Abstract
Binding of different regulatory subunits and methylation of the catalytic (C) subunit carboxy-terminal leucine 309 are two important mechanisms by which protein phosphatase 2A (PP2A) can be regulated. In this study, both genetic and biochemical approaches were used to investigate regulation of regulatory subunit binding by C subunit methylation. Monoclonal antibodies selectively recognizing unmethylated C subunit were used to quantitate the methylation status of wild-type and mutant C subunits. Analysis of 13 C subunit mutants showed that both carboxy-terminal and active site residues are important for maintaining methylation in vivo. Severe impairment of methylation invariably led to a dramatic decrease in Bα subunit binding but not of striatin, SG2NA, or polyomavirus middle tumor antigen (MT) binding. In fact, most unmethylated C subunit mutants showed enhanced binding to striatin and SG2NA. Certain carboxy-terminal mutations decreased Bα subunit binding without greatly affecting methylation, indicating that Bα subunit binding is not required for a high steady-state level of C subunit methylation. Demethylation of PP2A in cell lysates with recombinant PP2A methylesterase greatly decreased the amount of C subunit that could be coimmunoprecipitated via the Bα subunit but not the amount that could be coimmunoprecipitated with Aα subunit or MT. When C subunit methylation levels were greatly reduced in vivo, Bα subunits were found complexed exclusively to methylated C subunits, whereas striatin and SG2NA in the same cells bound both methylated and unmethylated C subunits. Thus, C subunit methylation is critical for assembly of PP2A heterotrimers containing Bα subunit but not for formation of heterotrimers containing MT, striatin, or SG2NA. These findings suggest that methylation may be able to selectively regulate the association of certain regulatory subunits with the A/C heterodimer.
INTRODUCTION
Although it is well established that phosphorylation of a single amino acid can regulate protein-protein interactions and enzyme activities in eukaryotes, the effect of methylation of a single residue is less well understood. Irreversible methylation on multiple arginines has been shown to inhibit certain protein-protein interactions (Bedford et al., 2000) and to affect cellular processes such as nuclear export (Shen et al., 1998) and transcription (Chen et al., 1999). However, mutational studies that would establish the role of a single arginine methylation have not been carried out to our knowledge. Even less is known about the role of carboxy methylation in eukaryotic systems. In the best understood case, neutralization of the processed CAAX carboxy terminus of Ras by a single carboxy methylation appears to promote hydrophobic interactions with membrane lipids (Farh et al., 1995), enhancing the localization to the plasma membrane (Hrycyna et al., 1991; Bergo et al., 2000). However, the effect of G protein carboxy methylation on protein-protein interactions is not well understood. The catalytic subunit of protein phosphatase 2A (PP2A) is also known to be carboxy methylated on its C-terminal residue (Rundell, 1987; Lee and Stock, 1993; Favre et al., 1994; Li and Damuni, 1994; Xie and Clarke, 1994), although the function of this methylation is presently unclear. PP2A methylation is reversible and PP2A methylesterase (Ogris et al., 1999a) and methyltransferase enzymes (De Baere et al., 1999) have been cloned, thus providing an interesting model system amenable to the study of the potential regulatory effects of reversible protein methylation.
PP2A is a major eukaryotic protein phosphatase that is highly conserved and is known to be involved in the regulation of multiple cellular events (reviewed in Cohen, 1989; Mumby and Walter, 1993; Hopkin, 1995), including transcription, translation, DNA replication, development, neuronal signaling, cell cycle progression, and cell division. To allow PP2A to function independently in these diverse roles, a variety of regulatory subunits exist that localize PP2A to specific cellular microenvironments and/or signaling pathways and modulate PP2A activity (for examples, see Cohen, 1989; Murata et al., 1997; Moreno et al., 2000). Although some regulatory subunits (e.g., alpha4; Murata et al., 1997) appear to bind and regulate only the PP2A catalytic subunit, most of these subunits associate with and regulate a heterodimeric A/C form of PP2A that consists of a 36-kDa catalytic (C) subunit and a 63-kDa constant regulatory (A) subunit (Usui et al., 1988).
The regulatory subunits that bind the A/C heterodimer can be divided into several families. The best characterized are the three major families of B-type subunits, B (or B55), B′ (or B56), and B′′ (or PR72/130). In addition, we recently described two members of a new family of PP2A regulatory subunits that bind the A/C heterodimer, striatin and SG2NA (Moreno et al., 2000). Moreover, a family of papovavirus PP2A regulatory subunits exists that includes simian virus 40 and polyomavirus small tumor antigens and polyomavirus middle tumor antigen (MT) (Pallas et al., 1990). In all these cases, the PP2A core A/C heterodimer is found complexed with the regulatory subunit, and the substrate specificity and activity of PP2A are modulated (Yang et al., 1991; Cayla et al., 1993; Cegielska et al., 1994; Kamibayashi et al., 1994; Mayer-Jaekel et al., 1994; Ogris et al., 1997; Moreno et al., 2000).
Little is known about how the association of the A/C heterodimer with the various regulatory subunits might be modulated. Differential expression of the various cellular regulatory subunits during development and differentiation may account for one level of regulation (for review, see (Mumby and Walter, 1993), but some evidence suggests the existence of a more dynamic means of altering PP2A complex composition (Chen et al., 1992; Ogris et al., 1997; Zhu et al., 1997). For example, tyrosine 307 of the catalytic subunit can be phosphorylated in vitro by pp60c-src, resulting in 90% inhibition of PP2A activity (Chen et al., 1992). Substitution of this same tyrosine with an acidic residue abolishes binding in vivo of the A/C heterodimer to B subunit, but not to MT (Ogris et al., 1997), suggesting that covalent modification of the C subunit carboxy terminus might selectively regulate association of certain regulatory subunits.
PP2A C subunit methylation has been shown to occur both in vivo and in cell lysates (Rundell, 1987; Lee and Stock, 1993; Favre et al., 1994; Li and Damuni, 1994; Xie and Clarke, 1994). The effect of methylation on PP2A activity is controversial, with some studies indicating up to a twofold increase in specific activity (Favre et al., 1994; Kowluru et al., 1996) and another finding no effect (De Baere et al., 1999). Although C subunit methylation appears to occur in vivo in a cell cycle-regulated manner (Floer and Stock, 1994; Turowski et al., 1995), the molecular mechanisms controlling such changes and the PP2A function(s) affected are not yet understood. In addition, those residues in the C subunit important for methylation and demethylation remain to be determined. Peptide substrate studies may not be helpful in their identification because earlier studies have shown that synthetic C subunit carboxy-terminal peptides functioned neither as substrates nor inhibitors of the PP2A methyltransferase (Xie and Clarke, 1994) and PP2A methylesterase (Lee et al., 1996).
The carboxy-terminal region of the PP2A C subunit seems to be a focal point for regulation of PP2A. In addition to containing the amino acids identified as the sites of tyrosine phosphorylation and methylation, this region contains residues essential for stable binding of the B regulatory subunit (Ogris et al., 1997). Deletion of the nine highly conserved C-subunit carboxy-terminal residues (amino acids 301–309) or nonconservative substitution of threonine 304 and tyrosine 307 abolished the C subunit's ability to form complexes with B subunit. However, these mutants could still form heterotrimers containing the viral regulatory subunit, MT (Ogris et al., 1997). Based on this finding, we proposed that covalent modification of the C subunit carboxy terminus might selectively regulate the association of certain PP2A regulatory subunits (Ogris et al., 1997). In this report we present four lines of evidence that methylation of leucine 309 is required for the association of the A/C heterodimer with Bα subunit but not with certain other cellular and viral PP2A regulatory subunits. Two genetic approaches used point mutants and deletion of leucine 309 to observe that loss of methylation results in loss of Bα subunit association but not of MT, SG2NA, or striatin. Two biochemical approaches decreased methylation of C subunit in vitro and in vivo to determine the effect on association of these regulatory subunits. In each case, methylation was essential for binding of Bα subunit but not for MT, SG2NA, or striatin. These results suggest that reversible carboxy methylation may be a mechanism for selectively regulating the formation of certain PP2A heterotrimers.
MATERIALS AND METHODS
Plasmids
Wild-type (wt) PP2A Aα and Bα subunit cDNAs (Hemmings et al., 1990; Pallas et al., 1992) and a cDNA encoding a mutant PP2A C subunit lacking the carboxy-terminal leucine were individually cloned into a dexamethasone-inducible vector, pGRE 5-2 (Mader and White, 1993), to try to minimize the potential deleterious effects of overexpression (if any) while the lines were being carried in culture. To accomplish this, wt PP2A Aα and Bα subunit cDNAs with an NcoI site containing the start ATG were first cloned into pcDNA I Amp (Invitrogen, San Diego, CA), together with a double-stranded oligonucleotide, which introduced at the 5′-end the coding sequence for a nine-amino acid peptide (TyrProTyrAspValProAspTyrAla) from influenza hemagglutinin (HA), followed by the thrombin recognition site (LeuValProArgGlySer). Then, the Aα and Bα subunit cDNAs, including the epitope tag sequence, were cloned into pGRE 5-2. Beginning with a previously described pGRE5-2 construct carrying epitope-tagged wt C subunit (Ogris et al., 1997), deletion of the C subunit carboxy-terminal leucine was performed by standard cloning techniques. This mutant cDNA, including the epitope tag sequence, was then cloned into pGRE 5-2.
Based on sequence homology to related crystallized phosphatases, four residues, which were predicted to be in the catalytic site of mammalian PP2A (histidine 59, aspartic acid 85, arginine 89, and histidine 118; Table 1) were individually mutated as described by Ogris et al. (1999a,b). PP2A activity was undetectable, confirming that these residues are indeed important for PP2A catalysis.
Table 1.
C subunit mutants used in this study
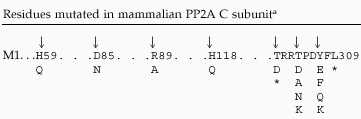 |
C subunit mutants are described in the text. Generally, the mutants are referred to by the position mutated preceded by the wt amino acid and followed by the introduced residue. Abbreviations for the amino acid residues are: A, Ala; D, Asp; E, Glu; F, Phe; H, His; K, Lys; L, Leu; N, Asn; Q, Gln; R, Arg; T, Thr; and Y, Tyr. Arrows indicate changed residues. Single amino acid substitutions introduced into the mammalian C subunit are shown below the corresponding amino acid of the wt sequence. The asterisk (*) indicates the replacement of a residue with a stop codon.
Production of Monoclonal Antibodies Sensitive to the C Subunit Methylation State
A 15-residue unmethylated PP2A C subunit carboxy-terminal peptide was conjugated to keyhole limpet hemocyanin via an added amino-terminal cysteine residue using a Imject conjugation kit (Pierce, Rockford, IL) . BALB/c female mice were immunized, boosted on d 7, 14, 21, and 28, and then euthanized by rapid asphyxiation on d 31. Splenocytes were fused to SP2/0 murine myeloma cells by using polyethylene glycol and standard techniques. The cells were then washed and plated into microtiter wells, which were screened after 10–14 d using an enzyme-linked immunosorbent assay in which wells were coated with the unconjugated peptide. A secondary screen was performed by immunoblotting NIH3T3 cell lysates, and positive clones were single-cell cloned by limiting dilution. Three monoclonal antibodies specific for the PP2A C subunit, 1d6, 4b7, and 4e1, were obtained.
Cells and Cell Culture
MT-transformed NIH3T3 cell lines expressing HA epitope-tagged wt or mutant PP2A C subunits (36wt [wt], 301stop, T301D, T304D, T304A, T304N, T304K, Y307E, Y307F, Y307Q, and Y307K) or empty vector (GREonly) were described previously (Ogris et al., 1997). Only ∼10% of PP2A C subunits are bound to MT in these cells (Haehnel and Pallas, unpublished data). These lines were maintained in DMEM/10% calf serum containing 150 μg/ml hygromycin B and 200 μg/ml geneticin. To generate cell lines individually expressing HA-tagged wt PP2A Bα subunit, wt PP2A Aα subunit, or C subunit lacking leucine 309 (L309Δ), NIH3T3 cells (MT-transformed in the case of L309Δ) were transfected with pGRE 5-2 vectors expressing the appropriate HA epitope-tagged proteins by the calcium phosphate precipitation method (Sambrook et al., 1989). Vector (20 μg) encoding the HA-tagged B, A, or mutant C subunit were cotransfected with 2 μg of a plasmid conferring resistance to hygromycin B. Selection medium contained 300 μg/ml hygromycin B. In each case, hundreds of individual clones were pooled and used for the experiments described herein. These lines were maintained in DMEM/10% calf serum containing 150 μg/ml hygromycin B and, in the case of L309Δ, 200 μg/ml geneticin as well. Although the inducible vector, pGRE5-2, was used to express these proteins, levels of HA-tagged Bα subunit, Aα subunit, and L309Δ proteins were substantial in the absence of dexamethasone. A similar result was found previously when the same vector was used to express wt and mutant PP2A C subunits in NIH3T3 cells (Ogris et al., 1997). However, dexamethasone treatment was used in most cases to obtain maximal expression. Control experiments showed that the dexamethasone treatment had no effect on PP2A methylation (data not shown).
Use of Methylation-sensitive C Subunit Monoclonal Antibodies to Assay PP2A Methylation
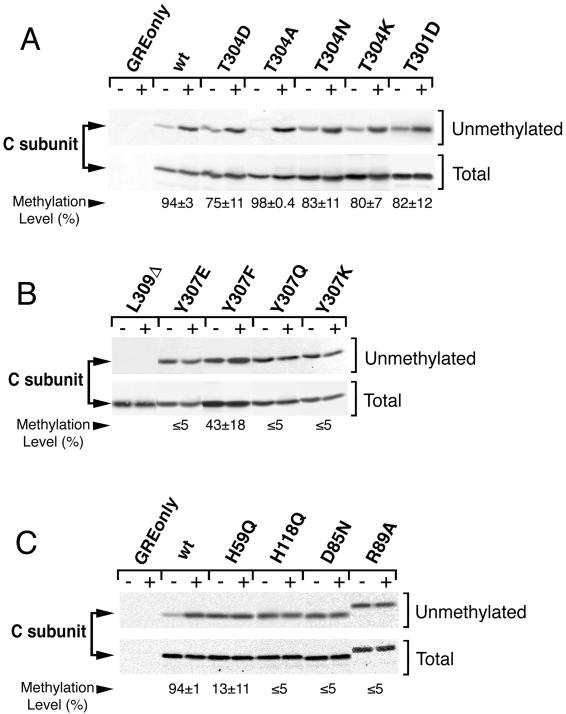
The primary methylation assay used in this study involved the use of methylation-sensitive antibodies to evaluate PP2A methylation (Turowski et al., 1995). It assays the steady-state methylation level of C subunit directly (labeling of PP2A methyl groups to steady-state may take many hours in a radioactive assay). In addition, this assay is not affected by variability in uptake of label between different cell lines, differences in turnover of the methyl group between different mutants, or effects of the inhibitor treatments necessary to prevent label incorporation into protein during the in vivo assay. The details of this assay are presented in the legend to Figure 3. A small, but consistent, decrease in signal was seen in the base-treated immunoprecipitate lanes using a methylation-insensitive anti-PP2A antibody (Transduction Laboratories, Lexington, KY) to probe for C subunit, indicating that there was some loss of C subunit epitopes during base treatment. An equivalent decrease was seen when mutants known to be unmethylated (as assayed by in vivo labeling) were blotted with 4b7. Quantitation of the nonspecific signal decrease caused by base treatment by using methylation-insenstive C subunit antibody facilitated correction for this nonspecific loss of C subunit signal (e.g., for data shown in Figure 3 the average correction for loss of C subunit signal was 11.5 ± 6.6%).
Figure 3.
Mutation of either carboxy terminal or catalytic site residues can dramatically reduce methylation. (A, B, and C) Steady-state in vivo methylation levels of wt and mutant C subunits. NIH3T3 cell lines expressing vector only, HA-tagged wt C subunit (wt), or the indicated HA-tagged mutant (see RESULTS and Table 1 for mutant definitions) were lysed as described by Moreno et al. (2000), except that lysis buffer contained 250 nM okadaic acid to prevent further methylation and demethylation (Li and Damuni, 1994). Antitag immunoprecipitates were prepared, and half of each immunoprecipitate (+) was demethylated by a brief base treatment (5-min incubation with 0.1 M NaOH at 4°C), followed by neutralization with an equivalent volume of HCl and 0.5 volume of 1 M Tris, pH 6.8. The untreated half of each immunoprecipitate (−) was combined with the same amount of preneutralized base solution as a control. After 10% SDS-PAGE, samples were sequentially immunobloted with the methylation-sensitive 4b7 monoclonal antibody (unmethylated) and a methylation-insensitive antibody (Total). The fraction (x) of unmethylated C subunit in the original sample is defined as the ratio (untreated to base treated) of C subunit signal quantitated with a Bio-Rad Fluor S-Max chemilumimager. The chemilumimager directly measures band intensities without the use of film via a supercooled charge-coupled device camera and provides quantitative data that is linear over 4.8 orders of magnitude. All measurements were well within this range. This method yielded highly reproducible results that did not vary with image capture times. The fraction (x) of unmethylated C subunit was used to calculate the steady-state in vivo methylation level (% = (100 × [1 − x]) for each mutant. The calculated % methylation ± SD is shown below each lane. All lanes in each panel are from the same experiment and were analyzed on the same gel, but different exposures of some pairs (−/+) of lanes are shown to facilitate visual comparison for the reader. R89A migrates slower on SDS-PAGE because of the mutation introduced; careful sequencing of the mutant cDNA revealed no other mutation (Ogris et al., 1999b). All mutants were analyzed in at least three separate experiments. The C subunits migrate as singlets in all experiments shown; whether double or single bands are seen can vary (see details in legend to Figure 1).
This assay can also be used to determine the methylation level of mutant C subunits in which the mutation partially impairs the binding of the methylation-sensitive antibody (although most in this study do not). The reason that such a mutant can still be accurately analyzed by this assay is that percentage methylation is determined by normalizing the 4b7 signal on an untreated sample with the 4b7 signal of an equivalent, unmethylated (base-treated) sample. This normalization corrects for any decrease in 4b7 signal due to mutation. The only problematic effect of a mutation would be 1) if 4b7 does not recognize a mutant at all (not a problem for the mutants we analyzed) or 2) if the mutation somehow strengthened the binding of 4b7 so that it bound even when the mutant was methylated. In the latter case, such mutants might appear as unmethylated or partially methylated when, in fact, they are fully methylated. For this reason, we have analyzed every mutant that appeared only partially methylated or unmethylated (see Figure 4) by a radioactive labeling assay to be sure that this is not the case in the present study.
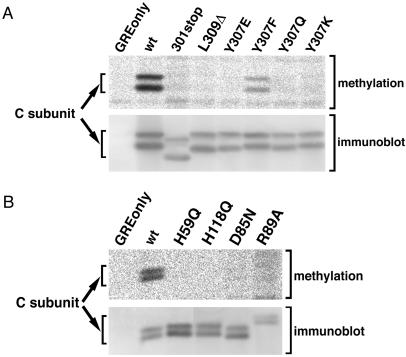
Figure 4.
Confirmation of the methylation-deficient status of C subunit mutants by an in vivo radiolabeling methylation assay. (A and B) In vivo 3H-methylation assays were performed as described in MATERIALS AND METHODS on the indicated C subunits (see RESULTS and Table 1 for mutant definitions). Phosphorimager (methylation) and fluorimager (immunoblot) images are shown. Similar results were obtained in at least two independent experiments. The C subunits migrate as doublets in these gels, but whether double or single bands are seen can vary (see details in legend to Figure 1). All lanes in each panel are from the same experiment and were analyzed on the same gel but were not all originally adjacent.
In Vivo Methylation Assay
The in vivo methylation assay was used as a backup method. Approximately 80% confluent dishes of cells were washed twice with DMEM lacking l-methionine and incubated 1 h in DMEM lacking l-methionine supplemented with 5% dialyzed fetal calf serum, 5 μm cycloheximide (Sigma, St. Louis, MO), and 30 μg/ml puromycin (Sigma). Then, l-[methyl-3H]methionine (Amersham Pharmacia Biotech, Piscataway, NJ) was added to a final concentration of 100 μCi/ml. Preincubation of the cells with 5 μM cycloheximide and 30 μg/ml puromycin prevents translational incorporation of subsequently added radiolabeled methionine into newly synthesized proteins (Favre et al., 1994; Kowluru et al., 1996); the fact that no radioactivity was detected in certain PP2A mutants indicates that protein synthesis was indeed completely inhibited. After labeling for 5 h, the cells were washed, scraped into 1 ml of ice-cold wash buffer, and centrifuged at 3000 × g for 1 min. The supernatant was discarded, and the cells were lysed at 4°C for 3 min in 55 mM Tris, pH 8.0, 55 mM NaCl, 0.2 mM DTT, 1.0 mM CaCl2, 1.0 mM MgCl2, and 0.55% Nonidet P-40 (Rundell, 1987) containing okadaic acid (100 nM final concentration). This concentration of okadaic acid prevents the loss of already incorporated radiolabeled methyl groups during subsequent immunoprecipitation by inhibiting the PP2A methylesterase (Lee et al., 1996). The lysates were cleared at 13,000 × g for 5 min, and the resulting supernatants were immunoprecipitated. Immunoprecipitates were analyzed by SDS-PAGE, proteins were transferred to nitrocellulose, and 3H-methyl incorporation and amount of C subunit protein were, respectively, visualized by a phosphorimager (Fuji, Tokyo, Japan) and a fluorimager (STORM, Molecular Dynamics, Sunnyvale, CA). For immunoblotting, methylation-insensitive mouse monoclonal anti-HA tag antibody (16B12; BAbCO, Richmond, CA) or anti-PP2A C subunit antibody (Transduction Laboratories), alkaline phosphatase-conjugated secondary antibody (Promega, Madison, WI), and Attophos substrate (JBL Scientific, Northridge, CA) were used.
In Vitro Demethylation with PP2A Methylesterase (PME-1)
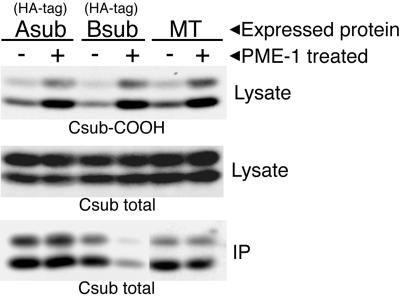
Two 15-cm dishes of each cell line expressing HA-tagged Aα or Bα subunits or MT were lysed as described by Moreno et al. (2000), except that the lysis buffer contained 55 mM Tris (pH 8.0), 55 mM NaCl, 0.5 mM dithiothreitol, 1 mM CaCl2, 1 mM MgCl2, and 0.55% Nonidet P-40. The cleared lysates were divided into two equal portions. PME-1 (2–16 μg/reaction, depending on the activity of the preparation), purified from bacteria expressing recombinant 6× His-tagged PME-1, was added to one portion of each lysate to demethylate C subunit and was incubated for 1–2 h at 31°C. Lysates were then further analyzed, as described in the legend to Figure 6.
Figure 6.
Addition of exogenous PME-1 to cell lysates demethylates C subunit and causes Bα subunit, but not MT or Aα subunit, to dissociate from C subunit. Cell lysates prepared from lines expressing HA-tagged Aα or Bα subunits or MT were divided into two equal portions. PME-1 was added to one portion of each lysate to demethylate C subunit as described in MATERIALS AND METHODS. A small amount of each lysate portion was analyzed by 10% SDS-PAGE and immunobloted with methylation-sensitive antibody (4b7; top) to show that C subunit had been highly demethylated in the PME-1-treated lysates (+ lanes) relative to the controls (− lanes). Subsequent reprobing of the lysates with a methylation-insensitive C subunit antibody (middle) shows that an equivalent total amount of C subunit was present in each pair of lanes. Epitope-tagged Aα or Bα subunit or MT was immunoprecipitated from the remainder of the lysates and the immunoprecipitates (IP) were analyzed by 10% SDS-PAGE and immunoblotted with a methylation-insensitive C subunit antibody (bottom) to determine the amount of C subunit coimmunoprecipitated in each case. The amount of C subunit coimmunoprecipitated with HA-tagged Bα was consistently decreased by ≥ 80% in lysates demethylated by PME-1. All lanes in each panel are from the same experiment and were analyzed on the same gel, but a longer exposure of the pair (−/+) of MT lanes in the bottom panel are shown to facilitate visual comparison for the reader. The C subunits migrated as doublets in these gels, but whether double or single bands are seen can vary (see details in legend to Figure 1).
Immunoprecipitations
Immunoprecipitation of PP2A complexes via HA-tagged wt or mutant PP2A A, B, and C subunits or via MT, striatin, or SG2NA was performed using anti-HA tag monoclonal antibody (12CA5; obtained from BAbCO, but now carried by Boehringer Mannheim, Indianapolis, IN) or anti-MT, striatin, or SG2NA polyclonal antibodies plus protein A-Sepharose beads (Pharmacia, Piscataway, NJ) (Moreno et al., 2000; Pallas et al., 1986). In some cases, covalently cross-linked 12CA5 antibody/bead complexes were used to reduce background at the position of Bα subunit and MT.
RESULTS
Association of Regulatory Subunits with a PP2A C Subunit Mutant Lacking Leucine 309
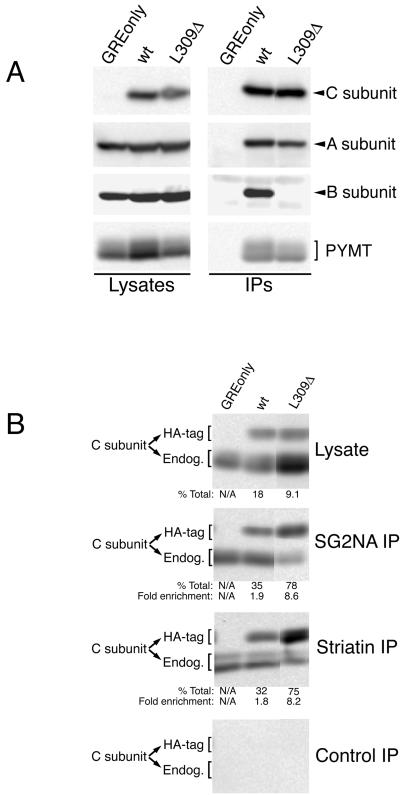
To investigate the possibility that methylation of C subunit leucine 309 might selectively regulate the association of certain regulatory subunits, we assayed whether deletion of this residue would have a different effect on the association of Bα subunit, MT, striatin, and SG2NA. Anti-HA tag immunoprecipitates were prepared from MT-transformed NIH3T3 cell lines individually expressing either HA-tagged wt C subunit (wt) or HA-tagged mutant C subunit lacking only leucine 309 (L309Δ), and the immunoprecipitates were then probed for the presence of A and Bα subunits and MT by immunoblotting (Figure 1A). L309Δ associated with substantial A subunit and MT, indicating that A/C/MT heterotrimers can form independently of this residue. However, even on long exposures, no Bα subunit could be seen in the L309Δ immunoprecipitate, indicating that loss of leucine 309 abrogates Bα subunit binding.
Figure 1.
Deletion of leucine 309 abolishes Bα subunit binding but not binding of MT, SG2NA, or striatin. (A) Lysates of NIH3T3 cells stably expressing MT antigen (PYMT) and HA-tagged wt C subunit, HA-tagged L309Δ C subunit, or empty vector (GREonly) were used to prepare anti-HA tag immunoprecipitates. Lysates and immunoprecipitates (IPs) were analyzed on the same 10% SDS-polyacrylamide gel and probed sequentially for epitope-tagged C subunit, A subunit, B subunit, and MT. The light bands above and below the position of B subunit in the immunoprecipitate lanes are antibody background bands. (B) Lysates of the same cell lines used in A were used to prepare SG2NA, striatin, and control immunoprecipitates (IPs) that were analyzed along with lysates by 10% SDS-PAGE and transferred to nitrocellulose. The membrane was probed with an anti-C subunit antibody (Transduction Laboratories) that recognizes both HA-tagged (HA-tag) and endogenous (Endog.) C subunits. Each band was quantitated using a Bio-Rad Fluor-S Max chemilumimager, which has a linear range of almost 5 orders of magnitude. The percentage of total C subunit that HA-tagged wt or L309Δ C subunit represent in lysates and in immunoprecipitates was calculated (% Total) and is shown beneath the respective panels. The ratio of % total in the immunoprecipitates to the % total in the respective lysates was calculated for each cell line as a measure of the efficiency of C subunit association with striatin or SG2NA and is indicated (Fold enrichment). The SDs for the fold enrichment values for SG2NA IPs were 0.4 and 1.7 for wt and L309Δ, respectively. For striatin IPs SDs were 0.4 and 1.1 for wt and L309Δ, respectively. All lanes in each panel were analyzed on the same gel but were not originally adjacent. The exposure times of the control, SG2NA, and striatin immunoprecipitate panels were equivalent. The C subunits migrate sometimes as singlets and sometimes as doublets; whether double or single bands are seen can vary for the same sample from gel to gel. This pattern of migration in SDS-PAGE has been noted previously for endogenous and epitope-tagged PP2A C subunits (Campbell et al., 1995; Turowski et al., 1995; Ogris et al., 1997) and does not appear to be due to degradation.
Epitope-tag immunoprecipitates of HA-tagged C subunit do not efficiently immunoprecipitate striatin and SG2NA (Yu, Du, Moreno and Pallas, unpublished data), presumably because binding of the epitope tag antibody is blocked. Therefore, to determine whether C subunit leucine 309 is important for striatin and SG2NA binding, striatin and SG2NA immunoprecipitates and lysates were probed for the presence of endogenous, HA-tagged wt, and L309Δ C subunits (Figure 1B). The amounts of these C subunits were quantitated using a Fluor S-Max chemilumimager (Bio-Rad, Richmond, CA), which directly measures band intensities without the use of film via a supercooled charge-coupled device camera that provides linear data over 4.8 orders of magnitude. This method yielded highly reproducible results that did not vary with image capture times. For both lysates and immunoprecipitates, the percentage of total C subunit that each HA-tagged C subunit constitutes was determined. The calculated percentage for each immunoprecipitate was divided by the calculated percentage for the corresponding lysate to determine the fold enrichment of wt and L309Δ in immune complexes. Addition of the HA-tag to wt C subunit increased its ability to compete with endogenous C subunit for binding to striatin and SG2NA approximately twofold. In contrast to the loss of binding observed for Bα subunit, L309Δ bound to striatin and SG2NA fourfold better than wt. Collectively, these results are consistent with the possibility that methylation of leucine 309 may selectively regulate binding of certain regulatory subunits.
Methylation-sensitive Monoclonal Antibody, 4b7, Recognizes Only Demethylated PP2A C Subunit
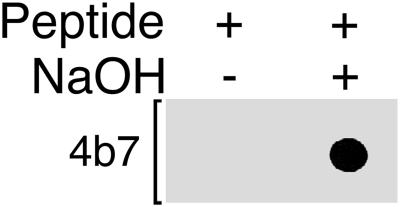
To develop a quantitative assay to measure the methylation levels of PP2A, monoclonal antibodies were raised to an unmethylated peptide corresponding to the last 15 amino acids of the catalytic subunit (see MATERIALS AND METHODS). To demonstrate the methylation sensitivity of these monoclonal antibodies, a methylated C subunit carboxy-terminal octomer peptide was synthesized and high-pressure liquid chromatography purified and used for a dot blot analysis. One aliquot of this peptide was demethylated by brief treatment with base and spotted onto nitrocellulose along side an equal amount of methylated peptide. Figure 2 shows the results of probing this membrane with 4b7, the antibody used here for methylation assays (see below). 4b7 strongly recognized the base-treated, unmethylated C subunit carboxy-terminal peptide but could not detect the methylated peptide, indicating that it is completely specific for unmethylated C subunit.
Figure 2.
Binding of the PP2A monoclonal antibody, 4b7, to the PP2A C subunit carboxy terminus is completely inhibited by methylation of leucine 309. A C subunit carboxy-terminal octopeptide carboxy methylated on leucine 309 was synthesized and high-pressure liquid chromatography purified. This peptide (0.25 μg) was demethylated by treatment with 0.1 N NaOH (+) for 5 min at 4°C and then neutralized while another 0.25 μg of peptide (−) was treated with an equivalent amount of preneutralized solution. The unmethylated and methylated aliquots of the peptide were then spotted onto nitrocellulose, and the membrane was probed with 4b7 monoclonal antibody. Even on long exposure, no reactivity with the methylated peptide could be detected, indicating that 4b7 is absolutely specific for unmethylated C subunit.
Steady-State Methylation Levels of C Subunit Mutants
A combination of methods was used to analyze the methylation status of various C subunit mutants to determine whether methylation correlated with their ability to bind various regulatory subunits. As with the L309Δ mutant analyzed above, all of these mutants retain substantial native structure, as evidenced by their ability to associate with A subunit and polyomavirus MT (Ogris et al., 1997, 1999a,b). We first measured the in vivo methylation level of the wt and mutant C subunits with an assay using a monoclonal antibody (4b7) specific for demethylated C subunit (see MATERIALS AND METHODS and Figure 3). In this assay, base treatment was used to demethylate half of each immunoprecipitated C subunit before immunoblotting with 4b7 to detect unmethylated C subunits in the treated and untreated samples (Figure 3).
Figure 3A shows the methylation status of wt C subunit, four mutant C subunits with substitutions at threonine 304, and one mutant C subunit with a substitution at threonine 301. Anti-HA tag immunoprecipitates prepared from control cells containing empty vector (GRE only lane) showed no immunoreactivity at the position of C subunit either without (−) or with (+) base treatment. Anti-HA tag immunoprecipitates of wt C subunit (wt), on the other hand, showed low immunoreactivity in the absence of base treatment and a large increase of immunoreactivity upon base-induced demethylation. Chemiluminescence quantitation determined that 94 ± 3% of HA-tagged wt C subunits are methylated in unsynchronized NIH3T3 cell populations. HA-tagged mutants in which threonine 304 is substituted with aspartate (T304D), alanine (T304A), asparagine (T304N), or lysine (T304K), or in which threonine 301 is substituted with aspartate (T301D), were also highly methylated, with methylation levels ranging from 75–98% (Figure 3A). These high levels of methylation indicate that threonines 301 and 304 are not essential for methylation of leucine 309.
Figure 3B shows an analysis of five other carboxy-terminal mutants altered at leucine 309 or tyrosine 307. Even though substantial L309Δ protein was immunoprecipitated, L309Δ was not recognized by 4b7, indicating that the epitope recognized by 4b7 includes unmethylated leucine 309. Substitution of tyrosine 307 with glutamic acid (Y307E), glutamine (Y307Q), or lysine (Y307K) nearly abolished methylation, indicating that this residue is very important for achieving a wt methylation level. Substitution of phenylalanine (Y307F) for tyrosine 307 resulted in an intermediate level of methylation (43 ± 18%), indicating that the tyrosine 307 hydroxyl group plays an important but not essential role, and that the aromatic ring of this tyrosine is important. Thus, tyrosine 307 is more important than threonines 301 and 304 for methylation of the C subunit in vivo.
A similar analysis of C subunit active site point mutants is shown in Figure 3C. The methylation status of these mutants was assayed because they had previously been shown to bind MT but not substantial Bα subunit (Ogris et al., 1999a,b). Although wt C subunit analyzed in parallel again showed a great increase in 4b7 reactivity upon base treatment, all four of these catalytically inactive mutants showed little change. Quantitation indicated that 94 ± 1% of HA-tagged wt C subunits were methylated, but only 3–13% of the catalytically inactive mutant C subunits were methylated, demonstrating that these active site residues are essential for obtaining a high steady-state level of C subunit methylation. Expression of these mutants did not affect the methylation status of endogenous, untagged wt C subunit (data not shown), indicating that these mutants do not have a general, indirect effect on PP2A methylation.
Figure 4A shows the results of an in vivo assay, based on incorporation of 3H-labeled methyl groups into C subunit, that was used to confirm the methylation status of the methylation-deficient mutants detected by the antibody assay (see MATERIALS AND METHODS). No protein synthesis occurs during the labeling period of this in vivo assay, so only methylation of pre-existing C subunits is seen. As expected, no [3H]C subunit was present in the control immunoprecipitates from cells expressing vector only (GRE only lane), whereas HA-tagged wt C subunit, the positive control, was efficiently methylated (wt lane). In agreement with the methylation-sensitive antibody assay results, Y307F was methylated at an intermediate level, but Y307E, Y307Q, and Y307K were essentially unmethylated. In addition, neither 301stop, a nine-amino acid C-terminal truncation mutant, nor L309Δ was detectably methylated even though a substantial amount of mutant protein was present in the assay (see immunoblot). Thus, the methyltransferase does not simply methylate the last residue of C subunit but specifically recognizes leucine 309.
Figure 4B shows the results of a similar in vivo labeling analysis in which the four catalytically inactive C subunit mutants along with a wt C subunit control (wt lane) were assayed. All four of the inactive mutants were highly defective in methylation, confirming the results obtained with the antibody assay (Figure 3C).
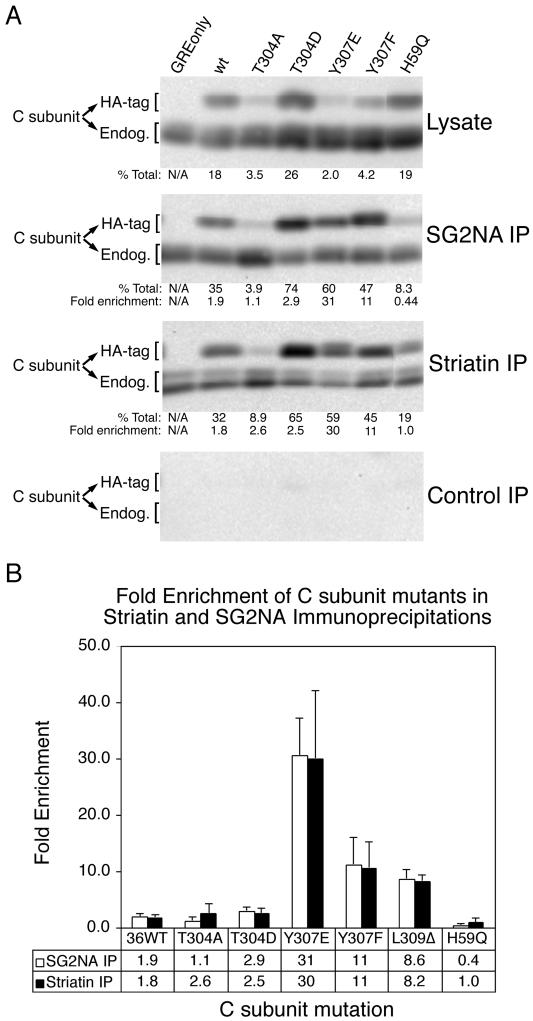
Unmethylated C Subunit Mutants Bind to Striatin and SG2NA
To analyze the binding of striatin and SG2NA to PP2A mutants, the ability of striatin and SG2NA antisera to coimmunoprecipitate these mutants relative to HA-tagged wt C subunit was determined. An essentially unmethylated catalytically inactive mutant (H59Q), an unmethylated C-terminal mutant (Y307E), a near-wt methylated mutant (T304D), an intermediately methylated mutant (Y307F), and a hypermethylated mutant (T304A) were analyzed along with wt C subunit (Figure 5A). For both lysates and immunoprecipitates, the percentage of total C subunit that each HA-tagged C subunit constitutes was determined. The calculated percentage for each immunoprecipitate was divided by the calculated percentage for the corresponding lysate to determine the fold enrichment of each mutant in immune complexes, which is shown graphically in Figure 5B. The fold enrichment was used as a measure of the efficiency of complex formation of various C subunit mutants to SG2NA and striatin. One caveat from this type of analysis is that the maximum fold enrichment is limited by the overall expression levels of each mutant, which varies among different cell lines. For example, a mutant that is expressed at 10% of total PP2A C subunit cannot be enriched more than 10-fold, whereas a mutant expressed at 2% of total C subunit could theoretically be enriched 50-fold. Nevertheless, none of the mutants were enriched to their theoretical maximum level.
Figure 5.
Striatin and SG2NA can bind unmethylated C subunit mutants. (A) Mutation of PP2A C subunit can dramatically affect association with SG2NA and striatin. Lysates of a subset of the cell lines used in Figure 3 were used to prepare SG2NA and striatin immunoprecipitates. Immunoprecipitates (IP) and lysates were analyzed by 10% SDS-PAGE, transferred to nitrocellulose, and probed with a methylation-insensitive monoclonal antibody to PP2A C subunit. Results from a typical immunoblot of lysates, SG2NA immunoprecipitates, striatin immunoprecipitates, and control immunoprecipitates are shown. The amount of HA-tagged (HA-tag) and endogenous (Endog.) C subunit in immunoprecipitates and lysates was quantitated by a Bio-Rad chemilumimager. The percentage of total (endogenous + HA-tagged) C subunit that HA-tagged C subunits represent in lysates and in immunoprecipitates was calculated (% Total) and is shown beneath the respective panels. The ratio of % total in the immunoprecipitates to the % total in the respective lysates was calculated for each cell line as a measure of the efficiency of C subunit association with striatin or SG2NA and is indicated (Fold enrichment). The average percentages and fold enrichments of at least three independent experiments are shown below each lane. The C subunits migrate sometimes as singlets and sometimes as doublets; whether double or single bands are seen can vary (see details in legend to Figure 1). (B) Graph of the fold enrichment (mean ± SD) of each HA-tagged wt or mutant PP2A C subunit present in striatin and SG2NA immunoprecipitations compared with expression levels in lysates. Values for the L309Δ mutant are also included for comparison.
The effects of these mutations on binding to striatin and SG2NA were dramatically different from those on Bα subunit binding. Compared with wt C subunit, Y307E and Y307F showed enhanced binding to striatin and SG2NA, T304D showed near-wt binding, and T304A showed near-wt or decreased binding. Not surprisingly, each mutation tended to have similar effects on binding to striatin and SG2NA. The differences among various mutants were not simply due to expression levels of the mutant C subunits, as shown by Y307E and T304A, which were both expressed at low levels (2 and 3.5%, respectively), but differed 30-fold in their ability to bind SG2NA. Moreover, T304A and T304D, which were expressed at very different levels (3.5 and 26%, respectively), showed no difference in their enrichment in striatin immunoprecipitates. Furthermore, methylation of C subunit is not required for striatin and SG2NA binding to the A/C heterodimer. In fact, the unmethylated mutants Y307E and L309Δ are two of the most highly enriched in striatin and SG2NA complexes.
Unmethylated C Subunit Mutants Bind to MT but Not to Bα Subunit
Table 2 shows a comparison of the steady-state methylation levels of the C subunit carboxy-terminal and active site mutants with their abilities to form complexes containing the different regulatory subunits. Data on the binding of these mutants to Bα subunit and MT are from published studies (Ogris et al., 1997, 1999a,b). Several major observations can be made. First, although C subunit methylation is required for Bα subunit binding, it is not required for binding of striatin, SG2NA, or MT. All point mutants highly defective in methylation (Y307E, Y307Q, Y307K, H59Q, H118Q, D85N, and R89A) fail to bind Bα subunit. The only mutants that still bind Bα subunit efficiently are methylated at medium (Y307F) to high (T304A) levels. These results are consistent with the hypothesis that methylation positively regulates Bα subunit association. MT, on the other hand, binds to all of these mutants, indicating that its association with the A/C heterodimer does not require methylation. The second major observation from Table 2 is that some mutations (T301D, T304D, T304N, and T304K) impair B binding without having a dramatic effect on leucine 309 methylation. Although all four of these mutants are methylated at a near-wt level, they all are partially (T301D) or completely (T304D, T304N, and T304K) defective in Bα subunit binding. Thus, Bα subunit binding is not important for maintaining a high methylation level on C subunit.
Table 2.
Methylation is required for A/C heterodimer binding to Bα subunit but not for its association with MT, striatin, or SG2NA
| C subunita | Methylation competenceb | Bα subunit bindingbc | Striatin bindingd | SG2NA bindingd | MT bindingcf |
|---|---|---|---|---|---|
| wt | +++ | +++ | +++ | +++ | wt |
| 301stop | − | − | +++e | +++e | wt |
| L309Δ | − | − | ++++ | ++++ | wt |
| T301D | +++ | + | n.a. | n.a. | wt |
| T304D | +++ | − | +++ | +++ | wt |
| T304A | +++ | ++++ | +++ | ++ | wt |
| T304N | +++ | − | n.a. | n.a. | wt |
| T304K | +++ | − | n.a. | n.a. | wt |
| Y307E | − | − | ++++ | ++++ | wt |
| Y307F | ++ | ++++ | ++++ | ++++ | wt |
| Y307Q | − | − | n.a. | n.a. | wt |
| Y307K | − | − | n.a. | n.a. | wt |
| H59Q | − | − | ++ | + | wt |
| H118Q | − | − | n.a. | n.a. | wt |
| D85N | − | − | n.a. | n.a. | wt |
| R89A | − | − | n.a. | n.a. | wt |
The description of the C subunit mutants and their nomenclature, as well as the abbreviations for the amino acid residues are the same as in the legend to Table 1 except that Δ indicates deletion of the specified residue. 301stop indicates the replacement of residue 301 with a stop codon.
For these assays: ++++, > wt level; +++, 71–100% of the wt level; ++, 41–70% of the wt level; +, 15–40% of the wt level; −, <15% of the wt level; n.a., not assayed.
Bα subunit and MT-binding results for mutants other than L309Δ were taken from Ogris et al. (1999a, 1999b, 1997) and were estimated from autoradiograms.
For these assays, the symbols are equivalent to footnote b except that ++++ indicates ≥twofold the wt level because lesser increases may not be significant.
These values were estimated from autoradiograms.
Because of the variability of MT expression in each of the MT transfected cell lines, only qualitative data were obtained. wt indicates that MT associated with each of these mutants at near-wt levels.
Demethylation of C Subunit by a PME-1 Results in Dissociation of Bα Subunit but Not MT or Aα Subunit
We recently reported the cloning and bacterial expression of PME-1, a PP2A methylesterase (Ogris et al., 1999a). We used PME-1 to demethylate C subunit in cell lysates and assayed by coimmunoprecipitation whether this treatment affected the amount of C subunit found in complex with Bα subunit, MT, or Aα subunit. The top panel of Figure 6 shows that treatment of lysates from cell lines expressing either HA-tagged Aα subunit (Asub), HA-tagged Bα subunit (B sub), or MT with PME-1 resulted in substantial demethylation of the C subunit in each of these lysates. Quantitation of percentage of methylation from four independent experiments was performed on a set of parallel immunoblots (not shown) using the method described for immunoprecipitates in the legend to Figure 3. This analysis determined that the methylation level of C subunit in PME-1-treated lysates from cells expressing HA-tagged Aα or Bα subunit or MT had been reduced to 21 ± 3, 20 ± 11, or 22 ± 4%, respectively. The bottom panel of this figure shows that demethylation of the C subunit did not greatly affect the amount of C subunit coimmunoprecipitated with Aα subunit or MT, but it dramatically reduced (≥5-fold) the amount of C subunit bound to Bα subunit. This result provides further evidence that the methylation state of the C subunit affects Bα subunit, but not MT, binding and also shows that A/C heterodimer formation is not greatly affected by the level of C subunit methylation. Moreover, these data show that PME-1 demethylates A/C/Bα complexes and induces dissociation of Bα from A/C, suggesting that it may perform a similar function in vivo.
Striatin and SG2NA, but Not Bα Subunit, Can Bind Unmethylated C Subunit in Cells with Reduced Methylation Levels
Typically, PP2A is at least 94% methylated in NIH3T3 cells (Figure 3). We next reduced the level of PP2A methylation in vivo and examined the effect on association of cellular regulatory subunits. Initial attempts to overexpress PME-1 were unable to decrease PP2A C subunit methylation (Du and Pallas, unpublished data). Okadaic acid treatment of cells has been reported to cause demethylation of PP2A (Favre et al., 1997), but prolonged treatment can cause apoptosis (Yan et al., 1997). Adenosine dialdehyde (AdOx), an inhibitor of S-adenosyl homocysteine hydrolase (Bartel and Borchardt, 1984), increases the cellular concentration of the S-adenosyl methionine (AdoMet)-dependent methylation byproduct, S-adenosyl homocysteine, thus reducing AdoMet-dependent protein methylation. Because PP2A is methylated by an AdoMet-dependent methyltransferase, AdOx treatment lowers PP2A methylation. We used sequential treatments with okadaic acid and AdOx to decrease PP2A methylation levels to ∼25%.
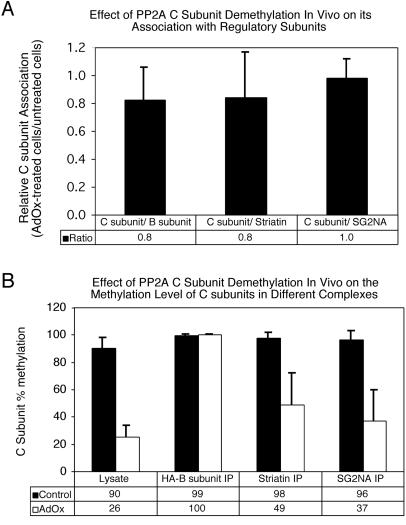
Analysis of PP2A complexes in control cells and in treated cells in which C subunit methylation was reduced showed no consistent decrease in the amount of C subunit coimmunoprecipitated with Bα subunit, striatin, or SG2NA (Figure 7A). However, striatin and SG2NA bound to both methylated and unmethylated C subunits in the treated cells, whereas Bα subunit associated exclusively with methylated C subunits (Figure 7B), suggesting that Bα subunit preferentially binds to the remaining population of methylated C subunits in the treated cells.
Figure 7.
Effect of PP2A C subunit demethylation in vivo on PP2A complexes. NIH3T3 cells stably expressing HA-tagged Bα subunit were treated sequentially with 100 nM okadaic acid for 14 h followed by 200 μM AdOx for 30 h to reduce C subunit methylation. Cell lysates were prepared from treated (AdOx) and untreated (Control) cells and immunoprecipitations were performed using HA-tag (12CA5), striatin, and SG2NA antibodies. Immunoprecipitates (IP) and lysates were analyzed by SDS-PAGE and probed with HA-tag, striatin, SG2NA, and methylation-sensitive and methylation-insensitive C subunit antibodies using enhanced chemiluminescence. A Bio-Rad Fluor S-Max chemilumimager was then used to calculate 1) the ratio of total C subunit (methylation-insensitive antibody) to regulatory subunit in each immune complex and 2) the methylation level of C subunit in the lysates and immune complexes. (A) Effect of PP2A C subunit demethylation on regulatory subunit association. C subunit association (C subunit/regulatory subunit) was calculated for each regulatory subunit for both control and treated cells. The ratio of C subunit association in treated cells to C subunit association in untreated cells is shown for each regulatory subunit. (B) Effect of PP2A C subunit demethylation on the methylation level of C subunits bound to different regulatory subunits. Shown is the percentage of methylation of C subunit in cell lysates and immunoprecipitations prepared from control and treated cells. The data shown represent the average and SD of at least three independent experiments. The error bars on C subunits associated with Bα are very small because the C subunit in these complexes was essentially 100% methylated in each experiment.
DISCUSSION
It has been known for several years that the PP2A C subunit is methylated at its carboxy terminus, but a biological function for this posttranslational modification has not yet been directly demonstrated. Here, we present four independent lines of evidence to show that methylation is essential for the binding of Bα subunit to the A/C heterodimer but not for association of MT, striatin, and SG2NA. First, deletion of leucine 309 abolished Bα subunit binding, whereas it enhanced binding to striatin and SG2NA and had no effect on MT association. Second, loss of C subunit methylation by mutation of other residues had effects similar to deletion of leucine 309. Third, demethylation of C subunit in vitro using purified PME-1 disrupted A/C/Bα complexes but not A/C/MT complexes. Finally, immunoprecipitations prepared from cells with reduced C subunit methylation showed that SG2NA and striatin can associate with unmethylated C subunit, whereas Bα subunits complexed exclusively with the remaining population of methylated C subunits. These findings provide evidence that reversible protein methylation at a single residue, much like phosphorylation, is capable of regulating protein-protein interactions and enzyme activity and demonstrate the first known function for PP2A methylation. In addition, the results of this study indicate the importance of specific C subunit carboxy-terminal residues and, more surprisingly, active site residues for efficient methylation. Two models consistent with these findings are either that the active site and the carboxy terminus are needed for recognition by methyltransferase and/or methylesterase enzymes or that mutations induce conformational changes elsewhere in PP2A that affect the interaction with these enzymes.
The complete loss of Bα subunit binding to L309Δ (as well as other C subunit mutants) cannot be due to competition between Bα and MT. MT is expressed only at ∼10% the level of PP2A in these cells (Haehnel and Pallas, unpublished data; Ulug et al., 1992), and overexpression of 10 times as much MT via an adenovirus does not cause complete dissociation of Bα subunit (Green and Pallas, unpublished). In addition, all MT in these cells is already bound to PP2A (Pallas et al., 1989); therefore, no excess MT is available to compete off additional Bα subunit. Our findings with L309Δ confirm that of Bryant et al. (1999), who showed that a single mutant (L309A) did not bind Bα subunit and would not incorporate methyl groups in vitro. We have also shown that leucine 309 is not important for several other viral and cellular PP2A regulatory subunits. Because the only mutant in the study of Bryant et al. changed leucine 309, they could not distinguish whether Bα subunit binding was affected by loss of methylation, mutation of the leucine residue, or both. Our analysis of multiple unmethylated mutants that retained leucine 309 together with several biochemical approaches have clearly demonstrated that methylation is required for binding of Bα subunit.
We have also developed a methylation-sensitive antibody assay that has the important advantage of measuring the steady-state methylation level of C subunit in vivo. Similar assays utilizing methylation-sensitive polyclonal antibodies have been used previously as a means of evaluating in vivo PP2A methylation levels (Favre et al., 1994, 1997; Turowski et al., 1995), but here we have used milder conditions and a chemilumimager to obtain quantitative data on methylation. The immunoprecipitated C subunit that was analyzed typically consisted of 15–20% of the total cellular C subunit, raising the possibility that it was not representative of the total pool of expressed protein. However, most HA-tagged C subunits were also analyzed directly in lysates for steady-state methylation level and gave similar results in every case (Yu, Du, Moreno, Green, and Pallas, unpublished data), indicating that the immunoprecipitation data are representative.
Mutational analysis of PP2A C subunit showed that loss of methylation induced by individual substitution of any one of five separate residues resulted in loss of Bα subunit binding but not of MT, striatin, or SG2NA binding. In fact, the binding characteristics of some of the mutants are consistent with the possibility that loss of C subunit methylation might increase striatin and SG2NA binding to the A/C heterodimer. Mutations in completely different regions of the C subunit protein primary sequence (positions 59, 85, 89, 118, and 307) were able to simultaneously affect methylation and Bα subunit binding, indicating a strong connection between the two events. Although two of the active site mutants (H59Q and H118Q) form a stable complex with PME-1 (Ogris et al., 1999a), the other two active site mutants do not, excluding the possibility that loss of Bα subunit binding is due to stable PME-1 association. The data also show that the dramatic loss of methylation seen with several mutants was not an indirect effect produced by loss of Bα subunit binding. Four separate C subunit mutants (T301D, T304D, T304N, and T304K) had very low or no Bα subunit binding and yet retained 80–89% of the wt level of methylation. Thus, Bα subunit binding is not necessary to maintain high methylation levels of C subunit. Taken together, these results strongly support the hypothesis that C subunit methylation positively regulates Bα subunit binding to the A/C heterodimer.
The B subunit antibody used to detect B subunit associating with the various mutant C subunits (Ogris et al., 1997, 1999a,b) was raised against a large portion of the Bα subunit containing extensive sequence identity with other B subunit isoforms (β, γ, and δ). Two-dimensional gel immunoblot analysis indicates that this antibody recognizes multiple isoforms of B subunit (Huehnel, Park, and Pallas, unpublished data). The fact that no B subunit could be detected by this antibody in immunoprecipitates of many unmethylated C subunit mutants (Ogris et al., 1997, 1999b) suggests that B subunit isoforms other than Bα also probably require methylation for efficient association with the A/C heterodimer.
We have recently obtained evidence showing that C subunit methylation is important for the efficient association of both B and B′ subunits in yeast (Wei et al., in press). We identified the major PP2A methyltransferase in S. cerevisiae as Ppm1p and found that deletion of the PPM1 gene resulted in almost complete loss of C subunit (Pph21p/Pph22p) methylation. Loss of methylation resulted in greatly decreased association of the B (Cdc55p) and B′ (Rts1p) subunits and, to a lesser degree, of A subunit (Tpd3p). Moreover, cells deleted for PPM1 exhibited nocodazole sensitivity, a known phenotype of CDC55 disruption, indicating that loss of methylation can affect PP2A function. Two other groups have published concurrent studies describing findings similar to ours in both yeast (Wu et al., 2000) and mammalian systems (Tolstykh et al., 2000). Wu et al. also identified Ppm1p as the major methyltransferase, found that methylation is important for association of Tpd3p and Cdc55p, and showed that deletion of PPM1 causes nocodazole sensitivity (Wu et al., 2000). However, in contrast to our data (Wei et al., in press), they observed a small amount of residual binding of Cdc55p in the absence of methylation, which may be the result of differences in experimental conditions. Consistent with our data demonstrating the importance of C subunit methylation for Bα association with A/C heterodimers in mammalian cells, Tolstykh et al. showed that methylation of A/C heterodimers enhanced their association with B subunit in vitro (Tolstykh et al., 2000). In addition, they presented data suggesting that C subunit methylation increases the affinity of the A/C heterodimer for B′ subunits in mammalian cells. Finally, the only other regulatory subunit of PP2A for which there is any evidence suggesting a possible effect of methylation is alpha 4, which unlike the regulatory subunits discussed above, binds to C subunit but not A subunit. Alpha 4 has been recently reported to have increased binding to a C subunit mutant altered at both Y307 and L309 (Chung et al., 1999), suggesting that methylation is not required for alpha 4 association with C subunit and actually may inhibit it. Consistent with this possibility, Wu et al. (Wu et al., 2000) found that association of C subunit with the yeast homolog of alpha 4 (Tap42) is enhanced ∼50% by disruption of PPM1.
Addition of the PP2A methylesterase, PME-1, to cell lysates caused C subunit demethylation and dissociation of C subunit from the Bα subunit-containing complexes but not from MT complexes or from Aα subunit. Thus, PME-1 demethylates A/C/Bα complexes and induces dissociation of Bα from A/C in vitro, suggesting that it may perform a similar function in vivo. Although it is possible that the exogenously added PME-1 may have physically competed off the Bα subunit from the A/C heterodimer, this seems unlikely because, unlike Bα subunit, PME-1 does not have high enough affinity for wt C subunit to stably complex with it (Ogris et al., 1999a). Furthermore, immunoblotting showed that the amount of added PME-1 represented as little as eightfold more than was present in the untreated lysates. Moreover, addition of inactive PME-1 at a concentration 64-fold above the endogenous level neither demethylated the C subunit nor displaced Bα subunit (McQuoid and Pallas, unpublished data). Because A/C/Bα heterotrimers exist in a dynamic equilibrium with A/C heterodimers, it is possible that, as Bα subunit naturally dissociates from the A/C heterodimer, exogenously added PME-1 could demethylate the A/C heterodimer, decreasing its affinity for Bα and preventing reassociation. This hypothesis would be consistent with the findings of Tolstykh et al. (Tolstykh et al., 2000), who showed that A/C/Bα complexes are demethylated by PME-1 at a much slower rate than A/C heterodimers.
Whereas a reduction in C subunit steady-state methylation in vivo resulted in a decrease in methylation of C subunits associated with striatin and SG2NA, Bα subunit-associated C subunits in the same cell remained essentially 100% methylated. This suggests that Bα subunit has a higher affinity than striatin and SG2NA for methylated C subunits and that methylation is essential for Bα subunit but not striatin and SG2NA binding to the A/C heterodimer. Because a substantial decrease in Bα subunit association with A/C heterodimer was not observed in the okadaic acid/AdOx-treated NIH3T3 cells, it can be inferred that Bα subunit associates with less than 26% of C subunit in these cells. It is possible that cells regulate Bα subunit association with the A/C heterodimer simply by reducing C subunit methylation in a particular cellular compartment below the level of associated Bα subunit. Alternatively, there may be a mechanism for specifically demethylating Bα subunit-associated C subunits by PME-1.
Earlier studies have shown that synthetic C subunit carboxy-terminal peptides functioned neither as substrates nor inhibitors of the PP2A methyltransferase (Xie and Clarke, 1994) and PP2A methylesterase (Lee et al., 1996), suggesting that carboxy-terminal residues might not be critical for interaction with these enzymes. The observations that L309Δ, 301stop, and several tyrosine 307 mutants abrogated methylation provide the first direct evidence that C subunit carboxy-terminal residues are important for recognition of PP2A by the methyltransferase and/or methylesterase enzymes in vivo. Taken together, these data suggest that proper recognition by the methyltransferase and/or methylesterase requires both C subunit carboxy-terminal residues and additional structure.
Four individual point mutations in the active site nearly abolished the methylation competence of the C subunit in a cis manner, suggesting that residues or coordinated metals in or near the active site may be part of the additional structure needed for recognition by methyltransferase and/or methylesterase enzymes. This hypothesis is supported by the recent finding that two of these mutants formed stable complexes with PME-1 that could be disrupted by PP2A inhibitors such as okadaic acid (Ogris et al., 1999a). Okadaic acid and microcystin-LR, whose binding sites on PP2A are thought to overlap (MacKintosh et al., 1990), may thus inhibit PP2A methylation and/or demethylation (Lee and Stock, 1993; Floer and Stock, 1994; Li and Damuni, 1994; Lee et al., 1996) because they overlap in binding with the methyltransferase and methylesterase enzymes. Microcystin has been shown to interact with multiple residues in the active site of the highly related phosphatase, PP1 (Goldberg et al., 1995), including a PP1 residue corresponding to one of the PP2A residues mutated in this study (arginine 89). An alternative hypothesis to a direct interaction between the active site and the methyltransferase and methylesterase enzymes is that mutations and inhibitors induce conformational changes elsewhere in PP2A that affect the interaction with these enzymes.
Protein phosphatase V, protein phosphatase G, and protein phosphatase X, several phosphatases >50% identical to PP2A, have the same three carboxy-terminal amino acids as PP2A, raising the possibility that the PP2A methyltransferase and methylesterase enzymes may also regulate these phosphatases. Whereas the methylation status of protein phosphatase V and protein phosphatase G has not been reported, protein phosphatase X is methylated on its C terminus (Kloeker et al., 1997). These phosphatases share two residues that have been shown in this study to be essential for PP2A methylation—a tyrosine analogous to PP2A tyrosine 307 and a carboxy-terminal leucine. Because these phosphatases share >50% identity with PP2A, including the active site residues mutated in this study, they may contain the other structural information necessary for interaction with the PP2A methyltransferase and/or methylesterase enzymes.
The observation that the unmethylated Y307E mutant was greatly enriched in striatin and SG2NA complexes suggests that phosphorylation of tyrosine 307 may enhance the affinity of C subunit for these regulatory subunits. In contrast, phosphorylation of this residue may inhibit Bα subunit binding (Ogris et al., 1997), perhaps indirectly by reducing methylation. Because the T304D mutant is highly methylated, it is unlikely that phosphorylation of threonine 304 regulates methylation. However, since this mutant cannot bind Bα subunit (Ogris et al., 1997), phosphorylation of this residue might directly regulate Bα association. Our previous data suggests that MT binding to the A/C heterodimer would be unaffected by phosphorylation of threonine 304 or tyrosine 307 (Ogris et al., 1997). The current results indicate that MT binding to PP2A is also independent of C subunit methylation. These two results may have implications for how MT may circumvent normal cellular regulation of PP2A.
On first consideration, the fact that Y307F has a twofold decrease in methylation compared with wt C subunit and yet binds Bα subunit as well or better than wt is puzzling. In the context of the other results in this study, we would suggest that this mutation simultaneously reduces recognition by a PP2A methyltransferase while strengthening Bα subunit binding in an independent manner. This result, in combination with the requirement for leucine 309 methylation for Bα subunit binding, supports a model in which hydrophobic interactions between the carboxy terminus of C subunit and Bα subunit play a critical role in stabilizing the A/B/C heterotrimer. Also consistent with this model is the fact that the T304A mutant exhibits enhanced Bα subunit binding. Alternatively, methylation of the C subunit carboxy terminus could induce a conformational change that affects its interaction with different regulatory proteins. Structural studies will be necessary to resolve these possibilities and to help elucidate the mechanisms by which multiple signals and events involving the C subunit carboxy terminus affect PP2A activity.
ACKNOWLEDGMENTS
We thank Carroll Weaver, Danielle McKelton, Danita Ashby, and Marie Kozel for excellent technical assistance, Brian Hemmings for the C subunit cDNA, John White for the pGRE 5-2 vector, and Anita Corbett, Gerald Shadel, Xiaodong Cheng, Shirish Shenolikar, Cori Beychok, and Tatiana Mendez for critical reading of the manuscript. Under agreements between Upstate Biotechnology Inc. and Emory University and between Calbiochem and Emory University, David Pallas is entitled to a share of sales royalty received by the University from these companies. In addition, this same author serves as a consultant to Upstate Biotechnology Inc. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict of interest policies.
Abbreviations used:
- AdOx
adenosine dialdehyde
- AdoMet
S-adenosyl methionine
- C subunit
catalytic subunit
- HA
hemagglutinin
- MT
middle tumor antigen
- PME-1
protein phosphatase methylesterase-1
- PP2A
protein phosphatase 2A
- wt
wild-type
REFERENCES
- Bartel RL, Borchardt RT. Effects of adenosine dialdehyde on S-adenosylhomocysteine hydrolase and S-adenosylmethionine-dependent transmethylations in mouse L929 cells. Mol Pharmacol. 1984;25:418–424. [PubMed] [Google Scholar]
- Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Young SG. Targeted. inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J Biol Chem. 2000;275:17605–17610. doi: 10.1074/jbc.C000079200. [DOI] [PubMed] [Google Scholar]
- Bryant JC, Westphal RS, Wadzinski BE. Methylated C-terminal leucine residue of PP2A catalytic subunit is important for binding of regulatory Balpha subunit. Biochem J. 1999;339:241–246. [PMC free article] [PubMed] [Google Scholar]
- Campbell KS, Auger KR, Hemmings BA, Roberts TM, Pallas DC. Identification of regions in polyomavirus middle T and small t antigens important for association with protein phosphatase 2A. J Virol. 1995;69:3721–3728. doi: 10.1128/jvi.69.6.3721-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayla X, Ballmer-Hofer K, Merlevede W, Goris J. Phosphatase 2A associated with polyomavirus small-T or middle-T antigen is an okadaic acid-sensitive tyrosyl phosphatase. Eur J Biochem. 1993;214:281–286. doi: 10.1111/j.1432-1033.1993.tb17922.x. [DOI] [PubMed] [Google Scholar]
- Cegielska A, Shaffer S, Derua R, Goris J, Virshup DM. Different oligomeric forms of protein phosphatase 2A activate and inhibit simian virus 40 DNA replication. Mol Cell Biol. 1994;14:4616–4623. doi: 10.1128/mcb.14.7.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Chen J, Martin BL, Brautigan DL. Regulation of protein serine-threonine phosphatase type-2A by tyrosine phosphorylation. Science. 1992;257:1261–1264. doi: 10.1126/science.1325671. [DOI] [PubMed] [Google Scholar]
- Chung H, Nairn AC, Murata K, Brautigan DL. Mutation of Tyr307 and Leu309 in the protein phosphatase 2A catalytic subunit favors association with the alpha 4 subunit which promotes dephosphorylation of elongation factor-2. Biochemistry. 1999;38:10371–10376. doi: 10.1021/bi990902g. [DOI] [PubMed] [Google Scholar]
- Cohen P. The structure and regulation of protein phosphatases. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- De Baere I, Derua R, Janssens V, Van Hoof C, Waelkens E, Merlevede W, Goris J. Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the human homologue. Biochemistry. 1999;38:16539–16547. doi: 10.1021/bi991646a. [DOI] [PubMed] [Google Scholar]
- Farh L, Mitchell DA, Deschenes RJ. Farnesylation and proteolysis are sequential, but distinct steps in the CaaX box modification pathway. Arch Biochem Biophys. 1995;318:113–121. doi: 10.1006/abbi.1995.1211. [DOI] [PubMed] [Google Scholar]
- Favre B, Turowski P, Hemmings BA. Differential inhibition and posttranslational modification of protein phosphatase 1 and 2A in MCF7 cells treated with calyculin-A, okadaic acid, and tautomycin. J Biol Chem. 1997;272:13856–13863. doi: 10.1074/jbc.272.21.13856. [DOI] [PubMed] [Google Scholar]
- Favre B, Zolnierowicz S, Turowski P, Hemmings BA. The catalytic subunit of protein phosphatase 2A is carboxyl-methylated in vivo. J Biol Chem. 1994;269:16311–16317. [PubMed] [Google Scholar]
- Floer M, Stock J. Carboxyl methylation of protein phosphatase 2A from Xenopus eggs is stimulated by cAMP and inhibited by okadaic acid. Biochem Biophys Res Commun. 1994;198:372–379. doi: 10.1006/bbrc.1994.1052. [DOI] [PubMed] [Google Scholar]
- Goldberg J, Huang HB, Kwon YG, Greengard P, Nairn AC, Kuriyan J. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature. 1995;376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- Hemmings BA, Adams-Pearson C, Maurer F, Muller P, Goris J, Merlevede W, Hofsteenge J, Stone SR. alpha- and beta-forms of the 65-kDa subunit of protein phosphatase 2A have a similar 39 amino acid repeating structure. Biochemistry. 1990;29:3166–3173. doi: 10.1021/bi00465a002. [DOI] [PubMed] [Google Scholar]
- Hopkin K. Phosphatases: new respect for cellular second banana. J NIH Res. 1995;7:27–30. [Google Scholar]
- Hrycyna CA, Sapperstein SK, Clarke S, Michaelis S. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J. 1991;10:1699–1709. doi: 10.1002/j.1460-2075.1991.tb07694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamibayashi C, Estes R, Lickteig RL, Yang SI, Craft C, Mumby MC. Comparison of heterotrimeric protein phosphatase 2A containing different B subunits. J Biol Chem. 1994;269:20139–20148. [PubMed] [Google Scholar]
- Kloeker S, Bryant JC, Strack S, Colbran RJ, Wadzinski BE. Carboxymethylation of nuclear protein serine/threonine phosphatase X. Biochem J. 1997;327:481–486. doi: 10.1042/bj3270481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowluru A, Seavey SE, Rabaglia ME, Nesher R, Metz SA. Carboxylmethylation of the catalytic subunit of protein phosphatase 2A in insulin-secreting cells: evidence for functional consequences on enzyme activity and insulin secretion. Endocrinology. 1996;137:2315–2323. doi: 10.1210/endo.137.6.8641181. [DOI] [PubMed] [Google Scholar]
- Lee J, Chen Y, Tolstykh T, Stock J. A specific protein carboxyl methylesterase that demethylates phosphoprotein phosphatase 2A in bovine brain. Proc Natl Acad Sci USA. 1996;93:6043–6047. doi: 10.1073/pnas.93.12.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Stock J. Protein phosphatase 2A catalytic subunit is methyl-esterified at its carboxyl terminus by a novel methyltransferase. J Biol Chem. 1993;268:19192–19195. [PubMed] [Google Scholar]
- Li M, Damuni Z. Okadaic acid and microcystin-LR directly inhibit the methylation of protein phosphatase 2A by its specific methyltransferase. Biochem Biophys Res Commun. 1994;202:1023–1030. doi: 10.1006/bbrc.1994.2031. [DOI] [PubMed] [Google Scholar]
- MacKintosh C, Beattie KA, Klumpp S, Cohen P, Codd GA. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Lett. 1990;264:187–192. doi: 10.1016/0014-5793(90)80245-e. [DOI] [PubMed] [Google Scholar]
- Mader S, White JH. A steroid-inducible promoter for the controlled overexpression of cloned genes in eukaryotic cells. Proc Natl Acad Sci USA. 1993;90:5603–5607. doi: 10.1073/pnas.90.12.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Jaekel RE, Ohkura H, Ferrigno P, Andjelkovic N, Shiomi K, Uemura T, Glover DM, Hemmings BA. Drosophila mutants in the 55 kDa regulatory subunit of protein phosphatase 2A show strongly reduced ability to dephosphorylate substrates of p34cdc2. J Cell Sci. 1994;107:2609–2618. doi: 10.1242/jcs.107.9.2609. [DOI] [PubMed] [Google Scholar]
- Moreno CS, Park S, Nelson K, Ashby DG, Hubalek F, Lane WS, Pallas DC. The WD40 repeat proteins striatin and SG2NA are members of a novel family of calmodulin-binding proteins that associate with PP2A. J Biol Chem. 2000;275:5257–5263. doi: 10.1074/jbc.275.8.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumby MC, Walter G. Protein serine/threonine phosphatases: structure, regulation, and functions in cell growth. Physiol Rev. 1993;73:673–699. doi: 10.1152/physrev.1993.73.4.673. [DOI] [PubMed] [Google Scholar]
- Murata K, Wu J, Brautigan DL. B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc Natl Acad Sci USA. 1997;94:10624–10629. doi: 10.1073/pnas.94.20.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogris E, Du X, Nelson KC, Mak EK, Yu XX, Lane WS, Pallas DC. A protein phosphatase methylesterase (PME-1) is one of several novel proteins stably associating with two inactive mutants of protein phosphatase 2A. J Biol Chem. 1999a;274:14382–14391. doi: 10.1074/jbc.274.20.14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogris E, Gibson DM, Pallas DC. Protein phosphatase 2A subunit assembly: the catalytic subunit carboxy terminus is important for binding cellular B subunit but not polyomavirus middle tumor antigen. Oncogene. 1997;15:911–917. doi: 10.1038/sj.onc.1201259. [DOI] [PubMed] [Google Scholar]
- Ogris E, Mudrak I, Mak E, Gibson D, Pallas DC. Catalytically inactive protein phosphatase 2A can bind to polyomavirus middle tumor antigen and support complex formation with pp60c-src. J Virol. 1999b;73:7390–7398. doi: 10.1128/jvi.73.9.7390-7398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas DC, Morgan W, Roberts TM. The cellular proteins which can associate specifically with polyomavirus middle T antigen in human 293 cells include the major human 70-kilodalton heat shock proteins. J Virol. 1989;63:4533–4539. doi: 10.1128/jvi.63.11.4533-4539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas DC, Schley C, Mahoney M, Harlow E, Schaffhausen BS, Roberts TM. Polyomavirus small t antigen: overproduction in bacteria, purification, and utilization for monoclonal and polyclonal antibody production. J Virol. 1986;60:1075–1084. doi: 10.1128/jvi.60.3.1075-1084.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- Pallas DC, Weller W, Jaspers S, Miller TB, Lane WS, Roberts TM. The third subunit of protein phosphatase 2A (PP2A), a 55-kilodalton protein which is apparently substituted for by T antigens in complexes with the 36- and 63-kilodalton PP2A subunits, bears little resemblance to T antigens. J Virol. 1992;66:886–893. doi: 10.1128/jvi.66.2.886-893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell K. Complete interaction of cellular 56,000- and 32,000-Mr proteins with simian virus 40 small-t antigen in productively infected cells. J Virol. 1987;61:1240–1243. doi: 10.1128/jvi.61.4.1240-1243.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstykh T, Lee J, Vafai S, Stock JB. Carboxyl methylation regulates phosphoprotein phosphatase 2A by controlling the association of regulatory B subunits. Embo J. 2000;19:5682–5691. doi: 10.1093/emboj/19.21.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski P, Fernandez A, Favre B, Lamb NJ, Hemmings BA. Differential methylation and altered conformation of cytoplasmic and nuclear forms of protein phosphatase 2A during cell cycle progression. J Cell Biol. 1995;129:397–410. doi: 10.1083/jcb.129.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulug ET, Cartwright AJ, Courtneidge SA. Characterization of the interaction of polyomavirus middle T antigen with type 2A protein phosphatase. J Virol. 1992;66:1458–1467. doi: 10.1128/jvi.66.3.1458-1467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui H, Imazu M, Maeta K, Tsukamoto H, Azuma K, Takeda M. Three distinct forms of type 2A protein phosphatase in human erythrocyte cytosol. J Biol Chem. 1988;263:3752–3761. [PubMed] [Google Scholar]
- Wei, H., Ashby, D.G., Moreno, C.S., Ogris, E., Yeong, F.M., Corbett, A.H., and Pallas, D.C. (2000). Carboxy-methylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits, Cdc55p and Rts1p. J. Biol. Chem. (in press). [DOI] [PMC free article] [PubMed]
- Wu J, Tolstykh T, Lee J, Boyd K, Stock JB, Broach JR. Carboxyl methylation of the phosphoprotein phosphatase 2A catalytic subunit promotes its functional association with regulatory subunits in vivo. Embo J. 2000;19:5672–5681. doi: 10.1093/emboj/19.21.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Clarke S. Protein phosphatase 2A is reversibly modified by methyl esterification at its C-terminal leucine residue in bovine brain. J Biol Chem. 1994;269:1981–1984. [PubMed] [Google Scholar]
- Yan Y, Shay JW, Wright WE, Mumby MC. Inhibition of protein phosphatase activity induces p53-dependent apoptosis in the absence of p53 transactivation. J Biol Chem. 1997;272:15220–15226. doi: 10.1074/jbc.272.24.15220. [DOI] [PubMed] [Google Scholar]
- Yang SI, Lickteig RL, Estes R, Rundell K, Walter G, Mumby MC. Control of protein phosphatase 2A by simian virus 40 small-t antigen. Mol Cell Biol. 1991;11:1988–1995. doi: 10.1128/mcb.11.4.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Matsuzawa S, Mizuno Y, Kamibayashi C, Mumby MC, Andjelkovic N, Hemmings BA, Onoe K, Kikuchi K. The interconversion of protein phosphatase 2A between PP2A1 and PP2A0 during retinoic acid-induced granulocytic differentiation and a modification on the catalytic subunit in S phase of HL-60 cells. Arch Biochem Biophys. 1997;339:210–217. doi: 10.1006/abbi.1996.9835. [DOI] [PubMed] [Google Scholar]