The human Ccr4-Not complex has two types of deadenylase subunits that shorten the polyadenylate tail of cytoplasmic mRNA. The authors present evidence for novel roles of the highly related Ccr4a/Ccr4b deadenylases in preventing cell death and senescence and show that they have distinct roles as compared with the Caf1a/Caf1b deadenylases.
Abstract
A key step in cytoplasmic mRNA degradation is the shortening of the poly(A) tail, which involves several deadenylase enzymes. Relatively little is known about the importance of these enzymes for the cellular physiology. Here we focused on the role of the highly similar Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the Ccr4–Not complex. In addition to a role in cell proliferation, Ccr4a and Ccr4b play a role in cell survival, in contrast to the Caf1a (CNOT7) and Caf1b (CNOT8) deadenylase subunits or the CNOT1 and CNOT3 noncatalytic subunits of the Ccr4–Not complex. Underscoring the differential contributions of the deadenylase subunits, we found that knockdown of Caf1a/Caf1b or Ccr4a/Ccr4b differentially affects the formation of cytoplasmic foci by processing-body components. Furthermore, we demonstrated that the amino-terminal leucine-rich repeat (LRR) domain of Ccr4b influenced its subcellular localization but was not required for the deadenylase activity of Ccr4b. Moreover, overexpression of Ccr4b lacking the LRR domain interfered with cell cycle progression but not with cell viability. Finally, gene expression profiling indicated that distinct gene sets are regulated by Caf1a/Caf1b and Ccr4a/Ccr4b and identified Ccr4a/Ccr4b as a key regulator of insulin-like growth factor–binding protein 5, which mediates cell cycle arrest and senescence via a p53-dependent pathway.
INTRODUCTION
Accurate regulation of gene expression requires appropriate control of mRNA levels, which are determined by the relative rates of pre-mRNA synthesis, nuclear processing, and cytoplasmic mRNA turnover. A key step in mRNA degradation is the shortening of the poly(A) tail, which involves several deadenylases containing ribonucleolytic activity (Parker and Song, 2004; Garneau et al., 2007). Approximately 10 deadenylase enzymes have been identified in human cells, which can be divided into two classes: those with a conserved DEDD domain and others belonging to the endonuclease–exonuclease–phosphatase (EEP) superfamily (Goldstrohm and Wickens, 2008). The shortening and removal of the poly(A) tail by deadenylase enzymes exposes the 3′ mRNA end to the cytoplasmic form of the exosome nuclease complex and facilitates decapping by the Dcp1–Dcp2 dimer, which renders the mRNA susceptible to 5′–3′ exonucleolytic degradation by the Xrn1 nuclease (Parker and Song, 2004; Garneau et al., 2007; Goldstrohm and Wickens, 2008). Many of the factors involved in these processes, as well as mRNA degradation intermediates, are enriched in cytoplasmic processing P-bodies (Sheth and Parker, 2003; Cougot et al., 2004).
Pioneering work in the yeast Saccharomyces cerevisiae identified the Ccr4–Not complex as the major deadenylase (Tucker et al., 2001). Although this factor contains two deadenylase subunits, Caf1 (Pop2; DEDD type) and Ccr4 (EEP), the major deadenylase activity is associated with the Ccr4 subunit (Chen et al., 2002; Tucker et al., 2002). By contrast, while Caf1 is an active deadenylase in S. pombe (Takahashi et al., 2007), it is less clear whether the ribonucleolytic activity of Caf1 is required for mRNA turnover in S. cerevisiae (Daugeron et al., 2001; Viswanathan et al., 2004). An important role for Caf1, however, is the recruitment of the Ccr4 subunit to the Ccr4–Not complex, which involves protein–protein interactions between Caf1 and residues in the leucine-rich repeat (LRR) domain of Ccr4 (Clark et al., 2004). In addition, the Ccr4–Not complex contains a number of additional noncatalytic subunits (Collart, 2003; Collart and Timmers, 2004; Denis and Chen, 2003).
The role of the Ccr4–Not complex in mRNA decay is conserved in metazoans, including Drosophila and humans (Temme et al., 2004; Temme et al., 2010; Yamashita et al., 2005). Interestingly, multiple homologues of Caf1 and Ccr4 have been identified in human cells. The paralogues Caf1a (CNOT7) and Caf1b (CNOT8) are components of the human Ccr4–Not complex, while a third, more distant homologue, Caf1z/TOE1, forms a separate nuclear complex involved in mRNA metabolism (Wagner et al., 2007). In HTGM5 fibrosarcoma cells, combined knockdown of Caf1a/Caf1b results in a global increase in the length of poly(A) tails, in contrast to combined knockdown of Ccr4a/Ccr4b, suggesting that Caf1a/Caf1b may have a greater contribution in global deadenylation in mammalian cells (Schwede et al., 2008). At the cellular level, knockdown of Caf1a and/or Caf1b results in a cellular proliferation defect, which depends in part on their catalytic activity (Aslam et al., 2009).
Five Ccr4 homologues have been identified in human cells, but only Ccr4a (CNOT6) and Ccr4b (CNOT6L) contain an amino-terminal LRR required for interactions with Caf1a/Caf1b (Dupressoir et al., 2001). Ccr4b, but not its paralogue Ccr4a, influences cell cycle progression by regulating p27/Kip1 mRNA levels in mouse 3T3 fibroblasts (Morita et al., 2007). Furthermore, Ccr4a is a component of P-bodies and required for foci formation by various P-body components in HeLa cells (Cougot et al., 2004; Andrei et al., 2005).
The exact mechanism of how the Ccr4–Not complex is recruited to mRNA targets remains unclear. In yeast, the PUF family of RNA-binding proteins acts as adapter proteins that mediate interactions with the Ccr4–Not complex to stimulate deadenylation of certain mRNAs (Goldstrohm et al., 2006). Members of the BTG/Tob family of antiproliferative proteins may also contribute to the recruitment of the Ccr4–Not complex (Winkler, 2010). Both Tob and BTG2 interact with the Caf1a/Caf1b subunits and enhance global deadenylation (Ezzeddine et al., 2007; Mauxion et al., 2008). It has been proposed that during translation termination, binding of Tob and PABPC1 may result in recruitment of the Ccr4–Not deadenylase (Ezzeddine et al., 2007; Funakoshi et al., 2007). In addition, several recent studies in Drosophila and mammalian cells have shown that microRNA-mediated gene repression is associated with deadenylation and mRNA decay (Behm-Ansmant et al., 2006; Wu et al., 2006) and implicates the recruitment of the Ccr4–Not complex by components of the microRNA machinery (Chen et al., 2009; Fabian et al., 2009; Zekri et al., 2009; Piao et al., 2010). Furthermore, the activity of the Ccr4–Not deadenylase subunits may be regulated after their recruitment to mRNA (Morozov et al., 2010).
In addition to a role in mRNA turnover, and consistent with its functions in yeast, a number of protein–protein interactions point to a separate role of the Ccr4–Not complex in transcription in human cells. Particularly, several subunits are reported to regulate the activity of nuclear receptors (Prevot et al., 2001; Hiroi et al., 2002; Morel et al., 2003; Winkler et al., 2006; Garapaty et al., 2008; Govindan et al., 2009).
Here we investigated the role of the Ccr4a/Ccr4b subunits of the Ccr4–Not complex in the regulation of cellular functions using MCF7 cells. We showed that the Ccr4 paralogues have distinct roles in mediating cell survival, the formation of P-bodies, and regulating gene expression as compared with the Caf1a/Caf1b deadenylase subunits. Furthermore, we found that the LRR domain was required for proper localization of Ccr4b and that expression of Ccr4b lacking this domain reduced cell proliferation but did not affect cell survival. Finally, we identified Ccr4a/Ccr4b as a key regulator of insulin-like growth factor–binding protein 5 (IGFBP5), which mediates cell cycle arrest and senescence via a p53-dependent pathway (Kim et al., 2007).
RESULTS
Knockdown of Ccr4a and/or Ccr4b results in reduced cell proliferation and decreased cell survival
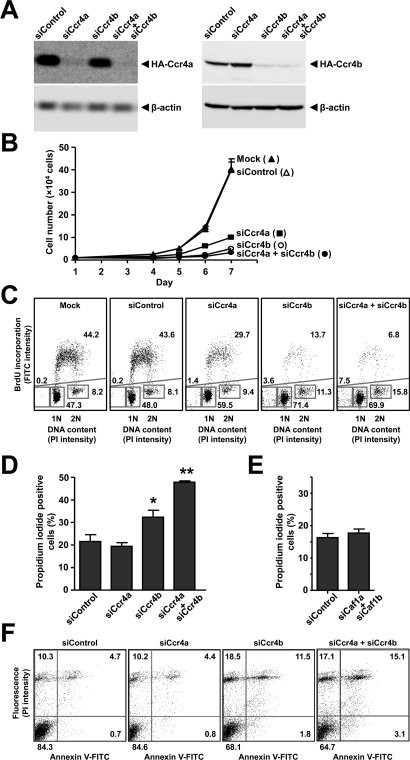
To study the cellular role(s) of the highly related Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4–Not complex in MCF7 breast cancer cells, we used small interfering RNA (siRNA)–mediated knockdown. Using different siRNA duplexes targeting distinct regions in the mRNAs, we obtained efficient knockdown (Figure 1A). At the mRNA level, we routinely achieved a knockdown efficiency of >80% (unpublished data). In agreement with earlier observations by Morita et al. (2007), we noted a strong effect on cell proliferation upon knockdown of Ccr4b (Figure 1B). Interestingly, however, we also observed a significant effect on MCF7 cell proliferation upon knockdown of Ccr4a (Figure 1B), which has no effect on cell proliferation of NIH 3T3 mouse fibroblasts (Morita et al., 2007). The effects on cell proliferation upon knockdown of Ccr4a and/or Ccr4b were confirmed by cell cycle profiling using flow cytometry (Figure 1C). In addition to a decreased fraction of cells in S phase and a concomitant increase in G1, we surprisingly also found a significant fraction of cells with sub-G1 DNA content upon Ccr4a/Ccr4b knockdown (Figure 1C), indicating reduced cell viability (Galluzzi et al., 2009). In agreement with the latter observation, we confirmed decreased cell viability upon knockdown of Ccr4b and particularly upon combined knockdown of Ccr4a and Ccr4b using propidium iodide exclusion in combination with flow cytometry (Figure 1D). Previously, we had not observed evidence for decreased cell viability upon (combined) knockdown of Caf1a (CNOT7) and Caf1b (CNOT8) using cell cycle profiling by flow cytometry (Aslam et al., 2009). This was confirmed using propidium iodide exclusion combined with flow cytometry (Figure 1E), suggesting unique functions for the Ccr4a/Ccr4b deadenylases as compared with the Caf1a/Caf1b catalytic subunits in mediating cell survival.
FIGURE 1:
Knockdown of Ccr4a and/or Ccr4b results in reduced cell proliferation and decreased cell viability. (A) Knockdown of Ccr4a and Ccr4b in MCF7 cells. Following transfection of vectors expressing HA-Ccr4a or HA-Ccr4b, cells were treated with siRNA targeting Ccr4a and/or Ccr4b. After 3 d, total lysates were analyzed by immunoblotting. Antibodies recognizing β-actin were used to assess equal loading. (B) Inhibition of cell proliferation upon knockdown of Ccr4a and/or Ccr4b. Following siRNA transfection, adherent cells were counted in a hemocytometer at 24 h intervals. (C) Cell cycle profiling of MCF7 cells transfected with siRNA targeting Ccr4a and/or Ccr4b. Dot plots of bivariate flow cytometry using propidium iodide fluorescence to determine DNA content (horizontal) and anti-BrdU fluorescence (FITC) to identify BrdU incorporation to label cells in S phase (vertical) are shown. Cells (n = 20,000) were analyzed per condition 72 h after transfection. The percentages of cells in (sub-)G1, S, and G2/M phase are indicated. (D) Decreased cell viability upon knockdown of Ccr4a and/or Ccr4b. Cell viability (n = 10,000) was monitored by propidium iodide exclusion as detected by flow cytometry at 72 h after transfection. The percentage of (nonviable) cells labeled by propidium iodide is indicated. *p < 0.05, **p < 0.01 (compared with nontargeting control siRNA). (E) Cell viability is not decreased upon combined knockdown of Caf1a and Caf1b. Cell viability was measured by propidium iodide exclusion as detailed in (D). (F) Increased apoptosis upon knockdown of Ccr4b. Apoptosis was monitored using annexin V binding (horizontal) and propidium iodide exclusion (vertical). Cells (n = 20,000) were analyzed 72 h after transfection. The percentage of cells present in each quadrant is indicated.
To investigate whether reduced survival was due to increased apoptosis or alternative mechanisms, we carried out bivariate flow cytometry using annexin V and propidium iodide staining (Vermes et al., 1995) (Figure 1F). The fraction of early and late apoptotic cells (bottom right and top right quadrant, respectively) was increased following Ccr4b knockdown and more pronounced upon combined knockdown of Ccr4a and Ccr4b (Figure 1F). In addition, the fraction of nonviable cells that did not bind annexin V was also increased in Ccr4b and combined Ccr4a/Ccr4b knockdown cells (top left quadrant). Taken together, these results indicate that decreased survival in Ccr4a/Ccr4b knockdown cells is due to both apoptosis-dependent and ‑independent mechanisms.
The CNOT1 and CNOT3 subunits of the Ccr4–Not complex are required for cell proliferation but do not contribute to cell survival
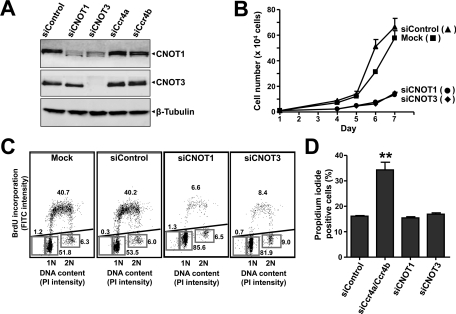
To investigate whether the Ccr4a/Ccr4b deadenylase subunits carry out a unique role, or whether other Ccr4–Not components also contribute to cell survival and proliferation, we used siRNA to knock down the CNOT1 and CNOT3 subunits (Figure 2A). These subunits do not have paralogues in human cells, and knockdown may result in disruption of the complex. Interestingly, we reproducibly observed a reduction of CNOT1 protein levels upon knockdown of CNOT3, which may indicate that CNOT3 is required for stability of the complex (Figure 2A). As was observed in Ccr4a and Ccr4b knockdown cells, as well as in Caf1a/Caf1b knockdown cells (Aslam et al., 2009), knockdown of CNOT1 and CNOT3 resulted in a strong reduction of cell proliferation (Figure 2B). Cell cycle profiling confirmed the roles for these subunits in cell cycle progression as a decreased fraction of cells in S phase, and a concomitant increase of cells in G1 was observed (Figure 2C). Remarkably, no significant increase in the percentage of cells with sub-G1 DNA content was found. This was confirmed by measurement of propidium iodide exclusion by flow cytometry, which did not show increased uptake of propidium iodide upon knockdown of CNOT1 or CNOT3 as compared with nontargeting control siRNA (Figure 2D). Thus these results indicate that CNOT1 and CNOT3 are required for cell proliferation of MCF7 cells but—in contrast to the Ccr4a/Ccr4b deadenylase subunits—are not required for cell survival.
FIGURE 2:
Knockdown of CNOT1 and CNOT3 results in reduced cell proliferation but does not affect cell viability. (A) Knockdown of CNOT1 and CNOT3 in MCF7 cells. Cells were transfected with the indicated siRNA, and protein levels were analyzed after 72 h using the indicated antibodies. (B) Inhibition of cell proliferation upon knockdown of CNOT1 and CNOT3. After siRNA transfection, adherent cells were counted in a hemocytometer at 24-h intervals. (C) Cell cycle analysis of MCF7 cells upon knockdown of CNOT1 and CNOT3. Cells were mock treated or transfected with nontargeting control siRNA or siRNA targeting CNOT1 or CNOT3. Dot plots show bivariate flow cytometry using propidium iodide fluorescence to determine DNA content (horizontal) and anti-BrdU fluorescence (FITC) to identify BrdU incorporation to label cells in S phase (vertical). Cells (n = 20,000) were analyzed per condition 72 h after transfection. The percentages of cells in (sub-)G1, S, and G2/M phase are indicated. (D) Cell viability is not decreased upon knockdown of CNOT1 or CNOT3. Cell viability was measured by propidium iodide exclusion as detailed in (Figure 1D). **p < 0.01 (compared with nontargeting control siRNA).
The deadenylase components Caf1a/Caf1b and Ccr4a/Ccr4b differentially affect foci formation of P-body markers
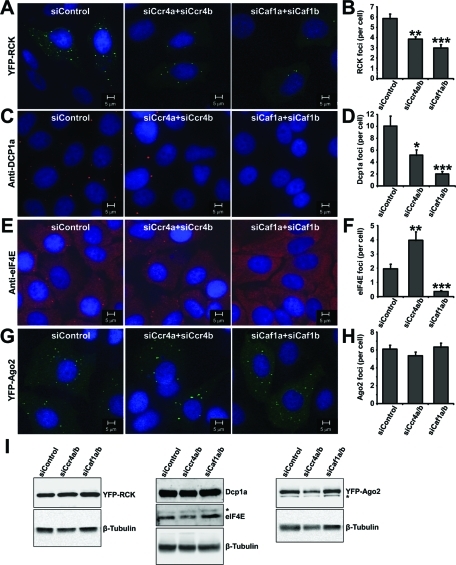
Because knockdown of the Caf1a/Cafb deadenylases did not affect cell viability, in contrast to combined knockdown of Ccr4a and Ccr4b, we characterized the differential requirements for the Ccr4a/Ccr4b versus the Caf1a/Caf1b deadenylases further by analyzing cytoplasmic foci formation of the P-body components p54/Rck/Ddx6, Dcp1a, eIF4E, and Ago2 (Figure 3). The average number of foci per cell formed upon expression of yellow fluorescent protein (YFP)–p54/Rck/Ddx6 was decreased upon combined Ccr4a/Ccr4b knockdown cells, which was unexpectedly more pronounced after combined knockdown of Caf1a/Caf1b (Figure 3, A and B). Similar observations were made using staining of Dcp1a with antibodies recognizing endogenous protein (Figure 3, C and D). By contrast, when antibodies recognizing endogenous eIF4E were used, the average number of foci per cell was increased upon combined knockdown of Ccr4a/Ccr4b, while a reduction was observed upon combined knockdown of Caf1a/Caf1b (Figure 3, E and F). No change in the number of foci formed after expression of YFP-Ago2 was observed upon knockdown of either Ccr4a/Ccr4b or Caf1a/Caf1b (Figure 3, G and H). No stress bodies were induced upon siRNA transfection as assessed by eIF3 foci formation (unpublished data). Furthermore, the effects on foci formation were not caused by altered expression of the respective proteins as determined by immunoblotting (Figure 3I). Together these results show that the accumulation of RCK, Dcp1a, and eIF4E foci is particularly sensitive to knockdown of Caf1a/Caf1b and support the notion that the Ccr4a/Ccr4b deadenylases have differential roles as compared with the Caf1a/Caf1b catalytic subunits of the Ccr4–Not complex.
FIGURE 3:
Distinct roles for the deadenylase components Caf1a/Caf1b and Ccr4a/Ccr4b in foci formation of P-body components. (A, B) Reduced foci formation of YFP-RCK upon knockdown of Ccr4a/Ccr4b and Caf1a/Caf1b. (C, D) Decreased formation of Dcp1a foci upon knockdown of Ccr4a/Ccr4b and Caf1a/Caf1b. (E, F) Increased foci formation by eIF4E in MCF7 cells upon knockdown of Ccr4a/Ccr4b but dramatically decreased foci formation upon knockdown of Caf1a/b. (G, H) Formation of YFP-Ago2 foci is not affected upon knockdown of Ccr4a/Ccr4b and Caf1a/Caf1b. Cells were transfected with the indicated siRNA and processed for immunofluorescence microscopy using antibodies recognizing endogenous eIF4E (C, D) or Dcp1a (E, F). p54/Rck (A, B) and Ago2 (G, H) were detected 24 h after transfection of the respective YFP-fusion proteins. *p < 0.05, **p < 0.01, ***p < 0.001 (compared with nontargeting control siRNA). (I) Protein levels of YFP-RCK, Dcp1a, eIF4E, and YFP-Ago2 were unaffected upon knockdown of Caf1a/Caf1b or Ccr4a/Ccr4b. Protein lysates from cells transfected with the indicated siRNA were subjected to immunoblotting. YFP-RCK and YFP-Ago2 were detected with antibodies recognizing YFP, whereas eIF4E and Dcp1a were detected with antibodies recognizing the endogenous proteins. Cross-reactive bands are indicated with an asterisk.
Interactions mediated by the LRR domain impact the subcellular localization of Ccr4b
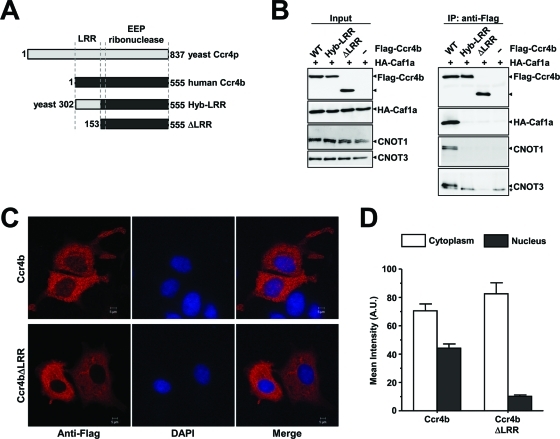
To further explore the relationship between Caf1a and Ccr4b, we next focused on the role of the N-terminal LRR domain of the Ccr4a/Ccr4b subunits (Figure 4A). As expected, Flag-Ccr4b can bind to HA-Caf1a. This interaction depends on the presence of the LRR domain (Figure 4B). Interestingly, replacing the LRR domain of human Ccr4b with the LRR of yeast Ccr4 disrupted the interaction with Caf1a, demonstrating that the human domain cannot be changed for the yeast residues. Furthermore, Flag-Ccr4b did coimmunoprecipitate the Ccr4–Not subunits CNOT1 and CNOT3 in addition to HA-CNOT7 (Figure 4B). However, this was not observed when the LRR residues were deleted, indicating that Ccr4b interacts with the CNOT1 and CNOT3 subunits via Caf1a protein in agreement with observations made in yeast (Clark et al., 2004).
FIGURE 4:
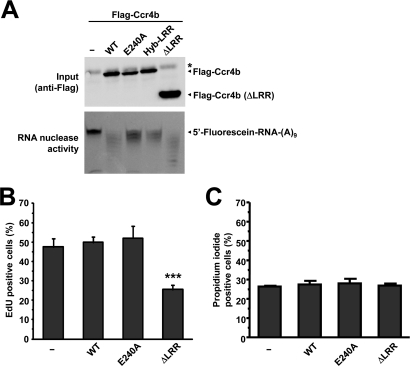
The LRR domain of Ccr4b is required for interactions with Caf1a, incorporation into the Ccr4–Not complex, and subcellular localization. (A) Schematic diagrams the Ccr4b constructs. Indicated are the S. cerevisiae Ccr4p protein (light gray), the human Ccr4b homologue of Ccr4p (black), and the locations of the LRR and EEP ribonuclease domain. (B) The LRR domain of Ccr4b interacts with CNOT1 and CNOT3 via Caf1a. Plasmids pCMV5-HA–CNOT7 (or control plasmid) and vectors expressing wild-type Flag-Ccr4b, Ccr4b (Hyb-LRR), or Cr4b (ΔLRR) were transiently cotransfected into HEK293 cells. Total lysates (left) and anti-Flag immunoprecipitates (right) were analyzed using the indicated antibodies. (C) The LRR domain of Ccr4b is required for its localization to the nucleus. Following transfection with the indicated cDNA expression vectors, MCF7 cells were processed for immunofluorescence using anti-Flag antibodies after 24 h. Flag-Ccr4b was found to localize to both the nucleus and cytoplasm (top) whereas the Flag-Ccr4b lacking the LRR domain (ΔLRR) was exclusively cytoplasmic (bottom). (D) Quantification of the nucleocytoplasmic distributions of Ccr4b and Ccr4bΔLRR. The mean fluorescence intensity was determined from cells expressing Flag-Ccr4b (n = 50) and Flag-Ccr4bΔLRR (n = 49) using three identical regions of interest per cell. The mean intensity is plotted. Error bars represent the standard error of the mean (SEM).
Surprisingly, the LRR domain influenced the subcellular localization of Ccr4b. On expression of Flag-Ccr4b, the majority of Ccr4b was detected in the cytoplasm, although an appreciable amount was also found in the nucleus (Figure 4C, top) (Cougot et al., 2004; Andrei et al., 2005). Remarkably, deletion of the LRR domain of Flag-Ccr4b resulted in an almost exclusive cytoplasmic localization (Figure 4C, bottom, and 4D).
Requirements of the LRR domain of Ccr4b for deadenylase activity and cell proliferation
To characterize the role of the LRR domain further, we expressed and immunopurified several Flag-Ccr4b variants from human embryonic kidney (HEK) 293 cells. Subsequently, the deadenylase activity of wild-type Flag-Ccr4b, enzymatically inactive Flag-Ccr4b, Flag-Ccr4b containing the yeast LRR domain, or Flag-Ccr4b lacking the LRR domain was measured using a fluorescently labeled oligonucleotide substrate. As expected, immunopurified Flag-Ccr4b was able to degrade the oligonucleotide substrate. The activity was severely reduced when Flag-Ccr4 containing the amino acid substitution E240A, or Flag-Ccr4b containing the yeast LRR domain, was used (Figure 5A). Interestingly, Flag-Ccr4b lacking the LRR domain was an active deadenylase enzyme, thereby demonstrating that the LRR domain is not absolutely required for the deadenylase activity of human Ccr4b in contrast to the yeast enzyme (Clark et al., 2004).
FIGURE 5:
The role of the LRR domain of Ccr4a/Ccr4b in deadenylation and cell proliferation. (A) The LRR domain of Ccr4b is dispensable for deadenylase activity. Cells were transfected with empty vector or the indicated Flag-Ccr4b expression plasmids. After immunoprecipitation with anti-Flag antibodies, bound proteins were eluted and assayed for deadenylase activity using a 5′ Flc-labeled RNA substrate. The asterisk indicates the presence of a cross-reactive band. (B) Expression of Ccr4b lacking the LRR domain inhibits cell proliferation. Cells were transfected with empty vector or the indicated Flag-Ccr4b expression plasmids. The percentage of cells in S phase as a measure of cell proliferation was determined using the thymidine analogue EdU and fluorescence microscopy. ***p < 0.001 (compared with empty vector control). (C) Expression of Ccr4b does not interfere with cell viability. Cells were transfected with empty vector or the indicated Flag-Ccr4b expression plasmids. Cell viability was determined using propidium iodide exclusion and flow cytometry.
To examine whether the LRR domain of Ccr4b was involved in the regulation of cell proliferation, we next overexpressed wild-type Flag-Ccr4b, an enzymatically inactive version (E240A), as well as Flag-Ccr4b lacking the LRR domain in MCF7 cells. A pulse labeling with the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU) was used to identify cells in S phase as a measure for cell proliferation. Interestingly, while overexpression of Flag-Ccr4b or an enzymatically inactive form did not affect cell proliferation, overexpression of Flag-Ccr4b lacking the LRR domain caused a significant drop in EdU-positive cells (Figure 5B). By contrast, overexpression of (enzymatically inactive) Ccr4b, or Ccr4b lacking the LRR domain, did not affect cell viability (Figure 5C). Together, these data suggest that interactions with Caf1a/Caf1b mediated by the LRR domain of Ccr4b are important for cell cycle progression.
Ccr4a/Ccr4b and Caf1a/Caf1b regulate distinct gene sets
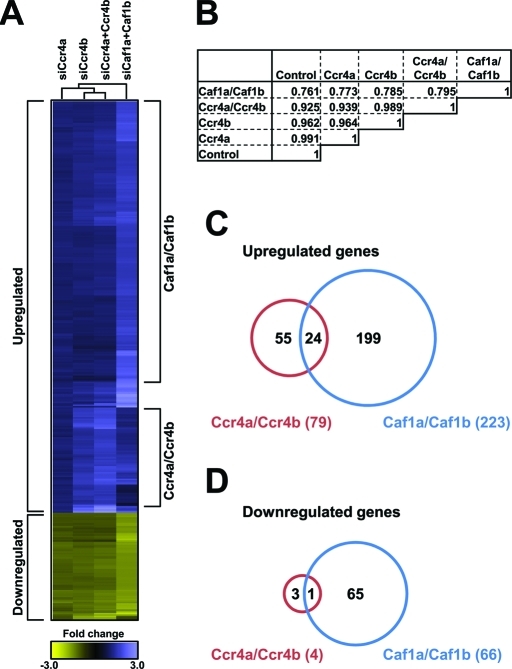
Next, we used gene expression profiling to understand in more detail the mechanism by which the Ccr4a/Ccr4b deadenylases regulate cell proliferation and survival. Thus MCF7 cells were treated with siRNA pools targeting Ccr4a and/or Ccr4b, Caf1a/Caf1b, and a nontargeting control pool. Total RNA was isolated and analyzed using Affymetrix Human Gene 1.0 ST Array GeneChips. In combined Ccr4a/Ccr4b knockdown cells, 79 genes were found to be up-regulated, whereas only 4 genes were down-regulated (fold change > 1.50, p < 0.050), which is expected based on their function as deadenylase enzymes. No differentially expressed genes were identified upon knockdown of Ccr4a. However, the expression profile upon knockdown of Ccr4b was enhanced when combined with knockdown of Ccr4a, indicating that knockdown of Ccr4a can be (partially) compensated for by Ccr4b. Interestingly, combined knockdown of Caf1a/Caf1b resulted in a larger number of differentially expressed genes (223 up and 66 down) with limited overlap with the differentially expressed gene set upon Ccr4a/Ccr4b knockdown (Figure 6, A–D). Together, these results demonstrate that Ccr4a/Ccr4b and Caf1a/Caf1b largely regulate distinct gene sets with limited overlap.
FIGURE 6:
Ccr4a/Ccr4b and Caf1a/Caf1b regulate distinct gene sets. (A) Diagram of hierarchical clustering of gene expression profiles of MCF7 cells treated with the indicated siRNA pools. Probes are represented vertically, while conditions are shown horizontally. The subset of probes was selected on the basis of the expression profiles (>1.50-fold differential expression compared with control nontargeting siRNA pool, p-value < 0.050). Hierarchical clustering was carried out using CARMAweb (https://carmaweb.genome.tugraz.at/carma). (B) Matrix of Pearson’s correlation coefficients of gene expression profiles as indicated. (C) Venn diagram showing limited overlap between the set of up-regulated genes in Ccr4a/Ccr4b and Caf1a/Caf1b knockdown cells. (D) Venn diagram showing limited overlap between the set of down-regulated genes in Ccr4a/Ccr4b and Caf1a/Caf1b knockdown cells.
Identification of Ccr4a/Ccr4b target genes
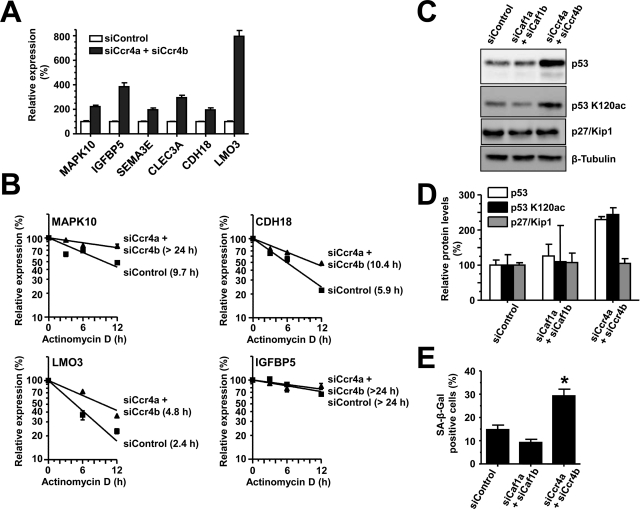
A number of genes, identified from gene expression profiling, were subsequently validated by reverse transcriptase–quantitative PCR (RT-qPCR) using gene-specific primers. We confirmed enhanced expression of IGFBP5 (approximately threefold), CLEC3A (approximately threefold), SEMA3E (approximately twofold), MAPK10 (approximately twofold), CDH18 (approximately twofold), and LMO3 (approximately eightfold) upon Ccr4a/Ccr4b knockdown (Figure 7A). To determine whether the enhanced expression of the genes was due to increased transcript stability following loss of Ccr4a/Ccr4b, we used the transcriptional inhibitor actinomycin D in combination with RT-qPCR to measure mRNA stability. Of the six genes identified, MAPK10, CDH18, and LMO3 mRNA transcripts were significantly more stable after Ccr4a/Ccr4b knockdown compared with control siRNA treatment (Figure 7B). The mRNAs of IGFBP5, SEMA3E, and CLEC3A were stable under normal conditions, precluding the assessment of increased mRNA half-lives of the mRNAs of these genes (Figure 7B and unpublished data).
FIGURE 7:
Identification of Ccr4a/Ccr4b target genes. (A) Confirmation of mRNA target genes of Ccr4a/Ccr4b. mRNA levels of the indicated genes were detected using RT-qPCR with GAPDH as a reference gene. All assays were carried out in triplicate. (B) Measurement of mRNA stability of Ccr4a/Ccr4b target genes. Actinomycin D was added (72 h after siRNA transfection), and total mRNA was isolated at 0, 3, 6, and 12 h after treatment. mRNA transcript levels were determined by RT-qPCR using GAPDH as a reference gene. mRNA half-lives were derived from the slope of the fitted line [mRNA] = 100 · e(−k · t), where [mRNA] = 100% at t = 0, k is the decay constant, and the half-life t1/2 is given by t1/2 = ln(2) / k. (C) Increased protein levels of p53 and p53 K120ac in MCF7 cells upon Ccr4a/Ccr4b knockdown. MCF7 cells were transfected with control, Caf1a/Caf1b, or Ccr4a/Ccr4b siRNA and analyzed by immunoblotting. (D) Quantitative immunoblotting of p53, p53 K120ac, and p27/Kip1 protein levels relative to β-tubulin. Signal intensities were measured (duplicates) and analyzed in ImageJ. Error bars indicate SEM. (E) Increased cellular senescence upon knockdown of Ccr4a/Ccr4b. MCF7 cells were treated with control, Caf1a/Caf1b, or Ccr4a/Ccr4b siRNA and stained for β-galactosidase activity. *p < 0.05. The error bars represent SEM.
IGFBP5 overexpression is associated with cellular senescence via a p53-dependent pathway in human umbilical vein endothelial cells (HUVEC) (Kim et al., 2007). Consistent with IGFBP5 up-regulation, p53 protein levels were increased upon Ccr4a/Ccr4b knockdown, although no change in mRNA levels was observed in the expression profiling data. Because activation of p53 residue at Lys-120 by acetylation is indispensable for p53-dependent growth arrest and apoptosis (Tang et al., 2008), we also determined the acetylation of p53 at this residue in Ccr4a/b and Caf1a/b knockdown MCF7 cells. Indeed, acetylation of Lys-120 was significantly induced in Ccr4a/Ccr4b knockdown cells as compared with control or Caf1a/Caf1b knockdown cells (Figure 7C). In contrast, we did not observe increased p27/Kip1 mRNA or protein levels in MCF7 cells (Figure 7C). These results were confirmed by quantitative immunoblotting. Interestingly, while the overall levels of both total p53 as well as p53 acetylated at Lys-120 were significantly increased in Ccr4a/Ccr4b knockdown cells, the fraction of p53 acetylated at Lys-120 was not increased (Figure 7D).
Finally, we looked at the senescence phenotype by β-galactosidase staining (Dimri et al., 1995). Consistent with the role of IGFBP5, knockdown of Ccr4a/Ccr4b caused a significant increase in senescence-associated β-galactosidase staining as compared with control or Caf1a/Caf1b knockdown (Figure 7E). Taken together, these results suggest that knockdown of Ccr4a/Ccr4b in MCF7 cells causes up-regulation of IGFBP5, which may mediate inhibition of cellular proliferation and induce senescence via a p53-dependent pathway.
DISCUSSION
Distinct roles for the human Ccr4a/Ccr4b and Caf1a/Caf1b deadenylases
In this report we show that the human deadenylase subunits associated with the Ccr4–Not complex have distinct roles based on three criteria: 1) The Ccr4 paralogues mediate cell survival and inhibit cellular senescence in contrast to the Caf1a/Caf1b subunits; 2) the accumulation of foci by P-body components is particularly dependent on Caf1a/Caf1b and less sensitive to knockdown of the Ccr4 paralogues; 3) distinct gene sets are regulated by Ccr4a/Ccr4b as compared with Caf1a/Caf1b as shown by genome-wide expression profiling; and 4) (acetylated) p53 is selectively induced upon knockdown of Ccr4a/Ccr4b as compared with the Caf1 paralogues. These phenotypes are not due to “off-target” effects, as different siRNA duplexes targeting different regions of the mRNAs yielded similar results. Moreover, the phenotypic differences are not merely quantitative, for instance, due to varying knockdown efficiencies. For example, while the effect on cell proliferation is less pronounced and no effect on cell viability is observed upon knockdown of Caf1a/Caf1b, the effect on foci formation of YFP-RCK and Dcp1a is quantitatively more significant as compared with combined knockdown of Ccr4a/Ccr4b. Furthermore, there are qualitative effects on foci formation by eIF4A upon knockdown of Ccr4a/Ccr4b and Caf1a/Caf1b, respectively. Third, using genome-wide expression profiling, qualitative differences were found upon knockdown of the Ccr4-type and Caf1 paralogues. The expression of more genes was affected upon knockdown of Caf1a/Caf1b, and little overlap was observed with the Ccr4a/Ccr4b knockdown profile.
Further evidence for unique roles for the Ccr4a/Ccr4b subunits is provided by the analysis of the cellular phenotype upon knockdown of the noncatalytic subunits CNOT1 and CNOT3. While the effect on the percentage of cells in S phase was more pronounced upon knockdown of these subunits as compared with knockdown of Ccr4b (and comparable to combined knockdown of Ccr4a/Ccr4b), no effect on cell viability was observed.
Ccr4a and Ccr4b are required for cell cycle progression and prevent cell death and senescence
Consistent with findings reported by Morita et al. (2007), we found that the Ccr4b deadenylase is important in controlling cell proliferation of MCF7 breast cancer cells. However, while up-regulation of p27/Kip1 is implicated in reduced cell cycle progression of NIH3T3 cells (Morita et al., 2007), we did not observe up-regulation at either mRNA or protein level of this cell cycle inhibitor in MCF7 cells. We extend these observations further by showing that the Ccr4a deadenylase as well as two noncatalytic subunits of the Ccr4–Not complex, the large subunit CNOT1 and CNOT3, are also important for efficient cell proliferation. Finally, we reveal two additional roles of the Ccr4a/Ccr4b proteins in mediating cell survival and preventing cellular senescence. We believe that the most likely explanation for the observed differences is provided by assuming cell type–specific roles of the Ccr4a/Ccr4b deadenylases, although other explanations cannot be excluded.
Distinct roles for Caf1a/Caf1b and Ccr4a/Ccr4b in foci formation by P-body components
While Ccr4 can be found in P-bodies in some cell types (Cougot et al., 2004; Andrei et al., 2005), we did not find Caf1a or Ccr4b to localize to P-bodies in MCF7 cells. Because the accumulation of several P-body components depends on active deadenylation and is reduced upon knockdown of Ccr4a (Andrei et al., 2005; Zheng et al., 2008), we looked at foci formation of the P-body components RCK, Dcp1a, eIF4E, and Ago2 to investigate in more detail the distinct roles for the Caf1a/Caf1b and Ccr4a/Ccr4b deadenylases. Specific dependencies in P-body assembly have been described (Teixeira and Parker, 2007). Whereas quantitative differences were observed when the accumulation of RCK and Dcp1a was analyzed, qualitative differences were notable upon analysis of eIF4E foci accumulation. The accumulation of eIF4E foci upon knockdown of Ccr4a/Ccr4b was not due to the formation of stress bodies as eIF3 foci were observed only after treatment with sodium arsenite and were absent upon combined knockdown of Ccr4a/Ccr4b (unpublished data). Thus these data suggest that, compared with Caf1a/Caf1b, the Ccr4a/Ccr4b proteins act at a different stage of the deadenylation process and therefore differentially affect the accumulation of P-body components. Alternatively, the structural contributions to P-body formation via protein–protein interactions by Caf1a/Caf1b may be more significant as compared with Ccr4a/Ccr4b. Interestingly, differential roles for Caf1 and Ccr4 on P-body formation and deadenylation have also been described in Aspergillus nidulans (Morozov et al., 2010).
Mechanistic insight: the role of the LRR domain
In yeast, the LRR domain of Ccr4 is critical for its function in vivo as well as its deadenylase activity (Clark et al., 2004). Furthermore, the LRR of Ccr4 mediates its interaction with Caf1, thereby linking Ccr4 to the Ccr4–Not complex (Dupressoir et al., 2001; Clark et al., 2004). In agreement with these studies, we find that deleting the LRR of Ccr4b abolishes its interaction with Caf1a and other components of the Ccr4–Not complex. However, in contrast to yeast Ccr4, the LRR of Ccr4b is not critical for its deadenylase activity. This is in agreement with recent reports showing that purified Ccr4b lacking the LRR domain is enzymatically active and the fact that Nocturnin, a Ccr4-type deadenylase lacking an LRR domain, is active in vitro (Baggs and Green, 2003; Wang et al., 2010).
We previously identified the Caf1a and Caf1b deadenylase subunits of the human Ccr4–Not complex as mediators of efficient proliferation of MCF7 cells, partially compensating each other’s function (Aslam et al., 2009). Because the Caf1a/Caf1b subunits are required for recruitment of the Ccr4a/Ccr4b subunits to the complex, we hypothesized that deregulated Ccr4 activity contributes to the phenotype. In agreement with this model, we found that overexpression of Ccr4b lacking the LRR domain resulted in aberrant localization and reduced cell proliferation, while cell viability remained unaffected. This is consistent with a unique role for the Ccr4-type deadenylases in mediating cell survival, which is independent of interactions with Caf1a/Caf1b. However, other explanations may also be possible; for example, the LRR domain may contribute to mRNA interactions required for regulated mRNA decay during cell cycle progression.
Mechanistic insight from gene expression profiling
Most of the differentially expressed genes upon knockdown of Ccr4a/Ccr4b were up-regulated, consistent with their role in mRNA turnover. In support of this notion, we tested transcript stability of six putative target genes, whose expression was significantly altered in the genome-wide analysis. The mRNA stability of LMO3, CDH18, and MAPK10 were significantly increased following Ccr4a/Ccr4b knockdown, consistent with their role in mRNA degradation. IGFBP5, SEMA3E, and CLEC3A were stable transcripts, which precluded the use of actinomycin D to accurately determine their stability. Thus these data suggest that at least a significant fraction of the genes identified in the gene expression profiling experiment appear to be direct targets as their up-regulation correlates with increased transcript stability. Interestingly, CLEC3A, SEMA3E, MAPK10, and IGFBP5 are thought to be involved in reduced breast cancer cell proliferation, apoptosis, and inhibition of tumor development (Bogoyevitch, 2006; Kigel et al., 2008; Tsunezumi et al., 2009). IGFBP5 is one of six members of the IGFBP protein family and is an important component of the IGF axis (Beattie et al., 2006). In breast cancer cells, IGFBP5 binds to IGF I/II and blocks the activation of IGF signaling. Reduction or cleavage of IGFBP5 is then followed by the release of IGF, which reduces apoptosis and activates cell proliferation (Beattie et al., 2006; Akkiprik et al., 2008). Consistent with this, previous reports have identified IGFBP5 as a key regulator of cell proliferation and apoptosis in breast cancer cell lines (Butt et al., 2003; Butt et al., 2005). Our data suggest that IGFBP5 may indeed exert its apoptotic effects via a p53-dependent mechanism, in support of Kim and coworkers (2007), who show similar data in HUVEC. The precise contribution of IGFBP5 and other direct or indirect target genes of Ccr4a/Ccr4b to the observed phenotype will be the focus of future studies.
MATERIALS AND METHODS
Plasmids, mutagenesis, and siRNA
The open reading frames of CNOT6 and CNOT6L were obtained by RT-PCR using MCF7 NKI total RNA as a template. After cloning the respective cDNAs into the SmaI site of pBluescript II KS(+) and sequence verification, the cDNAs were subcloned in pcDNA3-FLAG (BamHI-EcoRV) and pCMV5-HA (XhoI-digested cDNA ligated into SalI-digested vector). Site-directed mutations to inactivate the active sites (E240A) were introduced using standard protocols (QuikChange, Stratagene, La Jolla, CA). The hybrid cDNA encoding the yeast leucine-rich region (yeast residues 302–453) fused to the human CNOT6 (amino acids 148–557) and CNOT6L (amino acids 153–555) regions was constructed using overlap PCR techniques. Primer sequences are available upon request. Deletion of the leucine-rich regions in CNOT6 (residues 2–147) and CNOT6L (2–152) were also obtained by standard PCR techniques. See Aslam et al. (2009) for a description of plasmid pCMV5-HA–CNOT7.
In addition to those described before (Winkler et al., 2006; Aslam et al., 2009), the following siRNA duplexes were used (Dharmacon, Lafayette, CO): CNOT6 (D-019101–01 and On-Target plus SMARTpool L-019101–00), CNOT6L (D-016411–02 and On-Target plus SMARTpool L-016411–00).
Cell culture and transfection
MCF7 NKI and HEK293 cells were routinely maintained and transfected as described previously (Aslam et al., 2009).
Western blotting and immunoprecipitation
Proteins were separated by SDS–PAGE and transferred to nitrocellulose membranes. Proteins were visualized using an enhanced chemiluminescence detection kit (Pierce, Rockford, IL) and a Fujifilm LAS-4000 imager. Primary antibodies used were YFP (sc-8336, 1:1000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA), eIF4E (ab-1126, 1:1000 dilution; Abcam, Cambridge, MA), Dcp1a (1:1000 dilution) (Lykke-Andersen and Wagner, 2005), p53 (DO-1, sc-126, 1:1000 dilution; Santa Cruz) and tubulin (sc-7396, 1:1000 dilution; Santa Cruz), p27/Kip1 (2552, 1:1000 dilution; Cell Signaling Technology, Danvers, MA), p53-K120ac antibody (ab78316, 1:500 dilution; Abcam), and β-actin (AC-15, 1:1000; Sigma, St. Louis, MO). All horseradish peroxidase–conjugated secondary antibodies were purchased from Santa Cruz Biotechnology and used in 1:2500 dilutions. Other antibodies for immunoprecipitations and Western blotting were as used before (Winkler et al., 2006; Aslam et al., 2009). Chemiluminescent signals were visualized using a Fujifilm LAS-4000 imager.
Flow cytometry
MCF-7 NKI cells (400,000 cells in a T25 flask) were transfected with siRNA as described (Aslam et al., 2009). After 24 h, medium was removed and fresh medium was added to the cells. For cell cycle profiling, cells were labeled after an additional 46 h for 2 h in the presence of 1 μM 5-bromo-2′-deoxyuridine (BrdU) and prepared for bivariate flow cytometry using propidium iodide and fluorescein isothiocyanate (FITC)–conjugated anti-BrdU antibody 3D4 (BD PharMingen, San Jose, CA).
For cell viability analysis, transfected cells (150,000 per T25 flask) were harvested by trypsin treatment. After inactivation of trypsin by the addition of the supernatant medium, cells were washed and resuspended in phosphate-buffered saline (PBS), incubated with propidium iodide (1.0 μg/ml), and subjected to flow cytometry.
For annexin V binding, transfected cells (200,000 per T25 flask) were harvested by trypsin treatment. After inactivation of trypsin by the addition of the supernatant medium, cells were washed and resuspended in annexin V binding buffer (Invitrogen, San Jose, CA; 0.01 M HEPES-NaOH [pH 7.4], 0.14 M NaCl, 2.5 mM CaCl2), incubated with propidium iodide and FITC-labeled annexin V (BD Biosciences; according to the manufacturer’s instructions), and subjected to flow cytometry. Analysis was carried out using a FACS Aria flow cytometer, FACSDiva software (BD Biosciences, San Jose, CA), and the WinMDI package.
Fluorescence microscopy
To assess cell proliferation by fluorescence microscopy, MCF7 cells were transfected using GeneJuice (150,000 cells per well of a six-well plate containing a coverslip) following the manufacturer’s instructions (Merck, Darmstadt, Germany). After 46 h, cells were labeled for 2 h with 10 μM of the thymidine analogue EdU and processed using Click-iT reagents (Invitrogen). The percentage of EdU-labeled cells was determined in triplicate.
To stain for the endogenous P-body markers, eIF4E and Dcp1a, 60,000 MCF7-NKI cells were seeded into each well of six-well plates containing glass coverslips. After 72 h of siRNA transfection, the cells were washed twice with PBS, fixed with 4% paraformaldehyde in PBS for 10 min, and permeabilized with cold 0.5% Triton X-100 for 10 min. After blocking for 20 min in PBS containing 3% bovine serum albumin (BSA), the cells were stained with primary antibody diluted in PBS containing 3% BSA for 1 h at 37ºC. Primary antibodies used were eIF4E (1:500 dilution, rabbit polyclonal antibody [ab] 1126; Abcam) and hDcp1a (1:200 dilution) (Lykke-Andersen and Wagner, 2005). After washing with PBS, the cells were incubated for 1 h at 37ºC with Alexa Fluor–conjugated secondary antibodies (Molecular Probes, Carlsbad, CA), counterstained with Hoechst 33258 (0.5 μg/ml), and mounted on glass slides for microscopy.
To detect Rck/p54/DDX6 and Ago2, the cells were transfected with siRNA for 48 h and subsequently transfected with their respective YFP constructs (a kind gift from Martin Bushell, Leicester, UK). After 24 h of plasmid transfection, the cells were washed twice with PBS, fixed with 4% paraformaldehyde, counterstained with Hoechst 33258, and mounted on glass slides for microscopy.
Cell imaging was carried out using a Zeiss LSM510 Meta confocal laser scanning microscope. Images were processed and merged using the LSM Image Browser (Zeiss, Thornwood, NY) and the Paint.net package (www.getpaint.net).
To study the subcellular localization of Ccr4b, MCF7 cells were transfected with plasmids expressing either Flag-Ccr4b or Flag-Ccr4bΔLRR using JetPEI (Polyplus, New York, NY). Immunofluorescence using an anti-Flag antibody (F1804, 1:500 dilution; Sigma) was carried out as described above at 24 h after transfection. For quantification of the nucleocytoplasmic distribution, images were captured using identical laser settings. For each cell analyzed, the mean pixel intensity of three equal regions of interest in the nucleus and cytoplasm was determined in the appropriate channel using ImageJ (http://rsbweb.nih.gov/ij). Fluorescence intensities were obtained from MCF7 cells transfected with either Flag-Ccr4b (n = 50) or Flag-Ccr4bΔLRR (n = 49), and the mean intensity of nuclear and cytoplasmic regions of interest was calculated.
Gene expression profiling
MCF7 NKI cells (1.0 × 106 cells in a 100-mm culture dish) were transfected with 5 nM siRNA pools targeting Ccr4a(CNOT6), Ccr4b(CNOT6L), Ccr4a/Ccr4b, or Caf1a(CNOT7)/Caf1b(CNOT8) and/or a nontargeting control pool (Dharmacon On-Target Plus SMARTpool; total siRNA concentration was 10 nM). DNA-free total RNA of biological triplicates was isolated (EZNA total RNA kit, including on-column DNAse digestion, Omega, Norcross, GA; using RNase-free DNAse I, Qiagen, West Sussex, UK), subjected to quality control using an Agilent 2100 Bioanalyzer, and processed using Affymetrix Human Gene 1.0 ST Array GeneChips, the manufacturer’s labeling protocols, fluidic station, and scanner (Nottingham Arabidopsis Stock Centre’s International Affymetrix Service, Santa Clara, CA). Data were normalized using the RMA protocol with the Affymetrix Gene Console package and analyzed using Excel 2007 (Microsoft, Redmond, WA). Differentially expressed genes were identified on the basis of the following criteria: signal intensity (untransformed value) > 50.0, fold change > 1.50, and p-value < 0.050. Hierarchical clustering was carried out using the comprehensive R-based microarray analysis tool CARMAweb (Rainer et al., 2006). The microarray data have been deposited in the ArrayExpress database (European Bioinformatics Institute, Cambridge, UK, accession number E-MEXP-2926).
RTq-PCR
Total RNA was isolated using the Omega EZNA total RNA kit, and cDNA was prepared using an anchored oligo(dT) primer with 75–100 ng total RNA in a 10-μl reaction (Superscript III, Invitrogen). After 1:5 dilution of the cDNA reaction with Tris-EDTA, 1 μl diluted cDNA was analyzed in triplicates by quantitative PCR (10-μl reaction volume, SensiMix Low-Rox SYBR Green mix; Bioline, Taunton, MA) using a Stratagene MX3005p cycler. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-actin was used as a reference gene. Analysis was carried out using the Stratagene MXpro package. Analysis of mRNA stability was as described before (Aslam et al., 2009).
Deadenylase assay
HEK293 cells (60% confluent, 6-cm standard cell culture dish) were transfected with Flag-Ccr4b expression plasmids using GeneJuice following the manufacturer’s protocol (Merck). After 48 h, the cells were lysed in 500 μl lysis buffer (50 mM HEPES-NaOH, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 5% glycerol, 1 mM dithiothreitol [DTT], and protease inhibitors). Flag-Ccr4b proteins were immunoprecipitated using anti-Flag antibodies (2 μg antibody coupled to 20 μl protein G–agarose beads) overnight at 4°C. After three washes with lysis buffer, immunoprecipitates were washed twice in deadenylation buffer (50 mM HEPES-NaOH, pH 8.0, 150 mM NaCl, 2 mM MgCl2, 10% glycerol, 1 mM DTT, and protease inhibitors). To elute bound proteins by peptide competition, the flag resin was incubated with 20 μl deadenylase buffer containing 0.3 mg/ml 3× Flag peptide (Sigma) for 60 min at 37°C with occasional mixing. Then 1 μl 5′ fluorescein (Flc)–labeled substrate (Sigma, Flc-5′-CCUUUCCAAAAAAAAA-3′; final concentration: 0.1 μM) was added to 9.0 μl Flag eluate and incubated for 60 min at 37°C. Reactions were stopped by the addition of 12 μl RNA loading buffer (95% formamide, 0.025% bromophenol blue, 0.025% xylene cyanol FF, 0.025% SDS, and 5 mM EDTA) and heated 3 min at 85°C. RNA was analyzed by denaturing PAGE using a 20% acrylamide:bisacrylamide (19:1) gel containing 8.3 M urea. Flc-labeled RNA was visualized using a Fujifilm LAS-4000 imager.
Senescence-associated β-galactosidase assay
To assess senescence, 60,000 MCF7 NKI cells were seeded onto each well of a six-well dish and transfected in triplicate with control, Caf1a/Caf1b, or Ccr4a/Ccr4b siRNA using INTERFERin (Polyplus). After 72 h of transfection, the cells were fixed and stained for β-galactosidase activity using the Senescence β-Galactosidase Staining Kit (New England Biolabs, Ipswich, MA) according to the manufacturer’s instructions. The cells were visualized using a Leica DM2000 light microscope at 200× magnification. Approximately 750 cells were counted from each well, and the percentage of blue cells (exhibiting senescence) was calculated.
Supplementary Material
Acknowledgments
This work was supported by grants to G.S.W. from the Biotechnology and Biological Sciences Research Council (BBSRC grant BB/002338X/1) and the Association for International Cancer Research (AICR grant 07-0494). We are grateful to David Heery, members of the Gene Regulation Group, and Cornelia de Moor and Keith Spriggs for critical reading of the manuscript and invaluable support. Jens Lykke-Anderson (University of California, San Diego) is acknowledged for his generous gift of Dcp1a antibody.
Abbreviations used:
- BrdU
5-bromo-2′-deoxyuridine
- BSA
bovine serum albumin
- DTT
dithiothreitol
- EdU
5-ethynyl-2′-deoxyuridine
- EEP
endonuclease–exonuclease–phosphatase
- FITC
fluorescein isothiocyanate
- Flc
fluorescein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HA
hemagglutinin
- HEK
human embryonic kidney
- HUVEC
human umbilical vein endothelial cells
- IGFBP5
insulin-like growth factor–binding protein 5
- LRR
leucine-rich repeat
- P-body
processing body
- PBS
phosphate-buffered saline
- pre-mRNA
precursor mRNA
- RT-qPCR
reverse transcriptase–quantitative PCR
- SEM
standard error of the mean
- siRNA
small interfering RNA
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-11-0898) on January 13, 2011.
REFERENCES
- Akkiprik M, Feng Y, Wang H, Chen K, Hu L, Sahin A, Krishnamurthy S, Ozer A, Hao X, Zhang W. Multifunctional roles of insulin-like growth factor binding protein 5 in breast cancer. Breast Cancer Res. 2008;10:212–225. doi: 10.1186/bcr2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam A, Mittal S, Koch F, Andrau JC, Winkler GS. The Ccr4–Not deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol Biol Cell. 2009;20:3840–3850. doi: 10.1091/mbc.E09-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs JE, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- Beattie J, Allan GJ, Lochrie JD, Flint DJ. Insulin-like growth factor-binding protein-5 (IGFBP-5): a critical member of the IGF axis. Biochem J. 2006;395:1–19. doi: 10.1042/BJ20060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch MA. The isoform-specific functions of the c-Jun N-terminal kinases (JNKs): differences revealed by gene targeting. Bioessays. 2006;28:923–934. doi: 10.1002/bies.20458. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Dickson KA, Jambazov S, Baxter RC. Enhancement of tumor necrosis factor-α-induced growth inhibition by insulin-like growth factor-binding protein-5 (IGFBP-5), but not IGFBP-3 in human breast cancer cells. Endocrinology. 2005;146:3113–3122. doi: 10.1210/en.2004-1408. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Dickson KA, McDougall F, Baxter RC. Insulin-like growth factor-binding protein-5 inhibits the growth of human breast cancer cells in vitro and in vivo. J Biol Chem. 2003;278:29676–29685. doi: 10.1074/jbc.M301965200. [DOI] [PubMed] [Google Scholar]
- Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LB, Viswanathan P, Quigley G, Chiang YC, McMahon JS, Yao G, Chen J, Nelsbach A, Denis CL. Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J Biol Chem. 2004;279:13616–13623. doi: 10.1074/jbc.M313202200. [DOI] [PubMed] [Google Scholar]
- Collart MA. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. doi: 10.1016/s0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- Collart MA, Timmers HT. The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron MC, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis CL, Chen J. The CCR4–NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, Heidmann T. Identification of four families of yCCR4− and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, Yamashita Y, Zheng D, Shyu AB. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol. 2007;27:7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 2009;16:1093–1107. doi: 10.1038/cdd.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapaty SR, Mahajan MA, Samuels HH. Components of the CCR4-NOT complex function as nuclear hormone receptor coactivators via association with the NRC interacting factor, NIF-1. J Biol Chem. 2008;283:6806–6816. doi: 10.1074/jbc.M706986200. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- Govindan M, Meng X, Denis CL, Webb P, Baxter JD, Walfish PG. Identification of CCR4 and other essential thyroid hormone receptor coactivators by modified yeast synthetic genetic array analysis. Proc Natl Acad Sci USA. 2009;106:19854–19859. doi: 10.1073/pnas.0910134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N, Ito T, Yamamoto H, Ochiya T, Jinno S, Okayama H. Mammalian Rcd1 is a novel transcriptional cofactor that mediates retinoic acid-induced cell differentiation. EMBO J. 2002;21:5235–5244. doi: 10.1093/emboj/cdf521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigel B, Varshavsky A, Kessler O, Neufeld G. Successful inhibition of tumor development by specific class-3 semaphorins is associated with expression of appropriate semaphorin receptors by tumor cells. PLoS One. 2008;3:e3287. doi: 10.1371/journal.pone.0003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Seu YB, Baek SH, Kim MJ, Kim KJ, Kim JH, Kim JR. Induction of cellular senescence by insulin-like growth factor binding protein-5 through a p53-dependent mechanism. Mol Biol Cell. 2007;18:4543–4552. doi: 10.1091/mbc.E07-03-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauxion F, Faux C, Seraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel AP, Sentis S, Bianchin C, Le Romancer M, Jonard L, Rostan MC, Rimokh R, Corbo L. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J Cell Sci. 2003;116:2929–2936. doi: 10.1242/jcs.00480. [DOI] [PubMed] [Google Scholar]
- Morita M, Suzuki T, Nakamura T, Yokoyama K, Miyasaka T, Yamamoto T. Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol Cell Biol. 2007;27:4980–4990. doi: 10.1128/MCB.02304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov IY, Jones MG, Spiller DG, Rigden DJ, Dattenbock C, Novotny R, Strauss J, Caddick MX. Distinct roles for Caf1, Ccr4, Edc3 and CutA in the co-ordination of transcript deadenylation, decapping and P-body formation in Aspergillus nidulans. Mol Microbiol. 2010;76:503–516. doi: 10.1111/j.1365-2958.2010.07118.x. [DOI] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol. 2010;30:1486–1494. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevot D, Morel AP, Voeltzel T, Rostan MC, Rimokh R, Magaud JP, Corbo L. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor α signaling pathway. J Biol Chem. 2001;276:9640–9648. doi: 10.1074/jbc.M008201200. [DOI] [PubMed] [Google Scholar]
- Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36:3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Kontani K, Araki Y, Katada T. Caf1 regulates translocation of ribonucleotide reductase by releasing nucleoplasmic Spd1-Suc22 assembly. Nucleic Acids Res. 2007;35:1187–1197. doi: 10.1093/nar/gkm015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–1370. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunezumi J, Higashi S, Miyazaki K. Matrilysin (MMP-7) cleaves C-type lectin domain family 3 member A (CLEC3A) on tumor cell surface and modulates its cell adhesion activity. J Cell Biochem. 2009;106:693–702. doi: 10.1002/jcb.22062. [DOI] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- Viswanathan P, Ohn T, Chiang YC, Chen J, Denis CL. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J Biol Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- Wagner E, Clement SL, Lykke-Andersen J. An unconventional human Ccr4-Caf1 deadenylase complex in nuclear Cajal bodies. Mol Cell Biol. 2007;27:1686–1695. doi: 10.1128/MCB.01483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J. 2010;29:2566–2576. doi: 10.1038/emboj.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GS. The mammalian anti-proliferative BTG/Tob protein family. J Cell Physiol. 2010;222:66–72. doi: 10.1002/jcp.21919. [DOI] [PubMed] [Google Scholar]
- Winkler GS, Mulder KW, Bardwell VJ, Kalkhoven E, Timmers HT. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 2006;25:3089–3099. doi: 10.1038/sj.emboj.7601194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.