Abstract
A novel sucrose hydrolase (SUH) from Xanthomonas axonopodis pv. glycines, a causative agent of bacterial pustule disease on soybeans, was studied at the functional and molecular levels. SUH was shown to act rather specifically on sucrose (Km = 2.5 mM) but not on sucrose-6-phosphate. Protein analysis of purified SUH revealed that, in this monomeric enzyme with an estimated molecular mass of 70,223 ± 12 Da, amino acid sequences determined for several segments have corresponding nucleotide sequences in XAC3490, a protein-coding gene found in the genome of X. axonopodis pv. citri. Based on this information, the SUH gene, consisting of an open reading frame of 1,935 bp, was cloned by screening a genomic library of X. axonopodis pv. glycines 8ra. Database searches and sequence comparison revealed that SUH has significant homology to some family 13 enzymes, with all of the crucial invariant residues involved in the catalytic mechanism conserved, but it shows no similarity to known invertases belonging to family 32. suh expression in X. axonopodis pv. glycines requires sucrose induction, and insertional mutagenesis resulted in an absence of sucrose-inducible sucrose hydrolase activity in crude protein extracts and a sucrose-negative phenotype. Recombinant SUH, overproduced in Escherichia coli and purified, was shown to have the same enzymatic characteristics in terms of kinetic parameters.
Plant-pathogenic bacteria grow in the intercellular spaces of plant tissues, relying on nutrients available there. In higher plants, sucrose (α-d-glucopyranosyl β-d-fructofuranoside) is the major transportable product of photosynthesis that flows from the source organs to the sink organs. The process of the source-sink flow involves phloem loading, for which sucrose has to exit from the mesophyll cell, and from the apoplasm, it enters the phloem. Sucrose is naturally the predominant form of carbohydrate found in the intercellular spaces of photosynthetically active tissues. Pathogens with their habitats in mature leaves, therefore, may utilize sucrose as the main, if not the only, source for carbon and energy and possess systems for a sucrose utilization pathway. Nevertheless, information on such systems in plant-associated bacteria is rather scanty. To our knowledge, Erwinia amylovora, the causative agent for fire blight of rosaceous plants, is the only phytopathogen that has been studied at the molecular level in relation to sucrose utilization (4).
Many sucrose-positive bacteria have a phosphoenolpyruvate-dependent, sucrose-specific phosphotransferase system (PTS) that promotes sucrose translocation across the cytoplasmic membrane with concomitant phosphorylation (27, 28). The resulting intracellular sucrose-6-phosphate (S-6-P) is then hydrolyzed by S-6-P hydrolase to yield d-glucose-6-phosphate and d-fructose. In some bacteria, however, sucrose itself can be transported into the cytoplasm without phosphorylation via a pathway independent of PTS. For instance, the Escherichia coli strain EC3132, which is able to grow on sucrose, bears a chromosomally located regulon with structural genes for a sucrose hydrolase, a fructokinase, and a transporter, regulated by a sucrose-specific repressor (3, 13). Similar pathways involving facilitated diffusion or active sucrose-ion symport and intracellular invertase have also been invoked for several other bacteria (11, 14, 35, 38). Certain bacteria secrete enzymes acting on sucrose, such as hexosyltransferase, levansucrase, and levanase, which split sucrose in the culture medium (22, 25, 40): these microbes utilize sucrose as an energy source for growth, as well as a substrate for oligosaccharide synthesis. Some bacteria show more than one sucrose hydrolysis activity (18, 25).
The genus Xanthomonas is arguably one of the most ubiquitous groups of plant-associated bacteria. Members of this group have been shown to infect at least 124 monocotyledonous and 268 dicotyledonous plants (6). To gain insight into sucrose utilization by Xanthomonas pathogens, a novel sucrose hydrolase from Xanthomonas axonopodis was isolated and characterized at the functional and molecular levels in the present study. This protein, designated SUH, appears to be essential for sucrose metabolism in X. axonopodis pv. glycines, a causative agent of bacterial pustule disease on soybean. As a sucrose-specific enzyme, SUH is unique in that not only does it have no structural similarity to invertases (EC 3.2.1.26), the typical sucrose hydrolases, but it also shows no significant sequence homology to α-glucosidases (EC 3.2.1.20) with known functions.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Two strains of X. axonopodis pv. glycines, the rifampin-resistant wild-type strain 8ra, obtained from E. J. Braun of the University of Illinois at Urbana-Champaign, and a mutant constructed from strain 8ra, were cultured at 28°C in either Luria-Bertani (LB) broth or minimal medium. The basal composition of minimal medium was 20 mM NaCl, 10 mM (NH4)2SO4, 5 mM MgSO4, 1 mM CaCl2, 0.32 mM K2HPO4, 0.16 mM KH2PO4, 0.01 mM FeSO4, 1% tryptone (Difco), and 1 to 10 mM sucrose. When necessary, sucrose was replaced by glucose or fructose, with tryptone deleted from the medium. E. coli DH5α (31) and E. coli BL21(DE3)/pLysS (Novagen) were used as the hosts for plasmids and for suh gene expression, respectively, and were grown at 37°C in LB broth. Antibiotics were added to give the following concentrations: rifampin, 100 μg/ml; tetracycline, 25 μg/ml; kanamycin, 50 μg/ml; ampicillin, 50 μg/ml.
Enzyme purification.
Cells of strain 8ra were cultured in minimal medium with 10 mM sucrose and 1% tryptone and harvested by centrifugation. The biomass (∼26 g [wet weight]) was suspended in 200 ml of a suspension buffer (pH 7.5) containing 50 mM K-phosphate, 1 mM dithiothreitol (DTT), and 1.5 mM EDTA; sonicated on ice; and centrifuged at 20,000 × g for 20 min. The supernatant, hereafter referred to as crude protein extract, was subjected to ammonium sulfate (35 to 50% saturation) fractionation, and the precipitate was solubilized in 50 ml of elution buffer A (20 mM Tris-HCl [pH 7.5], 1 mM DTT, and 1% sucrose) and dialyzed against the same buffer. The dialysate was applied to an anion-exchange column (2 by 10 cm) packed with DEAE-Sepharose Fast Flow gel and equilibrated with elution buffer A. The bound proteins were eluted with a linear gradient of NaCl (0 to 0.3 M) in the same buffer. The fractions showing sucrose-hydrolyzing activity were pooled and dialyzed against elution buffer B (20 mM K-phosphate buffer [pH 7.5], 1 mM DTT, and 1% sucrose), which was then loaded onto a hydroxyapatite column (2.5 by 2 cm) equilibrated with the same buffer, and the flowthrough fraction was retrieved. This was concentrated by precipitating the protein with ammonium sulfate (35 to 50% saturation) and redissolving it in 1 ml of elution buffer B and was then applied to a Sephacryl S-100 HR gel permeation column (2 by 100 cm) equilibrated with elution buffer B supplemented with 0.25 M NaCl. The active fractions from the column were pooled, and ammonium sulfate was added to the pool to a final concentration of 0.8 M. This was then loaded onto a hydrophobic interaction column (1 by 17 cm) filled with Butyl Sepharose Fast Flow gel and equilibrated with elution buffer B containing 0.8 M (NH4)2SO4. Protein elution was done with a reverse linear gradient of (NH4)2SO4 (0.8 to 0.3 M) in the same buffer. The active fractions were pooled, dialyzed against elution buffer A devoid of sucrose, and applied to a Mono Q HR 5/5 column for fast protein liquid chromatography (FPLC) as the final step of SUH purification. The elution was carried out with a linear gradient of NaCl (0 to 0.3 M) in the same buffer. All packing materials for the chromatographic columns and the factory-packed FPLC column were purchased from Amersham Bioscience. The purification procedures were performed at 4°C.
Recombinant SUH (rSUH) with a histidine tag at the C terminus was isolated from transformed E. coli cells. Harvested cells (∼1g [wet weight]) were suspended in 30 ml of a saline phosphate buffer (50 mM Na-phosphate [pH 8.0] and 300 mM NaCl) and disrupted by freezing and thawing. After a 30-min incubation at 30°C with occasional shaking, the cell lysate was cleared by centrifugation, and the supernatant was loaded onto a Ni-nitrilotriacetic acid agarose (Qiagen) column. The column was washed thoroughly with the saline phosphate buffer containing 20 mM imidazole, and then the bound proteins were eluted with the same buffer containing 0.25 M imidazole. The eluate was mixed with an equal volume of 1.6 M (NH4)2SO4. From this, rSUH was further purified by employing the same hydrophobic interaction (Butyl Sepharose Fast Flow) chromatography and ion-exchange (Mono Q HR 5/5) FPLC used for SUH purification from the wild-type cells.
Measurements of enzyme activity.
Sucrose reactions catalyzed by SUH or rSUH in 20 mM K-phosphate buffer (pH 7.5) at 30°C were assayed primarily by the dinitrosalicylic acid (DNSA) method (23), which is based on color development as a result of the reaction of DNSA with reducing groups produced. The DNSA color reaction was calibrated under the assay conditions with defined amounts of glucose and fructose (the same amounts of these monosaccharides resulted in almost identical increases in the 575-nm absorption). One unit of enzyme activity was defined as the amount of protein hydrolyzing 1 μmol of sucrose per min. In cases where direct sugar analysis was necessary, either high-performance liquid chromatography (HPLC) or thin-layer chromatography (TLC) was performed to simultaneously measure the changes in both sucrose and the reaction products in enzyme reaction mixtures. For the HPLC analysis, a polyamine column (YMC Europe GmbH) was used with acetonitrile-water (3/1 [vol/vol]) as the mobile phase, and sugars were detected by differential refractive index and quantified from the respective peak areas. TLC was conducted using three ascents of the solvent system (ethylacetate-pyridine-H2O [100/35/25 {vol/vol/vol}]) on Merck silica 60 F254 gel plates as described by Liebl et al. (20), and sugars were visualized by dipping the plates into 5% (vol/vol) H2SO4 in ethanol containing 0.5% (wt/vol) α-naphthol, followed by heating them at 110°C for 10 min.
Protein analysis.
The homogeneity of the enzyme preparations was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on acrylamide (10%) gels in the discontinuous buffer system of Laemmli (16). The molecular mass of SUH was determined by matrix-assisted laser desorption- ionization mass spectrometry using a Perspective Biosystems Voyager-DE STR mass spectrometer in reflector mode with sinapinic acid as the matrix. The isoelectric point of SUH was measured by isoelectric focusing on polyacrylamide gels with a pH gradient developed by Ampholine carrier ampholites (Amersham Bioscience). The N-terminal amino acid sequence was determined using an Applied Biosystems Procise 491 automatic sequencer. For this, SUH on an SDS-PAGE gel was electroblotted onto a polyvinyldifluoride membrane in CAPS (3-cyclohexylamino-1-propanesulfonic acid) buffer and stained with Coomassie brilliant blue, and then the transferred protein band was excised and subjected to N-terminal sequencing based on Edman degradation. Meanwhile, the internal sequencing was done by electrospray ionization tandem mass spectrometry. For this, the stained SUH band on SDS-PAGE was excised and treated with trypsin for in-gel protein digestion. The resulting tryptic peptides were extracted and then subjected to amino acid sequencing in a Micromass Q-Tof2 mass spectrometer.
Gene identification.
Homology searches in the GenBank database using the BLAST algorithm (1) led to the finding that amino acid sequences determined for four segments of SUH match four nucleotide sequence segments of a protein-coding gene, XAC3490, found in the X. axonopodis pv. citri genome, which has recently been sequenced (7). Furthermore, the measured molecular mass of SUH was found to be almost identical to the predicted molecular mass of the XAC3490 gene product with an amino acid sequence deduced from the open reading frame. These findings allowed us to assume that the genes suh and XAC3490 are virtually the same. Based on the nucleotide sequence of XAC3490 compared with the partial amino acid sequences of SUH, a pair of oligonucleotide primers (forward, 5′-AGCACCTGCCCGATCGATTC-3′, and reverse, 5′-CCGCTGCTCTGGAAACTCTC-3′) were designed. These were used to amplify a 1.3-kb DNA by PCR (94°C for 1 min, 59°C for 2 min, 72°C for 3 min; 35 cycles) with genomic DNA from X. axonopodis pv. glycines 8ra as the template. The PCR product was cloned into pGEM-T Easy (Promega), yielding a plasmid called pGSC1. The insert in this plasmid was then used as a probe to screen an X. axonopodis pv. glycines genomic library that was constructed with the broad-host-range cosmid vector pLAFR3 (39) by cloning of total DNA partially digested with Sau3AI and transformed into E. coli DH5α. The positive recombinant clone from this library was subcloned into the Ampr-carrying vector pBluescript II SK(−) (Stratagene) through EcoRI-BamHI treatment. One of the resulting plasmids, pBSEB3, harboring a 3.7-kb EcoRI-BamHI fragment, was determined to contain suh by Southern hybridization using the insert DNA in pGSC1 as a probe, followed by DNA sequencing.
The full-length suh gene was amplified by PCR (94°C for 1 min, 67.5°C for 2 min, 72°C for 3 min; 25 cycles) using DNA from pBSEB3 as the template. For this, a pair of oligonucleotide primers (direct primer, 5′-ATATCGGACATATGAGCACCTGCCCGATC-3′, with the NdeI site in boldface and the authentic start codon underlined, and reverse primer, 5′-CTCACTCGAGTTCAGGCGCATGCTCAGT-3′, with the XhoI site in boldface) were designed. The reaction was carried out in the presence of 10% dimethyl sulfoxide to avoid nonspecific hybridization of the primers. The PCR product was cut with NdeI and XhoI and ligated to the corresponding sites of the Kanr-carrying expression vector pET41b (Novagen), yielding a plasmid called pETSC3. The insert in pETSC3 was found to contain no mutations.
The nucleotide sequences of the constructed plasmids were determined according to the dideoxy chain termination method of Sanger et al. (32) using a Prism dye primer cycle-sequencing kit and a Prism model 3700 automated DNA sequencer (Applied Biosystems). Sequencing was done for the entire length of both strands.
SUH expression in E. coli.
The plasmid pETSC3 was introduced into E. coli BL21(DE3)/pLysS. The transformed cells were grown in LB medium at 37°C to an optical density at 600 nm (OD600) of 0.6, and protein expression was induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 20°C for 12 h. The cultured bacteria were harvested and used to purify rSUH.
Construction of suh mutant.
The plasmid pBSEB3 was mutagenized by using an in vitro Tn5 (Kanr) transposition system, EZ::TN <KAN-2> (Epicentre), following the protocol of the manufacturer. The transposon insertion site was identified by sequence determination using the KAN-2 FP-1 and KAN-2 RP-1 primers (Epicentre). The insertion derivative was then introduced into X. axonopodis pv. glycines 8ra by electroporation, and selection was performed on LB plates supplemented with rifampin, ampicillin, and kanamycin. Cells of the transformant were cycled more than four times in LB broth containing rifampin and kanamycin to maintain selection for the presence of the transposon. A putative marker exchange mutant was screened by testing kanamycin resistance and ampicillin sensitivity and verified by Southern hybridization.
Measurement of sucrose uptake by cells.
The wild-type strain 8ra and the suh mutant were cultured in minimal medium containing 1 mM sucrose and 1% tryptone, harvested at mid-exponential phase, washed with 100 mM K-phosphate buffer (pH 6.6), and suspended in the same buffer to an OD600 of 3. To 1.0 ml of these suspensions, 0.5 ml of sucrose solution (3 mM) charged with 0.3 μCi of [U-14C]sucrose was added with shaking. Then, aliquots (0.2 ml each) were taken from the suspensions at 30-s intervals and filtered rapidly through Millipore membrane filters. The filters with collected cells were placed on paper towels to soak up the remaining solution, dried under a hot air stream, and then subjected to radioactivity counting in a liquid scintillation counter.
Bacterial inoculation and measurement of multiplication in planta.
Soybean plants grown in a greenhouse were inoculated with X. axonopodis pv. glycines by infiltrating cell suspensions into the fully expanded trifoliate leaves using a 1-ml syringe (the inoculation was successful when it was performed on rainy days). Cell suspensions to ∼105 CFU in 10 mM MgCl2 were prepared from 2-day-old bacterial plates. Six leaf disks (6-mm diameter) were taken at 1-day intervals after inoculation and ground in a microtube containing 200 μl of 10 mM MgCl2. Sample aliquots (100 μl each) were serially diluted and spread on LB agar plates. Colonies were counted after the plates were incubated for 48 h at 28°C.
Nucleotide sequence accession number.
The nucleotide sequence of suh has been submitted to the GenBank database under accession number AY359289.
RESULTS
General and enzymatic properties.
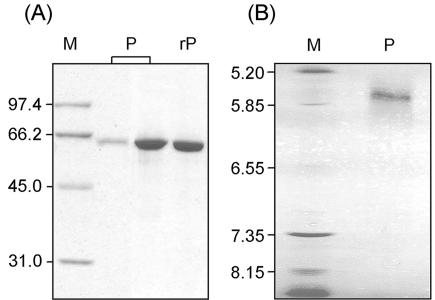
The overall purification schemes for SUH and rSUH are compiled in Table 1 (the average production levels of the enzyme in the Xanthomonas and Escherichia cells were ∼20 and 1,500 U/liter of culture, respectively, with crude protein extracts). The isolated SUH showed a single band on SDS-PAGE corresponding to ∼64 kDa, as well as a single protein peak corresponding to ∼54 kDa on size exclusion chromatography under nondenaturing conditions, indicating that the functioning SUH is a monomeric protein, probably with a highly compact structure. These estimated molecular mass values appeared considerably smaller than the molecular mass of 70,216 Da predicted from the deduced amino acid sequence. However, matrix-assisted laser desorption-ionization mass spectrometry of purified SUH revealed a molecular mass of 70,223 ± 12 Da (an average of three measurements), which agrees well with the predicted molecular mass. A measured pI of ca. 5.7 for SUH is also close to the theoretical pI of 5.5. Together with these observations (Fig. 1), the partial sequencing data from the natural SUH, i.e., STCPIDPPAL (positions 1 to 10), YEATLGQV (positions 213 to 220), AEAIVP (positions 320 to 325), and GESFQSSG (positions 431 to 438), strongly suggest that the Xanthomonas sucrose hydrolase is indeed encoded by suh.
TABLE 1.
Purification of natural and recombinant SUH
| Enzyme | Stepa | Protein (mg) | Total activity (U) | Sp act (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|---|
| SUH | CPX | 2,270 | 180 | 0.079 | 1 | 100 |
| ASF | 840 | 158 | 0.19 | 2.4 | 88 | |
| AEC | 320 | 123 | 0.38 | 4.8 | 68 | |
| HAC | 99 | 91.7 | 0.93 | 12 | 51 | |
| GPC | 9.6 | 40.9 | 43 | 54 | 23 | |
| HIC | 0.76 | 20.2 | 27 | 342 | 11 | |
| FPLC | 0.097 | 6.1 | 64 | 810 | 3.4 | |
| rSUH | CPX | 68 | 387 | 5.7 | 1 | 100 |
| Ni-NTA | 31 | 372 | 12 | 2.1 | 96 | |
| HIC | 8.8 | 253 | 29 | 5.1 | 65 | |
| FPLC | 2.2 | 148 | 67 | 12 | 38 |
CPX, crude protein extract; ASF, ammonium sulfate fractionation; AEC, anion-exchange chromatography; HAC, hydroxyapatite chromatography; GPC, gel permeation chromatography; HIC, hydrophobic interaction chromatography; Ni-NTA, Ni-nitrilotriacetic acid affinity chromatography.
FIG. 1.
SDS-PAGE (A) and isoelectric focusing (B) of SUH. Lanes P, natural protein; lane rP, recombinant protein; lanes M, markers for molecular size (A) and pI (B). In SDS-PAGE, the natural protein was loaded at high and low levels, ca. 0.4 and 3 μg, respectively, while the recombinant protein was loaded at a high level only.
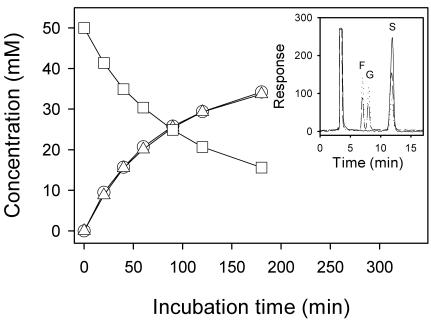
Upon incubation of sucrose at various concentrations (10 to 200 mM) with the preparations of either natural or recombinant SUH in reaction buffer, glucose and fructose were released at equal molar ratios, and the amounts of sucrose consumed appeared identical with the amounts of the respective monosaccharides produced at molar base (Fig. 2). Consistent with this, no production of other saccharides, such as smaller maltosaccharides and sucrose isoforms, was detected on HPLC. The presence of high concentrations (up to 10 mg/ml) of glycogen or soluble starch affected neither the rate of sucrose consumption nor the ratio of glucose to fructose produced. Furthermore, under the same reaction conditions (in terms of enzyme and sugar concentrations, buffer composition and pH, and reaction temperature) as for sucrose hydrolysis (see the legend to Fig. 2), no hydrolytic reaction products were detected by the HPLC and DNSA methods in 24-h incubations of SUH with the following sugars: disaccharides having an α-d-glucopyranosyl moiety, such as maltose [α-d-glucopyranosyl-(1→4)-d-glucopyranose], isomaltose [α-d-glucopyranosyl-(1→6)-d-glucopyranose], and trehalose (α-d-glucopyranosyl α-d-glucopyranoside), and the typical substrates for fructofuranosidases, such as raffinose [α-d-galactopyranosyl-(1→6)-α-d-glucopyranosyl-(1↔ 2)-β-d-fructofuranoside] andstachyose [α-d-galactopyranosyl-(1→6)-α-d-galactopyranosyl-(1→6)-α-d-glucopyranosyl-(1↔2)-β-d-fructofuranoside]. These results may indicate that among reactions involving sugars, sucrose hydrolysis is virtually the only reaction catalyzed by SUH, at least, under our assay conditions. Some bacterial sucrose hydrolases have been shown to hydrolyze not only sucrose but also S-6-P (4, 43). However, SUH apparently did not act on S-6-P, as no reducing group was released from S-6-P even in a prolonged (>2-day) incubation in the presence of the enzyme under optimal reaction conditions. Despite the inability to hydrolyze the α-d-glucopyranosyl-containing disaccharides tested, except for sucrose, SUH was thought to be α-glucosidase recognizing the glucose moiety, because p-nitrophenyl α-d-glucopyranoside, a typical chromogenic substrate for α-glucosidases, was shown to undergo SUH-catalyzed hydrolytic cleavage, as monitored by the absorbance of p-nitrophenolate at 405 nm, although the reaction rate was very low. We estimated that the rate of p-nitrophenolate production by SUH was lower by at least 3 orders of magnitudes than that by brewer's yeast α-glucosidase under the same reaction conditions.
FIG. 2.
Time courses of sucrose consumption and glucose-fructose production by SUH. To 1.0 ml of 50 mM sucrose in 20 mM K-phosphate buffer (pH 7.5) kept at 30°C, 10 μl of the enzyme preparation (1.2 μg/μl) in the same buffer was added, and the reaction mixture was incubated at 30°C. Aliquots (0.1 ml each) were drawn at the time intervals shown, immediately boiled for 2 min, filtered through Millipore membranes (0.2-μm pore size), and then subjected to HPLC analysis. Squares, sucrose; triangles, glucose; circles, fructose. The inset shows the representative HPLC chromatograms of samples taken 0 (solid line), 60 (broken line), and 180 (dotted line) min after the start of the reaction, respectively. The peaks are fructose (F), glucose (G), and sucrose (S) following the solvent peak.
The initial rates of sucrose hydrolysis were determined by measuring the amounts of reducing group produced for the first 2 min of reaction using the DNSA method. The double-reciprocal plot revealed that sucrose hydrolysis by SUH follows the Michaelis-Menten kinetics, with a Km of 2.4 mM and a Vmax of 65 μmol min−1 mg of protein−1 at 30°C and pH 7.5. Virtually the same values for enzyme kinetic parameters were determined for the reaction catalyzed by rSUH. The enzyme remained active over a neutral pH (6.5 to 8.5) range, with a pH optimum of 7.5. The temperature-activity profile, constructed from measurements of the initial rates, showed the maximal activity at ca. 40°C. At this temperature, however, the enzyme was thermally inactivated with a rather short halftime, i.e., ∼3.5 h. Therefore, it would be practical to work with SUH at ca. 30°C, at which the enzyme was shown to retain as much as 85% of the maximal activity with a thermal inactivation halftime of ∼14 h.
Sequence comparison of SUH with related enzymes.
From database searches, the deduced sequence of SUH with 644 amino acid residues (or 643 residues with the terminal Met deleted, as confirmed by the N-terminal sequencing data and molecular mass determination) was found to be, among structurally characterized proteins, the most similar to amylosucrase from Neisseria polysaccharea (NpAS), with 36% identity and 57% similarity. In addition, the genomes of bacteria from different genera, such as Caulobacter crescentus (26), Deinococcus radiodurans (45), and Pirellula sp. (10), predicted the possible existence of proteins that also show high degrees of sequence homology (36 to 43% identity) to SUH. These proteins, designated RB5196, CC1135, and DR0933, whose expression and functions have yet to be tested (although their functions were assigned as α-amylase or amylosucrase, amylosucrase, and α-amylase, respectively), have been placed in family 13, together with NpAS (http://afmb.cnrs-mrs.fr/CAZY/). Since the Xanthomonas enzyme was regarded as α-glucosidase, known-function α-glucosidases available from databases were subjected to a similarity test, revealing that the 31 different sequences tested have only low degrees of sequence homology to SUH, with identities ranging from 9 to 19%.
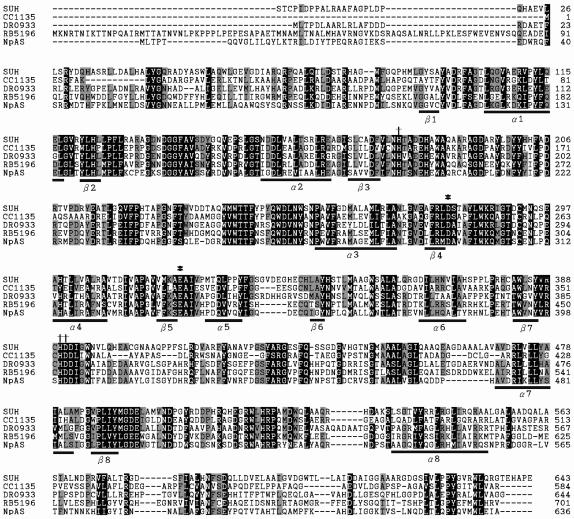
Family 13 enzymes share a common catalytic domain in the form of the (β/α)8 barrel (8, 41). This characteristic domain was predicted in the primary structure of SUH by using Pfam (http://pfam.wustl.edu/hmmsearch.shtml). Multiple sequence alignment using the CLUSTAL W program version 1.8 (42) (http://www.ebi.ac.uk/clustalw/) revealed that two catalytic carboxylic (aspartate and glutamate) residues and three other active-site residues (i.e., the second aspartate and two histidines), identified in NpAS (30, 33, 37) and long-recognized as being conserved in family 13 proteins (21), are all conserved in the sequence of SUH (Fig. 3). That is, D279 and E321 of SUH correspond to the critical residues D294 (the catalytic nucleophile) and E336 (the proton donor) situated right after the ends of the fourth and fifth β-sheet strands in NpAS, respectively, while H179, H390, and D391 of SUH correspond to the active-site residues H195, H400, and D401 situated near the ends of the third (for H195) and seventh (for H400 and D401) β-sheet strands in NpAS. The short sequence segments involving these invariant residues appeared to be rather highly conserved in the family 13 proteins examined.
FIG. 3.
Sequence alignment for SUH and related proteins. The residues in solid boxes are similar in all five sequences, whereas the residues in shaded boxes are similar to those of SUH in at least three of the four other sequences. β1 to β8 and α1 to α8 indicate the positions of the (β/α)8 barrel elements as determined for NpAS (37). Two catalytic carboxylic (aspartate and glutamate) residues (*) and three active-site (the second aspartate and two histidine) residues (†) conserved in family 13 enzymes are indicated. The sequences are those of X. axonopodis SUH (accession number AY359289), a putative C. crescentus amylosucrase (CC1135; accession number AAK23119), a putative D. radiodurans α-amylase (DR0933; accession number AAF10510), a putative Pirellula sp. α-amylase or amylosucrase (RB5196; accession number CAD78342), and NpAS (accession number CAA09772).
Bacterial multiplication in culture media and in planta.
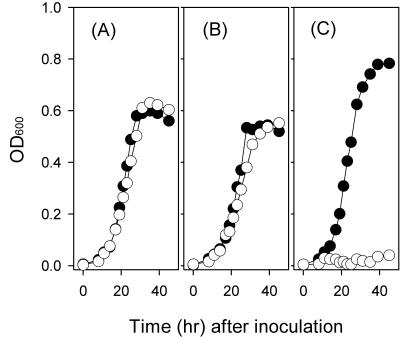
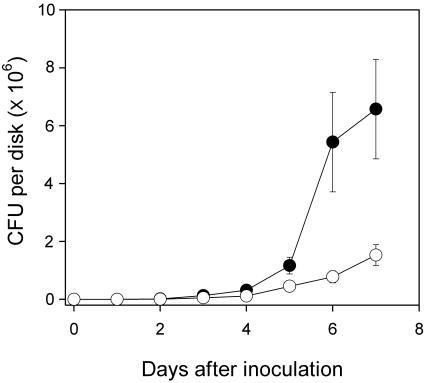
Strain 8ra and the suh mutant were cultured in minimal medium with tryptone deleted and with different carbon sources. When glucose or fructose was provided, the mutant grew as well as did 8ra, with doubling times only slightly different from each other, indicating that the glucose- and fructose-utilizing systems of strain 8ra were not affected by the mutation. In the presence of sucrose as the sole carbon source, however, the suh mutant, unlike the wild-type strain, was not able to grow (Fig. 4). These results appeared to be consistent with in vivo bacterial multiplication data: that is, the population size of the suh mutant in the inoculated soybean leaves was only about one-fourth of that of the wild-type strain when estimated on the seventh day after inoculation (Fig. 5). The multiplication of the mutant in soybean plants could be due to the presence of some carbon sources other than sucrose in the intercellular spaces of leaf tissues, the natural habitats of the phytopathogen. The mutant was shown to be pathogenic, and yet disease symptom development on leaves was delayed to some extent (estimated roughly, by 1 day).
FIG. 4.
Growth of X. axonopodis pv. glycines on different sugars. Cells of strain 8ra (solid circles) and the suh mutant (open circles) were pregrown in LB broth, washed with 100 mM K-phosphate buffer (pH 6.6), and then inoculated into minimal medium with tryptone deleted containing 20 mM glucose (A), 20 mM fructose (B), or 10 mM sucrose (C) as the sole carbon source. Cell multiplication was monitored by measuring the OD600. The data presented are from one representative experiment, which was confirmed twice in additional independent experiments.
FIG. 5.
Multiplication of X. axonopodis pv. glycines in planta. Cells of strain 8ra (solid circles) and the suh mutant (open circles) were infiltrated into fully matured soybean leaves. Leaf disks taken at 1-day intervals after inoculation were ground in 10 mM MgCl2, serially diluted, and spread on LB plates. Colonies were counted after the plates were incubated for 48 h at 28°C. The data are means of triplicate measurements with standard deviations shown as vertical bars.
Sucrose uptake and hydrolysis by cells.
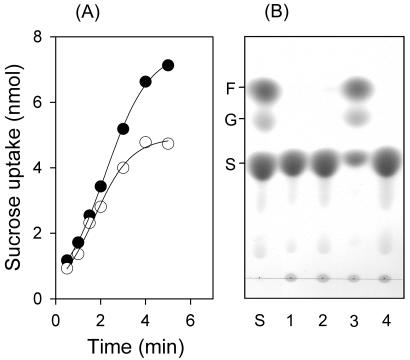
When harvested cells of X. axonopodis pv. glycines were exposed to sucrose labeled with a radioisotope, rapid sucrose uptake took place. The initial uptake rates were more or less the same for both strain 8ra and the suh mutant, indicating that the sucrose transport activity of the bacterial membrane remained intact in the suh mutant. Therefore, the inability of the mutant to grow on sucrose was thought to be due to an essential lack of intracellular metabolic activity for sucrose. Such an inference appeared to conform to the observation that, when crude protein extracts of X. axonopodis pv. glycines cells were incubated with sucrose, the production of glucose and fructose was detected in incubations containing the extracts of the wild-type cells only, but not those of the mutant cells. A lower sucrose saturation level in the suh mutant than in the 8ra strain, as noted from the sucrose uptake kinetics, may also be connected to the absence of intracellular sucrose metabolism in the mutant. Intriguingly, no significant sucrose hydrolase activity was seen in crude protein extracts from the wild-type cells when they had been cultured in medium containing carbon sources other than sucrose, such as glucose, fructose, and succinate. This may be taken as an indication that suh gene expression in X. axonopodis requires sucrose induction. The relevant data are presented in Fig. 6.
FIG. 6.
Uptake and hydrolysis of sucrose by X. axonopodis pv. glycines. (A) Time courses of sucrose uptake by strain 8ra (solid circles) and the suh mutant (open circles). Sucrose uptake was monitored by measuring the radioactivity of cells exposed to [U-14C]sucrose at ambient temperature. The data are expressed as amounts of sucrose (in nanomoles) taken up by cells in 0.2 ml of suspension at an OD600 of 2. (B) TLC profiles of sugars contained in 18-h incubations of sucrose (50 mM) with crude protein extracts (0.6 mg of protein/ml) at 30°C. The extracts were from the 8ra cells grown on glucose (lane 1), fructose (lane 2), and sucrose (lane 3) and from suh mutant cells grown on sucrose (lane 4). A mixture of sugar standards (F, fructose; G, glucose; S, sucrose; 0.1 μmol each) was also developed (lane S).
DISCUSSION
Through protein purification and analysis, gene identification and characterization, mutant construction and phenotype examination, and recombinant-protein production, we have described a new enzyme, SUH, that is solely responsible for intracellular sucrose hydrolysis and thus is essential for growth of X. axonopodis on sucrose. There can be little doubt that SUH is rather specific for sucrose, promoting hydrolytic cleavage of the disaccharide probably as the only reaction, and yet the enzyme is not structurally similar to any known sucrose hydrolase. Most of the typical sucrose hydrolases are invertases that are β-fructosidases, although some α-glucosidases also possess sucrose-hydrolyzing activity (2, 15, 36). The invertases whose primary structures have been characterized, such as those from Klebsiella pneumoniae (44), Bacillus stearothermophilus (19), Bacillus subtilis (9), Zymomonas mobilis (11), Streptococcus mutans (34), and Thermotoga maritima (20), have been classified into glycoside hydrolase family 32 (12) (http://afmb.cnrs-mrs.fr/CAZY/). The sequences of these proteins show strong similarity to each other in several regions. The amino acid residues are particularly conserved in the N terminus, with an identical segment of five residues, NDPNG, found throughout family 32 enzymes. The so-called “sucrose box,” which has been suggested to be important for catalysis of the transfer of fructose from sucrose (34), is also conserved. Such primary structural characteristics of those invertases, however, were not seen in SUH. Rather, a strong structural similarity to SUH is seen in NpAS and several hypothetical proteins, all of which are categorized in family 13. Such sequence homology with similar lengths (seen, in particular, between NpAS and SUH) and carrying many conserved sequences, especially the crucial invariant residues believed to be involved in catalytic activity, leads to a conclusion that SUH, together with NpAS, may be placed in the same family.
Although SUH is similar to amylosucrase in terms of not only the primary structure but also substrate specificity, the main function of the latter is glucosyltransferase (EC 2.4.1.4) activity that catalyzes the transfer of an α-d-glucopyranosyl moiety from sucrose to an acceptor molecule, synthesizing α-1,4-linked glucan (5, 29). Nevertheless, it is not unusual that amylosucrase can promote sucrose hydrolysis to a certain extent when sucrose is the sole substrate in reaction systems. The hydrolytic products, glucose and fructose, by themselves may then be used as the glucosyl acceptors for the glucosyl transfer reaction, resulting in production of di- and trisaccharides, such as maltose, maltotriose, trehalose, and turanose, as seen in NpAS (29). Sucrose hydrolysis by amylosucrase may be a natural consequence of the catalytic reaction that has been proposed to proceed via a double-displacement mechanism, in which a covalent glucosyl enzyme intermediate (Gl-Ez) is first formed (24). This glucosyl moiety could then be transferred either onto a water molecule, resulting in sucrose hydrolysis, or onto a hydroxyl group of a saccharide molecule, finally leading to transglucosylation. Between these two reactions that share Gl-Ez as the common reactant there would be competition. In most cases, the Gl-Ez reaction with glucosyl acceptors prevails over the reaction with water, as amylosucrase activity is principally manifested by transglucosylation. However, it would not be unreasonable to hypothesize that in certain amylosucrases a water molecule might compete strongly with an acceptor molecule for the reaction with Gl-Ez. An extreme case could be that the rate of the “Gl transfer to water” reaction far exceeds the rate of the “Gl transfer to acceptor” reaction; SUH might be such a case. To our knowledge, if hydrolytic activity toward sugars is considered the major function, the Xanthomonas enzyme is the first enzyme found to be specific for sucrose among the proteins in family 13, to which it belongs.
It is unlikely that PTS is involved in the process of sucrose uptake by X. axonopodis cells. If sucrose is transported across the bacterial membranes by the action of a sucrose-specific PTS, the transported form of sucrose is S-6-P, which should then be cleaved into glucose-6-phosphate and fructose to be metabolized in cells. However, the observed inability of SUH to hydrolyze S-6-P despite its indispensability for sucrose utilization by the bacterial cells conforms to the notion that sucrose itself is transported by a system independent of PTS. In this regard, it is noteworthy that X. axonopodis pv. citri and X. campestris pv. campestris, two Xanthomonas pathogens whose genomes have been sequenced (7), carry a structural gene designated suc1 and annotated as a sugar transporter gene. The nomenclature SUC1 was originally given to a transmembrane sucrose carrier found in Arabidopsis thaliana. Many plant species have also been shown to contain sucrose transporters homologous to SUC1, with different nomenclatures, in their plasma membranes (17). Because the predicted gene products of suc1 from X. axonopodis pv. citri and X. campestris pv. campestris show sequence homology to some extent (19 and 20% identity, respectively) to plant SUC1 and because no sucrose-specific PTS genes are found in the genomes of those bacteria, Xanthomonas suc1 could be the PTS-independent transporter responsible for sucrose uptake by the Xanthomonas cells. This concept seems to be supported by our preliminary results from a study in progress of the sucrose utilization pathway in Xanthomonas bacteria. Briefly, we cloned an X. axonopodis pv. glycines gene extensively similar to suc1 of X. axonopodis pv. citri, constructed a knockout mutant by insertional mutagenesis targeting that gene, and tested the mutant for its phenotype on sugars. It turned out that the mutant suffered a complete loss of sucrose uptake activity, being unable to grow on sucrose, while it grew on other carbon sources like glucose and fructose as well as did the wild-type strain.
The SUH gene of X. axonopodis pv. glycines is nearly identical with a structural gene, XAC3490, on the chromosome of X. axonopodis pv. citri. In the amino acid sequences of both gene products, deduced from open reading frames with the same length of 1,935 bp, differences are found only in 11 positions that are significantly separated from each other. That is, the residues 7P, 16G, 88P, 209V, 275A, 296S, 505D, 533K, 574A, 591L, and 601V in SUH are replaced by 7S, 16A, 88Q, 209L, 275T, 296P, 505H, 533N, 574V, 591P, and 601I in the XAC3490 product. The chromosome of X. campestris pv. campestris bears a structural gene, XCC3359, coding for a protein with a similar length (637 amino acid residues) that also appears extensively similar (77% identical) to SUH. In this context, it might be suggestive that SUH is a homologous protein promoting intracellular sucrose hydrolysis throughout Xanthomonas bacteria.
Acknowledgments
This work was supported in part by the Brain Korea21 project.
We thank Choonki Sung for technical assistance in protein purification work.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tools. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Badr, H. R., K. A. Sims, and M. W. W. Adams. 1994. Purification and characterization of sucrose α-glucohydrolase (invertase) from the hyperthermophilic archaeon Pyrococcus furiosus. Syst. Appl. Microbiol. 17:1-6. [Google Scholar]
- 3.Bockmann, J., H. Heuel, and J. W. Lengeler. 1992. Characterization of a chromosomally encoded, non-PTS metabolic pathway for sucrose utilization in Escherichia coli EC3132. Mol. Gen. Genet. 235:22-32. [DOI] [PubMed] [Google Scholar]
- 4.Bogs, J., and K. Geider. 2000. Molecular analysis of sucrose metabolism of Erwinia amylovora and influence on bacterial virulence. J. Bacteriol. 182:5351-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Büttcher, V., T. Welsh, L. Willmitzer, and J. Kossmann. 1997. Cloning and characterization of the gene for amylosucrase from Neisseria polysaccharea: production of a linear α-1,4-glucan. J. Bacteriol. 179:3324-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, J. W. Y. F., and P. H. Goodwin. 1999. The molecular genetics of virulence of Xanthomonas campestris. Biotechnol. Adv. 17:489-508. [DOI] [PubMed] [Google Scholar]
- 7.da Silva, A. C. R., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. C. Alves, A. M. do Amaral, M. C. Bertolini, L. E. A. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. B. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. S. Ferreira, R. C. C. Ferreira, M. I. T. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. M. Lemos, M. V. F. Lemos, E. C. Locali, M. A. Machado, A. M. B. N. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. M. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. M. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. D. Sena, C. Silva, R. F. de Souza, L. A. F. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. D. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 8.Davies, G., and B. Henrissat. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3:853-859. [DOI] [PubMed] [Google Scholar]
- 9.Fouet, A., A. Klier, and G. Rapoport. 1986. Nucleotide sequence of the sucrase gene of Bacillus subtilis. Gene 45:221-225. [DOI] [PubMed] [Google Scholar]
- 10.Glöckner, F. O., M. Kube, M. Bauer, H. Teeling, T. Lombardot, W. Ludwig, D. Gade, A. Beck, K. Borzym, K. Heitmann, R. Rabus, H. Schlesner, R. Amann, and R. Reinhardt. 2003. Complete genome sequence of the marine Planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 100:8298-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunasekaran, P., T. Karunakaran, B. Cami, A. G. Mukundan, L. Preziosi, and J. Baratti. 1990. Cloning and sequencing of the sacA gene: characterization of a sucrase from Zymomonas mobilis. J. Bacteriol. 172:6727-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jahreis, K., L. Bentler, J. Bockmann, S. Hans, A. Meyer, J. Siepelmeyer, and J. W. Lengeler. 2002. Adaptation of sucrose metabolism in the Escherichia coli wild-type strain EC3132. J. Bacteriol. 184:5307-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kakinuma, Y., and T. Unemoto. 1985. Sucrose uptake is driven by the Na+ electrochemical potential in the marine bacterium Vibrio alginolyticus. J. Bacteriol. 163:1293-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly, C. T., and W. M. Fogarty. 1983. Microbial α-glucosidases. Process Biochem. 18:6-12. [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lemoine, R. 2000. Sucrose transporters in plants: update on function and structure. Biochim. Biophys. Acta 1465:246-262. [DOI] [PubMed] [Google Scholar]
- 18.Lepesant, J. A., F. Kunst, J. Lepesant-Kejzlarova, and R. Dedonder. 1972. Chromosomal location of mutations affecting sucrose metabolism in Bacillus subtilis Marburg. Mol. Gen. Genet. 118:135-160. [DOI] [PubMed] [Google Scholar]
- 19.Li, Y., and T. Ferenci. 1996. The Bacillus stearothermophilus NUB36 surA gene encodes a thermophilic sucrase related to Bacillus subtilis SacA. Microbiology 142:1651-1657. [DOI] [PubMed] [Google Scholar]
- 20.Liebl, W., D. Brem, and A. Gotschlich. 1998. Analysis of the gene for β-fructosidase (invertase, inulinase) of the hyperthermophilic bacterium Thermotoga maritima, and characterization of the enzyme expressed in Escherichia coli. Appl. Microbiol. Biotechnol. 50:55-64. [DOI] [PubMed] [Google Scholar]
- 21.MacGregor, E. A., Š. Janeèek, and B. Svensson. 2001. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta 1546:1-20. [DOI] [PubMed] [Google Scholar]
- 22.Martin, I., M. Débarbouillé, E. Ferrari, A. Klier, and G. Rapoport. 1987. Characterization of the levanase gene of Bacillus subtilis which shows homology to yeast invertase. Mol. Gen. Genet. 208:177-184. [DOI] [PubMed] [Google Scholar]
- 23.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 24.Mirza, O., L. K. Skov, M. Remaud-Simeon, G. Potocki de Montalk, C. Albenne, P. Monsan, and M. Gajhede. 2001. Crystal structures of amylosucrase from Neisseria polysaccharea in complex with d-glucose and the active site mutant Glu328Gln in complex with the natural substrate sucrose. Biochemistry 40:9032-9039. [DOI] [PubMed] [Google Scholar]
- 25.Munro, C., S. M. Michalek, and F. L. Macrina. 1991. Cariogenicity of Streptococcus mutans V403 glucosyltransferase and fructosyltransferase mutants constructed by allelic exchange. Infect. Immun. 59:2316-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1996. Phosphoenolpyruvate:carbohydrate phosphotransferase systems, p. 1149-1174. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington D.C.
- 29.Potocki de Montalk, G., M. Remaud-Simeon, R. M. Willemot, P. Sarçabal, V. Planchot, and P. Monsan. 2000. Amylosucrase from Neisseria polysaccharea: novel catalytic properties. FEBS Lett. 471:219-223. [DOI] [PubMed] [Google Scholar]
- 30.Potocki de Montalk, G., M. Remaud-Simeon, R. M. Willemot, V. Planchot, and P. Monsan. 1999. Sequence analysis of the gene encoding amylosucrase from Neisseria polysaccharea and characterization of the recombinant enzyme. J. Bacteriol. 181:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarçabal, P., M. Remaud-Simeon, R. M. Willemot, G. Potocki de Montalk, B. Svensson, and P. Monsan. 2000. Identification of key amino acid residues in Neisseria polysaccharea amylosucrase. FEBS Lett. 474:33-37. [DOI] [PubMed] [Google Scholar]
- 34.Sato, Y., and H. K. Kuramitsu. 1988. Sequence analysis of the Streptococcus mutans scrB gene. Infect. Immun. 56:1956-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scholle, R. R., V. E. Coyne, R. Maharaj, F. T. Robb, and D. R. Woods. 1987. Expression and regulation of a Vibrio alginolyticus sucrose utilization system cloned in Escherichia coli. J. Bacteriol. 169:2685-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schönert, S., T. Buder, and M. K. Dahl. 1998. Identification and enzymatic characterization of the maltose-inducible α-glucosidase MalL (sucrase-isomaltase-maltase) of Bacillus subtilis. J. Bacteriol. 180:2574-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skov, L. K., O. Mirza, A. Henriksen, G. Potocki De Montalk, M. Remaud-Simeon, P. Sarçabal, R. M. Willemot, P. Monsan, and M. Gajhede. 2001. Amylosucrase, a glucan-synthesizing enzyme from the α-amylase family. J. Biol. Chem. 276:25273-25278. [DOI] [PubMed] [Google Scholar]
- 38.Slee, A. M., and J. M. Tanzer. 1982. Sucrose transport by Streptococcus mutans, evidence for multiple transport systems. Biochim. Biophys. Acta 692:415-424. [DOI] [PubMed] [Google Scholar]
- 39.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinmetz, M., D. Le Coq, S. Aymerich, G. Gonzy-Tréboul, and P. Gay. 1985. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol. Gen. Genet. 200:220-228. [DOI] [PubMed] [Google Scholar]
- 41.Svensson, B. 1994. Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol. Biol. 25:141-157. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J., N. Y. Nguyen, D. L. Sackett, and J. A. Donkersloot. 1991. Transposon-encoded sucrose metabolism in Lactococcus lactis. Purification of sucrose-6-phosphate hydrolase and genetic linkage to N5-(L-1-carboxyethyl)-L-ornithine synthase in strain K1. J. Biol. Chem. 266:14573-14579. [PubMed] [Google Scholar]
- 44.Titgemeyer, F., K. Jahreis, R. Ebner, and J. W. Lengeler. 1996. Molecular analysis of the scrA and scrB genes from Klebsiella pneumoniae and plasmid pUR400, which encode the sucrose transport protein enzyme IIScr of the phosphotransferase system and a sucrose-5-phosphate invertase. Mol. Gen. Genet. 250:197-206. [DOI] [PubMed] [Google Scholar]
- 45.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]