The Gipie/GRP78 interaction serves as a molecular switch for the regulation of the apoptotic signaling pathway, which appears to contribute to the protection of endothelial cells from endoplasmic reticulum stress-induced apoptosis.
Abstract
Continued exposure of endothelial cells to mechanical/shear stress elicits the unfolded protein response (UPR), which enhances intracellular homeostasis and protect cells against the accumulation of improperly folded proteins. Cells commit to apoptosis when subjected to continuous and high endoplasmic reticulum (ER) stress unless homeostasis is maintained. It is unknown how endothelial cells differentially regulate the UPR. Here we show that a novel Girdin family protein, Gipie (78 kDa glucose-regulated protein [GRP78]-interacting protein induced by ER stress), is expressed in endothelial cells, where it interacts with GRP78, a master regulator of the UPR. Gipie stabilizes the interaction between GRP78 and the ER stress sensor inositol-requiring protein 1 (IRE1) at the ER, leading to the attenuation of IRE1-induced c-Jun N-terminal kinase (JNK) activation. Gipie expression is induced upon ER stress and suppresses the IRE1-JNK pathway and ER stress-induced apoptosis. Furthermore we found that Gipie expression is up-regulated in the neointima of carotid arteries after balloon injury in a rat model that is known to result in the induction of the UPR. Thus our data indicate that Gipie/GRP78 interaction controls the IRE1-JNK signaling pathway. That interaction appears to protect endothelial cells against ER stress-induced apoptosis in pathological contexts such as atherosclerosis and vascular endothelial dysfunction.
INTRODUCTION
The endothelial cells that surround the lumen of blood vessels are directly exposed to mechanical stresses, chemicals, pathogens, hypoxia, and repetitive inflammatory insults. Previous studies have identified the mechanisms by which these extracellular stresses are sensed and transduced into intracellular biochemical signals in endothelial cells (Bonetti et al., 2002; Helenius and Schumacker, 2002). Dysregulation of that system leads to pathological changes in conditions such as atherosclerosis and breakdown of tumor vessels (Shimokawa, 1999). In response to extracellular stresses, endothelial cells accumulate misfolded proteins, fail to posttranslationally modify secretory proteins, and alter calcium homeostasis, events that elicit the unfolded protein response (UPR), also known as the endoplasmic reticulum (ER) stress response (Shen et al., 2004).
The lumen of the ER provides a highly specialized environment for the production of secretory and membrane proteins (Lee, 2001; Ellgaard and Helenius, 2003). When cells are subjected to extracellular stresses coupled with other ER stress conditions (such as nutrient depletion or severe hypoxia), the acute increase of newly translated secretory proteins imposes a major problem for the cells because of the potential buildup of improperly folded proteins within the ER (Szegedi et al., 2006). The initial aim of the UPR is to adapt to changes in the environment and to reestablish the ER’s function. This process begins with three ER-resident transmembrane proteins: activating transcription factor 6 (ATF6), protein kinase regulated by RNA (PKR)-like ER kinase (PERK), and inositol-requiring protein 1 (IRE1) (Schröder and Kaufman, 2005). When unfolded proteins accumulate within the ER luminal domains, those three proteins act as sentinels, sensing the levels of free 78 kDa glucose-regulated protein (GRP78; also termed BiP), an ER-resident chaperone protein (Lee, 1992, 2001). On imposition of ER stress, the three proteins are released from GRP78 and activated, leading to either transduction of apoptotic signals or transcription of genes encoding molecular chaperones and proteins involved in the ER-associated degradation pathway (Römisch, 2005). Given evidence that impaired functioning of ER stress transducers contributes to the degeneration of endothelial cells and atherosclerosis (Zhou et al., 2005), understanding how endothelial cells survive ER stress is an important issue for vascular biology. At present, the molecular mechanisms that differentially control survival and apoptotic death of endothelial cells are unclear.
In this study, we identified a novel protein named GRP78-interacting protein induced by ER stress (Gipie) encoded by the CCDC88B gene (previously described FLJ00354) that is preferentially expressed in endothelial cells and macrophages. The primary structure of Gipie shows high sequence similarity to that of Girdin, which we have previously identified as a novel actin cytoskeleton-binding protein and Akt substrate that regulates cell migratory responses in various biological contexts (Enomoto et al., 2005, 2006, 2009; Jiang et al., 2008, 2009; Kitamura et al., 2008). Comparison of their sequences indicates only a remote homology between the carboxyl (C)-terminal domains of Gipie and that of Girdin, suggesting that Gipie has a function distinct from that of Girdin. We found that endothelial cell expression of Gipie, which localizes at the ER and Golgi apparatus, is up-regulated by ER stress conditions induced by chemical reagents and experimental vascular balloon injury. Our biochemical studies revealed that Gipie can stabilize the interaction between GRP78 and IRE1, reducing IRE1-induced c-Jun N-terminal kinase (JNK) activation and ER stress-induced apoptosis. Given these findings, we suggest that Gipie is a regulator of the UPR, protecting cells from apoptosis in the vascular system.
RESULTS
Gipie is a member of the Girdin family of proteins
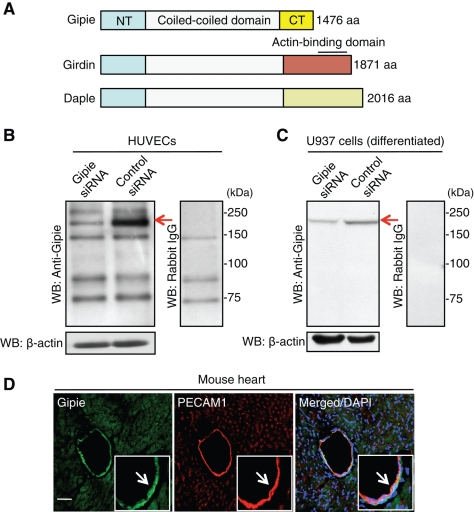
In a series of recent reports from our laboratory, we characterized the novel actin-binding protein Girdin (also termed APE, GIV, and HkRP1), which possesses a long coiled-coil domain that spans more than two-thirds of the middle of the protein (Figure 1A) (Enomoto et al., 2005, 2006). The coiled-coil domain of Girdin is flanked by unique amino (N)-terminal and C-terminal domains that lack similarity to any known functional domains. A BLAST search of the human genome and expressed sequence tag (EST) database for sequences related to Girdin identified two additional paralogues: dishevelled-associating protein with a high frequency of leucine residues (Daple) and a novel protein named Gipie. The full-length cDNA of Gipie, a transcript of the human CCDC88B gene, encodes a novel 1476-amino-acid protein, which is shorter than Girdin and Daple by 395 and 540 amino acid residues, respectively (Figure 1A). These three proteins share a conserved N-terminal domain and a central coiled-coil domain, but they diverge at the C-terminal domains, which may define their distinctive functions.
FIGURE 1:
Primary structure of Gipie and its expression in endothelial cells. (A) Schematic presentation of primary structures of Gipie and the Girdin family of proteins. Gipie can be divided into three domains. The N-terminal domain (NT) and central coiled-coil domain show high homologies (22.5 and 25.9%, 55.6 and 61.1%, respectively) with those of the Girdin family of proteins, Girdin and Daple. In contrast, the C-terminal domain (CT) of Gipie with unknown function shows remote similarities (3.4 and 5.7%) with those of the Girdin family of proteins. (B and C) Endogenous expression of Gipie in HUVECs (B) and U937 cells (C). Lysates from HUVECs or U937 cells transfected with either control or Gipie-specific siRNA were subjected to Western blot analysis (WB) using anti-Gipie antibody (left panels) or control rabbit IgG (right panels). Arrows denote the bands corresponding to Gipie. (D) Expression of Gipie in endothelial cells in vivo. A frozen-fixed section of mouse heart was stained with anti-Gipie (green) and anti–PECAM-1 (red) antibodies. Nuclei were visualized by DAPI staining (blue). Arrows denote the colocalization of Gipie and PECAM-1 in endothelial cells of the vessel. Scale bar: 100 μm.
Expression of Gipie in endothelial cells
Reverse transcription-PCR (RT-PCR) analyses using total RNA extracted from various mouse tissues indicated ubiquitous expression of Gipie in all postnatal (P2) and adult (P56) mouse tissues analyzed, with high expression in liver, spleen, and heart at P2 and in bone marrow and heart at P56 (Supplemental Figure S1A). To facilitate further investigations of Gipie, we generated a polyclonal antibody which was raised against its 18 C-terminal amino acids. Western blot analysis revealed that the anti-Gipie antibody recognized a 170 kDa band in total cell lysates from human umbilical vein endothelial cells (HUVECs) (Figure 1B). The 170 kDa band was less prominent in lysates from HUVECs transfected with Gipie-specific small interfering RNA (siRNA), indicating the specificity of the antibody (Figure 1B). We also screened multiple cell lines derived from various tissues and malignant tumors for expression of Gipie by both Western blot analysis and RT-PCR. These experiments showed that Gipie was also expressed in human myeloid and monocytoid leukemic cell lines U937 (Figure 1C), HL-60, and THP-1 (Supplemental Figure S1B). None of the epithelial and mesenchymal cell lines and none of the tumor cells that we examined (COS7, HEK293, HeLa [adenocarcinoma], HT1080 [fibrosarcoma], A549 [squamous cell carcinoma], TGW [neuroblastoma], and SW480 [adenocarcinoma] cells) expressed detectable levels of Gipie (Supplemental Figure S1B). Given the expression of Gipie in HUVECs (Figure 1B) and human coronary artery endothelial cells (HCAECs, Supplemental Figure S1B), we used immunofluorescent staining to examine the expression of Gipie in the endothelia of vessels in mouse heart sections (Figure 1D). Gipie expression was detected in the vascular endothelium, which also stained positively for platelet endothelial cell adhesion molecule-1 (PECAM-1, CD31, indicating that Gipie is normally expressed by vascular endothelial cells in vivo.
Gipie is localized in the Golgi apparatus and ER
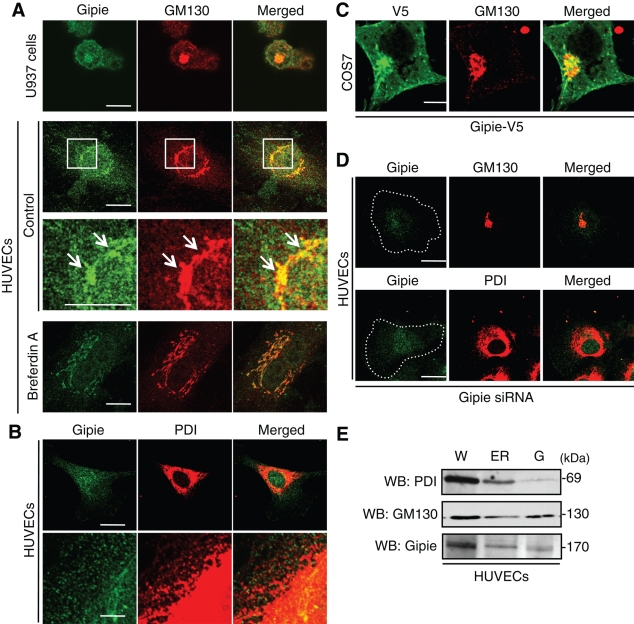
To elucidate the subcellular localization of Gipie, we performed fluorescence immunocytochemistry (Figure 2, A–C). In both HUVECs and U937 cells, anti-Gipie antibody yielded a perinuclear and a cytoplasmic lacelike staining pattern, suggesting that Gipie was expressed in both the Golgi apparatus and the ER (Figure 2, A and B). This subcellular localization of Gipie was further examined by double-staining, which revealed that the majority of Gipie was colocalized with GM130, a marker protein for the Golgi apparatus, and protein disulfide isomerase (PDI), a marker protein for ER (Figure 2, A and B). Whereas Gipie and GM130 were almost always colocalized, Gipie and PDI displayed a somewhat different staining pattern (Figure 2B). Coimmunoprecipitation experiments also failed to detect the association of Gipie with PDI in HUVECs (Supplemental Figure S2). These data indicate that Gipie does not interact with PDI directly and functions with other partners to form a protein complex in the ER.
FIGURE 2:
Subcellular localization of Gipie. (A) Localization of endogenous Gipie in the Golgi apparatus and the cytoplasm. U937 cells and HUVECs were fixed and stained with anti-Gipie (green) and anti-GM130 (red) antibodies, showing the localization of Gipie in GM130-positive Golgi apparatus (arrows) and the cytoplasm. The regions within the white boxes are shown at a higher magnification in adjacent lower panels. Shown in the bottom panels is the localization of Gipie in HUVECs treated with BFA at 30 nmol/l for 30 min. Scale bars: 10 μm. (B) Localization of endogenous Gipie in the ER in HUVECs. HUVECs were fixed and stained with anti-Gipie (green) and anti-PDI (red) antibodies, showing coincident or adjacent localization of Gipie and PDI in the ER. Scale bars: 10 μm (top panel), 1 μm (bottom panel). (C) Localization of exogenous Gipie in COS7 cells. COS7 cells, which lack endogenous expression of Gipie, were transfected with Gipie tagged with the V5 epitope, followed by immunostaining with anti-V5 (green) and GM130 (red) antibodies. Scale bar: 10 μm. (D) HUVECs were transfected with Gipie-specific siRNA and stained with the indicated antibodies. The staining of Gipie in the Golgi apparatus and ER was significantly decreased in Gipie-depleted HUVECs, showing the specificity of the anti-Gipie antibody. Scale bars: 10 μm. (E) Gipie expression in the fractions containing the ER and the Golgi apparatus. Equal amounts of proteins from each subcellular fraction were analyzed by Western blotting using the indicated antibodies. W, whole cell lysate; G, Golgi apparatus.
The specificity of Gipie staining was also shown by incubating HUVECs with brefeldin A (BFA), which specifically blocks protein transport from ER to the Golgi apparatus and causes the absorption of Golgi membranes into the ER (Lippincott-Schwartz et al., 1990). In cells treated with BFA (30 ng/ml), the Golgi apparatus displayed fragmentation and reorganization of the tubular networks around the nucleus, whereas Gipie signals merged with GM130 (bottom panels in Figure 2A), again confirming that Gipie is localized in the Golgi apparatus.
We observed that exogenously expressed Gipie tagged with the V5 epitope (Gipie-V5) also localized in the Golgi apparatus and the cytoplasm in COS7 cells (Figure 2C). Finally, when HUVECs were transfected with siRNA specific for human Gipie, the Gipie signals merged with the Golgi apparatus and ER in control cells almost disappeared (Figure 2D), indicating the specificity of the Gipie antibody.
To further confirm the localization of Gipie in the Golgi apparatus and ER, we analyzed the expression of Gipie in subcellular fractions obtained by differential centrifugation of HUVEC lysates. The ER and Golgi fractions were monitored using antibodies against PDI and GM130, respectively. Gipie expression was detected in both the ER and Golgi fractions (Figure 2E).
Identification of Gipie interacting proteins
Proteins specifically captured in Gipie immunoprecipitates were collected from U937 cell extracts and eluted with high salt buffer (NaCl at 0.5 mol/l) and analyzed by capillary liquid chromatography followed by tandem mass spectrometry (LC–MS/MS) after trypsin digestion. The majority of peptides identified unambiguously in the Gipie immunoprecipitates included GRP78, sorting nexin, lymphocyte-specific protein 1, nucleolin, and cathepsin Z.
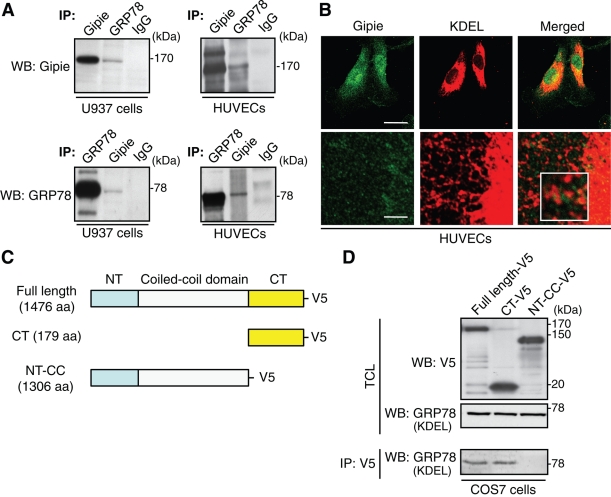
We focused further on the interaction between Gipie and GRP78, which is an ER chaperone that displays subcellular localization similar to Gipie and has pivotal roles in ER homeostasis (Liu et al., 1997). Validation of GRP78 as an interactant with Gipie was obtained by coimmunoprecipitation experiments using extracts from HUVECs as well as U937 cells which showed that endogenous Gipie interacts with GRP78 (Figure 3A). Other members of the Girdin family of proteins, Girdin and Daple, did not interact with GRP78, suggesting that the interaction was specific to Gipie (Supplemental Figure S3A). In immunofluorescence studies using HUVECs, cytoplasmic staining of Gipie partially overlapped with the signals obtained by anti-KDEL antibody. The latter recognizes the C-terminal ER retrieval sequence (Lys-Asp-Glu-Leu) and preferentially detects GRP78 and GRP94 (Figure 3B). These results imply that endogenous Gipie associates with endogenous GRP78 in the ER.
FIGURE 3:
Interaction of Gipie with GRP78. (A) Interaction of endogenous Gipie and GRP78. Lysates from U937 cells (left panels) and HUVECs (right panels) were immunoprecipitated with anti-Gipie, anti-GRP78, and normal rabbit IgG antibodies, followed by Western blot analysis using the indicated antibodies. (B) Colocalization of Gipie with GRP78 in HUVECs. HUVECs were fixed and stained with anti-Gipie (green) and anti-KDEL (red) antibodies, showing the localization of Gipie in GRP78-positive ER structures (white box). Scale bars: 10 μm (top panel), 1 μm (bottom panel). (C) Fragments of Gipie tagged with the V5 epitope used in this study. (D) The C-terminal domain (CT) of Gipie is required for interaction with GRP78. Lysates from COS7 cells transfected with fragments of Gipie were immunoprecipitated with anti-V5 antibody, followed by Western blot analysis with anti-KDEL antibody (bottom panels). Expression of Gipie and GRP78 fragments was monitored by Western blot analysis using the indicated antibodies (top and middle panels, respectively). TCL, total cell lysates. CT-V5, C-terminal fragment of Gipie. NT-CC-V5, the mutant lacking the C-terminal fragment of Gipie.
To identify the GRP78-binding domain in Gipie, we generated deletion mutants of Gipie fused with the V5 epitope (Figure 3C). Cotransfection experiments indicated that full-length Gipie and its C-terminal fragment (Gipie-CT-V5), but not the mutant lacking the C-terminal fragment (Gipie-NT-CC-V5), interact with GRP78, indicating that Gipie binds to GRP78 through its C-terminal domain (Figure 3D).
Expression of Gipie is induced by ER stress
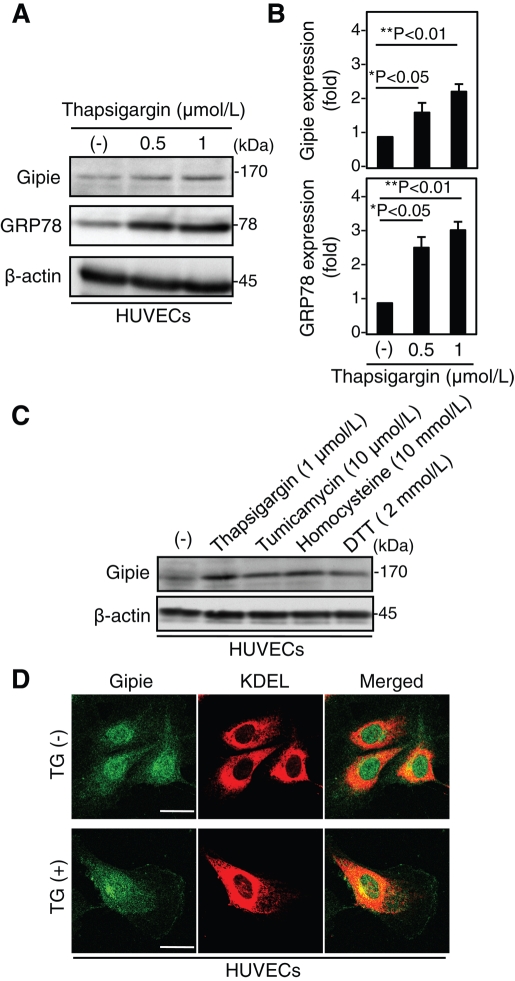
GRP78 was first identified as a eukaryotic cell protein which was up-regulated by depletion of glucose (Shiu et al., 1977). GRP78 functions as an ER molecular chaperone and as a calcium-binding protein, and it has been used extensively as a biomarker for activation of the UPR pathway (Hendershot, 2004). To examine whether the expression of Gipie is transcriptionally regulated in a similar way, we investigated the expression of Gipie following ER stress induced by chemical reagents. HUVECs were cultured for 8 h in the absence or presence of thapsigargin (TG), a specific blocker of ER calcium ATPase pumps, at 0.5 or 1 μmol/l. Following treatment with TG, the expression of Gipie was significantly increased (up to twofold or more) in parallel with the increase of GRP78 expression (Figure 4, A and B). The expression of Gipie was also moderately but reproducibly increased in HUVECs treated with other ER stressors, tunicamycin, homocysteine, and dithiothreitol (DTT), a reversible inhibitor of protein folding in the ER (Figure 4C). Thus Gipie expression seems to be regulated by ER stress through a mechanism yet to be identified. The TG treatment had no significant effects on subcellular localization of Gipie in HUVECs, with little increase in the expression of Gipie at the cell periphery (Figure 4D). Expression levels of Girdin and Daple were not significantly changed by TG-induced ER stress (Supplemental Figure S3, B and C).
FIGURE 4:
Induced expression of Gipie in HUVECs by ER stress. (A) Induction of Gipie expression in HUVECs by ER stress. HUVECs were incubated with TG at 0.5 or 1 μmol/l for 8 h. Total cell lysates were subjected to Western blot analysis with the indicated antibodies. (B) Expression levels of Gipie (top panel) and GRP78 (bottom panel) shown in (A) were quantified by densitometric scanning. The values are presented as the fold increase in expression relative to the control. Asterisks indicate significant differences (*p < 0.05; **p < 0.01, t test). (C) Induction of Gipie expression in HUVECs by various ER stressors. Total cell lysates from HUVECs untreated or treated with TG at 1 μmol/l, tunicamycin at 10 μmol/l, homocysteine at 10 mmol/l, or DTT at 2 mmol/l for 8 h were subjected to Western blot analysis with the indicated antibodies. (D) Negligible effects of TG on subcellular localization of Gipie in HUVECs. HUVECs incubated with or without TG at 1 μmol/l for 8 h were fixed and stained with anti-Gipie (green) and anti-KDEL (red) antibodies. Scale bars: 10 μm.
Gipie regulates the association between GRP78 and IRE1
In mammalian cells, the UPR pathway is initiated by three prototypical ER-localized stress sensors: IRE1, PERK, and ATF6. These sensor proteins bind to GRP78 under normal conditions but they dissociate when unfolded proteins accumulate during ER stress, which in turn activates various signaling cascades characteristic of the UPR pathway. After being activated, IRE1 recruits the adaptor protein tumor necrosis factor receptor–associated factor 2 (TRAF2) and apoptosis signal-regulating kinase 1 (ASK1) to the ER membrane, which subsequently activates ASK1 and the JNK pathway, leading to cell death (Urano et al., 2000). Other UPR pathway steps include the phosphorylation of serine-51 of eukaryotic initiation factor 2α (eIF2α) by active PERK, which results in the global inhibition of protein synthesis (Schröder and Kaufman, 2006). Also, proteolytic processing of activated ATF6 releases a 50 kDa cytosolic basic leucine zipper transcription factor that translocates into the nucleus to bind to several different promoter elements in various UPR genes (Haze et al., 1999).
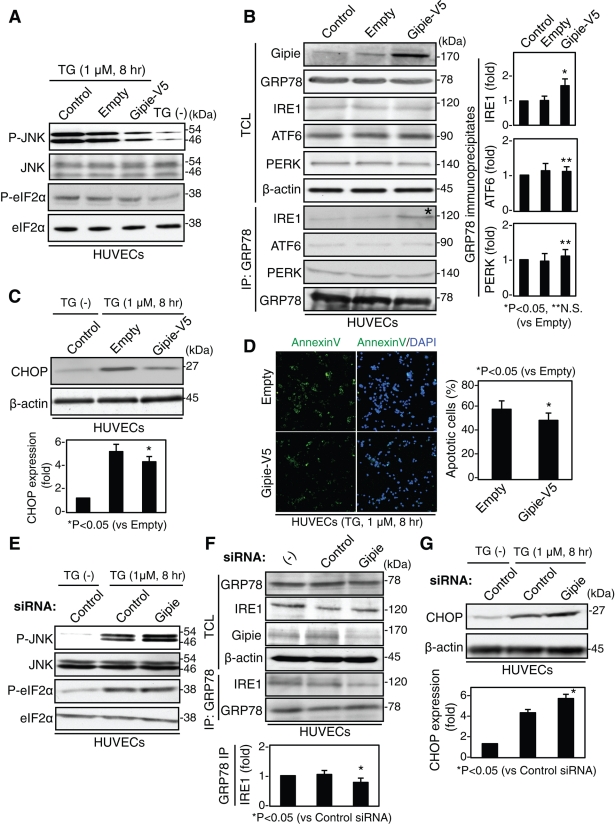
To test whether Gipie was involved in the UPR pathway, HUVECs were transfected with either Gipie or control vector and were then incubated with TG at 1 μmol/l. Phosphorylation of JNK and eIF2α was examined by Western blot analysis (Figure 5A). The phosphorylation level of eIF2α in Gipie-transfected cells was comparable to that in empty vector-transfected cells, whereas JNK phosphorylation was moderately attenuated in Gipie-expressing cells compared with empty vector-transfected cells (Figure 5A). These data show that Gipie opposes IRE1-mediated JNK activation in ER stress conditions. We could not test the role of Gipie in the cleavage of ATF6 in the UPR pathway in this study because antibody for the detection of cleaved ATF6 was not commercially available.
FIGURE 5:
Gipie regulates the interaction between GRP78 and IRE1, and also ER stress-induced apoptosis. (A) Gipie suppressed ER stress-induced phosphorylation of JNK but not that of elF2α. Nontransfected (control) HUVECs or cells transfected with either Gipie or V5 empty vector were treated with TG at 1 μmol/l for 8 h, and total cell lysates were subjected to Western blot analysis using the indicated antibodies. (B) Regulation of GRP78 interaction with IRE1 by Gipie. Total cell lysates and GRP78 immunoprecipitates from nontransfected HUVECs or cells transfected with either Gipie or V5 empty vector were subjected to Western blot analysis using the indicated antibodies. Note that the amount of IRE1 detected in the GRP78 immunoprecipitates was increased by the transfection of Gipie (asterisk). In the right panels, the amounts of IRE1, ATF6, and PERK detected in the GRP78 immunoprecipitates shown in the left panels were quantified by densitometric scanning, and the values are presented as the fold increase in expression relative to the control (*p < 0.05; **not significant [N.S.], t test). (C) Effects of Gipie on ER stress-induced expression of CHOP. Total cell lysates from HUVECs that had been transfected with either Gipie or V5 empty vector were subsequently treated with TG at 1 μmol/l for 8 h and analyzed by Western blotting using anti-CHOP antibody. In the bottom panel, the amounts of CHOP were quantified by densitometric scanning, and the values are presented as the fold increase in expression relative to the control. An asterisk indicates a significant difference (*p < 0.05, t test). (D) Effects of Gipie on ER stress-induced apoptosis. HUVECs transfected with either Gipie or V5 empty vector were incubated with TG at 1 μmol/l for 8 h, followed by annexin V staining (green) to detect apoptotic cells. In the right panel, the percentage of annexin V–positive cells was quantified. An asterisk indicates a statistically significant difference (*p < 0.05, t test). (E and F) Effects of endogenous Gipie depletion on JNK phosphorylation (E) and the interaction between GRP78 and IRE1 (F) Nontransfected HUVECs or cells transfected with either control or Gipie-specific siRNA were collected. Total cell lysates and GRP78 immunoprecipitates were subjected to Western blot analysis using the indicated antibodies. In the bottom panel of (F), the amounts of IRE1 detected in the GRP78 immunoprecipitates were quantified by densitometric scanning, and the values are presented as the fold increase in expression relative to the control. An asterisk indicates statistically significant difference (*p < 0.05, t test). (G) Effects of Gipie-depletion on ER stress-induced expression of CHOP. Total cell lysates from HUVECs that had been transfected with either control or Gipie-specific siRNA were analyzed. An asterisk indicates a statistically significant difference (*p < 0.05, t test).
It was of interest to determine how Gipie attenuated IRE1-mediated JNK activation. HUVECs were transfected with either Gipie or control vector, and immunoprecipitates generated with anti-GRP78 antibody were subjected to Western blot analysis. The results showed that the amount of IRE1 in the GRP78 immunoprecipitates from Gipie-transfected cells was significantly increased relative to that from control cells, without affecting the expression levels of IRE1, PERK, and ATF6 (Figure 5B). In contrast, the amounts of PERK and ATF6 in the GRP78 immunoprecipitates were comparable between Gipie-transfected and control cells. The effects of Gipie on IRE1-mediated JNK activation and IRE1/GRP78 interaction were also demonstrated in COS7 cells that overexpress exogenous Gipie (Supplemental Figure S4).
One possible mechanism for the increase in IRE1/GRP78 interaction is that Gipie regulates the turnover of IRE1, which has been shown to undergo proteasome-dependent degradation (Gao et al., 2008). We addressed this possibility by measuring IRE1 protein expression by pulse-chase analysis (Supplemental Figure S5). The exogenous expression of Gipie, however, had no significant effect on the half-life of IRE1, suggesting that Gipie is not involved in proteasome-dependent degradation of IRE1. These data indicate that Gipie, presumably through binding to GRP78, augments and/or stabilizes the GRP78/IRE1 protein complex by yet unidentified mechanisms that ultimately attenuate the release of IRE1 from GRP78 and the phosphorylation of JNK in the UPR pathway.
Effect of Gipie overexpression on ER stress-induced apoptosis
JNK activation downstream of the IRE1/TRAF2/ASK1 signaling pathway triggers proapoptotic signals during continuous ER stress (Urano et al., 2000). Therefore we asked whether Gipie had effects on ER stress-induced apoptosis. Active JNK increases the amount of transcription factor C/EBP homologous protein (CHOP) (also termed GADD153), a factor involved in ER stress-induced apoptosis (Wang and Ron, 1996; Marciniak et al., 2004). We found that transfection of Gipie into HUVECs (Figure 5C) and COS7 cells (Supplemental Figure S4C) decreased the expression of CHOP induced by ER stress. Consistent with this observation, the frequency of ER stress-induced apoptotic cells was lower in Gipie-transfected cells than in controls (Figure 5D and Supplemental Figure S4D). Expression of the NT-CC mutant of Gipie, which lacks the ability to bind to GRP78 (Figure 3D), did not affect CHOP expression or TG-induced apoptosis, indicating that the anti-apoptotic effect of Gipie is mediated by its interaction with GRP78 (Supplemental Figure S6). These data show that Gipie and its interaction with GRP78 regulate ER stress-induced apoptosis by modulating the IRE1/ASK1/JNK signaling pathway.
The function of endogenous Gipie in the UPR pathway was investigated by knock-down experiments in HUVECs. In Gipie-depleted HUVECs, ER stress-induced interaction of GRP78 with IRE1, JNK phosphorylation, and CHOP expression were dysregulated compared with control cells (Figure 5, E–G).
Expression of Gipie in nascent endothelial cells of the carotid artery after balloon injury
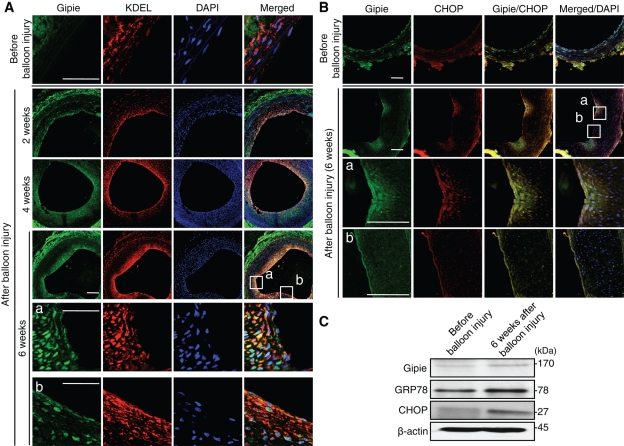
Given that endothelial cells of large arteries are subject to mechanical stretch and sheer stresses due to blood pressure and flow, it is plausible to consider the perturbation of the UPR pathway as a mechanism for atherosclerosis (Zhou et al., 2005). It was demonstrated that elevated GRP78 expressed in endothelial cells has a protective role in the pathogenesis of atherosclerosis (Zhou et al., 2004; Bhattacharjee et al., 2005; Feaver et al., 2008). Considering our findings mentioned earlier in the text, we investigated Gipie’s expression in rat carotid artery endothelial cells after balloon injury, a model of endothelial damage and restenosis (Clowes et al., 1983; Numaguchi et al., 1999). Tissue samples of the carotid arteries before and 2, 4, and 6 wk after balloon injury were stained with anti-Gipie, anti-KDEL, and anti–PECAM-1 antibodies (Figure 6A and Supplemental Figure S7A). The samples were also stained with anti–α-smooth muscle actin (SMA) antibody to show the proliferation of smooth muscle cells in the neointima (Supplemental Figure S7B). We found that Gipie expression increased in the cytoplasm of nascent endothelial cells and neointima 2 wk after balloon injury. The increased expression of Gipie persisted at least 6 wk after the injury (Figure 6A), which parallels GRP78 and CHOP expression (Figure 6, A and B).
FIGURE 6:
Expression of Gipie in nascent endothelial cells after balloon injury. (A) Paraffin-embedded sections from rat carotid arteries either before or 2, 4, or 6 wk after balloon injury were stained with anti-Gipie (green) and anti-KDEL (red) antibodies. Nuclei were visualized with DAPI staining (blue). The regions within the white boxes (a and b) are shown at a higher magnification in bottom panels. Note that Gipie expression is significantly increased in endothelial cells and neointima of the artery after the balloon injury. Scale bars: 200 μm. (B) Paraffin-embedded sections from rat carotid arteries either before or 6 wk after balloon injury were stained with anti-Gipie (green) and anti-CHOP (red) antibodies. The regions within the white boxes (a and b) are shown at a higher magnification in bottom panels. Three rats were analyzed in each group. Scale bars: 200 μm. (C) Protein extracts from carotid arteries either before or 6 wk after the balloon injury were subjected to Western blot analysis with anti-Gipie, anti-GRP78, anti-CHOP, and anti–β-actin antibodies.
After balloon injury (6 wk), the expression of Gipie was moderately but significantly higher in protein samples collected from carotid arteries than in those from control arteries, paralleling the increase of GRP78 and CHOP (Figure 6C). Those results agreed with immunofluorescence findings. Thus the data imply that induced GRP78 and Gipie have important roles in ER homeostasis and/or protecting endothelial cells against apoptosis during their regeneration after balloon injury.
Expression of Gipie in endothelial cells in the inner curvature of the aortic arch
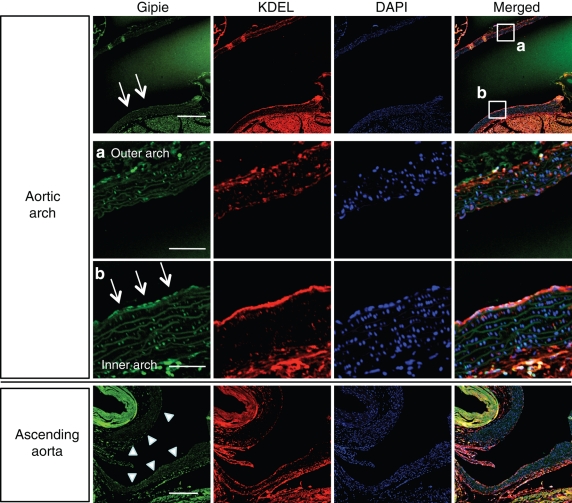
Hemodynamically induced shear stress generates various mechanical signals, which put endothelial cells at risk for developing atherosclerosis. Considering that GRP78 is up-regulated by mechanical shear stress and is expressed at greater levels at the inner curvature of the aortic arch with disturbed flow patterns than the outer curvature (Feaver et al., 2008), we stained aortic tissue samples from adult P56 mice with anti-Gipie and anti-KDEL antibodies (Figure 7). Consistent with the previous study, GRP78 was highly expressed in endothelial cells of the inner curvature but not the outer curvature of the aortic arch, which paralleled the increased expression of Gipie. In the ascending aorta, the increased expression of both Gipie and GRP78 was not evident in either of the lateral walls of the aorta (Figure 7). These data indicate that Gipie expression may be regulated by mechanical shear stress that induces the UPR in atheroprone regions of the vasculature in vivo.
FIGURE 7:
Expression of Gipie in the inner curvature of the aortic arch. Paraffin-embedded sections from the aortic arch (top panels) and the ascending aorta (bottom panel) of adult C56BL/6 P56 mice were stained with anti-Gipie (green) and anti-KDEL (red) antibodies. Nuclei were visualized with DAPI staining (blue). Scale bars: 200 μm. The regions within the white boxes in the aortic arch (a, outer curvature; b, inner curvature) are shown at a higher magnification in bottom panels. Three mice were analyzed. Scale bars: 20 μm (in a and b). Arrows in aortic arch indicate endothelial cells with high levels of Gipie expression. Arrowheads in ascending aorta indicate the lateral walls.
DISCUSSION
We describe here the identification and physiological role of Gipie, a member of the Girdin family of proteins. We showed that Gipie is present in the ER and the Golgi apparatus in endothelial cells where it associates with the ER chaperone GRP78 to regulate ER stress-induced apoptosis.
Of all the cells constituting the vascular system, endothelial cells have the greatest susceptibility to injury induced by mechanical forces, reactive oxygen species, and hypoxia, all of which are transduced into biological responses via the UPR pathway (VanderLaan et al., 2004). Cytoprotective responses of chaperone proteins such as GRP78 reduce cellular injury from misfolded proteins and promote cell survival. The mechanism by which endothelial cells differentially regulate cytoprotective or proapoptotic signaling responses, however, has been obscure (Xu et al., 2005). Our study showed that Gipie has a cytoprotective effect against ER stress conditions. This novel mechanism appears to protect endothelial cells against ER stress.
An intriguing observation presented here is that expression of Gipie is up-regulated by inducers of ER stress. An accompanying finding is that the up-regulation of Gipie expression may be implicated in the pathogenesis of restenosis after balloon injury of the carotid arteries (Figure 6) and atherogenic response occurring in atheroprone regions of the vasculature (Figure 7). At present, the regulatory mechanisms responsible for the induction of Gipie expression are unknown and require further investigation. It is possible that the up-regulation of Gipie gene expression is mediated by transmembrane integrin receptors and/or stretch-activated ion channels used by cells to sense mechanical changes and initiate intracellular responses (Shyy and Chien, 2002). In preliminary experiments using uniaxial cyclical mechanical stretch, we examined the levels of Gipie expression in HUVECs cultured on fibronectin-coated silicon membranes. No effects were apparent (Supplemental Figure S8), indicating that other mechanisms and factors are responsible for the induction of Gipie expression in the context of balloon injury and the inner curvature of the aortic arch.
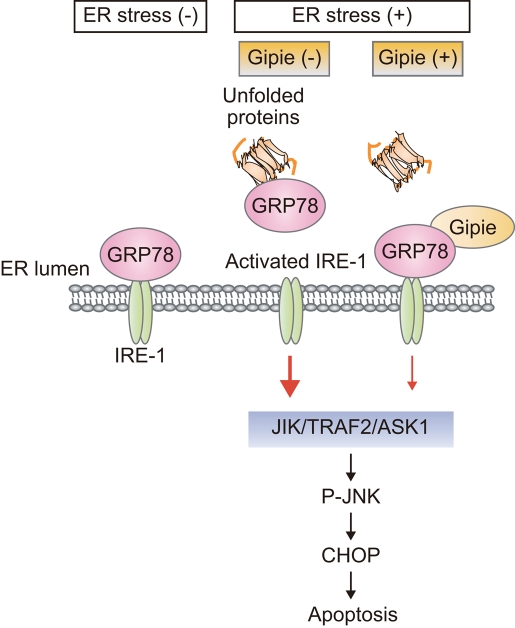
Our data showed that Gipie interacts with GRP78 and enhances the interaction between GRP78 and IRE1. This interaction attenuated the IRE1/ASK1/JNK signaling pathway and consequent expression of CHOP, leading to decreased apoptosis (Figure 8). In contrast, Gipie had no effect on the splicing of the transcription factor X-box binding protein 1 (XBP-1), which is an effector of IRE1 (Supplemental Figure S9) (Calfon et al., 2002). The mechanism by which Gipie regulates GRP78/IRE1 interaction remains unclear. In the current study, we were unable to detect an association of Gipie with IRE1 by coimmunoprecipitation experiments, indicating that Gipie may not directly interact with IRE1 in cells (unpublished data). It is possible that, upon binding with Gipie, GRP78 undergoes a conformational change that stabilizes its interaction with IRE1. In addition, Gipie did not significantly influence the interaction of GRP78 with PERK and ATF6, indicating that regulation of GRP78 function by Gipie appears to be more complex (Figure 5).
FIGURE 8:
Proposed model of Gipie-mediated modulation of the UPR pathway in endothelial cells. In cells exposed to persistent ER stresses, the expression of Gipie was induced to regulate the interaction of GRP78 and IRE1, resulting in a protective role against apoptotic cell death by regulating JNK activation and ER stress-induced expression of CHOP.
Expression of Gipie has been detected in all tissues examined, although at varying levels. Our histological data, however, revealed its limited expression in endothelial cells of the vessels. Considering that ER stress-mediated endothelial dysfunction is assumed to represent an early step in atherosclerosis, it is significant that endothelial cells’ expression of Gipie (as in the case of GRP78) was up-regulated in the context of balloon injury models and the native conditions of mechanical shear stress. Thus Gipie may be a potential target for pharmacological treatment of human atherosclerosis, although it is important to determine how induced expression of Gipie impacts the pathogenesis of atherosclerotic lesions. To address the question, its biological functions should be studied in in vivo contexts in which cell-signaling events play important roles. In this regard, generation of Gipie gene-deficient animals will prove crucial. This strategy will also improve our understanding of the function of Gipie in the hematopoietic system, where Gipie was found to be preferentially expressed based on the analysis of numerous EST clones in the GenBank database.
MATERIALS AND METHODS
Cloning of mouse Gipie
A cDNA encoding a full-length mouse Gipie (4100 base pairs) was isolated from an EST clone (MMM1013–7510332; GenBank accession no. BC038890) obtained from Open Biosystems (Huntsville, AL). The cDNA was amplified by PCR with introduction of appropriate restriction enzyme sites in the primers, and was introduced into the pGEM-11Zf cloning vector (Promega, Madison, WI). A V5 tag was fused to the carboxy terminus of the protein.
Plasmids and antibodies
The cDNA for mouse IRE1β was provided by H. Nishitoh and H. Ichijyo (Tokyo University, Tokyo, Japan). Rabbit anti-Gipie polyclonal antibody was developed against the 18 carboxyl-terminal amino acids of Gipie and affinity-purified with the immunopeptide. It was raised by immunizing rabbits with a keyhole limpet hemocyanin–conjugated peptide of Gipie. Antiserum was purified as a bound fraction of the peptide-conjugated column. Other antibodies used in this study include anti-GRP78 and anti-KDEL mouse monoclonal antibodies (mAbs) (Stressgene, Victoria, BC, Canada); anti-GRP78, anti-PERK, and anti-PDI rabbit polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA); anti-PDI and anti-CHOP mouse mAbs (Affinity BioReagents, Golden, CO); anti-GM130 and PECAM-1 mouse mAbs (BD Bioscience PharMingen, San Diego, CA); anti-IRE1 mouse mAb (MoBiTec GmbH, Gottingen, Germany); anti-ATF6 rabbit polyclonal antibody (LifeSpan Bioscience, Seattle, WA); anti-PERK and anti-PDI rabbit polyclonal antibodies (Santa Cruz Biotechnology); anti-eIF2α mouse mAb, anti–phospho-eIF2α (Ser51) rabbit mAb, anti-JNK, anti–phospho-JNK (Thr183/Tyr185) rabbit polyclonal antibodies, and anti-CHOP mouse mAb (Cell Signaling Technology, Beverly, MA); anti–α-SMA and anti–β-actin mouse mAbs (Sigma-Aldrich, St. Louis, MO); and anti-HaloTag rabbit polyclonal antibody (Promega).
Culture of cell lines and primary endothelial cells
COS7 and U937 cells, purchased from the American Type Culture Collection (ATCC; Rockville, MD), were cultured at 37ºC in a humidified atmosphere of 5% CO2. COS7 cells were grown in DMEM supplemented with 5% fetal bovine serum (FBS), whereas U937 cells were cultured in RPMI 1640 supplemented with 10% FBS. HUVECs obtained from Lonza (Basel, Switzerland) were cultured in endothelial medium-2 (EBM2, Lonza) supplemented with EGM-2-MV-SingleQuots (Lonza) containing vascular endothelial growth factor A on plates coated with 2% (wt/vol) gelatin in a humidified 37°C incubator with 5% CO2. HUVECs from passages three to five were used in this study.
Semiquantitative RT-PCR
For semiquantitative analyses of Gipie and β-actin transcripts, total RNAs from normal tissues of P2 and P56 C57BL/6 mice were isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany). cDNA transcripts were then generated using Superscript II reverse transcriptase (Invitrogen, San Diego, CA). RT-PCR was performed with primers specific to mouse Gipie (sense, 5′-ACAAGGTGAAGAGGCTCATTCG -3′; antisense, 5′-ACAGAATCACTGGCTGAGGGAA-3′), human Gipie (sense, 5′-AGACGGAGCTTCCTGAGGGCAG-3′; antisense, 5′-ACGGTGTCACTGGCTGAGGGA -3′), human XBP-1(sense, 5′-CCTTGTAGTTGAGAACCAGG-3′; antisense, 5′-GGGGCTTGGTATATATG-TGG-3′), mouse β-actin (sense, 5′-TGGAATCCTGTGGCATCCATGAAAC-3′; antisense, 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′), human β-actin (sense, 5′-ACTCTTCCAGCCTTCCTTCC-3′; antisense, 5′-CGTCATACTCCTGCTTGCTG -3′).
Western blot analysis
Cells cultured under various conditions were washed with phosphate-buffered saline (PBS) (sodium phosphate at 10 mmol/l, NaCl at 150 mmol/l, pH 7.4). After washing, cells were lysed with SDS sample buffer (Tris-HCl at 10 mmol/l, 2% SDS, DTT at 50 mmol/l, EDTA at 2 mmol/l, 0.02% bromophenol blue, 6% glycerol, pH 6.8) and treated at 97ºC for 5 min. Samples were separated by SDS–PAGE. Proteins were transferred to polyvinylidene fluoride membranes (Immobilon; Millipore, Bedford, MA), blocked in 5% milk in PBS containing 0.05% Tween 20, incubated with primary antibodies, and detected by horseradish peroxidase–conjugated secondary antibodies (Dako, Carpinteria, CA). In some experiments, the primary antibodies were diluted with Can-Get-Signal Solution 1 (TOYOBO, Osaka, Japan) to enhance antibody–antigen binding.
Immunoprecipitation
The cell lysates were immunoprecipitated with 5 to 10 μl of the indicated antibodies for 2 to 8 h at 4ºC, followed by incubation with 10% Protein A slurry (Sigma) for 4 h with intermittent mixing. All immunoprecipitates were washed five times with low salt buffer (NaCl at 0.15 mol/l, EDTA at 5 mmol/l, Tris-HCl at 20 mmol/l, pH 7.5) and then eluted with high salt buffer (NaCl at 0.5 mol/l, EDTA at 5 mmol/l, Tris-HCl at 20 mmol/l, pH 7.5). Eluates were analyzed by SDS–PAGE, followed by Western blot analysis.
Subcellular fractionation
Cultured HUVECs were collected in cold sucrose buffer (250 mM sucrose, 2 mM EDTA, 0.5 mM EGTA, 10 mM DTT, 10 mM Tris-HCl, pH 7.4) and homogenized in a Kontes Dounce tissue grinder. Differential centrifugation rounds were performed to separate the fractions of the nuclei (500 × g), ER (7600 × g), and Golgi apparatus (80,000 × g). The lysates of each fraction were analyzed by Western blot analysis using anti-PDI (a marker protein of ER), anti-GM130 (a marker protein of Golgi apparatus), and anti-Gipie antibodies.
RNA interference
siRNA-mediated knock down of Gipie protein expression was performed using a previously described method (Enomoto et al., 2005). The 21-nucleotide synthetic duplexes were purchased from Qiagen. The targeted sequence that effectively silenced human Gipie expression was 5′-TGGGCTGATCTTGAGCCTTAA-3′ (sense sequence). HUVECs were transfected with either the Gipie-specific siRNA or 21-nucleotide control siRNA (Qiagen) using Lipofectamine 2000 according to the manufacturer’s protocol.
Mass spectrometric analysis (LC/MS/MS)
After immunoprecipitation, proteins eluted with the high salt buffer were first precipitated using TCA to remove salts, dissolved in guanidine hydrochloride at 7 mol/l in Tris-HCl buffer (0.5 mol/l, pH 8.5), reduced with DTT at 10 mmol/l for 30 min at room temperature and alkylated with iodoacetamide at 55 mmol/l for 1 h at room temperature. The resulting protein solution was digested by addition of gold grade trypsin (Promega) at 10 ng/μl and overnight incubation at 37ºC. Peptide samples were analyzed by nanoflow high performace liquid chromatography–microelectrospray ionization on an LTQ mass spectrometer (Thermo Fisher Scientific, Waltham, MA).
Fluorescence immunocytochemistry
HUVECs and COS7 cells grown on glass base dishes were fixed in 4% (wt/vol) paraformaldehyde, permeabilized with PBS containing 0.05% (vol/vol) Triton X-100, incubated with antibodies, and then stained with Alexa 488/594-conjugated goat anti–mouse or anti–rabbit immunoglobulin (Ig) G (Invitrogen). After washing in PBS, fluorescence was visualized with a confocal laser scanning microscope (FLUOVIEW FV500; Olympus, Tokyo, Japan).
Apoptosis assay
HUVECs or COS7 cells transfected with either control or Gipie-V5 were treated with 1 mmol/l TG (Sigma) for 8 h, and the induction of apoptosis was analyzed by Alexa 488–conjugated annexin V (Invitrogen) according to the protocol provided by the manufacturer. In the assay, annexin V, a calcium-dependent phospholipid-binding protein with a high affinity for phosphatidylserine, was used to detect early-stage apoptosis. The nuclei of the cells were detected by staining with the DNA fluorochrome 4′-6-diamidine-2-phenyl indole (DAPI). Determinations were made in three separate experiments, and the frequency of apoptosis per ∼100 cells was measured.
Pulse-chase assay
HaloTag interchangeable labeling technology (Promega) was used to assess the stability of IRE1 protein. IRE1β cDNA was introduced into the pFc14K HaloTag CMV Flexi vector (Promega). COS7 cells transfected with IRE1β-HaloTag vector and either Gipie-V5 vector or V5 empty vector were pulse-labeled with HaloTag TMR (tetramethylrhodamine) ligand at 5 μmol/l (Promega) for 10 min. After washing three times with PBS at 37ºC, cells were chased in DMEM containing 8% FBS in the presence or absence of 10 μM MG132 for the indicated time. Cells were lysed in SDS sample buffer and subjected to SDS–PAGE. The HaloTag TMR ligand-labeled IRE1β was visualized with a fluorescence image analyzer (LAS 4010; GE Healthcare UK, Little Chalfont, UK).
Carotid balloon injury in rats
Six-month-old male Wistar rats were maintained on a 12-h light/dark cycle at 24ºC. All procedures were performed according to protocols approved by the Institutional Committee for the Care and Use of Laboratory Animals, Nagoya University School of Medicine. Rats underwent anesthesia with thiopental, and carotid injury was generated by a 2 French Fogarty balloon catheter. The catheter was inserted through an incision made in the external carotid artery and advanced along the length of the carotid artery to the aortic arch. The balloon was inflated and passed three times. Then the balloon catheter was removed, and the external carotid artery was permanently ligated. Six weeks after balloon injury, animals were killed and the carotid was collected free of adventitia. For immunohistological analysis, tissue fragments were fixed in 4% buffered formaldehyde, embedded in paraffin. The sections were stained with anti-Gipie, anti-KDEL, anti–α-SMA, or anti–PECAM-1 antibodies and analyzed by confocal laser scanning microscopy. In these experiments, tissue sections of the carotid arteries from three rats before and after the balloon injury were evaluated, and representative images from three independent immunohistological experiments are shown. For the measurements of protein levels of Gipie, GRP78, and CHOP in total extracts from the carotid artery, the arteries were excised and immediately homogenized in ice-cold buffer (HEPES at 5 mmol/l, pH 7.9, containing 26% [vol/vol] glycerol, MgCl2 at 1.5 mmol/l, EDTA at 0.2 mmol/l, DTT at 0.5 mmol/l, phenylmethylsulfonyl fluoride at 0.5 mmol/l, NaCl at 300 mmol/l) and incubated on ice for 30 min. After centrifugation at 100,000 g at 4ºC for 20 min, the supernatant was mixed with SDS sample buffer and subjected to Western blot analysis.
Data analysis
Data are presented as the means ± SE. Statistical significance was evaluated with Student’s t test.
Supplementary Material
Acknowledgments
We thank M. Amano, T. Nishioka, Y. Yamakawa, and K. Taki (Nagoya University) for helpful instruction in the mass spectrometric analysis; S. Ishigaki (Nagoya University) for helpful discussions; and K. Ushida for technical assistance. This work was supported by Grants-in-Aid for Global Center of Excellence (GCOE) Research, Scientific Research (A), and Scientific Research on Innovative Areas (to M.T.), a Grant-in-Aid for Exploratory Research (to N.A.), and Program for Improvement of Research Environment for Young Researchers from Special Coordination Funds for Promoting Science and Technology (SCF) (to A.E.) commissioned by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Abbreviations used:
- ASK1
apoptosis signal-regulating kinase 1
- ATF6
activating transcription factor 6
- BFA
brefeldin A
- CHOP
C/EBP homologous protein
- DAPI
DNA fluorochrome 4′-6-diamidine-2-phenyl indole
- Daple
dishevelled-associating protein with a high frequency of leucine residues
- DTT
dithiothreitol
- eIF2α
eukaryotic initiation factor 2α
- ER
endoplasmic reticulum
- EST
expressed sequence tag
- Gipie
GRP78-interacting protein induced by ER stress
- GRP78
78 kDa glucose-regulated protein
- HUVECs
human umbilical vein endothelial cells
- IRE1
inositol-requiring protein 1
- JNK
c-Jun N-terminal kinase
- LC–MS/MS
liquid chromatography followed by tandem mass spectrometry
- mAb
monoclonal antibody
- PDI
protein disulfide isomerase
- PERK
PKR-like ER kinase
- PKR
protein kinase regulated by RNA
- RT-PCR
reverse transcription-PCR
- siRNA
small interfering RNA
- SMA
smooth muscle actin
- TG
thapsigargin
- TRAF2
tumor necrosis factor receptor–associated factor 2
- UPR
unfolded protein response
- XBP-1
X-box binding protein 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0724) on February 2, 2011.
REFERENCES
- Bhattacharjee G, Ahamed J, Pedersen B, El-Sheikh A, Mackman N, Ruf W, Liu C, Edgington TS. Regulation of tissue factor-mediated initiation of the coagulation cascade by cell surface Grp78. Arterioscler Thromb Vasc Biol. 2005;25:1737–1743. doi: 10.1161/01.ATV.0000173419.31242.56. [DOI] [PubMed] [Google Scholar]
- Bonetti PO, Lerman LO, Larman A. Endothelial dysfunction: a marker of atherosclerotic risk. Arterioscler Thromb Vasc Biol. 2002;22:1065–1074. doi: 10.1161/01.atv.0000051384.43104.fc. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding PH, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Clowes AW, Reidy MA, Clowes MM. Mechanisms of stenosis after arterial injury. Lab Invest. 1983;49:208–215. [PubMed] [Google Scholar]
- Ellgaard L, Helenius AM. Quality control in the endoplasmic reticulum. Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Murakami H, Asai N, Morone N, Watanabe T, Kawai K, Murakumo Y, Usukura J, Kaibuchi K, Takahashi M. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev Cell. 2005;9:389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Jiang P, Takahashi M. Girdin, a novel actin-binding protein, and its family of proteins possess versatile function in the Akt and Wnt signaling pathways. Ann NY Acad Sci. 2006;1086:169–184. doi: 10.1196/annals.1377.016. [DOI] [PubMed] [Google Scholar]
- Enomoto A, et al. Roles of disrupted-In-schizophrenia 1-interacting protein Girdin in postnatal development of the dentate gyrus. Neuron. 2009;63:774–787. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Feaver RE, Hastings NE, Pryor A, Blackman BR. GRP78 Upregulation by atheroprone shear stress via p38-, α2β1-dependent mechanism in endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:1534–1541. doi: 10.1161/ATVBAHA.108.167999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Lee S-M, Chen A, Zhang J, Zhang DD, Kannan K, Ortmann RA, Fang D. Synoviolin promotes IRE1 ubiquitination and degradation in synovial fibroblasts from mice with collagen-induced arthritis. EMBO Rep. 2008;9:480–485. doi: 10.1038/embor.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by protein in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius AM, Schumacker PT. Endothelial responses to mechanical stress: where is the mechanosensor? Crit Care Med. 2002;30:198–206. doi: 10.1097/00003246-200205001-00005. [DOI] [PubMed] [Google Scholar]
- Hendershot LM. The ER chaperone Bip is a master regulator of ER function. Mt Sinai J Med. 2004;5:289–297. [PubMed] [Google Scholar]
- Jiang P, Enomoto A, Jijiwa M, Kato T, Hasegawa T, Ishida M, Sato T, Asai N, Murakumo Y, Takahashi M. An actin-binding protein Girdin regulates the motility of breast cancer cells. Cancer Res. 2008;68:1310–1318. doi: 10.1158/0008-5472.CAN-07-5111. [DOI] [PubMed] [Google Scholar]
- Jiang P, Enomoto A, Takahashi M. Cell biology of the movement of breast cancer cells: Intracellular signaling and the actin cytoskeleton. Cancer Lett. 2009;284:122–130. doi: 10.1016/j.canlet.2009.02.034. [DOI] [PubMed] [Google Scholar]
- Kitamura T, et al. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nat Cell Biol. 2008;10:329–337. doi: 10.1038/ncb1695. [DOI] [PubMed] [Google Scholar]
- Lee AS. Mammalian stress response: induction of the glucose-regulated protein family. Curr Opin Cell Biol. 1992;4:267–273. doi: 10.1016/0955-0674(92)90042-b. [DOI] [PubMed] [Google Scholar]
- Lee AS. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci. 2001;8:504–510. doi: 10.1016/s0968-0004(01)01908-9. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Donaldson LG, Schweizer A, Berger EG, Hauri H-P, Yuan LC, Klausner RD. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- Liu H, Bowes III RC, van de Water B, Sillence C, Nagelkerke JF, Stevens JL. Endoplasmic reticulum chaperones GRP78 and calreticulin prevent oxidative stress, Ca2+ disturbances, and cell death in renal epithelial cells. J Biol Chem. 1997;272:21751–21759. doi: 10.1074/jbc.272.35.21751. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP increase death by promoting protein synthesis and oxidation in the stressed endothelial reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numaguchi Y, et al. Prostacyclin synthase gene transfer accelerates reendothelialization and inhibits neointimal formation in rat carotid arteries after balloon injury. Arterioscler Thromb Vasc Biol. 1999;19:727–733. doi: 10.1161/01.atv.19.3.727. [DOI] [PubMed] [Google Scholar]
- Römisch K. Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Schröder M, Kaufman RJ. ER stress and the unfolded protein response. Mutant Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Schröder M, Kaufman RJ. Divergent roles of IRE1α and PERK in the unfolded protein response. Curr Mol Med. 2006;6:5–36. doi: 10.2174/156652406775574569. [DOI] [PubMed] [Google Scholar]
- Shen X, Zhang K, Kaufman RJ. The unfolded protein response—a stress pathway of the endoplasmic reticulum. J Chem Neuroanat. 2004;28:79–92. doi: 10.1016/j.jchemneu.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Shimokawa H. Primary endothelial dysfunction: atherosclerosis. J Mol Cell Cardio. 1999;31:23–37. doi: 10.1006/jmcc.1998.0841. [DOI] [PubMed] [Google Scholar]
- Shiu RP, Pouyssegur J, Pastan I. Glucose dependent accounts for the induction of two transformation-sensitive membrane proteins in Rous sarcoma virus-transformed chick embryo fibroblasts. Proc Natl Acad Sci USA. 1977;79:3840–3844. doi: 10.1073/pnas.74.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyy JY-J, Chien S. Role of integrins in endothelial mechanosensing of shear stress. Circ Res. 2002;91:769–775. doi: 10.1161/01.res.0000038487.19924.18. [DOI] [PubMed] [Google Scholar]
- Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang XZ, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of jnk protein kinases by transmembrane protein kinase IRe1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- VanderLaan PA, Reardon CA, Gets GS. Site specificity of atherosclerosis site-selective response to atherosclerotic modulators. Arterioscler Thromb Vasc Biol. 2004;24:12–22. doi: 10.1161/01.ATV.0000105054.43931.f0. [DOI] [PubMed] [Google Scholar]
- Wang XZ, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP(GADD153) by p38 MAP kinase. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, et al. Association of multiple cellular stress pathways with accelerated atherosclerosis in hyperhomocysteinemic apolipoprotein E-deficient Mice. Circulation. 2004;110:207–213. doi: 10.1161/01.CIR.0000134487.51510.97. [DOI] [PubMed] [Google Scholar]
- Zhou J, Lhotak S, Hilditch BA, Austin RC. Activation of the unfolded protein response occurs at all stages of atherosclerotic lesion development in apolipoprotein E-deficient mice. Circulation. 2005;111:1814–1829. doi: 10.1161/01.CIR.0000160864.31351.C1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.