which were completely nonfluorescent and avoided mutants which exhibited weak fluorescence. It is possible that, in some members of the latter class, the residual fluorescence was contributed either by a fluorescent intermediate in the pyochelin biosynthetic pathway, such as Dha, or some other unrelated sulfur-containing secondary metabolite. Thus, in effect, we may have been looking for mutants which failed to produce both pyochelin and one or more additional fluorescent compounds which require an adequate supply of sulfur for their biosynthesis.
Acknowledgments
We are grateful to the University of Sheffield School of Medicine and Biomedical Sciences for the award of a postgraduate studentship to Kate Farmer.
REFERENCES
- 1.Abe, M., M. Tsuda, M. Kimoto, S. Inouye, A. Nakazawa, and T. Nakazawa. 1996. A genetic analysis system of Burkholderia cepacia: construction of mobilizable transposons and a cloning vector. Gene 174:191-194. [DOI] [PubMed] [Google Scholar]
- 2.Ankenbauer, R. G., T. Toyokuni, A. Staley, K. L. Rinehart, Jr., and C. D. Cox. 1988. Synthesis and biological activity of pyochelin, a siderophore of Pseudomonas aeruginosa. J. Bacteriol. 170:5344-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmi, R., S. Carmeli, E. Levy, and F. J. Gough. 1994. (+)-(S)-Dihydroaeruginoic acid, an inhibitor of Septoria tritici and other phytopathogenic fungi and bacteria, produced by Pseudomonas fluorescens. J. Nat. Prod. 57:1200-1205. [DOI] [PubMed] [Google Scholar]
- 4.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in E. coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 5.Clowes, R. C., and W. Hayes. 1968. Experiments in microbial genetics. Blackwell Scientific Publications, Oxford, United Kingdom.
- 6.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, C. D. 1980. Iron uptake with ferripyochelin and ferric citrate by Pseudomonas aeruginosa. J. Bacteriol. 142:581-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, C. D., and R. Graham. 1979. Isolation of an iron-binding compound from Pseudomonas aeruginosa. J. Bacteriol. 137:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, C. D., K. L. Rinehart, M. L. Moore, and J. C. Cook. 1981. Pyochelin: novel structure of an iron-chelating growth promoter for Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 78:4256-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darling, P., M. Chan, A. D. Cox, and P. A. Sokol. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun. 66:874-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 13.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeShazer, D., P. J. Brett, R. Carlyon, and D. E. Woods. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeShazer, D., and D. E. Woods. 1996. Broad-host-range cloning and cassette vectors based on the R388 trimethoprim resistance gene. BioTechniques 20:762-764. [DOI] [PubMed] [Google Scholar]
- 16.Farmer, K. L. 1998. Ph.D. thesis. University of Sheffield, Sheffield, United Kingdom.
- 17.Gough, J. A., and N. E. Murray. 1983. Sequence diversity among related strains for recognition of specific targets in DNA molecules. J. Mol. Biol. 166:1-19. [DOI] [PubMed] [Google Scholar]
- 18.Heinrichs, D. E., and K. Poole. 1996. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and a repressor. J. Bacteriol. 178:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hryniewicz, M., A. Sirko, A. Palucha, A. Bock, and D. Hulanicka. 1990. Sulfate and thiosulfate transport in Escherichia coli K-12: identification of a gene encoding a novel protein involved in thiosulfate binding. J. Bacteriol. 172:3358-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hummerjohann, J., E. Kuttel, M. Quadroni, J. Ragaller, T. Leisinger, and M. A. Kertesz. 1998. Regulation of the sulfate starvation response of Pseudomonas aeruginosa: role of cysteine biosynthetic intermediates. Microbiology 144:1375-1386. [DOI] [PubMed] [Google Scholar]
- 22.Iwanicka-Nowicka, R., and M. M. Hryniewicz. 1995. A new gene, cbl, encoding a member of the LysR family of transcriptional regulators belongs to Escherichia coli cys regulon. Gene 166:11-17. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 24.Kertesz, M. A. 1999. Riding the sulfur cycle: metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 25.Kertesz, M. A. 2001. Bacterial transporters for sulfate and organosulfur compounds. Res. Microbiol. 152:279-290. [DOI] [PubMed] [Google Scholar]
- 26.Koedam, N., E. Wittouck, A. Gaballa, A. Gillis, M. Hofte, and P. Cornelis. 1994. Detection and differentiation of microbial siderophores by isoelectric focusing and chrome azurol S overlay. BioMetals 7:287-291. [DOI] [PubMed] [Google Scholar]
- 27.Kredich, N. M. 1992. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol. Microbiol. 6:2747-2753. [DOI] [PubMed] [Google Scholar]
- 28.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss III, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 29.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 30.Levesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 31.Lewenza, S., and P. A. Sokol. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 183:2212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 33.LiPuma, J. J. 1998. Burkholderia cepacia-management issues and new insights. Clin. Chest Med. 19:473-486. [DOI] [PubMed] [Google Scholar]
- 34.Lochowska, A., R. Iwanicka-Novicka, D. Plochocka, and M. M. Hryniewicz. 2001. Functional dissection of the LysR-type CysB transcriptional regulator. J. Biol. Chem. 276:2098-2107. [DOI] [PubMed] [Google Scholar]
- 35.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533-538. [DOI] [PubMed] [Google Scholar]
- 36.McKevitt, A. I., S. Bajaksouzian, J. D. Klinger, and D. E. Woods. 1989. Purification and characterization of an extracellular protease from Pseudomonas cepacia. Infect. Immun. 57:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metcalf, W. W., W. Jiang, and B. L. Wanner. 1994. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6Kγ origin plasmids at different copy numbers. Gene 138:1-7. [DOI] [PubMed] [Google Scholar]
- 38.Meyer, J-M., P. Azelvandre, and C. Georges. 1992. Iron metabolism in Pseudomonas: salicylic acid, a siderophore of Pseudomonas fluorescens CHA0. BioFactors 4:23-27. [PubMed] [Google Scholar]
- 39.Meyer, J-M., V. T. Van, A. Stintzi, O. Berge, and G. Winkelmann. 1995. Ornibactin production and transport properties in strains of Burkholderia vietnamiensis and Burkholderia cepacia (formerly Pseudomonas cepacia). BioMetals 8:309-317. [DOI] [PubMed] [Google Scholar]
- 40.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quadri, L. E. N. 2000. Assembly of aryl-capped siderophores by modular peptide synthetases and polyketide synthases. Mol. Microbiol. 37:1-12. [DOI] [PubMed] [Google Scholar]
- 42.Quadri, L. E. N., T. A. Keating, H. M. Patel, and C. T. Walsh. 1999. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: in vitro reconstitution of aryl-4,2-bisthiazoline synthetase activity from PchD, PchE and PchF. Biochemistry 38:14941-14954. [DOI] [PubMed] [Google Scholar]
- 43.Reimmann, C., H. M. Patel, L. Serino, M. Barone, C. T. Walsh, and D. Haas. 2001. Essential PchG-dependent reduction in pyochelin biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 183:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reimmann, C., L. Serino, M. Beyeler, and D. Haas. 1998. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology 144:3135-3148. [DOI] [PubMed] [Google Scholar]
- 45.Rinehart, K. L., A. L. Staley, S. R. Wilson, R. G. Ankenbauer, and C. D. Cox. 1995. Stereochemical assignment of the pyochelins. J. Org. Chem. 60:2786-2791. [Google Scholar]
- 46.Rodley, P. D., U. Romling, and B. Tummler. 1995. A physical genome map of the Burkholderia cepacia type strain. Mol. Microbiol. 17:57-67. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22:1-20. [DOI] [PubMed] [Google Scholar]
- 49.Serino, L., C. Reimmann, H. Baur, M. Beyeler, P. Visca, and D. Haas. 1995. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol. Gen. Genet. 249:217-228. [DOI] [PubMed] [Google Scholar]
- 50.Serino, L., C. Reimmann, P. Visca, M. Beyeler, V. Della Chiesa, and D. Haas. 1997. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J. Bacteriol. 179:248-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seth, A. 1984. A new method for linker ligation. Gene Anal. Techn. 1:99-103. [Google Scholar]
- 52.Sirko, A., M. Zatyka, and D. Hulanicka. 1987. Identification of the Escherichia coli cysM gene encoding O-acetylserine sulfhydrylase B by cloning with mini-Mu-lac containing a plasmid replicon. J. Gen. Microbiol. 133:2719-2725. [DOI] [PubMed] [Google Scholar]
- 53.Sirko, A., M. Hryniewicz, D. Hulanicka, and A. Bock. 1990. Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of the cysTWAM gene cluster. J. Bacteriol. 172:3351-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sirko, A., M. Zatyka, E. Sadowy, and D. Hulanicka. 1995. Sulfate and thiosulfate transport in Escherichia coli K-12: evidence for a functional overlapping of sulfate- and thiosulfate-binding proteins. J. Bacteriol. 177:4134-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sokol, P. A. 1984. Production of the ferripyochelin outer-membrane receptor by Pseudomonas species. FEMS Microbiol. Lett. 23:313-317. [Google Scholar]
- 56.Sokol, P. A. 1986. Production and utilization of pyochelin by clinical isolates of Pseudomonas cepacia. J. Clin. Microbiol. 23:560-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sokol, P. A., P. Darling, S. Lewenza, C. R. Corbett, and C. D. Kooi. 2000. Identification of a siderophore receptor required for ferric ornibactin uptake in Burkholderia cepacia. Infect. Immun. 68:6554-6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sokol, P. A., C. J. Lewis, and J. J. Dennis. 1992. Isolation of a novel siderophore from Pseudomonas cepacia. J. Med. Microbiol. 36:184-189. [DOI] [PubMed] [Google Scholar]
- 59.Stephan, H., S. Freund, W. Beck, G. Jung, J.-M. Meyer, and G. Winkelmann. 1993. Ornibactins—a new family of siderophores from Pseudomonas cepacia. BioMetals 6:93-100. [DOI] [PubMed] [Google Scholar]
- 60.Stewart, G. S. A. B., S. Lubinsky-Mink, C. G. Jackson, A. Cassel, and J. Kuhn. 1986. A pBR322 copy number derivative of pUC18 for cloning and expression. Plasmid 15:172-181. [DOI] [PubMed] [Google Scholar]
- 61.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 62.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Y. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 63.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. W. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:188-1200. [DOI] [PubMed] [Google Scholar]
- 64.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. W. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 65.van der Ploeg, J. R., R. Iwanicka-Novicka, M. A. Kertesz, T. Leisinger, and M. M. Hryniewicz. 1997. Involvement of CysB and Cbl regulatory proteins in expression of the tauABCD operon and other sulfate starvation-inducible genes in Escherichia coli. J. Bacteriol. 179:7671-7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vermeij, P., and M. A. Kertesz. 1999. Pathways of assimilative sulfur metabolism in Pseudomonas putida. J. Bacteriol. 181:5833-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Visca, P., G. Colotti, L. Serino, D. Verzili, N. Orsi, and E. Chiancone. 1992. Metal regulation of siderophore synthesis in Pseudomonas aeruginosa and functional effects of siderophore-metal complexes. Appl. Environ. Microbiol. 58:2886-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Visca, P., A. Ciervo, V. Sanfilippo, and N. Orsi. 1993. Iron-regulated salicylate synthesis by Pseudomonas spp. J. Gen. Microbiol. 139:1995-2001. [DOI] [PubMed] [Google Scholar]
- 69.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha-subunits and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilkinson, S. G., and T. L. Pitt. 1995. Burkholderia (Pseudomonas) cepacia: pathogenicity and resistance. Rev. Med. Microbiol. 6:10-17. [Google Scholar]
- 71.Wooff, E., S. L. I. Michell, S. V. Gordon, M. A. Chamber, S. Bardarov, W. R. Jacobs, Jr., R. G. Hewinson, and P. R. Wheeler. 2002. Functional genomics reveals the sole sulphate transporter of the Mycobacterium tuberculosis complex and its relevance to the acquisition of sulphur in vivo. Mol. Microbiol. 43:653-663. [DOI] [PubMed] [Google Scholar]
- 72.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]