Abstract
FtsI (also called PBP3) of Escherichia coli is a transpeptidase required for synthesis of peptidoglycan in the division septum and is one of several proteins that localize to the septal ring. FtsI comprises a small cytoplasmic domain, a transmembrane helix, a noncatalytic domain of unknown function, and a catalytic (transpeptidase) domain. The last two domains reside in the periplasm. We used PCR to randomly mutagenize ftsI, ligated the products into a green fluorescent protein fusion vector, and screened ∼7,500 transformants for gfp-ftsI alleles that failed to complement an ftsI null mutant. Western blotting and penicillin-binding assays were then used to weed out proteins that were unstable, failed to insert into the cytoplasmic membrane, or were defective in catalysis. The remaining candidates were tested for septal localization and ability to recruit another division protein, FtsN, to the septal ring. Mutant proteins severely defective in localization to the septal ring all had lesions in one of three amino acids—R23, L39, or Q46—that are in or near the transmembrane helix and implicate this region of FtsI in septal localization. Mutant FtsI proteins defective in recruitment of FtsN all had lesions in one of eight residues in the noncatalytic domain. The most interesting of these mutants had lesions at G57, S61, L62, or R210. Although separated by ∼150 residues in the primary sequence, these amino acids are close together in the folded protein and might constitute a site of FtsI-FtsN interaction.
Cell division in Escherichia coli requires approximately a dozen proteins, all of which localize to a ring structure at the division site (Fig. 1A) (for a recent review, see reference 15). The division proteins localize in a defined order, which starts with assembly of the tubulin homolog FtsZ into a contractile ring at the midcell. Subsequent recruitment of the other division proteins is thought to result in the assembly of a complex that mediates inward growth of the cell envelope. According to the current model, the last four proteins recruited to the septal ring are FtsW, FtsI, FtsN, and AmiC (in that order) (4). The precise roles of FtsW and FtsN in septum assembly are not yet known. FtsI is a transpeptidase that introduces peptide cross-linking into the peptidoglycan cell wall in the division septum (6, 37). AmiC is a periplasmic amidase that hydrolyzes peptide cross-links and contributes to the separation of daughter cells after division (25).
FIG. 1.
(A) Recruitment of proteins to the septal ring of E. coli. Assembly of the ring starts with formation of the FtsZ ring at the midcell. The remaining proteins are then recruited in the order indicated, with AmiC being the last. (B) Domain structure of FtsI. FtsI contains a cytoplasmic domain (CD), a single transmembrane helix (TM), a noncatalytic domain, and a transpeptidase catalytic domain. The last two domains reside in the periplasm. The catalytic domain contains a serine (S307) that becomes transiently acylated during transpeptidation and forms a long-lived covalent adduct with β-lactams. Amino acid substitutions characterized in this study are shown. Those above the protein severely impair localization, while those below the protein do not. (C) Model showing the locations of amino acid substitutions that impair localization (red balls) and recruitment of FtsN (black balls). Two structures of PBP2x determined by X-ray crystallography were combined to generate this model, as described in Materials and Methods. The α-carbon backbone of PBP2x obtained from 1PMD is shown, with the catalytic domain in purple and the noncatalytic domain in green. Twelve residues at the N terminus that are not resolved in the 1PMD structure but are present in the 1K25 structure are shown in red. The cytoplasmic domain, the transmembrane helix, and a 20-amino-acid portion of the noncatalytic domain are of unknown structure and have been drawn in for reference only; they are not to scale. Serine 307 in the catalytic site is represented by a green ball. The amino acids represented by yellow balls were also found to be important for FtsI function, but interpretation of lesions at these sites is complicated by decreased protein stability (see Discussion). R141 cannot be precisely located because the corresponding region in PBP2x is a 30-residue loop (shown in yellow) that is much shorter in FtsI of E. coli.
The ftsI gene encodes a protein of 588 amino acids, but a proteolytic processing event removes 11 residues from the C terminus (32). Processing is not required for FtsI function (22). The membrane topology of FtsI has been studied with protein fusions, which revealed that the protein can be divided into three domains: an amino-terminal cytoplasmic domain (23 amino acids), a single transmembrane helix (17 residues), and a large periplasmic domain (537 residues) (7). The periplasmic domain appears to comprise two parts, a noncatalytic domain of unknown function and a catalytic domain that is directly responsible for introducing cross-links into septal peptidoglycan (reviewed in reference 33). A cartoon of the domain structure of FtsI is shown in Fig. 1B.
Several reports have implicated the cytoplasmic domain and membrane-spanning segment in targeting of FtsI to the septal ring (12, 17, 20, 40). The most direct evidence comes from analysis of hybrid proteins in which these domains have been replaced with the corresponding parts of other membrane proteins. Such hybrid proteins fail to complement temperature-sensitive and null alleles of ftsI and also fail to localize to the septal ring (20, 40).
The catalytic domain extends from residues 237 to 577 and is the site of the transpeptidase activity responsible for cross-linking the peptidoglycan cell wall (1). The catalytic domain exhibits homology to other transpeptidases involved in peptidoglycan metabolism (18). In particular, the active site contains a universally conserved serine residue (S307) that forms a covalent bond with the peptide substrate during transpeptidation, a reaction that proceeds via an acyl enzyme intermediate. The catalytic domain also binds β-lactam antibiotics, which mimic a transpeptidase substrate and serve as suicide inhibitors by forming a long-lived covalent adduct with the catalytic serine (28, 34). Thus, FtsI is also known as penicillin-binding protein 3 (PBP3).
The noncatalytic domain extends from residues 41 to 236. This domain was first recognized as a distinct structural element when sequence comparisons revealed that it was conserved among a subset of transpeptidases called the class B high-molecular-mass penicillin-binding proteins (16). It has been proposed that the noncatalytic domain interacts with other proteins of the septal ring and that these interactions are important for septal localization of FtsI, recruitment of downstream proteins such as FtsN, and regulation of the transpeptidase activity of FtsI (33). Marrec-Fairley and coworkers have characterized several mutants with properties consistent with these roles. Most notably, they have suggested that changing D58 to V prevents localization of FtsI to the septal ring and changing both R210 and R213 to Q (a double mutation) interferes with a protein-protein interaction but not septal localization per se (29). These interpretations were based on the recessive nature of the D58V lesion and the dominant nature of the R210Q-R213Q lesions. The abilities of the mutant proteins to localize and/or interact with other division proteins were not tested directly.
The three-dimensional structure of an FtsI homolog, PBP2x from Streptococcus pneumoniae, has been solved by X-ray crystallography (Fig. 1C) (13, 35). The catalytic domain looks like a typical penicillin-binding domain, except that the active site is at the center of a long groove that is not seen in other penicillin-binding enzymes. This groove presumably accommodates the two peptides to be joined in the cross-linking reaction. The noncatalytic domain has a strikingly elongated shape. One end fits into a pocket in the catalytic domain, and two long arms extend into solution, creating a large central cavity that could accommodate another protein in vivo. This arrangement suggests that the noncatalytic domain regulates the catalytic domain and that the arms might engage in protein-protein interactions (35). This arrangement also suggests why the noncatalytic domain appears to be required for folding of the catalytic domain (17); these domains interact so extensively that neither is likely to be stable alone.
To identify sequences in FtsI that are needed for localization to the septal ring and for interaction with other division proteins, we have isolated and characterized mutant forms of the protein that fail to support cell division but appear to be normal for insertion into the membrane and catalytic activity (penicillin binding). These mutants are described in this paper. Single-amino-acid substitutions that severely impaired localization invariably mapped to the membrane-spanning segment rather than the noncatalytic domain. We also found several mutant proteins that were defective in recruitment of FtsN to the septal ring. These proteins had single-amino-acid substitutions in the noncatalytic domain and might define a site of FtsI-FtsN interaction. Finally, we found one lesion in the noncatalytic domain that renders FtsI incapable of supporting cell division, but the protein appears essentially normal in all of our assays.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strain construction was done by generalized transduction with P1 (31), specialized transduction with lambda InCh (9), or transformation with integration vectors (21).
The E. coli strains used were MG1655, DHB4 [F′ lacIq pro/λ− ΔlacX74 galE galK thi rpsL phoR ΔphoA(PvuII) ΔmalF3], EC295 [MG1655 ftsI23(Ts) leu::Tn10], EC812 [MG1655 ftsI::cat Δ(λattL-lom)::bla lacIq PBAD-ftsI], and EC1137 [MG1655 attL::pDSW539 (Kanr lacIq P209-gfp-ftsI)]. MG1655 and DHB4 have been described elsewhere (5, 8). EC295 was constructed by transducing MG1655 to Tetr with P1 grown on LMG64 (20) and screening for temperature sensitivity. EC812 was constructed in several steps. First, an arabinose-controlled ftsI gene was introduced into the chromosome of MC4100 from plasmid pDSW172 (pBAD24-ftsI) using λ InCh1 (19). The PBAD-ftsI was then moved into MG1655 by transduction with P1 to make EC811. Finally, an ftsI::cat allele from JE7947 (23) was transduced into EC811 to make EC812. Strain EC1137 was constructed by integrating pDSW539 into the λ att site (21). Derivatives that express mutant ftsI alleles were constructed similarly from plasmids pDSW546 to -559. These strains were EC1138 (R23H), EC1139 (L39P), EC1140 (Q46H), EC1141 (G57D), EC1142 (S61P), EC1143 (L62P), EC1144 (V86E), EC1145 (R141C), EC1146 (H177R), EC1147 (G188E), EC1148 (R210C), EC1149 (R210H), EC1150 (S61F), and EC1155 (R23C).
The plasmid pDSW287 was constructed by replacing the 1.3-kb promoter-bearing EcoRV-EcoRI fragment in pDSW254 with the corresponding fragment carrying a weaker promoter, P206, from pDSW209 (40). The construction of pDSW478 (pBAD18-Kan-ftsI) has been described elsewhere (14). Derivatives that express mutant alleles of ftsI were constructed by replacing a 1.5-kb EcoRI-KpnI fragment that carries most of ftsI. The resulting plasmids were pDSW479 (R23H), pDSW480 (L39P), pDSW481 (G57D), pDSW484 (Q46H), pDSW486 (S61P), pDSW487 (L62P), pDSW488 (V86E), pDSW489 (R141C), pDSW490 (H177R), pDSW492 (G188E), pDSW494 (R210C), pDSW495 (R210H), pDSW496 (R23C), and pDSW498 (S61F).
The plasmid pDSW521 (P206-gfp-ftsI) was constructed by PCR amplifying the gfp-ftsI fusion gene from pDSW287 using primers P324 (5′-CACGATATCAGCCAACAGACCATGAGTAAAGGAG-3′) and P304 (5′-GTGAAGCTTACGATCTGCCACCTGTCCC-3′). The 2.5-kb product was digested with EcoRV and HindIII (the sites are underlined) and ligated into pTH18-kr (24) that had been cut with EcoRI, made blunt ended by filling it with T4 DNA polymerase, and then cut with HindIII. Derivatives that carry mutant alleles of ftsI were obtained directly during the screen or constructed by subcloning from pDSW287 derivatives by replacing a 560-bp EcoRI-XmnI fragment in pDSW521 with the corresponding fragment from the pDSW287 derivatives. The resulting plasmids were pDSW562 (R23C), pDSW563 (R23H), pDSW566 (L39P), pDSW567 (Q46H), pDSW568 (G57D), pDSW569 (S61F), pDSW570 (S61P), pDSW571 (L62P), pDSW572 (V86E), pDSW573 (R141C), pDSW574 (H177R), pDSW575 (G188E), pDSW576 (R210C), and pDSW577 (R210H).
The vectors used to integrate gfp-ftsI fusion genes into the chromosome were constructed in several steps. The gene for gfp was amplified from pDSW207 (40) using the primers P459 (CAAGGATCCAGGAGGATGAATATGAAAGGAGAAGAACTTTTC) and P460 (CGTGAGCTCTCAGAATTCTTTGTATAGTTCATCCATGCC). The 0.8-kb product was cut with BamHI and SacI (the sites are underlined) and ligated into the same sites of pAH144 (21) to create pDSW499. Then, a 1.7-kb SphI-NdeI restriction fragment that carries lacIq and P206-gfp′ from pDSW287 was ligated into pDSW499 that had been cut with SphI and NdeI. The resulting construct was designated pDSW535. This plasmid was digested with SphI and NheI, and the 2.5-kb fragment carrying lacIq, P209-gfp, and tL3 was ligated into the same sites of pAH120 (21) to create pDSW537. Finally, ftsI was cloned into pDSW537 to create pDSW539. This was accomplished by PCR amplifying ftsI from pDSW287 with the primers GFP666-F (GAGACCACATGGTCCTTCTTGAG) and P500 (ACGGACAATTGAAGCTTCTAGAATTACGATCTGCCACCTGTCC). The 1.8-kb product was cut with EcoRI (a site in the pDSW287 template plasmid) and MfeI (the site is underlined) and ligated into the EcoRI site of pDSW537 to yield pDSW539. Derivatives that express mutant alleles of ftsI were constructed by replacing a 1.5-kb EcoRI-KpnI fragment that carries most of ftsI. The resulting plasmids were pDSW546 (R23C), pDSW547 (R23H), pDSW548 (L39P), pDSW549 (Q46H), pDSW550 (G57D), pDSW551 (S61F), pDSW552 (S61P), pDSW553 (L62P), pDSW554 (V86E), pDSW555 (R141C), pDSW556 (H177R), pDSW557 (G188E), pDSW558 (R210C), and pDSW559 (R210H).
Media and reagents.
The medium was Luria broth (LB) (10 g of tryptone, 5 g of yeast extract, 10 g of NaCl per liter, with 15 g of agar/liter for plates). l-Arabinose and d-glucose were used to modulate the expression of genes under the control of the PBAD promoter (19), and IPTG (isopropyl-β-d-thiogalactoside) was used to induce expression from lac promoters. Antibiotics were used at various concentrations as indicated in the description of each experiment.
Molecular biological procedures.
Standard protocols for cloning and analysis of DNA, PCR, and electroporation were used (3). Restriction enzymes were from New England Biolabs (Beverly, Mass.). PCR was performed with SuperMix from Life Technologies (Rockville, Md.). Oligonucleotides were from Integrated DNA Technologies (Coralville, Iowa). DNA was sequenced at the DNA Core Facility of the University of Iowa using dye-terminator cycle-sequencing chemistry.
Isolation of mutant ftsI alleles.
Random mutagenesis of the ftsI gene was performed by PCR with SuperMix, which contains Taq DNA polymerase. Amplification reaction mixtures had a volume of 100 μl and included 10 ng of pDSW235 plasmid template, which carries a wild-type gfp-ftsI fusion gene. The primers used were 5′-GAGACCACATGGTCCTTCTTGAG-3′, which anneals in gfp, and 5′-TGCTCTAGATTACGATCTGCCACCTGT-3′, which anneals to the 3′ end of ftsI. Amplification was done for 25 cycles with denaturation at 94°C, annealing at 52°C, and extension at 72°C. The 1.8-kb amplification product was digested with EcoRI and XbaI (sites at the gfp-ftsI junction and in the primer [underlined], respectively). The digested fragment was ligated into the same sites of pDSW287 or pDSW521. The resulting plasmid pools were introduced into EC812 by electroporation, selecting for kanamycin resistance on LB plates containing 40 μg of kanamycin/ml, 25 μg of ampicillin/ml, and 0.2% arabinose at 30°C. The colonies were then patched in a grid pattern onto similar plates that contained 0.0002% arabinose to predeplete the cells of wild-type ftsI expressed from the chromosome. Colonies that grew on the low-arabinose plates were tested for complementation by replica plating them onto the same medium with either 0.2% arabinose or 0.2% glucose. EC812 derivatives carrying an empty vector (pDSW286 or pTK18kr) and gfp-ftsI (pDSW286 or pDSW521) served as negative and positive controls, respectively.
Assays for expression, membrane insertion, catalytic activity, and stability of mutant FtsI proteins by Western blotting and penicillin binding.
Western blotting was done as described previously (40). Briefly, proteins were separated on sodium dodecyl sulfate (SDS)-10% polyacrylamide gels and transferred to nitrocellulose. Then, FtsI and green fluorescent protein (GFP)-FtsI were probed with a polyclonal anti-FtsI serum, followed by a secondary antibody conjugated to horseradish peroxidase, which in turn was detected with a chemiluminescent substrate. The blots were visualized with an LAS-1000 luminescent imager from Fuji (Stamford, Conn.). Penicillin binding was assayed with Bocillin-FL from Molecular Probes (Eugene, Oreg.) essentially as described previously (14, 20). For initial screening of expression, catalytic activity, and membrane insertion, we assayed penicillin binding in spheroplasts using a procedure similar to one previously used to demonstrate catalytic activity and membrane insertion for FtsI “swap” proteins (20). Cells harboring plasmids that expressed gfp-ftsI fusions were induced with 100 μM IPTG for 5 h and then converted to spheroplasts. The spheroplasts were incubated with 20 μM Bocillin FL for 20 min at 30°C. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and the fluorescent label incorporated into GFP-FtsI was visualized and quantified with a Typhoon 8600 imager from Amersham (Piscataway, N.J.). A penicillin-binding assay was also used to determine thermostability (14). Cells harboring pBAD18-Kan derivatives that expressed ftsI without the gfp moiety were induced for 2 h with 0.2% arabinose. Then, the membranes were isolated by extraction of soluble proteins with a nonionic detergent (14). The isolated membranes were incubated at 47 or 33°C for different lengths of time, and residual penicillin-binding activity was assayed with Bocillin FL at 30°C.
Localization of GFP-FtsI in merodiploids.
To visualize GFP-FtsI in merodiploids, strains were grown overnight at 37°C in LB containing 10 μg of kanamycin/ml. The next morning, the cultures were diluted 1:2,000 in LB containing 5 mM IPTG and were grown at 30°C to an optical density at 600 nm of ∼0.3, at which time they were fixed and analyzed by fluorescence microscopy with a GFP filter set as described previously (30). The typical exposure time was 12 s. Digital images were captured as 12-bit images, converted to 8-bit TIFF format, and imported to Adobe Photoshop and Canvas for the preparation of figures. At the time the cells were fixed for microscopy, the cells from 1 ml of culture were collected by centrifugation and resuspended in 100 μl of loading buffer for Western blotting.
Localization of GFP-FtsI and FtsN in filaments depleted of FtsI.
Strains were grown overnight at 37°C in LB containing 40 μg of kanamycin/ml, 10 μg of chloramphenicol/ml, 25 μg of ampicillin/ml, and 0.2% arabinose. The next morning, the cultures were diluted 1:50 in LB containing 40 μg of kanamycin/ml, 0.02% arabinose, and 100 μM IPTG. These cultures were grown at 30°C to an optical density at 600 nm of ∼0.3, and then the arabinose was removed by pelleting 1 ml of culture in a microcentrifuge tube and resuspending it in 1 ml of LB. The cells were washed in this manner three times before being diluted 1:250 in LB containing 40 μg of kanamycin/ml, 100 μM IPTG, and 0.2% arabinose or 0.2% glucose. Growth was continued for ∼4.5 to 5 h, at which time the glucose-grown cells had become filaments ∼20 to 30 μm long, except in the case of strains that expressed GFP-FtsI fusions that complemented the ftsI null mutation. The cells were fixed with cross-linking agents directly in growth medium and permeabilized by treatment with lysozyme as described previously (36). Anti-FtsN serum was used at a dilution of 1:200 overnight at 4°C. The secondary antibody was goat anti-rabbit conjugated to Texas Red (Molecular Probes) at 1:400 for 2 h at room temperature. For photography of Texas Red, the filter set was from Chroma Technology Corp. (Brattleburo, Vt.) and comprised a 555- to 560-nm-wavelength excitation filter, a 595-nm dichroic mirror (long pass), and a 645- to 675-nm emission filter. The typical exposure time was 2 s. Images were captured as 8-bit images, converted to TIFF format, and exported to Adobe Photoshop and Canvas for the preparation of figures. GFP-FtsI was also visualized in these filaments as described above for merodiploids. GFP-FtsI was photographed first to minimize bleaching. Staining with DAPI (4′,6′-diamidino-2-phenylindole) was done to visualize nucleoids and to verify proper segregation (not shown). At the time the cells were fixed for microscopy, the cells from 1 ml of culture were collected by centrifugation and resuspended in 100 μl of loading buffer for Western blotting.
Quantitation of fluorescence intensity.
Localization of GFP-FtsI and FtsN was quantified using the line profile tool in Image-Pro Plus version 4.1 from Media Cybernetics (Silver Spring, Md.). Transects ≥3 μm long were centered on the brightest GFP-FtsI bands (green channel). An average transect was then calculated after centering ≥24 transects on their brightest pixel. The average transects for wild-type and the various mutant proteins could not be compared directly owing to substantial day-to-day variation in the absolute fluorescence intensities in our images. However, relative fluorescence intensities were consistent between experiments, so to compare mutants to the wild type, we scaled the average transects as follows. First, the background fluorescence intensity, meaning the intensity away from the septal band, was determined by averaging the intensity values in 1-μm regions to the left (−1.5 to −0.5 μm) and right (+0.5 to +1.5 μm) of the peak. Values along each transect were then divided by the background intensity to produce a normalized transect. Finally, the baseline was set to zero by subtracting 1 from each pixel. This procedure was then repeated for FtsN after the transects were automatically replicated from the green channel to the corresponding position in the red channel.
Antiserum for detection of FtsN by immunofluorescence microscopy.
Polyclonal antibodies against the periplasmic domain of FtsN were raised in New Zealand White rabbits (Covance, Denver, Pa). The protein used as the antigen was obtained as follows. The periplasmic domain of FtsN (residues 56 to 319) was fused to the maltose-binding protein (MBP) in pMAL-c2 and purified by affinity chromatography on amylose-agarose as described previously (3). The final preparation was ∼95% pure, and the yield was ∼3 mg/liter of culture. Characterization of the antiserum by Western blotting revealed two bands corresponding to FtsN and MBP. Antibodies against MBP were depleted by incubating the serum with MBP-agarose that was made by incubating purified MBP2* from New England Biolabs with AminoLink coupling gel from Pierce Biotechnology, Inc. (Rockford, Ill.). One milliliter of antiserum was diluted with 1 ml of phosphate-buffered saline and passed over a column containing 200 μl of immobilized MBP slurry. Subsequent Western blotting revealed only a single protein. This band was present in a malE mutant but disappeared upon depletion of FtsN. The specificity of the antibody in immunofluorescence microscopy was verified by detection of FtsN as a band of fluorescence at the midcell in about half of the cells from a wild-type population and by showing that this signal was lost when the cells were depleted of FtsN (data not shown).
Model of FtsI.
The program O (27) was used to model the three-dimensional structure of FtsI based on the Cα coordinates for PBP2x deposited in the Brookhaven Protein Data Bank (PDB) under the PDB identification codes 1PMD and 1K25. First, the B subunit of 1K25 was superimposed on 1PMD. Then, the alpha carbon backbone for amino acids 76 to 633 of 1PMD and amino acids 66 to 77 of the B subunit of 1K25 were combined into a single PDB file.
RESULTS
Isolation of mutants.
We screened for ftsI mutants that failed to support division and were either recessive to wild type or exhibited dominant-negative phenotypes only at high levels of expression. To isolate such mutants, we constructed strain EC812, in which ftsI was expressed from an arabinose-inducible promoter integrated into the chromosome at the λ attachment site. This strain grew normally on media containing arabinose but failed to form colonies on media containing glucose. The growth of EC812 on media containing glucose was rescued by a plasmid that expresses a bright variant of gfp (gfpmut2 [11]) fused to the wild-type ftsI gene. We used two plasmids in this study, both of which direct expression of gfp-ftsI fusions under the control of IPTG-regulated promoters, although IPTG was not present in the plates during the initial screen to ensure low levels of expression. Plasmid pDSW287 has a copy number of ∼20 and overproduces GFP-FtsI ∼5-fold in the absence of induction. The other plasmid, pDSW521 has a copy number of ∼5 and produces GFP-FtsI at roughly the normal chromosomal level in the absence of induction.
To randomly mutagenize ftsI, we amplified the gene by PCR with Taq DNA polymerase, which resulted in ∼1 mutation per kb (data not shown). Since the ftsI gene is ∼1.8 kb long, most of the amplified copies had an amino acid substitution, even though some mutations are silent. The PCR products were ligated into a GFP fusion vector. The resulting plasmids were transformed into EC812 on LB medium containing arabinose. The transformants were then tested for failure to grow on LB medium containing glucose, when division depends on the GFP-FtsI protein produced from the plasmid. Noncomplementing plasmids were recovered from DNA minipreps, analyzed by restriction digestion to verify the presence of a gfp-ftsI fusion gene, and transformed back into EC812 to confirm the mutant phenotype. The candidates were then screened with a combination of Western blotting and penicillin-binding assays to weed out those that expressed proteins which were truncated, unstable, or catalytically inactive or failed to insert into the membrane. In particular, it should be noted that the penicillin-binding assay requires that FtsI perform catalysis on a β-lactam substrate and that this substrate is not able to diffuse across the cytoplasmic membrane. Thus, a positive result in this assay, which was done with spheroplasts, implies that the mutant protein in question retained catalytic activity and inserted into the membrane. Candidates that passed these tests were examined by fluorescence microscopy to determine whether the respective GFP-FtsI fusion protein localized to the septal ring, as indicated by a fluorescent band at the midcell. In parallel, DNA sequencing was used to identify the amino acid substitution(s) in the products of these alleles. If multiple substitutions were found, subcloning was used to identify the relevant lesion. Proteins whose defects could not be attributed to a single lesion were not studied further.
We started with ∼7,500 transformants generated from 15 independent PCRs. About 5% of the transformants recovered on arabinose exhibited poor complementation on glucose. Most of these produced proteins that were markedly unstable or that lacked penicillin-binding activity. Ultimately, we found only 23 plasmids that passed all of the tests. Of these, eight had striking defects in septal localization, i.e., ≤5% of the cells had a fluorescent band at the midcell compared to ∼50% for wild-type GFP-FtsI. The remaining 15 fusion proteins exhibited near-normal levels of septal localization.
Sequence changes.
Although mature FtsI is 577 amino acids long, the 23 mutant proteins were found to have lesions in only 11 amino acids (Table 1). The fact that most of the targeted amino acids were altered multiple times in independent mutants indicates that our analysis was reasonably thorough. Lesions in the eight localization-defective proteins mapped to three residues clustered in a 23-amino-acid region from R23 to Q46. These amino acids are in or near the membrane-spanning segment, implying that the membrane-spanning segment is important for septal localization. In the 15 mutant proteins that appeared to be proficient in localization, the lesions mapped to 8 amino acids in a 153-amino-acid region extending from Gly 57 to Arg 210. All of these amino acid substitutions are in the noncatalytic domain. Apparently this domain is important for something other than septal localization.
TABLE 1.
Amino acid changes and localization phenotypes of ftsI mutants
| Amino acid substitution | No. of times isolateda | Localization of GFP-FtsI fusions to septal ring
|
||
|---|---|---|---|---|
| In ftsI/gfp-ftsI merodiploids (% of cells with ringb) | In cells (filaments) depleted of FtsI
|
|||
| % of cells with ring(s) | Spacing of ringsd | |||
| Wild type | 40 | 47 | 9.5 (714) | |
| R23C | 2 | 2 | 71 | 12 (96) |
| R23H | 2 | 3 | 90 | 15 (82) |
| L39P | 1 | 0 | 26c | 88 (54) |
| Q46H | 3 | 5 | 67 | 11 (151) |
| G57D | 3 | 33 | 86 | 19 (63) |
| S61F | 1 | 40 | 88 | 14 (111) |
| S61P | 1 | 40 | 96 | 16 (44) |
| L62P | 3 | 29 | 98 | 14 (87) |
| V86E | 1 | 21 | 98 | 15 (94) |
| R141C | 1 | 30 | 96 | 17 (69) |
| H177R | 1 | 17 | 99 | 13 (90) |
| G188E | 2 | 40 | 93 | 13 (45) |
| R210C | 1 | 32 | 91 | 14 (45) |
| R210H | 1 | 36 | 93 | 13 (84) |
From independent PCRs.
Value reported is mean of ≥2 independent experiments in which ≥200 cells were scored.
Rings were faint.
The spacing is a measure of the frequency of rings per unit of cell mass and is calculated by dividing the total number of rings into the total length of cells or filaments scored. The number of filaments scored is indicated in parentheses.
Localization of the mutant FtsI proteins.
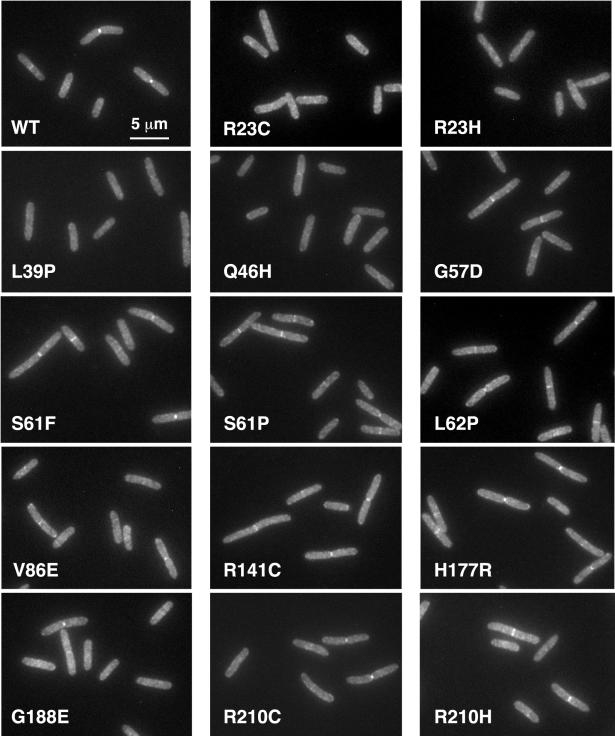
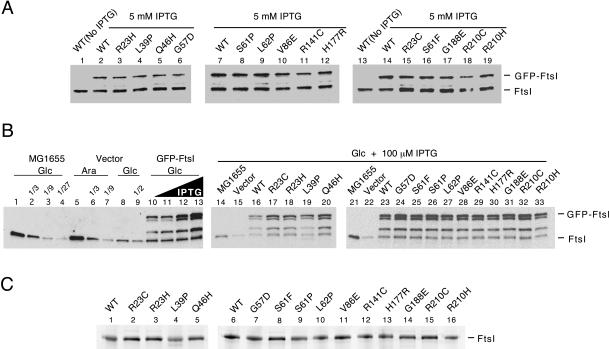
To study localization more carefully, each of the mutant gfp-ftsI alleles was integrated into the chromosome in single copy at the λ attachment site (21). The resulting strains were merodiploids, because a wild-type copy of ftsI was present at the normal chromosomal locus. Expression of gfp-ftsI was driven from a weak IPTG-inducible promoter, P206 (40). Cells were grown to mid-exponential phase in LB containing 5 mM IPTG, fixed with cross-linking agents, and examined by fluorescence microscopy. About 40% of the cells exhibited localization of wild-type GFP-FtsI to the septal ring (Fig. 2 and Table 1). The frequencies of localization for various mutant proteins ranged from 0 to 40%. Proteins with lesions in or near the membrane-spanning segment had localization frequencies ranging from 0% for L39P to 5% for Q46H. In contrast, proteins with lesions in the noncatalytic domain had localization frequencies ranging from 17 to 40%, close to wild type. (Formally, Q46 is considered to be in the noncatalytic domain, but just barely—the membrane-spanning segment probably ends at R40.) Western blotting indicated that the various mutant gfp-ftsI fusions were expressed at levels similar to those of the wild-type gfp-ftsI fusion and that this level was about half that of authentic ftsI expressed from its normal chromosomal locus (Fig. 3A). Thus, there was no reason to suspect that high expression levels were masking a mild localization defect in the cases of the mutant FtsI proteins with lesions in the noncatalytic domain.
FIG. 2.
Localization of GFP-FtsI mutant proteins in cells that also express wild-type ftsI from the chromosome. Cells in exponential growth were fixed, and GFP was visualized by fluorescence microscopy. The levels of FtsI and GFP-FtsI in these strains are shown in Fig. 3A. The strains shown are EC1137 (WT), EC1155 (R23C), EC1138 (R23H), EC1139 (L39P), EC1140 (Q46H), EC1141 (G57D), EC1150 (S61F), EC1142 (S61P), EC1143 (L62P), EC1144 (V86E), EC1145 (R141C), EC1146 (H177R), EC1147 (G188E), EC1148 (R210C), and EC1149 (R210H).
FIG. 3.
Expression and penicillin-binding activity of mutant FtsI proteins. (A) Steady-state levels of FtsI and GFP-FtsI proteins in merodiploid strains as determined by Western blotting. The strains used are listed in the legend to Fig. 2. They have a wild-type (WT) copy of ftsI at the normal chromosomal locus and a gfp-ftsI fusion (wild type or mutant) integrated into the chromosome at the λ attachment site. Expression of the gfp-ftsI fusion is under IPTG control. Amino acid substitutions in GFP-FtsI proteins and the amount of IPTG used for induction are indicated above the lanes. The positions of FtsI and GFP-FtsI are shown on the right. (B) Levels of FtsI and GFP-FtsI proteins in depletion strains as determined by Western blotting. The FtsI depletion strain EC812 was transformed with low-copy-number plasmids that express gfp-ftsI alleles. In this experiment, expression of wild-type ftsI from the chromosome of the host strain is controlled by the addition of 0.2% arabinose (Ara; induced) or 0.2% glucose (Glc; repressed), while expression of the plasmidborne gfp-ftsI alleles to be tested is controlled by the addition of 0 to 100 μM IPTG. Where indicated (Glc), strains were grown in the presence of glucose for 5 h (∼9 doublings) to deplete FtsI before samples were taken for Western blotting. Lanes 1 to 4, FtsI in wild-type strain MG1655 for purposes of comparison only. Lanes 5 to 9, FtsI in the depletion strain EC812/pTH18-kr (empty vector). Lanes 10 to 13, FtsI and GFP-FtsI in EC812/pDSW521 (gfp-ftsI). IPTG was present at 0, 2.5, 10, and 100 μM. Note that when GFP-FtsI is overproduced, several prominent breakdown products accumulate, including one that comigrates with authentic FtsI and makes it appear that the chromosomally encoded protein was not effectively depleted. Lanes 14 to 33, MG1655, EC812/pTH18-kr (empty vector), and EC812/pDSW521 derivatives that express wild-type and mutant GFP-FtsI proteins. Equivalent amounts of cell material were loaded, except for lanes 2 to 4, 6, 7, and 9, which were diluted as indicated. (C) Penicillin-binding activities of mutant FtsI proteins. Membranes isolated from DHB4/pDSW478 (PBAD-ftsI) derivatives were incubated with a fluorescent β-lactam, and then proteins were separated by SDS-PAGE and labeled FtsI was detected with a fluorescence imager.
We also assayed localization in cells (filaments) depleted of wild-type FtsI. These studies were done with EC812 derivatives transformed with low-copy-number plasmids that express gfp-ftsI fusions under the control of a lac promoter. Note that this is the genetic configuration used to isolate the mutants. Characterization of this system by Western blotting demonstrated that (i) EC812 produced normal amounts of FtsI when the inducer arabinose was present; (ii) FtsI levels were reduced ∼10-fold after several hours of growth in the presence of glucose, at which time the cells had become filaments ∼20 μm long; (iii) in the absence of IPTG, the level of GFP-FtsI fusion protein produced from the plasmid was similar to what is normal for FtsI from the chromosome; and (iv) induction with 100 μM IPTG results in severalfold overproduction of the GFP-FtsI fusion protein and the appearance of significant amounts of proteolytic breakdown products (Fig. 3B).
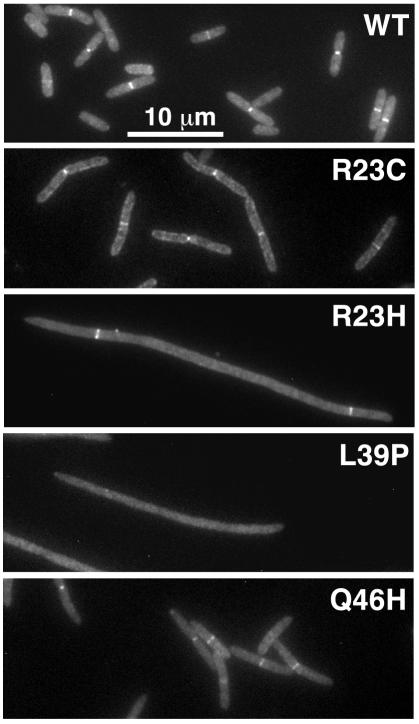
Results of experiments to test the effect of depleting FtsI on localization of the “localization-defective” mutants are shown in Fig. 4 and Table 2. Depletion of FtsI had no effect on the morphology of cells that express a wild-type gfp-ftsI fusion from the plasmid, because the protein localizes well and supports cell division. Interestingly, localization of the R23C, R23H, and Q46H proteins improved greatly upon depletion of the wild-type protein. Only the L39P protein appeared to have a severe localization defect after depletion of the wild-type protein, although even in that case, localization improved somewhat. Figure 4 also shows that the R23C and Q46H proteins support division reasonably well in broth when produced at this level (100 μM IPTG). We later found that all of the localization-defective proteins could rescue EC812 on plates when induced with enough IPTG (see below). One interpretation of these findings is that FtsI localization involves recognition of a limiting factor at the septal ring, and the localization-defective mutants compete poorly for this factor, which might be FtsW (30). Chen et al. made a similar observation in their study of localization-defective mutants of ftsQ and also suggested that this phenomenon reflects competition for a limiting component of the septal ring (10). An alternative interpretation is that the lesions impair dimerization of FtsI. The oligomeric state of FtsI in vivo is not yet known.
FIG. 4.
Localization of GFP-FtsI mutant proteins in cells (filaments) depleted of wild-type FtsI. In the experiment shown, chromosomally encoded wild-type (WT) FtsI was depleted for 5 h, and plasmidborne GFP-FtsI (wild type or mutant) was induced with 100 μM IPTG. Cells depleted of FtsI were fixed, and GFP was visualized by fluorescence microscopy. DAPI staining verified proper nucleoid segregation (not shown). The levels of FtsI and GFP-FtsI proteins in these cells are shown in Fig. 3B.
TABLE 2.
Abilities of mutant FtsI proteins to recruit FtsN
| Amino acid substitution | No. of transectsa | Normalized fluorescence (arbitrary units)b
|
Red/green ratioc | % of wild-type recruitment activity | |
|---|---|---|---|---|---|
| GFP (FtsI) | Texas Red (FtsN) | ||||
| Wild type | 29 | 0.7 | 1.6 | 2.3 | 100 |
| G57D | 27 | 0.6 | 0.4 | 0.7 | 30 |
| S61F | 39 | 0.8 | 0.5 | 0.6 | 36 |
| S61P | 24 | 0.7 | 0.3 | 0.4 | 17 |
| L62P | 53 | 0.7 | 0.3 | 0.4 | 17 |
| V86E | 48 | 0.6 | 0.3 | 0.5 | 22 |
| R141C | 58 | 0.6 | 0.3 | 0.5 | 22 |
| H177R | 49 | 0.5 | 1.0 | 2.0 | 87 |
| G188E | 55 | 0.6 | 0.5 | 0.8 | 35 |
| R210C | 27 | 0.8 | 0.5 | 0.6 | 26 |
| R210H | 43 | 0.6 | 0.7 | 1.2 | 52 |
The number of transects through the strong GFP-FtsI bands that were averaged to quantitate fluorescence intensity. Each transect was replicated to the corresponding position in the red (FtsN) channel.
Highest value in transect.
Similar signal ratios were obtained using the area under the transect peak, but this was harder to calculate than peak height.
All of the mutant GFP-FtsI proteins that localized well in merodiploids also localized well when wild-type FtsI was depleted (Fig. 5, left, and Table 2). This result appears to exclude the formal possibility that localization defects were masked in the merodiploids because the wild-type FtsI recruited the mutant proteins to the septal ring, perhaps by forming a dimer or some other oligomer. Rather, the mutant FtsI proteins with lesions in the domain of unknown function really are proficient in septal localization, even though they fail to support cell division. In the experiment shown, 100 μM IPTG was used to induce gfp-ftsI alleles from the plasmid. Similar results were obtained with depletion filaments grown in the presence of 10 μM IPTG, which resulted in GFP-FtsI levels essentially the same as those of FtsI (not shown).
FIG. 5.
Localization of GFP-FtsI mutant proteins and FtsN in cells (filaments) depleted of wild-type (WT) FtsI. In the experiment shown, chromosomally encoded wild-type FtsI was depleted for 5 h, and plasmidborne GFP-FtsI (wild type or mutant) was induced with 100 μM IPTG. Cells were fixed, and FtsI (green) and FtsN (red) were visualized by fluorescence microscopy. The transects on the right quantitate fluorescence signal as described in Materials and Methods. All of the micrographs are at the same magnification, except for G57D and S61F, which are slightly smaller. In some panels (S61F, S61P, L62P, H177R, and R210C), wild-type cells of strain MG1655 are shown together with the filaments. These short cells served as positive controls for detection of FtsN by immunofluorescence microscopy but do not express gfp-ftsI, so they are not readily apparent in the green images. DAPI staining verified proper nucleoid segregation (not shown). The levels of FtsI and GFP-FtsI proteins in these cells are shown in Fig. 3B. Note that the use of high levels of IPTG in this experiment results in high background fluorescence in the green channel and the accumulation of proteolytic breakdown products as assessed by Western blotting (Fig. 3B).
Localization of FtsN.
Because localization of FtsN to the septal ring depends upon FtsI (2), we determined the abilities of the mutant forms of FtsI to recruit FtsN. To do this, we used indirect immunofluorescence microscopy to visualize FtsN in filamentous cells depleted of wild-type FtsI but containing GFP-FtsI fusion proteins that were produced from a plasmid. FtsN was detected with a polyclonal primary antibody followed by a secondary antibody conjugated to Texas Red. Filaments were photographed using filters appropriate for green (GFP-FtsI) and red (FtsN) light.
At the resolution afforded by light microscopy, FtsN colocalized with wild-type GFP-FtsI to the septal ring (Fig. 5, middle). In the filamentous cells that expressed GFP fusions to mutant forms of FtsI, the results were more complex. In most cases, FtsN appeared to localize poorly in the filaments, but some of the mutant proteins, such as the H177R protein, appeared to recruit FtsN rather well. Nevertheless, we found it difficult to make simple yes-no calls as to whether FtsN localized to a particular site, largely because many of these sites had weak accumulations of fluorescence that were too bright to ignore. To score FtsN localization more objectively, we used computer software to draw transects across the strongest GFP-FtsI bands and let the computer replicate these transects in the same position in the red (FtsN) image. The fluorescence intensities in these transects were then measured, normalized, and averaged over at least 24 GFP-FtsI bands as described in Materials and Methods. The results are shown in Fig. 5 (right) and Table 2 and agree with more subjective impressions of localization. With wild-type GFP-FtsI, which recruited FtsN very well, the red fluorescence was about twice as bright as the green. The H177R mutant protein also recruited FtsN well—the ratio of red to green was 87% of that for the wild type. At the other extreme, the S61F and L62P mutant proteins appeared to have only 17% of the wild-type recruitment activity.
Western blot analysis showed that the different GFP-FtsI proteins were produced at similar levels (Fig. 3B) and that these levels were ∼4-fold above what is normal for FtsI expressed from the chromosome. This level of expression should be adequate for FtsN recruitment, if a particular mutant protein were proficient in this function. The same blots also revealed that the filaments contained residual FtsI. While some of this FtsI could be mutant protein derived from the fusions by proteolysis, residual FtsI was also seen in an EC812 derivative carrying a plasmid that did not express a gfp-ftsI fusion. These observations raise the possibility that residual wild-type FtsI, rather than mutant GFP-FtsI, is responsible for the weak localization of FtsN observed in some cases. If so, then mutant proteins such as S61F and L62P are probably more defective for recruitment of FtsN than they appear.
Complementation and dominance.
Mutant FtsI proteins that localize efficiently to the septal ring but fail to recruit FtsN ought to have dominant-negative effects. Indeed, merodiploids expressing gfp fusions to localization-proficient mutant proteins were slightly elongated (Fig. 2). Still, the fact that we recovered such mutants implies that any dominant-negative effects are mild. Conversely, elevated production of some localization-defective proteins appeared to rescue cell division (Fig. 4). These observations were confirmed by testing for complementation and dominant-negative effects in various ftsI mutant backgrounds (Table 3). With no IPTG, the gfp-ftsI fusions are expressed at normal levels, and the localization-defective proteins failed to complement either a depletion strain or a temperature-sensitive mutant. The presence of 100 μM IPTG resulted in ∼4-fold overproduction and some complementation by each mutant allele. In contrast, the set of proteins that exhibited fairly normal localization did not complement at either level of production. These proteins were not very toxic in the depletion strain, even when 100 μM IPTG was used for induction. However, in the temperature-sensitive background, which has ∼10% as much FtsI protein as the wild type even at the permissive temperature (data not shown), there was considerable toxicity. In fact, most of the localization-proficient alleles could not be transformed into the temperature-sensitive background, and those that we did recover were not genetically stable upon restreaking. Fusion genes that express localization-defective proteins, however, were not toxic in this background and even complemented slightly when mildly overproduced.
TABLE 3.
Complementation and dominance of mutant gfp-ftsI alleles as assessed by growth (colony formation) in FtsI depeletion and ftsI(Ts) backgrounds
| Amino acid substitution | Growth in FtsI depletion straina
|
Growth in ftsI(Ts) strainb
|
||||||
|---|---|---|---|---|---|---|---|---|
| Arabinose
|
Glucose
|
30°C
|
42°C
|
|||||
| −IPTG | +IPTG | −IPTG | +IPTG | −IPTG | +IPTG | −IPTG | +IPTG | |
| Vector | +++ | +++ | − | − | +++ | +++ | − | − |
| WT | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| R23C | +++ | +++ | − | ++ | +++ | +++ | + | + |
| R23H | +++ | +++ | − | ++ | +++ | +++ | ± | ± |
| L39P | +++ | +++ | − | ++ | +++ | +++ | ± | ± |
| Q46H | +++ | +++ | + | +++ | +++ | +++ | ± | + |
| G57D | +++ | +++c | − | − | −d | NDe | ND | ND |
| S61F | +++ | +++c | − | − | −d | ND | ND | ND |
| S61P | +++ | +++c | − | − | −d | ND | ND | ND |
| L62P | +++ | +++c | − | − | −d | ND | ND | ND |
| V86E | +++ | +++c | − | − | −d | ND | ND | ND |
| R141C | +++ | +++c | − | − | −d | ND | ND | ND |
| H177R | +++ | +++c | − | − | + | − | − | − |
| G188E | +++ | +++c | − | − | −d | ND | ND | ND |
| R210C | +++ | +++c | − | − | + | − | ± | ± |
| R210H | +++ | +++c | − | − | + | − | ± | ± |
The FtsI depletion strain EC812 was transformed with low-copy-number plasmids that express gfp-ftsI alleles. In this experiment, expression of wild-type ftsI from the chromosome of the host strain is controlled by the addition of 0.2% arabinose (induced) or 0.2% glucose (repressed), while expression of the plasmidborne gfp-ftsI alleles to be tested is controlled by the addition of 0 or 100 μM IPTG. Growth was scored on the basis of colony size. Scoring ranged from good growth (+++) if the isolated colonies were equivalent in size to those produced by transformants harboring a gfp-ftsI wild-type plasmid to no growth (−) if isolated colonies did not form, although there sometimes was growth in the heavy streak in these cases. ±, a few tiny colonies were observed. The plasmids used were pTH18-kr (vector with no gfp or ftsI), pDSW521 (WT), pDSW562 (R23C), pDSW563 (R23H), pDSW566 (L39P), pDSW567 (Q46H), pDSW568 (G57D), pDSW569 (S61F), pDSW570 (S61P), pDSW571 (L62P), pDSW572 (V86E), pDSW573 (R141C), pDSW574 (H177R), pDSW575 (G188E), pDSW576 (R210C), and pDSW577 (R210H).
Transformants of the ftsI(Ts) strain EC295 were scored for growth as described in footnote a, except that temperature rather than arabinose or glucose was used to control the FtsI produced from the host's chromosome.
Colonies were normal in size but contained some filamentous cells, indicating that the mutant alleles behave as mild dominant negatives when overproduced.
Plasmids expressing these alleles could not be transformed into EC295, presumably because they were too toxic (dominant negative) in a strain that has reduced amounts of functional FtsI even under permissive conditions.
ND, not determined.
Penicillin-binding activity and thermostability.
Our initial screen included a penicillin-binding assay done with spheroplasts isolated from cells that overproduced GFP-FtsI fusion proteins. We therefore anticipated that the mutant proteins described here would fractionate with the membranes and exhibit essentially normal reactivity toward a β-lactam. To verify this expectation, cell envelopes were isolated from strains that overproduced wild-type and mutant forms of FtsI (without the GFP). Cell envelopes were incubated at 30°C with a fluorescent β-lactam, and then the proteins were separated by SDS-PAGE and the gel was visualized with a fluorescence imager. As expected, wild-type and mutant proteins bound similar amounts of the β-lactam (Fig. 3C).
We also used a penicillin-binding assay to determine the half-lives of wild-type and mutant forms of FtsI at 47°C as described previously (14). Briefly, cell envelopes were incubated at 47°C for various lengths of time and then transferred to 30°C and assayed for binding of a fluorescent β-lactam. The stability assay was done at 47°C because at that temperature wild-type FtsI has a half-life of ∼5 min, so we anticipated that even mild stability problems would be reflected in very short half-lives for the mutant proteins. As summarized in Table 4, 10 of the mutant proteins (R23C, R23H, L39P, Q46H, G57D, S61F, S61P, L62P, R210C, and R210H) had half-lives similar to that of the wild type, indicating that the lesions do not cause a folding defect. The remaining mutants (V86E, R141C, H177R, and G188E) had distinctly shorter half-lives. For these mutants, we also determined the half-life at 33°C. At the lower temperature, the wild-type, V86E, and R141C proteins had half-lives of ∼33 h, while the H177R and G188E proteins had half-lives of ∼5 h.
TABLE 4.
Half-lives of wild-type and mutant FtsI proteins
| Amino acid substitution | Half-life (min)a
|
|
|---|---|---|
| 47°C | 33°C | |
| Wild type | 5.1 ± 0.8 (7) | ∼2,000 (3)b |
| R23C | 5.9 ± 1.3 (3) | |
| R23H | 6.0 ± 1.4 (2) | |
| L39P | 5.8 ± 0.7 (2) | |
| Q46H | 5.0 ± 0.8 (3) | |
| G57D | 5.0 ± 0.9 (3) | |
| S61F | 4.6 ± 0.9 (3) | |
| S61P | 5.1 ± 0.9 (2) | |
| L62P | 6.0 ± 0.5 (3) | |
| V86E | 2.9 ± 0.6 (2) | ∼2,000 (3)b |
| R141C | 2.4 ± 0.9 (2) | ∼1,800 (3)b |
| H177R | 1.4 (1) | 330 ± 90 (4) |
| G188E | 1.0 ± 0.3 (2) | 330 ± 50 (3) |
| R210C | 4.4 ± 1.4 (3) | |
| R210H | 6.3 ± 1.7 (2) | |
Mean ± standard deviation (number of independent determinations).
The time course examined was too short to calculate precise half-lives for these proteins.
DISCUSSION
Amino acid substitutions that impair septal localization fall in or near the membrane-spanning segment.
Our results indicate that amino acids 23, 39, and 46 of E. coli FtsI are important for localization to the septal ring. The amino acid substitutions recovered at these sites reduced localization by a factor of 8 or more but did not prevent insertion of the protein into the membrane, impair catalytic activity as judged by a penicillin-binding assay, or reduce the stability of the protein. Interestingly, changes at R23 (to C and H) were recovered four times, and changes at Q46 (all to H) were recovered three times. Given that mature FtsI is 577 amino acids long and that the mutants were obtained after random mutagenesis, these statistics imply that if other parts of FtsI are important for localization, a different approach will be needed to find them.
Residues R23, L39, and Q46 fall within or near the transmembrane helix, defined operationally as extending from R23 to R40 (7). Several other observations implicate the membrane-spanning segment in septal localization. An FtsI protein, constructed by site-directed mutagenesis, that has lesions at both ends of the membrane-spanning segment was previously described (20). Although this form of FtsI supports division, it localizes poorly (41). It was also shown that an FtsI protein with an extra leucine in the transmembrane helix does not support cell division (20) and does not localize to the septal ring (M. Wissel and D. Weiss, unpublished data).
Our results provide little support for previous speculation that the noncatalytic domain, which resides in the periplasm, engages in protein-protein interactions involved in recruiting FtsI to the septal ring. Otherwise, we should have found localization-defective mutants with lesions in this domain. We did, however, find several amino acid substitutions in the domain that had modest effects on septal localization and that may hint at a role for the domain in targeting FtsI to the septal ring. A previous report suggested that the D58V amino acid substitution prevented FtsI from localizing to the septal ring (29). This conclusion was based on the recessive nature of the mutation rather than on a direct assay of the ability of the mutant protein to localize. We have constructed the D58V protein by site-directed mutagenesis and have confirmed that it does not support cell division, but in our hands it localizes almost as well as the wild type (J. Wendt and D. Weiss, unpublished data). In these respects, the D58V protein is similar to the G57D protein that we found by random mutagenesis.
How the membrane-spanning segment of FtsI targets the protein to the septal ring is a matter of speculation. It might serve as a binding site for another protein, such as FtsI itself (if the protein is a dimer) or FtsW, which localizes immediately ahead of FtsI and is needed to recruit FtsI to the septal ring (30). Alternatively, changes in the membrane-spanning segment might subtly perturb the positioning of the periplasmic domain and thus impair dimerization or interaction with another protein. However, this hypothesis suggests that the real localization determinants might be in the noncatalytic domain, despite our inability to find lesions in that domain that prevent septal localization (other than Q46H at the beginning of the domain). Perhaps no single amino acid substitution in the noncatalytic domain impairs the putative interaction sufficiently to prevent complementation without causing additional defects.
Amino acid substitutions that impair recruitment of FtsN affect a domain of unknown function.
Our results indicate that amino acids 57, 61, 62, and 210 of E. coli FtsI are important for recruitment of FtsN to the septal ring but are not critical for septal localization of FtsI, insertion of the protein into the membrane, catalytic activity as judged by a penicillin-binding assay, or stability. Substitutions at these positions were isolated repeatedly: changes at G57 (all to D) were recovered three times, changes at S61 (to F and P) were recovered twice, changes at L62 (all to P) were recovered three times, and changes at R210 (to C and H) were recovered twice. Since these mutants were obtained after random mutagenesis, it is remarkable that lesions conferring a relatively specific defect in FtsN recruitment repeatedly mapped to the same four residues. The four amino acids targeted by our mutant hunt are in the noncatalytic domain, which we now conclude is needed for recruitment of FtsN to the septal ring. This inference is consistent with previous evidence that the periplasmic domain of FtsN directs that protein to the septum (2, 12). Our finding that R210 is important for recruitment of FtsN confirms and extends findings of Marrec-Fairley and coworkers, who reported that an R210Q-R213Q double-mutant protein was toxic and inferred that it probably localizes to the septal ring but fails to interact properly with another protein (29).
The three-dimensional structure of FtsI is not known, but the structure of a homolog, PBP2x from S. pneumoniae, has been determined by X-ray diffraction (13, 35). We used an alignment of the two proteins to identify the amino acids in PBP2x that correspond to G57, S61, L62, and R210 of FtsI. Despite being separated by 150 residues in the primary sequence, these amino acids appear close together in the folded protein (Fig. 1C). They cluster near the end of one of the arms that were suggested on the basis of the structure to be involved in protein-protein interactions (35). Our results suggest that one of these interactions is with FtsN, although biochemical evidence will be needed to prove this point. Because the resolution in the PBP2x structure is low, it is not possible to say whether any of these amino acids have solvent-exposed side chains that could interact with FtsN directly.
Whether the defects in FtsN recruitment account for the failure of these mutant proteins to support division is not yet known. There is enough residual localization of FtsN (17 to 30% of wild-type activity) that, if this were the only defect, we would expect the mutant ftsI alleles to complement to some extent. But the mutant proteins might be more defective than they appear, because Western blotting revealed some residual wild-type FtsI protein in the filaments used to assay FtsN recruitment.
Other lesions in the domain of unknown function.
Our screen identified several other critical amino acids in the noncatalytic domain: V86, R141, H177, and G188. Although lesions at these sites resulted in a failure to complement, further characterization indicated that the defects in these proteins are complex. Each mutant protein was somewhat less stable than the wild type. This instability does not by itself account for the failure to complement, because the steady-state level of each of the mutant proteins is similar to that of the wild type. Still, the instability suggests that parts of FtsI distant from the lesions might have been disrupted. One of the lesions, G188E, is at the start of a block of sequence, G188IEGVEKSFD, that is highly conserved among class B penicillin-binding proteins (18) and that has previously been shown to be important for the folding and stability of FtsI (17). The V86E and H177R proteins localize about half as well as the wild type, suggesting that the domain of unknown function has a role in septal localization, even though our best localization-defective mutants mapped to the transmembrane helix. The V86E, R141C, and G188E proteins did not recruit FtsN very well and lend further support to the notion that the noncatalytic domain is needed for recruitment of FtsN. The H177R protein is in some ways the most intriguing one to come out of the study. Although it does not support cell division, it is essentially normal in everything we can measure—septal localization, recruitment of FtsN, and penicillin-binding activity. Perhaps FtsI has functions that we do not know of. Alternatively, the H177R protein might point to the limitations of our assays. For example, the H177R lesion might alter the orientation of FtsI with respect to the peptidoglycan wall so that transpeptidation is not possible, but because the active site is not disrupted, penicillin binding is not impaired. Finally, Höltje and coworkers have used affinity chromatography to recover complexes that contain a host of murein enzymes, including FtsI, Slt70, MltA, MltB, PBP1b, PBP1c, PBP2, PBP4, PBP7, and AmiC (26, 38, 39). Thus, the H177R mutant (and possibly others too) might be defective in formation of a proposed “holoenzyme” for peptidoglycan synthesis.
The lesions in these mutant proteins are scattered throughout the domain of unknown function. In PBP2x, the amino acids that correspond to V86, H177, and G188 are near the interface between the noncatalytic and catalytic domains (Fig. 1C). This may explain why lesions at these sites reduce stability. R141 of FtsI is difficult to map onto the structure of PBP2x because it falls in a region where PBP2x has an ∼30-amino-acid insertion that is not present in the E. coli protein. Nevertheless, it appears safe to infer that R141 is in one of the arms of the noncatalytic domain, opposite the arm that contains residues implicated in FtsN recruitment.
Acknowledgments
We thank Ryan Arends, Jennie Blair, Calista Mitchell, Kari Schmidt, Ryan Seipke, Victor Torres, James Urton, Jennifer Wendt, and Brian Wulff for help with experiments and Subramanian Ramiswamy for help producing Fig. 1C.
This work was supported by a grant from the National Institutes of Health (GM59893) to D.S.W. M.C.W. was supported in part by a National Science Foundation Research Training Grant (DBI9602247). The DNA facility is supported by the Diabetes and Endocrinology Research Center with National Institutes of Health grant DK25295 and by the School of Medicine.
REFERENCES
- 1.Adam, M., C. Fraipont, N. Rhazi, M. Nguyen-Disteche, B. Lakaye, J. M. Frere, B. Devreese, J. Van Beeumen, Y. van Heijenoort, J. van Heijenoort, and J. M. Ghuysen. 1997. The bimodular G57-V577 polypeptide chain of the class B penicillin-binding protein 3 of Escherichia coli catalyzes peptide bond formation from thiolesters and does not catalyze glycan chain polymerization from the lipid II intermediate. J. Bacteriol. 179:6005-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25:303-309. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Bernhardt, T. G., and P. A. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Botta, G. A., and J. T. Park. 1981. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J. Bacteriol. 145:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowler, L. D., and B. G. Spratt. 1989. Membrane topology of penicillin-binding protein 3 of Escherichia coli. Mol. Microbiol. 3:1277-1286. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, D., C. Manoil, and J. Beckwith. 1987. Determinants of membrane protein topology. Proc. Natl. Acad. Sci. USA 84:8525-8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J. C., M. Minev, and J. Beckwith. 2002. Analysis of ftsQ mutant alleles in Escherichia coli: complementation, septal localization, and recruitment of downstream cell division proteins. J. Bacteriol. 184:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 12.Dai, K., Y. Xu, and J. Lutkenhaus. 1996. Topological characterization of the essential Escherichia coli cell division protein FtsN. J. Bacteriol. 178:1328-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dessen, A., N. Mouz, E. Gordon, J. Hopkins, and O. Dideberg. 2001. Crystal structure of PBP2x from a highly penicillin-resistant Streptococcus pneumoniae clinical isolate: a mosaic framework containing 83 mutations. J. Biol. Chem. 276:45106-45112. [DOI] [PubMed] [Google Scholar]
- 14.Eberhardt, C., L. Kuerschner, and D. S. Weiss. 2003. Probing the catalytic activity of a cell division-specific transpeptidase in vivo with beta-lactams. J. Bacteriol. 185:3726-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghuysen, J. M. 1991. Serine beta-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 45:37-67. [DOI] [PubMed] [Google Scholar]
- 17.Goffin, C., C. Fraipont, J. Ayala, M. Terrak, M. Nguyen-Disteche, and J. M. Ghuysen. 1996. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J. Bacteriol. 178:5402-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. S. Weiss, and J. Beckwith. 1997. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J. Bacteriol. 179:5094-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara, H., Y. Nishimura, J. Kato, H. Suzuki, H. Nagasawa, A. Suzuki, and Y. Hirota. 1989. Genetic analyses of processing involving C-terminal cleavage in penicillin-binding protein 3 of Escherichia coli. J. Bacteriol. 171:5882-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara, H., S. Yasuda, K. Horiuchi, and J. T. Park. 1997. A promoter for the first nine genes of the Escherichia coli mra cluster of cell division and cell envelope biosynthesis genes, including ftsI andftsW. J. Bacteriol. 179:5802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto-Gotoh, T., M. Yamaguchi, K. Yasojima, A. Tsujimura, Y. Wakabayashi, and Y. Watanabe. 2000. A set of temperature sensitive-replication/-segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene 241:185-191. [DOI] [PubMed] [Google Scholar]
- 25.Heidrich, C., M. F. Templin, A. Ursinus, M. Merdanovic, J. Berger, H. Schwarz, M. A. de Pedro, and J. V. Holtje. 2001. Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167-178. [DOI] [PubMed] [Google Scholar]
- 26.Höltje, J. V. 1996. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology 142:1911-1918. [DOI] [PubMed] [Google Scholar]
- 27.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 28.Keck, W., B. Glauner, U. Schwarz, J. K. Broome-Smith, and B. G. Spratt. 1985. Sequences of the active-site peptides of three of the high-Mr penicillin-binding proteins of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 82:1999-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrec-Fairley, M., A. Piette, X. Gallet, R. Brasseur, H. Hara, C. Fraipont, J. M. Ghuysen, and M. Nguyen-Distèche. 2000. Differential functionalities of amphiphilic peptide segments of the cell-septation penicillin-binding protein 3 of Escherichia coli. Mol. Microbiol. 37:1019-1031. [DOI] [PubMed] [Google Scholar]
- 30.Mercer, K. L., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 32.Nagasawa, H., Y. Sakagami, A. Suzuki, H. Suzuki, H. Hara, and Y. Hirota. 1989. Determination of the cleavage site involved in C-terminal processing of penicillin-binding protein 3 of Escherichia coli. J. Bacteriol. 171:5890-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen-Disteche, M., C. Fraipont, N. Buddelmeijer, and N. Nanninga. 1998. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol. Life Sci. 54:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholas, R. A., J. L. Strominger, H. Suzuki, and Y. Hirota. 1985. Identification of the active site in penicillin-binding protein 3 of Escherichia coli. J. Bacteriol. 164:456-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pares, S., N. Mouz, Y. Petillot, R. Hakenbeck, and O. Dideberg. 1996. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 3:284-289. [DOI] [PubMed] [Google Scholar]
- 36.Pogliano, J., K. Pogliano, D. S. Weiss, R. Losick, and J. Beckwith. 1997. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. USA 94:559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vollmer, W., and J. V. Höltje. 2001. Morphogenesis of Escherichia coli. Curr. Opin. Microbiol. 4:625-633. [DOI] [PubMed] [Google Scholar]
- 39.von Rechenberg, M., A. Ursinus, and J. V. Höltje. 1996. Affinity chromatography as a means to study multienzyme complexes involved in murein synthesis. Microb. Drug Resist. 2:155-157. [DOI] [PubMed] [Google Scholar]
- 40.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss, D. S., K. Pogliano, M. Carson, L. M. Guzman, C. Fraipont, M. Nguyen-Disteche, R. Losick, and J. Beckwith. 1997. Localization of the Escherichia coli cell division protein Ftsl (PBP3) to the division site and cell pole. Mol. Microbiol. 25:671-681. [DOI] [PubMed] [Google Scholar]