Relatively constant diameters of Caenorhabditis elegans mitochondria and their cristae are disrupted by mutations in a novel mitochondrial outer membrane protein, MOMA-1, and by mutations in a mitofilin homologue, which is anchored in the inner membrane. Genetic data suggest that these proteins act in the same pathway but localize to different membranes.
Abstract
Three proteins with similar effects on mitochondrial morphology were identified in an RNA interference (RNAi) screen for mitochondrial abnormalities in Caenorhabditis elegans. One of these is the novel mitochondrial outer membrane protein MOMA-1. The second is the CHCHD3 homologue, CHCH-3, a small intermembrane space protein that may act as a chaperone. The third is a mitofilin homologue, IMMT-1. Mitofilins are inner membrane proteins that control the shapes of cristae. RNAi or mutations in each of these genes change the relatively constant diameters of mitochondria into highly variable diameters, ranging from thin tubes to localized swellings. Neither growth nor brood size of the moma-1, chch-3, or immt-1 single mutants is affected, suggesting that their metabolic functions are normal. However, growth of moma-1 or immt-1 mutants on chch-3(RNAi) leads to withered gonads, a lack of mitochondrial staining, and a dramatic reduction in fecundity, while moma-1; immt-1 double mutants are indistinguishable from single mutants. Mutations in moma-1 and immt-1 also have similar effects on cristae morphology. We conclude that MOMA-1 and IMMT-1 act in the same pathway. It is likely that the observed effects on mitochondrial diameter are an indirect effect of disrupting cristae morphology.
INTRODUCTION
Mitochondria are tubular organelles with relatively constant diameters but widely varying lengths and numbers of connections with other mitochondria. This architecture is important for a range of cellular activities such as ATP production, Ca2+ buffering, biogenesis of Fe-S clusters, synaptic function, and apoptosis (Tsang and Lemire, 2003; Mannella, 2008). In the past decade, many proteins that control mitochondrial shape and connectivity have been uncovered.
Most attention on mitochondrial dynamics has been given to proteins that regulate mitochondrial fission and fusion. Mitochondrial fission and fusion are mediated by specific dynamin family members and their accessory proteins (van der Bliek, 1999; Griparic et al., 2004; Okamoto and Shaw, 2005; Chan, 2006; Hoppins and Nunnari, 2009). These fission and fusion processes are the primary determinants of mitochondrial lengths; the actions of fission proteins produce shorter mitochondria, while the actions of fusion proteins produce longer and more interconnected mitochondria.
In addition to fission and fusion, mitochondrial networks are influenced by connections to the cytoskeleton. Mitochondria of budding yeast are connected to the actin cytoskeleton, while mitochondria of fission yeast and other organisms are often connected to microtubules through the actions of motor proteins and their adaptors (Boldogh and Pon, 2006; Frederick and Shaw, 2007). Treatment with nocodazole or fixation of cells for electron microscopy (EM), although affecting the overall organization of the mitochondrial network, does not alter the tubular shape of mitochondria (Munn, 1968; Smirnova et al., 2001). These pieces of data suggest that the main role for cytoskeletal-mitochondrial interactions is in mitochondrial intracellular transport.
Fission, fusion, and cytoskeletal attachment control the connectivity and distribution of mitochondria. The other major parameter of mitochondria, tubular shape, is predominantly specified by mitochondrial resident proteins. These proteins are localized to one or more of the mitochondrial subcompartments: the outer (OM) or inner membranes (IM), the intermembrane space (IMS), or the matrix (Mannella, 2006; Zick et al., 2009). The IM is usually divided into two subcompartments: the inner boundary membrane (IBM) and cristae. The IBM is the region of the IM that closely opposes the mitochondrial OM and is separated by ∼10 nm of IMS. Cristae are balloon-like and/or sheetlike infoldings of the IM that project into the matrix space (Zick et al., 2009).
Recently, the maintenance of one of these mitochondrial subcompartments, the IM, has become an area of active research. Unlike bacteria, mitochondria lack actin- or tubulin-like molecules that could provide the necessary rigidity or scaffolding that might regulate IM morphology (Margolin, 2009). Instead, a handful of eukaryotic specific IM and IMS proteins have been implicated in the maintenance of IM morphology. Mitofilin has been characterized in mammalian cells and, more recently, in the model organisms Saccharomyces cerevisiae and Caenorhabditis elegans (John et al., 2005; Rabl et al., 2009; Mun et al., 2010). In these reports, mitofilin has been tied to maintenance of cristae morphology. A model emerges in which the ATP synthase F0 subunit e, F0 subunit g, and mitofilin modulate cristae curvature by antagonistically regulating ATP synthase dimerization (Rabl et al., 2009; Velours et al., 2009). In turn, it has recently been demonstrated that knockdown of CHCHD3 affects the levels of mitofilin in the IM and, correspondingly, affects cristae morphology (Darshi et al., 2011). The number of proteins that directly affect the mitochondrial IM is, nevertheless, still limited.
In an effort to identify novel proteins that affect mitochondrial morphology, we tested the effects of 232 mitochondrial proteins with unknown mitochondrial functions using C. elegans feeding RNA interference (RNAi). We identified three genes, moma-1, chch-3, and immt-1, that have similar effects on mitochondrial morphology. The mitochondria in these mutants show localized swellings and constriction as if they have lost their ability to control tubular shape. Single mutations in the corresponding genes have little to no effect on growth or viability of the worms, suggesting that these mutations do not severely disrupt mitochondrial function. Here we describe genetic interactions and some initial biochemical characterizations that lead us to a model in which MOMA-1 and IMMT-1 function in the control of cristae morphology and overall tubular shape.
RESULTS
Proteins of unknown function that affect mitochondrial morphology
We compiled a list of 232 proteins that were previously localized to mitochondria (Da Cruz et al., 2003; Mootha et al., 2003; Sickmann et al., 2003; Taylor et al., 2003), but with unknown or unresolved mitochondrial functions. To analyze the effects of these proteins, C. elegans were grown for one or two generations on bacteria that express double-stranded RNA targeting each of the 232 genes (Kamath et al., 2003). Mitochondrial morphologies were observed by expressing matrix-targeted green fluorescent protein (GFP) in muscle cells. RNAi that gave rise to abnormal mitochondrial morphologies were retested with mitochondrial GFP in hypodermal cells. Our survey led to the identification of three genes with similar effects on mitochondrial morphology (T14G11.3, M176.3, and K02F3.10). RNAi for these genes changes the relatively constant diameters of mitochondria into localized swellings and thin tubular extensions (Figure 1 and Supplemental Figure S4). Other RNAis produced mitochondrial morphology aberrations that were different from these three genes, suggesting that the morphological effects of T14G11.3, M176.3, and K02F3.10 knockdown were not an indirect response to disruption of other mitochondrial processes. Results of this screen are summarized in Supplemental Table 1, and examples of mitochondrial abnormalities observed with knockdown of common mitochondrial proteins and with several other novel proteins identified in this screen are shown in Supplemental Figures S1 and S2.
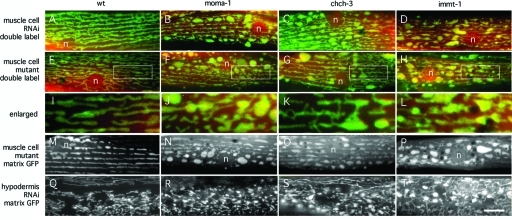
FIGURE 1:
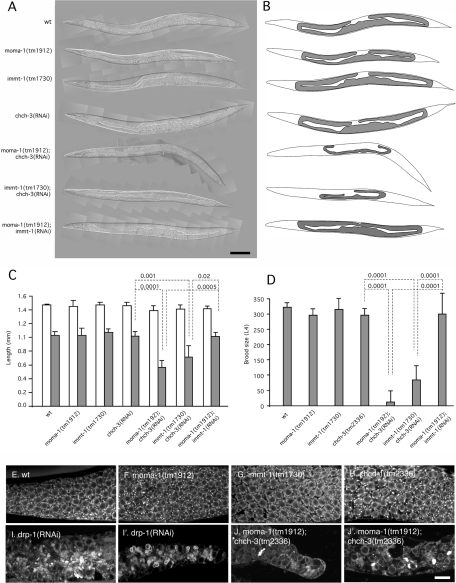
Three different proteins with similar effects on mitochondrial morphology. (A–D and Q–T) Effects of feeding RNAi (A, Q, no insert; B, R, moma-1 K02F3.10; C, S, chch-3 M176.3; D, T, immt-1 T14G11.3 RNAi) on muscle and hypodermal cell mitochondria. (E–P) Effects of chromosomal deletions (E, I, M, wild-type; F, J, N, moma-1[tm1912]; G, K, O, chch-3[tm2336]; H, L, P, immt-1[tm1730]) on muscle cell mitochondria. Muscle cell mitochondria were detected with the matrix marker Pmyo-3::mls::CFP (red) and the outer membrane marker Pmyo-3::Tom70::YFP (green) in A–L and with the matrix marker Pmyo-3::mls::GFP in M–P. Hypodermal cells mitochondria were detected with the matrix marker Pcol-12::mls::GFP in Q–T. Positions of nuclei are indicated (n), and the scale bar is 10 μm, except for I–L, which show threefold enlargements of the boxed areas in E–H.
The T14G11.3 gene is named immt-1. This gene encodes a C. elegans mitofilin homologue (Mun et al., 2010). In yeast and mammalian cells, mitofilin is anchored in the mitochondrial IM with a large part of the protein exposed to the mitochondrial IMS (Gieffers et al., 1997; John et al., 2005; Rabl et al., 2009). Fluorescence and immunoelectron microscopy show staining along the rims of mitochondria, indicating that a substantial fraction of mitofilin is localized to the IBM (Odgren et al., 1996; John et al., 2005). Mitofilin small interfering RNA (siRNA) disrupts cristae in mammalian cells, suggesting that mitofilin is involved in cristae formation or maintenance (John et al., 2005). Mitochondrial cristae of a C. elegans immt-1 mutant are similarly disrupted (Mun et al., 2010). A recent article showed that yeast mitofilin is required for the formation of mitochondrial cristae junctions, where it was proposed to promote membrane curvature (Rabl et al., 2009).
The M176.3 gene encodes a homologue of the human CHCHD3 protein. CHCHD3 belongs to a family of proteins with a characteristic coiled-coil-helix–coiled-coil-helix domain (CHCHD) (Westerman et al., 2004). Proteins in this family lack mitochondrial leader sequences, but they have a twin CX9C motif embedded in their CHCHD. Twin CX9C motifs and CHCHDs are also present in 10 small yeast proteins that are imported into the mitochondrial IMS (Gabriel et al., 2007). The fixed position of CX9C within each helix suggests that the helices fold back to form a hairpin structure that is stabilized by disulfide bonds (Westerman et al., 2004). This structure may mediate protein targeting and protein folding, or it may be a functional domain in the IMS. The mouse CHCHD3 homologue has also been localized to mitochondria (Schauble et al., 2007), and human CHCHD3 is found in a complex with mitofilin, consistent with localization to the mitochondrial IMS (Xie et al., 2007). The M176.3 gene was named chch-3.
The K02F3.10 gene is named moma-1 for Mitochondrial Outer Membrane Abnormal. It encodes a protein that is similar to two proteins in humans (Supplemental Figure S3). The human proteins have alternatively been named my025 and CXORF33 or apolipoprotein O and Apo-O-like because of weak similarity to apolipoproteins (Lamant et al., 2006). Yeast also has two similar proteins (YNL100W/Aim37 and YGR235C), which were shown with GFP tags to localize to mitochondria (Huh et al., 2003). Neither of the MOMA-1 homologues has a mitochondrial leader sequence, but each has hydrophobic sequences consistent with a transmembrane segment.
Chromosomal deletions in immt-1, chch-3, and moma-1 genes were generated by S. Mitani (National BioResource Project [NBRP], Shinjuku-ku, Japan). The mutant alleles are immt-1(tm1730), chch-3(tm2336), and moma-1(tm1912). The mutant genes encode highly truncated proteins and are therefore likely null alleles. Western blot analyses show that moma-1(tm1912) animals have no MOMA-1 protein (see Figure 3M later in the paper). Morphological abnormalities of mitochondria in the muscle cells of these strains are similar to those obtained with feeding RNAi, confirming that the phenotypes are due to loss of function of the intended gene and not to off-target effects on other genes (Figure 1). These mutations had little or no effect on growth or brood size (Supplemental Figure S4), nor were they more sensitive to stress, as suggested by the lack of nuclear localization of DAF-16::GFP (unpublished data), which is a reporter for elevated levels of reactive oxygen species (ROS) in C. elegans (Henderson and Johnson, 2001). The effects of these deletions on growth, fecundity, and ROS production therefore appear minimal.
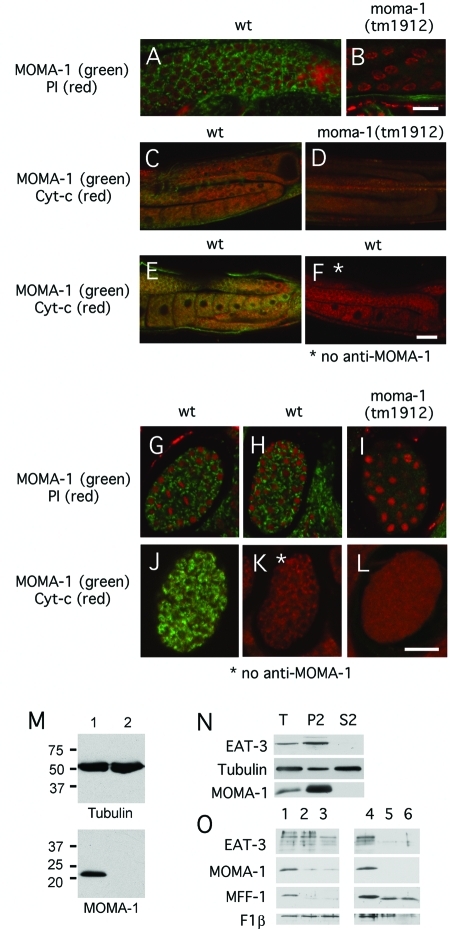
FIGURE 3:
Localization of MOMA-1 to the mitochondrial outer membrane. (A–F) Staining of C. elegans gonads. (G–L) Staining of C. elegans embryos with MOMA-1 antibody (green) and propidium iodide (red in A, B, and G–I) or cytochrome c antibody (red in C–F and J–L). Strains are indicated above the panels, and the combinations of stains are shown on the left, except for F and K, which were stained with cytochrome c antibody but not with MOMA-1 antibody to control for bleed-through. The scale bar is 10 μm for A and B, 20 μm for C–F, and 10 μm for G–L. (M) Western blots of extracts from wild-type (lane 1) and moma-1(tm1912) (lane 2) animals probed with antibodies against tubulin or MOMA-1. Size markers are in kDa. (N) Subcellular distribution of MOMA-1 determined by differential centrifugation. Lanes were loaded with total extracts (T), with 14,000 × g pellets (P2), and with 14,000 × g supernatants (S2; P2 and S2 fractions were volume equivalents). Blots were probed with EAT-3, tubulin, and MOMA-1 antibodies as indicated. The distributions were quantified with densitometry of P2 and S2 fractions: 88% of EAT-3, 12% of tubulin, and 98% of MOMA-1 is in the mitochondrial pellet. (O) Protease protection experiment to determine the submitochondrial localization of MOMA-1. Mitochondria were isolated by differential centrifugation and treated with 0 (lanes 1 and 4), 30 (lanes 2 and 5), and 60 (lanes 3 and 6) μg/ml proteinase K, without (lanes 1–3) or with (lanes 4–6) 1% Triton X-100. Blots were probed with antibodies for EAT-3 (an inner membrane marker), MOMA-1, MFF-1 (an outer membrane marker), and F1β subunit of the ATP synthase complex (a mitochondrial matrix marker). MOMA-1 is digested when no detergent is added, like MFF-1, while EAT-3 and F1β are protease protected. For unknown reasons, an intermediate cleavage product of MFF-1 is detected when Triton is added to the protease digestion but not when Triton is omitted.
C. elegans has a second mitofilin that is functionally distinct from IMMT-1
The C. elegans W06H3.1 gene, named immt-2, encodes a second mitofilin homologue. Each of the two C. elegans mitofilin homologues has a predicted amino terminal mitochondrial leader sequence, followed by a single transmembrane segment and a series of coiled-coil segments, which are presumably exposed to the mitochondrial IMS, similar to yeast and mammalian mitofilin proteins. A strain with a deletion in the immt-2 gene (tm2366) was kindly provided by S. Mitani (NBRP). The immt-2(tm2366) mutants have smaller brood sizes than wild-type or immt-1(tm1730) animals (Supplemental Figure S4). The immt-2(tm2366) deletion and immt-2(RNAi) also severely disrupt the morphologies of mitochondria in C. elegans hypodermal and muscle cells (Figure 2). Because immt-1 and immt-2 RNAi and mutations each cause phenotypes in the same somatic cells, we conclude that these genes are coexpressed. There are, however, noticeable differences between mitochondrial morphologies observed with immt-1 and immt-2 defects, whether introduced with RNAi or with chromosomal deletions. Defects in immt-1 yield mitochondria with localized swellings and thin tubular extensions, while immt-2 defects give rise to mitochondria that are generally thinner and more connected than mitochondria in wild-type cells or cells with immt-1 defects (Figure 2 and Supplemental Figure S4). The immt-2 phenotype is more similar to mitochondrial morphologies observed with RNAi for adenine nucleotide translocator and Sam50 homologues (Supplemental Figure S1). Different effects on brood size and mitochondrial morphology show that immt-1 and immt-2 have different effects on mitochondria, even though they are both clearly mitofilin homologues.
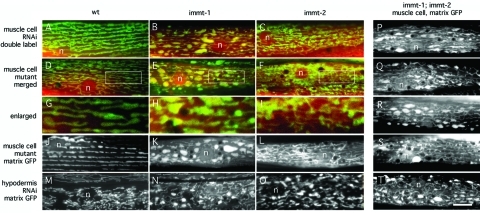
FIGURE 2:
Different effects of IMMT-1 and IMMT-2 on mitochondrial morphology. (A–C and M–O) Effects of feeding RNAi (A, M, no insert; B, N, immt-1 T14G11.3; C, O, immt-2 W06H3.1) on the morphologies of muscle cell and hypodermal mitochondria. (D–L and P–T) Effects of chromosomal deletions (D, G, J, wild-type; E, H, K, immt-1[tm1730]; F, I, L, immt-2[tm2366]; and P–T, immt-1[tm1730]; immt-2[tm2366]) on muscle cell mitochondria. Muscle cell mitochondria were detected with the matrix marker Pmyo-3::mls::CFP (red) and the outer membrane marker Pmyo-3::Tom70::YFP (green) in A–I and with the matrix marker Pmyo-3::mls::GFP in J–L and P–T. Hypodermal cells mitochondria were detected with the matrix marker Pcol-12::mls::GFP in M–O. Positions of nuclei are indicated (n), and the scale bar is 10 μm, except for G–I, which show threefold enlargements of the boxed areas in D–F.
To further test whether IMMT-1 and IMMT-2 affect different processes within mitochondria, we generated an immt-1(tm1730); immt-2(tm2366) double mutant strain. This strain grows slower and has brood sizes that are reduced when compared with mutations in either gene alone (Supplemental Figure S4). Images of mitochondrial morphologies in double mutants show phenotypes that appear to be a combination of IMMT-1 and IMMT-2 defects (Figure 2). The extent of the differences between single and double mutant strains suggests that the effects of mutations in immt-1 and immt-2 are additive. Similar results were obtained in a recent independent study of immt-1 and immt-2 mutants (Mun et al., 2010). We conclude that immt-1 and immt-2 are expressed in the same cells but there are differences in their effects on mitochondria.
MOMA-1 is localized to the mitochondrial OM
Submitochondrial localizations of C. elegans mitofilins and of the CHCH-3 protein are likely similar to the localizations of homologous proteins in other organisms (IM and IMS, respectively), but the localization of MOMA-1 had not been determined. To investigate this localization, we raised polyclonal antibodies against bacterially expressed C. elegans MOMA-1 protein. These antibodies detect a protein of the expected size (22 kDa) on Western blots of whole-worm extracts. This band was not detected in moma-1(tm1912) animals, which have a large deletion in the moma-1 gene, confirming that this is indeed the MOMA-1 protein (Figure 3M). Differential centrifugation shows that MOMA-1 is present in the P2 fraction, which contains mitochondria (Figure 3N). Immunofluorescence shows colocalization with cytochrome c in C. elegans embryos (Figure 3). Human and yeast homologues of MOMA-1 are also localized to mitochondria, as shown with immunofluorescence and GFP-tagged versions of these proteins (unpublished data) (Huh et al., 2003; Barbe et al., 2008). We conclude that MOMA-1 is predominantly localized to mitochondria.
We used two approaches to determine the submitochondrial localization of MOMA-1. In the first, we generated MOMA-1 protein with an amino- or carboxy-terminal yellow fluorescent protein (YFP) tag under control of the myo-3 promoter. Both MOMA-1::YFP and YFP::MOMA-1 are able to restore normal mitochondrial morphology in muscle cells of moma-1(tm1912) animals, showing that the fusion proteins are functional (Supplemental Figure S5 and unpublished data). Fluorescence labeling showed a pattern similar to patterns observed with YFP fused to a bona fide mitochondrial OM protein (YFP::FIS-1), suggesting that MOMA-1 is localized to the mitochondrial OM (Supplemental Figure S5). We then grew wild-type worms expressing a mitochondrial OM marker or YFP::MOMA-1 on drp-1 RNAi bacteria. This treatment blocks mitochondrial OM fission, but IM fission and mitochondrial fusion still occur, leading to the formation of a contiguous network with thin tubules of mitochondrial OM connecting blebs formed by contracted matrix compartments (Labrousse et al., 1999). Clear separation between mitochondrial OMs and matrix compartments enables unambiguous localization of YFP::MOMA-1. In this experiment, we could detect YFP::MOMA-1 along all of the thin tubular connections, suggesting that the protein is anchored in the mitochondrial OM (Supplemental Figure S5).
Submitochondrial localization was further investigated with protease protection experiments using isolated mitochondria. Mitochondrial proteins that are exposed to the cytosol are digested by small amounts of protease while proteins that are protected, because they reside in an internal compartment, are digested only when detergents are used to solubilize the membranes. We used antibodies against a C. elegans Mff homologue, encoded by the F55F8.6 gene, as a control for proteins exposed to the cytosol (Gandre-Babbe and van der Bliek, 2008) and antibodies against the C. elegans Opa1 homologue EAT-3 (Kanazawa et al., 2008) as a control for proteins exposed to the IMS, along with antibodies against the F1β subunit of the ATP synthase complex as a control for proteins in the mitochondrial matrix. Our experiments show that MOMA-1 is affected by proteases at concentrations similar to those that digest the F55F8.6 protein. MOMA-1 and F55F8.6 proteins are both much more sensitive to proteases than EAT-3, even though EAT-3 is a much larger protein and readily digested when membranes are solubilized with detergents (Figure 3O). We conclude from these experiments that MOMA-1 is more sensitive to protease digestion than EAT-3 in intact mitochondria. We do note, however, that a small but reproducible fraction of MOMA-1 (∼10%, as determined by densitometry of four separate experiments) remained undigested, even at the highest concentrations of proteinase K. It therefore remains possible that a small fraction of MOMA-1 is localized to the mitochondrial IM. Together with the differential centrifugation, immunofluorescence, and GFP tagging results, these experiments do, however, show that the bulk of MOMA-1 is embedded in the mitochondrial OM with part of the protein exposed to the cytosol.
MOMA-1 does not affect mitochondrial fission in somatic cells
Mitochondrial abnormalities in moma-1, chch-3, and immt-1 mutants resemble those observed with mitochondrial fission mutants in C. elegans muscle cells. Both sets of mutants have mitochondria with localized swellings and thin tubular connections. However, the mitochondria of moma-1, chch-3, and immt-1 mutants are relatively short, in contrast with mitochondria of drp-1 mutants, which have very few or no loose ends and form a closed network (Figure 4A). Strong effects on mitochondrial morphology but not increased connectivity are also seen with RNAi for moma-1, chch-3, and immt-1 in an anc-1 mutant background (Supplemental Figure S6) where mitochondria are no longer attached to the actin cytoskeleton (Starr and Han, 2002), showing that these morphological abnormalities do not depend on cytoskeletal attachments. There is, however, some variability in the degree of connectivity in moma-1, chch-3, and immt-1 deletions strains, raising the possibility that these mutations have a modest or incompletely penetrant effect on fission.
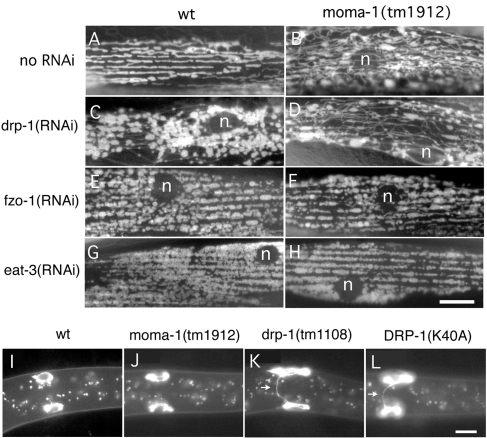
FIGURE 4:
Comparison of the MOMA-1 phenotype with mitochondrial fission and fusion defects. (A–H) C. elegans muscle cells labeled with the mitochondrial matrix marker Pmyo-3::mls::GFP. Wild-type animals (A, C, E, and G) or moma-1(tm1912) mutants (B, D, F, and H) were grown on bacteria with no RNAi (A and B), with drp-1 RNAi (C and D), with fzo-1 RNAi (E and F), or with eat-3 RNAi (G and H). The scale bar is 10 μm. (I–L) M cells after their first division in L1 larvae labeled with a mitochondrial outer membrane marker (Phlh-8::Tom70::GFP). Images show a wild-type animal (I), a moma-1(tm1912) mutant (J), a drp-1(tm1108) mutant (K), and a wild-type animal with the dominant negative construct Phlh-8::DRP-1(K40A) (L). Arrows point to mitochondrial tubules connecting the two daughter cells. These tubules persist in animals with a mitochondrial fission defect. The scale bar is 10 μm.
To test more thoroughly whether moma-1, chch-3, and immt-1 mutations affect mitochondrial fission, we knocked down mitochondrial fusion genes (fzo-1 and eat-3) in these backgrounds and determined their epistatic relationship. Strong defects in mitochondrial fusion produce cells with highly fragmented mitochondria. Mutations in drp-1 prevent the fragmentation of mitochondria induced by mutations in fzo-1 or eat-3, showing that fission mutants are epistatic to fusion mutants (Kanazawa et al., 2008). However, moma-1(tm1912) mutant animals grown with feeding RNAi for fzo-1 and eat-3 acquire a high percentage of cells with fragmented mitochondria (100% of cells examined, n = 41) similar to the mitochondria of fusion mutants, suggesting that fusion defects are epistatic to the effects of moma-1 on mitochondrial morphology (Figure 4A). Moreover, the mitochondria of moma-1 mutants that were grown on drp-1 RNAi gave rise to mitochondria that were indistinguishable from wild-type animals grown on drp-1 RNAi. Similar results were obtained with chch-3 and immt-1 mutant animals (unpublished data). We conclude that moma-1, chch-3, and immt-1 mutants do not strongly impact mitochondrial fission.
To further investigate a possible role of MOMA-1 in mitochondrial fission, we looked at the distribution of mitochondria during cell division. We showed previously that defects in mitochondrial fission are easy to detect during cell division because undivided mitochondria form bridges between daughter cells (Labrousse et al., 1999). The distributions of mitochondria during cell division were observed with a mitochondrial OM marker (TOM70::GFP) expressed from the hlh-8 promoter, which is specific for the M cell lineage (Harfe et al., 1998). The M cell lineage starts as a single mesodermal blast cell in early L1 larvae. This M cell divides a predetermined number of times to form body wall muscles, vulval muscles, and coelomocytes. When DRP-1(K40A) is expressed in the first M cell, this cell still divides, but the daughter cells remain connected by a tubule that can be detected with the mitochondrial OM marker (36% retain connections, SD = 25, n = 49), while 0% of wild-type cells retain connections (SD = 0, n = 80). This effect is also observed with drp-1(tm1108) mutant, which has a large deletion in the drp-1 gene (72% retain connections, SD = 22, n = 39). In contrast, only one of the moma-1(tm1912) animals (1.3% retain connections, SD = 4.8, n = 75) and none of the wild-type animals have cells that are visibly connected even though moma-1 is ubiquitously expressed (see Figure 4B for examples). We conclude that moma-1 has little or no effect on mitochondrial fission in this assay.
Next, we tested whether moma-1(tm1912) or immt-1(tm1730) mutations affect mitochondrial DNA (mtDNA) levels, as might be expected from mutations that affect mitochondrial fission or fusion. We did this with quantitative PCR (qPCR) using primers that were designed to compare levels of mitochondrial and nuclear DNA in C. elegans (Meyer et al., 2007). There were no statistically significant differences between mitochondrial to nuclear DNA ratios in wild-type (N2) and mutant animals (Student’s t test, n = 3 for each strain).
Last, we tested whether MOMA-1 affects the distributions of other mitochondrial OM proteins. We focused on representative mitochondrial fission proteins. C. elegans has two Fis1 homologues, encoded by fis-1 and fis-2, and two Mff homologues, encoded by mff-1(F11C1.2) and mff-2(F55F8.6). GFP-tagged versions of all four mitochondrial fission proteins, as well as cyan fluorescent protein (CFP)::DRP-1, were expressed in the muscle cells of moma-1(tm1912) mutant and wild-type animals. Single cell analysis of fluorescence intensity was used to determine whether the distributions within mitochondria and the distributions between mitochondria and cytosol were affected. Neither parameter was noticeably affected by the moma-1 mutations (unpublished data), suggesting that there is also no indirect effect on fission.
Synthetic interactions with chch-3, but not with each other, suggest that moma-1 and immt-1 function in the same pathway
Mutations in the three genes that we focus on here, immt-1, chch-3, and moma-1, have very similar effects on mitochondrial morphology and on brood size (Figure 1 and Supplemental Figure S4), suggesting that the proteins might act in the same pathway. To determine whether MOMA-1 protein levels are sensitive to loss of CHCH-3 or IMMT-1, we blotted whole-worm extracts of chch-3(tm2336) and immt-1(tm1730) mutant animals and processed them with MOMA-1 antibodies. MOMA-1 protein levels were not significantly altered in these worms nor were they altered by mutations in mitochondrial fission or fusion proteins (unpublished data). We also tested whether the effects of heterozygous deletions in moma-1 could be enhanced by heterozygous deletions in immt-1, but the muscle cell mitochondria in these transheterozygous animals almost all appeared wild type (unpublished data), suggesting that these genes are haplosufficient and therefore could not be not enhanced in this assay.
To test more thoroughly whether IMMT-1, CHCH-3, and MOMA-1 act in the same pathway, we asked whether RNAi of chch-3 or immt-1 could enhance the phenotypes of moma-1(tm1912) or immt-1(tm1730) homozygous animals. If IMMT-1, CHCH-3, and MOMA-1 are indeed components of a single pathway, then we would expect no additional effects when these gene knockdowns or knockouts are combined. To our surprise, the phenotypes of moma-1; chch-3(RNAi) and of immt-1; chch-3(RNAi) were much worse than the phenotypes of the single mutants. These animals have few or no brood, grow poorly, and have withered gonads (Figure 5, A–D). In contrast, moma-1(tm1912); immt-1(RNAi) animals show no additional mitochondrial abnormalities or growth defects (Figure 5, A–D). The effects of chch-3 on moma-1 were examined in more detail by dissecting the gonads of mutant animals and staining them with a mitochondrial dye. The gonads of single mutants have mitochondrial distributions that are similar to wild type, but the effects of the moma-1(tm1912); chch-3(tm2336) double mutant are as strong or stronger than those previously observed with drp-1(RNAi) (Labrousse et al., 1999). These gonads have very little mitochondrial staining, and what is left appears in clumps (Figure 5, E–J). Similarly strong effects on gonad morphology and brood size were previously observed with mutations in essential mitochondrial proteins, such as clk-1 or subunits of complex I (Jonassen et al., 2003; Curran et al., 2004; Grad and Lemire, 2004; Kawasaki et al., 2007). We conclude that IMMT-1 and MOMA-1 function in the same pathway, while CHCH-3 may have additional functions or it acts in a different pathway. Mutations in either pathway have relatively mild effects on growth, brood size, and mitochondrial morphology, but the combinations are lethal, suggesting that their combined actions are essential for mitochondrial function.
FIGURE 5:
Withered gonads caused by genetic interactions between chch-3 and moma-1 or immt-1. (A) Composite differential interference contrast images of adult worms with mutations and RNAi treatments as indicated on the left. (B) Tracings of these animals and their gonads. The scale bar is 100 μm. (C) Lengths of adult animals with the indicated genotypes and feeding RNAi (open bars). Closed bars show the lengths of gonads measured from the anterior bend to the posterior bend. Mean and SD were determined with five or more animals for each condition. P values were determined with an unpaired Student’s t test. (D) Brood size of animals with the indicated genotypes and feeding RNAi. Brood sizes were determined as the numbers of progeny that survive into the L4 larval stage. Mean and SD were determined with 10 or more animals for each condition. P values were determined with an unpaired Student’s t test. (E–J) Gonads of animals with the indicated genotypes and RNAi treatments. Gonads were dissected, stained with rhodamine 6G, and photographed with a confocal microscope. Scale bar is 10 μm.
EM shows similar disruptions of cristae in moma-1(tm1912) and immt-1(tm1730) mutant animals
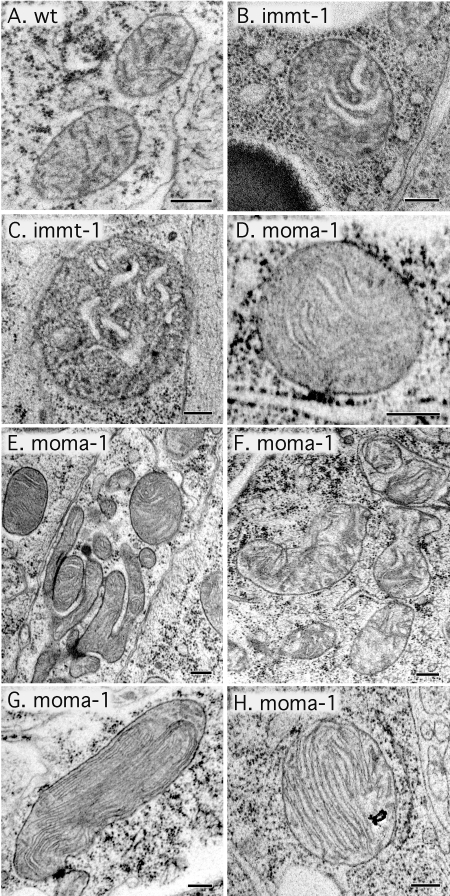
Due to the similarities between immt-1 and moma-1 mutants and the known role of mitofilins in the control of cristae morphology (Rabl et al., 2009; Mun et al., 2010), we asked whether MOMA-1 also affects cristae morphologies. Samples of wild-type (N2), immt-1(tm1730), and moma-1(tm1912) animals were prepared for EM with high-pressure freezing. Cristae are evident as thin tubules in mitochondria of wild-type animals (Figure 6A), similar to previously published reports (Mun et al., 2010). Cristae become distended in immt-1(tm1730) mutant animals, consistent with the role of mitofilins in maintaining cristae curvature and morphology (Figure 6, B and C). Similarly distended cristae were invariably observed in the mitochondria of moma-1(tm1912) mutant animals (Figure 6, D–F; none of the several hundred mutant mitochondria that were observed had thin tubular cristae). We did observe a few mitochondria in which cristae formed concentric whorls (Figure 6, G and H). Similar concentric whorls were previously observed in the mitochondria of mammalian cells upon transfection with mitofilin siRNA (John et al., 2005) and in yeast (Rabl et al., 2009; Mun et al., 2010). We conclude that moma-1(tm1912) mutant animals have disrupted mitochondrial cristae similar to the disrupted cristae of mitofilin mutants.
FIGURE 6:
Abnormal cristae observed by electron microscopy in immt-1 and moma-1 mutant animals. (A) Mitochondria in a wild-type (N2) animal. (B and C) Mitochondria in immt-1(tm1730) animals. (D–H) Mitochondria in moma-1(tm1912) animals. The thin tubular cristae of wild-type mitochondria (dashes seen in the mitochondrial matrix in panel A) are converted to swollen cristae (B and C for immt-1 and D–F for moma-1). Occasionally, the internal morphology of moma-1(tm1912) mitochondria is more severely disrupted, resulting in concentric whorls of cristae (G and H). Scale bar is 200 nm.
DISCUSSION
Over the years a number of screens for mitochondrial morphology defects have been performed (Hermann et al., 1997; Dimmer et al., 2002; Gandre-Babbe and van der Bliek, 2008). However, new genes are still being discovered, suggesting that this approach is not yet exhausted. The screen described here was limited to 232 mitochondrial proteins, but this was enough to find several proteins that were missed in previous screens. CHCH-3 may have been missed before because it is not found in yeast, while the specific effects of IMMT-1 described here may have been missed because other organisms have only one mitofilin, which could mask specialized functions that are divided between IMMT-1 and IMMT-2 in C. elegans. These proteins are nonredundant because mutations in either gene affect mitochondrial morphologies in the same cell, as shown here and by others (Mun et al., 2010). The opposite could have happened with MOMA-1. This protein may have been missed before because of redundancy caused by gene duplication: Mammals and yeast each have two MOMA-1 homologues while C. elegans has one. Knockdown of either mammalian homologue has little or no effect on mitochondrial morphology (unpublished data). Deletion of the yeast homologue Aim37/YNL100W does slow growth on a nonfermentable carbon source (Hess et al., 2009), but the other yeast homologue, YGR235C, has not been found in screens for mitochondrial defects. The effects described here for C. elegans are, nevertheless, striking and robust, suggesting that MOMA-1 plays an important and evolutionarily conserved role in mitochondria.
In this article, we show that MOMA-1 is a mitochondrial OM protein with mutant phenotypes that are very similar to the phenotypes caused by mutations in the CHCHD3 homologue CHCH-3 and the mitofilin homologue IMMT-1. Mutant mitochondria have numerous swellings or bulges, similar to those observed in mitochondrial fission mutants, but the mitochondria are not connected like fission mutants, suggesting that these proteins affect a different process. EM shows that moma-1 and immt-1 mutants both have numerous mitochondria with distended cristae. In addition, some of the moma-1 mutant mitochondria also have cristae in concentric whorls similar to those previously observed in other organisms (John et al., 2005). Genetic interactions indicate that MOMA-1 and IMMT-1 act in a single pathway that governs mitochondrial morphology while CHCH-3 acts in a different pathway with similar effects on mitochondrial morphology (Figure 7). Mutations in either pathway have relatively benign effects on mitochondrial function, but the combinations of moma-1 and chch-3 or immt-1 and chch-3 defects dramatically disrupt mitochondria in C. elegans gonads. It would appear that these gonads completely lack functional mitochondria. We conclude that MOMA-1 and IMMT-1 on one hand and CHCH-3 on the other hand play separate roles governing mitochondrial morphology but they also play redundant roles in an essential mitochondrial process.
FIGURE 7:
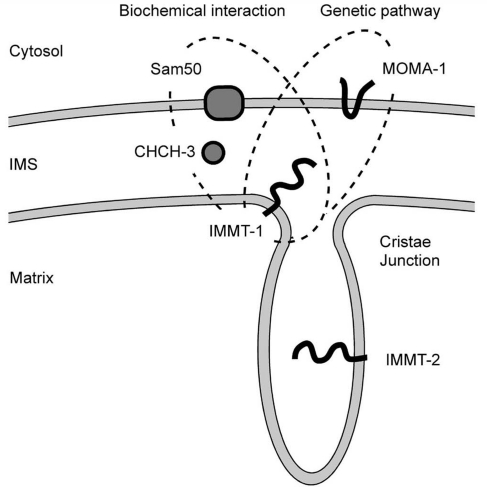
Model for interactions between MOMA-1, CHCH-3, and IMMT-1. Our genetic data show that MOMA-1 and IMMT-1 act in the same pathway even though they are on different membranes. In addition, mitofilin was previously shown to coimmunoprecipitate with Sam50 and the CHCH-3 homologue CHCHD3, but our data suggest that these proteins act in different pathways. Our data also show that the two C. elegans mitofilins (IMMT-1 and IMMT-2) are functionally distinct, possibly reflecting the two functions that mitofilins are known to have in mitochondria (formation of cristae junctions and control of membrane curvature within cristae) (Rabl et al., 2009) or a novel function.
CHCH-3 may contribute to protein import or assembly in the mitochondrial IMS because the mammalian CHCH-3 homologue CHCHD3 coimmunoprecipitates with other mitochondrial import proteins, such as Sam50 and metaxins (Xie et al., 2007), and although yeast does not have a CHCH-3 homologue, it does have several other proteins with CX9C motifs that act like chaperones (Gabriel et al., 2007). In addition, a new study by Darshi et al. (2011) shows that the levels of mammalian CHCHD3 control the levels of mitofilin, consistent with the proposed role of CHCHD3 in mitofilin biogenesis or stability. The synthetic effects that we observe between chch-3 and immt-1 mutations suggest that CHCH-3 and IMMT-1 do not have fully overlapping functions. They might have additional roles in other essential pathways or, perhaps more likely, CHCH-3 might also have other substrates that are important for mitochondrial function.
How might IMMT-1 and MOMA-1 proteins affect morphology? Previous submitochondrial localization and functional studies clearly show that mitofilin controls the diameters of cristae junctions and membrane curvature within cristae (Rabl et al., 2009). Effects on cristae morphology are also likely to have indirect effects on the overall diameter of mitochondria. Cristae morphologies could, for example, alter the ability of mitochondria to sequester IM within cristae. Cristae might also provide tensile resistance against mitochondrial swelling, as suggested by the observation that they often traverse the mitochondrial matrix with multiple connections to the IBM (Mannella, 2006). Regardless of mechanism, it seems likely that the effects of mitofilin on mitochondrial diameter are an indirect result of effects on cristae morphology.
Does MOMA-1 also indirectly affect mitochondrial diameter through a direct effect on cristae morphology? This would be consistent with the similar phenotypes of IMMT-1 and MOMA-1 mutants and their genetic interactions, but it is not immediately apparent how MOMA-1 and IMMT-1 would interact when they are on different membranes. There are a few examples of interactions between mitochondrial IM and OM proteins (Wong et al., 2003). In addition, a small but consistent amount of MOMA-1 was protease resistant, raising the possibility that this fraction is localized to the mitochondrial IM. It is therefore conceivable that MOMA-1 interacts directly with IMMT-1 to control cristae formation, even though direct interactions have not yet been detected in coimmunoprecipitation experiments (unpublished data). It is also possible that MOMA-1 affects IMMT-1 protein levels through effects on biogenesis or protein stability, similar to the effects of CHCHD3 on mitofilin in mammalian cells (Darshi et al., 2011). Further characterization of the interactions between IMMT-1 and MOMA-1 are therefore needed to determine how these two proteins affect mitochondrial diameter.
In summary, our experiments show the existence of a new mitochondrial morphology protein, MOMA-1, and a new pathway controlling mitochondrial cristae and diameter. More experiments are needed to test potential interactions between MOMA-1 and IMMT-1 and to define their effects on mitochondrial membranes.
MATERIALS AND METHODS
Molecular cloning
Full-length moma-1 cDNA was amplified from a C. elegans cDNA library (Invitrogen, Carlsbad, CA) and cloned into the NheI and XhoI sites of the pPD96.52 vector (kindly provided by A. Fire, J. Ahnn, G. Seydoux, and S. Xu, Carnegie Institution of Washington, Baltimore, MD). This vector has a myo-3 promoter for muscle cell–specific expression. Full-length fis-1 cDNA was similarly cloned into the NheI and EcoRI sites of pPD96.52. Full-length mff-1 and mff-2 cDNAs were cloned into the NheI and XhoI sites of pPD96.52. The fis-2 gene was amplified from C. elegans genomic DNA and cloned into the NheI and EcoRI sites of pPD96.52. These clones were used to create YFP::MOMA-1, YFP::FIS-1, YFP::FIS-2, CFP::MFF-1, and YFP::MFF-2 fusion constructs by inserting YFP or CFP coding sequences (Labrousse et al., 1999) into the NheI site of the pPD96.52 expression constructs. The MOMA-1::YFP fusion construct was made by PCR amplification of moma-1 coding sequences and cloning this into the KpnI and AgeI sites of the myo3::OuterMembrane/TOM-70::YFP plasmid (Labrousse et al., 1999). Hypodermal mitochondria were detected with a construct containing the 402 base pairs immediately upstream of the ATG site of col-12 cloned into the HindIII and KpnI sites of the pPD96.32 vector. This vector has a mitochondrial matrix–targeting sequence fused to GFP. Constructs were verified by sequencing. Mitochondrial OM and matrix markers under control of the myo-3 and the hlh-8 promoters were described previously (Labrousse et al., 1999). Feeding RNAi procedures were as described (Timmons et al., 2001). The bacterial RNAi library was provided by J. Ahringer (University of Cambridge, Cambridge, UK).
Worm strains
C. elegans moma-1(tm1912), chch-3(tm2336), immt-1(tm1730), immt-2(tm2366), and drp-1(tm1108) were provided by S. Mitani (NBRP and Tokyo Women’s Medical University School of Medicine, Tokyo, Japan). These strains were backcrossed four to six times with wild-type N2 animals to remove adventitious mutations. The C. elegans strains CB3335 anc-1(e1802), WM27 rde-1(ne219), CB6055 bus-8(e2698), and TJ356 Is(DAF-16::GFP) were obtained from the C. elegans stock center (Caenorhabditis Genetics Center, University of Minnesota, Minneapolis, MN). All strains were grown at 20°C.
Antibodies and other reagents
Rabbit polyclonal antibodies were generated against polypeptide fragments of C. elegans MOMA-1, EAT-3, and MFF-1 (F55F8.6) proteins. We used pET21d Novagen (Merck, Whitehouse Station, NJ) vectors encoding residues 1–119 of MOMA-1, 614–852 of EAT-3, and 1–138 of MFF-1. These His-tagged fragments were expressed in BL21 pLysS Escherichia coli, purified by column chromatography, and sent to Robert Sargeant (Ramona, CA) for antibody production. EAT-3 antibodies were blot purified; MOMA-1 and MFF-1 antibodies were not blot purified as they showed few cross-reacting bands on Western blots of whole-worm extracts. Cytochrome c and tubulin antibodies were purchased from Mitosciences (Eugene, OR) and Sigma (St. Louis, MO), respectively. Antibodies against ATP synthase F1β subunit were a gift from Carla Koehler (University of California, Los Angeles). Goat anti–mouse HRP– and goat anti–rabbit HRP–conjugated secondary antibodies were purchased from Pierce (Thermo-Fisher, Waltham, MA). Propidium iodide and rhodamine 6G were from Sigma-Aldrich (St. Louis, MO) and Molecular Probes (Eugene, OR), respectively.
Fluorescence microscopy and image analysis
Fluorescence images were captured using a Zeiss Axiovert 200M equipped with 40×/NA 1.3 Plan-Neofluar, 63×/NA 1.4 Plan-Apochromat, or 100×/NA 1.45 α-Plan-Fluar objectives and an ORCA ER-CCD camera (Hamamatsu, Shizuoka, Japan). Young adult worms (picked 1 d after L4) were used for imaging of muscle and hypodermal cell mitochondria. Gonad lengths were measured using ImageJ software (National Institutes of Health, Bethesda, MD), and gonad tracings were made with Canvas 8 software (ACD Systems, Victoria, BC, Canada). Embryos and gonad samples were processed for immunofluorescence as described (Epstein and Shakes, 1995). Rhodamine 6G–stained gonads and YFP- or CFP-tagged FIS-1, FIS-2, MFF-1, and MFF-2 in muscle cells were imaged with a Zeiss LSM 5 Pascal confocal microscope equipped with a 100×/NA 1.45 Plan-Fluar objective. CFP was excited with 458-nm, YFP with 514-nm, and rhodamine with 543-nm laser lines. Dissections for staining gonads were described previously (Labrousse et al., 1999). The degrees of colocalization of YFP and CFP labels were assessed with the Zeiss LSM 5 Confocal software, which assigned pixels to mitochondria in individual muscle cells. Thresholds were manually adjusted to distinguish mitochondria from surrounding cytoplasm.
Subcellular fractionation
Hypochlorite-synchronized bus-8(e2698) animals were grown to adulthood on nematode growth medium plates with OP50 bacteria and harvested by flotation in M9. Pooled worms were washed six times with M9, washed once with STEG pH 7.4 (Curran et al., 2004), and suspended in chilled STEG pH 7.4 with a protease inhibitor cocktail (Roche, Basel, Switzerland) and 1 mM phenylmethanesulfonyl fluoride (PMSF). All subsequent fractionation steps were performed on ice or at 4°C. Animals were homogenized in a Kontes (Vineland, NJ) ground glass tissue homogenizer using 75 strokes with a 1-min pause every 15 strokes. The resulting homogenate was centrifuged at 2500 × g for 20 min. The supernatant was passed through a nylon filter with 30-μ pores (Spectrum, Houston, TX) and centrifuged at 14,000 × g for 30 min. The P2 fraction (14,000 × g pellet) was washed twice with STEG and protease inhibitors. Protein concentrations were measured with a dye-based assay (Bio-Rad, Hercules, CA).
Protease protection experiment
A 60-μg fresh P2 fraction was pelleted and resuspended in 100 μl STEG or STEG with 1% Triton X-100. Proteinase K (Roche) was added to a final concentration of 0, 30, or 60 μg/ml and incubated for 15 min on ice. PMSF was then added to a final concentration of 2 mM, and samples were placed on ice for an additional 5 min. Laemmli sample buffer was added, and the samples were placed in boiling water for 8 min. Proteins were size fractionated by SDS–PAGE and transferred to nitrocellulose membrane (GE Healthcare, Little Chalfont, UK) by Western blotting. Membranes were probed with the indicated primary antibodies and developed with horseradish peroxidase–conjugated secondary antibodies and ECL Plus chemiluminescence (GE Healthcare). Where indicated, relative expression levels were quantified with a Personal Densitometer SI and ImageQuant software (GE Healthcare).
EM
Worms were synchronized by treatment with hypochlorite and grown to L2 or L3 larval stages. These worms were washed with M9, pelleted, and mixed with a concentrated suspension of E. coli OP50, which served as a cryoprotectant. This mixture was transferred to 1.5 mm × 0.2 mm sample holders and frozen in an EM PACT2-RTS High Pressure Freezer (Leica Microsystems, Bannockburn, IL). Frozen samples were transferred to 1.0% osmium tetroxide plus 0.1% uranyl acetate in acetone and placed in an EM AFS2 Automatic Freeze-Sub System (Leica Microsystems). The temperature was raised slowly from –144°C to –90°C >96 h, and from –90°C to –25°C >16 h, followed by more rapid warming to room temperature. Specimens were removed from the freeze-sub solution, washed with acetone, washed with propylene oxide, and embedded by baking in SPURR resin (Ted Pella, Redding, CA) for 24 h at 60°C. Sections of 75 nm were made with a UCT Ultramicrotome (Leica Microsystems) and poststained with uranyl acetate and lead citrate. Sections were examined using Titan S/TEM (FEI) and JEM1200-EX (JEOL, Tokyo, Japan) electron microscopes at 80 kV.
qPCR
L4 stage larvae were transferred to lysis buffer (1× thermophilus buffer [New England Biolabs, Ipswich, MA] with 1.2 μg/ml proteinase K [Roche]), frozen at –20°C, and incubated for 1 h at 65°C and for 15 min at 95°C. Each qPCR contained 2 μl lysate corresponding to two L4 worms. qPCR was performed with SYBR Green reagent, as recommended by the manufacturer (Invitrogen), with annealing and extension at 63°C in a Mx3000P Thermal Cycler (Agilent, Santa Clara, CA) followed by analysis with MxPro Software (Agilent). Primers to amplify a 195–base pair region of mtDNA and a 225–base pair region of polymerase ε subunit A in nuclear DNA were previously described (Meyer et al., 2007). Dissociation curves confirmed individual PCR products for mtDNA and two PCR products for nuclear DNA (nDNA). Ct values were converted to percentages, and mtDNA:nDNA ratios were calculated from these percentages.
Supplementary Material
Acknowledgments
We thank other members of the lab and Alison Frand for helpful discussions and critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health (NIH) (GM-051866) to A.V.D.B. B.P.H. received a stipend from an NIH training grant (T32-GM-07104).
Abbreviations used:
- CFP
cyan fluorescent protein
- CHCHD
coiled-coil-helix–coiled-coil-helix domain
- EM
electron microscopy
- GFP
green fluorescent protein
- IBM
inner boundary membrane
- IM
inner membrane
- IMS
intermembrane space
- MOMA-1
mitochondrial outer membrane abnormal protein
- mtDNA
mitochondrial DNA
- nDNA
nuclear DNA
- OM
outer membrane
- qPCR
quantitative PCR
- RNAi
RNA interference
- ROS
reactive oxygen species
- siRNA
small interfering RNA
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcgi/doi/10.1091/mbc.E10-07-0600) on January 19, 2011.
REFERENCES
- Barbe L, et al. Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics. 2008;7:499–508. doi: 10.1074/mcp.M700325-MCP200. [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta. 2006;1763:450–462. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- Curran SP, Leverich EP, Koehler CM, Larsen PL. Defective mitochondrial protein translocation precludes normal Caenorhabditis elegans development. J Biol Chem. 2004;279:54655–54662. doi: 10.1074/jbc.M409618200. [DOI] [PubMed] [Google Scholar]
- Da Cruz S, Xenarios I, Langridge J, Vilbois F, Parone PA, Martinou JC. Proteomic analysis of the mouse liver mitochondrial inner membrane. J Biol Chem. 2003;278:41566–41571. doi: 10.1074/jbc.M304940200. [DOI] [PubMed] [Google Scholar]
- Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining cristae integrity and mitochondrial function. J Biol Chem. 2011;286:2918–2932. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, Westermann B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Shakes DC, editors. San Diego, CA: Academic Press; 1995. Caenorhabditis elegans: Modern Biological Analysis of an Organism. [Google Scholar]
- Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel K, Milenkovic D, Chacinska A, Muller J, Guiard B, Pfanner N, Meisinger C. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J Mol Biol. 2007;365:612–620. doi: 10.1016/j.jmb.2006.10.038. [DOI] [PubMed] [Google Scholar]
- Gandre-Babbe S, Van Der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieffers C, Korioth F, Heimann P, Ungermann C, Frey J. Mitofilin is a transmembrane protein of the inner mitochondrial membrane expressed as two isoforms. Exp Cell Res. 1997;232:395–399. doi: 10.1006/excr.1997.3539. [DOI] [PubMed] [Google Scholar]
- Grad LI, Lemire BD. Mitochondrial complex I mutations in Caenorhabditis elegans produce cytochrome c oxidase deficiency, oxidative stress and vitamin-responsive lactic acidosis. Hum Mol Genet. 2004;13:303–314. doi: 10.1093/hmg/ddh027. [DOI] [PubMed] [Google Scholar]
- Griparic L, Head BP, Van Der Bliek AM. Mitochondrial division and fusion. Topics Curr Genet. 2004;8:227–249. [Google Scholar]
- Harfe BD, Gomes AV, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 1998;12:2623–2635. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, King EJ, Shaw JM. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J Cell Biol. 1997;137:141–153. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DC, et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Nunnari J. The molecular mechanism of mitochondrial fusion. Biochim Biophys Acta. 2009;1793:20–26. doi: 10.1016/j.bbamcr.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ, Zha J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell. 2005;16:1543–1554. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen T, Davis DE, Larsen PL, Clarke CF. Reproductive fitness and quinone content of Caenorhabditis elegans clk-1 mutants fed coenzyme Q isoforms of varying length. J Biol Chem. 2003;278:51735–51742. doi: 10.1074/jbc.M308760200. [DOI] [PubMed] [Google Scholar]
- Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kanazawa T, Zappaterra MD, Hasegawa A, Wright AP, Newman-Smith ED, Buttle KF, McDonald K, Mannella CA, Van Der Bliek AM. The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet. 2008;4:e1000022. doi: 10.1371/journal.pgen.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Hanazawa M, Gengyo-Ando K, Mitani S, Maruyama I, Iino Y. ASB-1, a germline-specific isoform of mitochondrial ATP synthase b subunit, is required to maintain the rate of germline development in Caenorhabditis elegans. Mech Dev. 2007;124:237–251. doi: 10.1016/j.mod.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zapaterra M, Rube DA, Van Der Bliek AM. C. elegans dynamin-related protein drp-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Lamant M, et al. ApoO, a novel apolipoprotein, is an original glycoprotein up-regulated by diabetes in human heart. J Biol Chem. 2006;281:36289–36302. doi: 10.1074/jbc.M510861200. [DOI] [PubMed] [Google Scholar]
- Mannella CA. Structure and dynamics of the mitochondrial inner membrane cristae. Biochim Biophys Acta. 2006;1763:542–548. doi: 10.1016/j.bbamcr.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Mannella CA. Structural diversity of mitochondria: functional implications. Ann NY Acad Sci. 2008;1147:171–179. doi: 10.1196/annals.1427.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Mun JY, Lee TH, Kim JH, Yoo BH, Bahk YY, Koo HS, Han SS. Caenorhabditis elegans mitofilin homologs control the morphology of mitochondrial cristae and influence reproduction and physiology. J Cell Physiol. 2010;224:748–756. doi: 10.1002/jcp.22177. [DOI] [PubMed] [Google Scholar]
- Munn EA. On the structure of mitochondria and the value of ammonium molybdate as a negative stain for osmotically sensitive structures. J Ultrastruct Res. 1968;25:362–380. doi: 10.1016/s0022-5320(68)80092-9. [DOI] [PubMed] [Google Scholar]
- Odgren PR, Toukatly G, Bangs PL, Gilmore R, Fey EG. Molecular characterization of mitofilin (HMP), a mitochondria-associated protein with predicted coiled coil and intermembrane space targeting domains. J Cell Sci. 1996;109:2253–2264. doi: 10.1242/jcs.109.9.2253. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Rabl R, et al. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauble S, King CC, Darshi M, Koller A, Shah K, Taylor SS. Identification of ChChd3 as a novel substrate of the cAMP-dependent protein kinase (PKA) using an analog-sensitive catalytic subunit. J Biol Chem. 2007;282:14952–14959. doi: 10.1074/jbc.M609221200. [DOI] [PubMed] [Google Scholar]
- Sickmann A, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- Taylor SW, et al. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tsang WY, Lemire BD. The role of mitochondria in the life of the nematode, Caenorhabditis elegans. Biochim Biophys Acta. 2003;1638:91–105. doi: 10.1016/s0925-4439(03)00079-6. [DOI] [PubMed] [Google Scholar]
- Van Der Bliek AM. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- Velours J, Dautant A, Salin B, Sagot I, Brèthes D. Mitochondrial F1F0-ATP synthase and organellar internal architecture. Int J Biochem Cell Biol. 2009;41:1783–1789. doi: 10.1016/j.biocel.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Westerman BA, Poutsma A, Steegers EA, Oudejans CB. C2360, a nuclear protein expressed in human proliferative cytotrophoblasts, is a representative member of a novel protein family with a conserved coiled coil-helix-coiled coil-helix domain. Genomics. 2004;83:1094–1104. doi: 10.1016/j.ygeno.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Wong ED, Wagner JA, Scott SV, Okreglak V, Holewinske TJ, Cassidy-Stone A, Nunnari J. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J Cell Biol. 2003;160:303–311. doi: 10.1083/jcb.200209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–3549. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793:5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.